Abstract
Chronic lymphocytic leukemia (CLL) is preceded by monoclonal B-cell lymphocytosis (MBL), a CLL precursor state with a prevalence of up to 12% in aged individuals; however, the duration of MBL and the mechanisms of its evolution to CLL remain largely unknown. In this study, we sequenced the B-cell receptor (BcR) immunoglobulin heavy chain (IGH) gene repertoire of 124 patients with CLL and 118 matched controls in blood samples taken up to 22 years prior to diagnosis. Significant skewing in the BcR IGH gene repertoire was detected in the majority of patients, even before the occurrence of lymphocytosis and irrespective of the clonotypic IGH variable gene somatic hypermutation status. Furthermore, we identified dominant clonotypes belonging to major stereotyped subsets associated with poor prognosis up to 16 years before diagnosis in 14 patients with CLL. In 22 patients with longitudinal samples, the skewing of the BcR IGH gene repertoire increased significantly over time to diagnosis or remained stable at high levels. For 14 of 16 patients with available samples at diagnosis, the CLL clonotype was already present in the prediagnostic samples. Overall, our data indicate that the preclinical phase of CLL could be longer than previously thought, even in adverse-prognostic cases.
Key Points
-
•
Significant skewing in the BcR IGH gene repertoire is an early event in CLL pathogenesis.
-
•
Even high-risk CLL subtypes may display a prolonged indolent preclinical stage.
Introduction
Since 2008, chronic lymphocytic leukemia (CLL) has been defined by the presence of a persisting monoclonal B-cell population ≥5 × 109 cells per liter, whereas monoclonal expansions below this limit are classified as monoclonal B-cell lymphocytosis (MBL).1 MBL has been detected up to 6 years before CLL diagnosis. 2, 3, 4, 5 Clinically, MBL is categorized as high count (HC) or low count (LC), based on a cutoff of 0.5 × 109 cells per liter.4 LC MBL can be detected in up to 12% of the elderly population and is found at increased prevalence in relatives of patients with CLL.6, 7, 8, 9 HC MBL cases progress to CLL requiring treatment at a rate of ∼1% per year.4
A key factor for risk stratification of patients with CLL is the somatic hypermutation status of the immunoglobulin heavy variable (IGHV) gene.10 CLL cases with mutated IGHV (M-CLL) genes are generally characterized by a more prolonged indolent disease course than cases of CLL with unmutated IGHV genes (U-CLL).11 Another prognostically relevant immunogenetic feature of CLL concerns the stereotypy of the B-cell receptor (BcR) immunoglobulins (IGs).12 Indeed, distinct stereotyped subsets can be defined by the expression of shared sequence motifs and are associated with particular presentations and outcomes.10, 12
In this context, we aimed to gain insight into the composition of the BcR IG repertoire during the early stages of CLL through the in-depth study of samples taken up to 22 years before diagnosis.
Study design
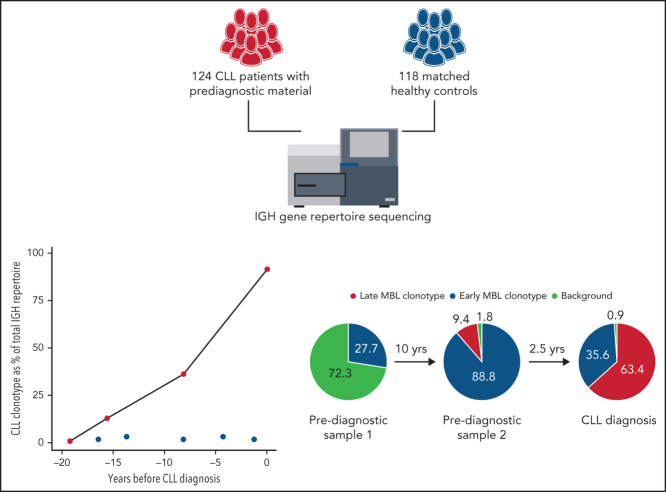
Study subjects were participants in the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort.13 For the current study, 124 healthy individuals who were later diagnosed with CLL or small lymphocytic leukemia along with 118 controls who were matched with regard to age, sex, blood draw date, and center were selected (supplemental Figure 1 and supplemental Table 1, available on the Blood Web site). Longitudinal samples were available for 22 of 124 patients. Postdiagnostic clinical data were available for 32 of 124 patients (supplemental Table 2). The timing of CLL diagnosis ranged from 1 month to 22 years after initial blood sampling. Genomic DNA was isolated from buffy coats obtained at blood sampling. Lymphocyte counts at the time of prediagnostic blood sampling were available for 28 patients and 28 matched controls. The immunoglobulin heavy chain (IGH) gene repertoire was sequenced using a leader-based IGH assay on an Illumina MiSeq (supplemental Table 6). The IGH repertoire was characterized using the ARResT/Interrogate immunoprofiler.14 Stereotyped subsets were annotated through the ARResT/AssignSubsets tool. For details, see supplemental Methods.
Results and discussion
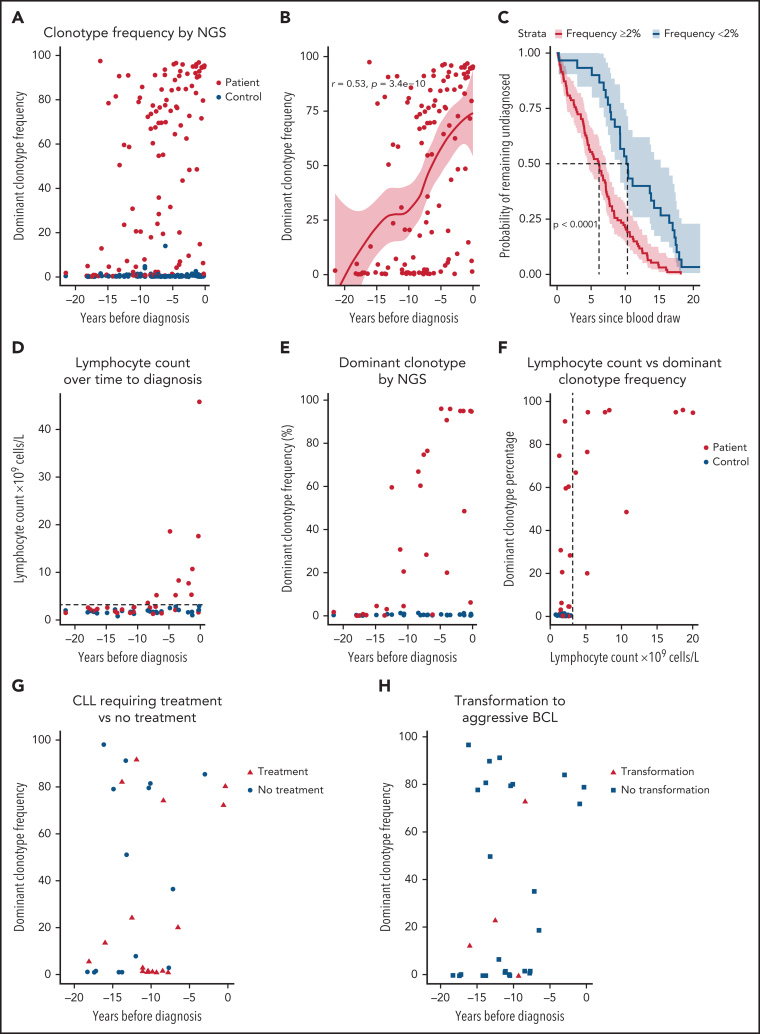
First, unsurprisingly, we observed a significant difference in the frequency of the dominant (most frequent) clonotype in patients with CLL vs controls (P < .0001): the median frequency was 54.9% (interquartile range, 2.2% to 84.4%) and 0.38% (interquartile range, 0.26-0.64%), respectively (Figure 1A). Patients with a shorter time to CLL diagnosis had a higher dominant clonotype frequency (P < .0001; Figure 1B; supplemental Table 3). Moreover, patients with a dominant clonotype frequency > 2% of the total repertoire exhibited a shorter interval from sampling to diagnosis (Figure 1C). Presence of a prediagnostic clonotype >2% of the IGH gene repertoire did not have a significant impact on overall survival of patients with CLL after diagnosis (supplemental Figure 2).
Figure 1.
Skewing of the BcR IGH gene repertoire is detectable by next-generation sequencing up to 16 years before CLL diagnosis. Dominant clonotype frequency represents the size of the largest (most frequent) productive clonotype as a percentage of the total productive IGH gene reads in each given sample. Time to diagnosis for controls reflects the time from sampling of the control to the diagnosis of the matched case. One sample was taken from each patient. (A) Dominant clonotype frequency for patients with CLL and matched controls over time to CLL diagnosis. (B) Positive correlation between time to diagnosis (TTD) and the dominant clonotype frequency as determined by Spearman correlation. The red line represents Loess regression, with 95% confidence intervals (CIs) marked around. (C) Kaplan-Meier (survival) analysis for TTD from prediagnostic sample collection stratified by clonotype frequency. TTD of patients with a dominant clonotype frequency ≥2% of the productive IGH gene repertoire is depicted in red, whereas TTD of patients with a dominant clonotype frequency <2% is depicted in blue. The 95% CI is marked for each line. Significance was determined by the log-rank test. (D) Lymphocyte counts in patients with future CLL and matched controls. (E) Dominant clonotype frequency for the same samples and individuals as in panel D. (F) Lymphocyte counts plotted against dominant clonotype frequency. The lymphocyte count of 1 outlier at 45 × 109/L was winsorized to 20 × 109/L to preserve visibility of low-level dynamics. Dashed line indicates cutoff for abnormal lymphocyte counts. (G) Skewing of the BcR IGH repertoire was detectable for CLL requiring treatment (red triangles) and indolent CLL during follow-up (blue circles), with a median follow-up period of 8.7 years. (H) Skewing of the IGH repertoire was detectable for patients with a transformation to an aggressive B-cell lymphoma and those without transformation during follow-up.
For 56 participants, lymphocytes counts were recorded at the time of baseline blood sampling (Figure 1D). Lymphocytosis (>3 × 109 lymphocytes per liter of blood) was evident in 10 of 28 patients up to 8 years before CLL diagnosis, suggesting undiagnosed instances of HC MBL or asymptomatic CLL. In contrast, next-generation sequencing results showed detectable skewing of the IGH gene repertoire in 21 of 28 patients up to 15 years before CLL diagnosis, often in the absence of elevated lymphocyte counts (Figure 1E-F). Selection mechanisms may operate long before lymphocyte counts become abnormal, leading to restriction of the IGH gene repertoire and clonal expansion. Remarkably, some patients with CLL requiring treatment and clinical transformation to an aggressive B-cell lymphoma displayed considerable skewing in the IGH gene repertoire as early as 16 years before CLL diagnosis (Figure 1G-H).
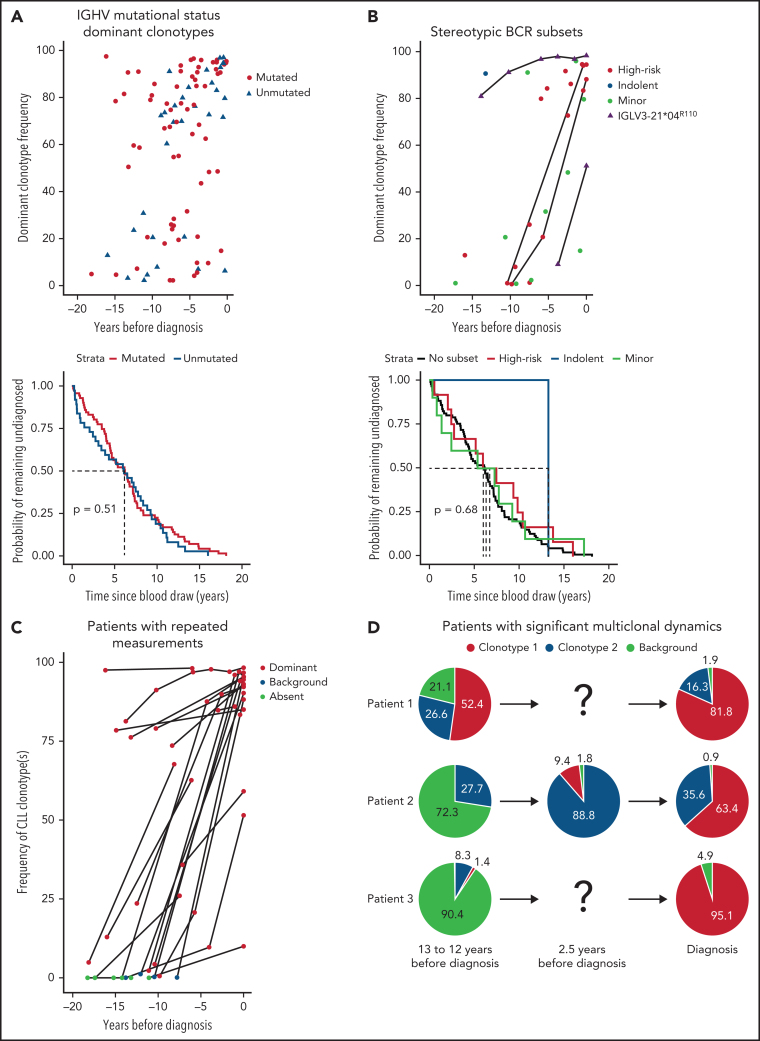
We determined the IGHV mutational status for all 100 future patients with CLL with a dominant clonotype frequency >2% (Figure 2A). We observed 68 mutated and 32 unmutated dominant clonotypes, in accordance with previous reports of an IGHV mutational status ratio of 2:1 in MBL.5 Regardless of the association of U-CLL with a poorer prognosis, there was no significant difference in the time to diagnosis between M-CLL and U-CLL. However, patients with a prediagnostic IGHV-unmutated dominant clonotype had a significantly shorter overall survival after CLL diagnosis compared with patients with an IGHV-mutated clonotype (supplemental Figure 2). Furthermore, at early time points (>10 years before diagnosis), patients with a high dominant clonotype frequency were more likely to be IGHV mutated, whereas this tendency was lost closer to diagnosis (Figure 2A), indicating that the prediagnostic phase may be even longer than 16 years for patients with M-CLL.
Figure 2.
Characterization of prediagnostic and diagnostic samples. (A) IGHV mutational status determination of all samples from patients with future CLL with a dominant clonotype frequency >2%. If no clonotype > 2% dominant clonotype frequency was present in the earliest sample of a patient, dominant clonotypes from any later measurements were used. Mutated clonotypes are shown as red circles, whereas unmutated clonotypes (IGHV mutational status >98% germline sequence identity) are shown as blue triangles (upper panel). A Kaplan-Meier (survival) curve for time until CLL diagnosis stratified by IGHV mutational status, indicating that the log-rank test does not find any significant difference (lower panel). (B) Overview of all CLL subsets identified in the data; major subsets were divided by subsets associated with aggressive or indolent disease course. Minor subsets are shown separately in light green. Samples in which the K16 and YDSD motifs and the R110 mutation of light chain subset 2L were confirmed are shown as purple triangles. Patients with repeated samples are connected by a black line (upper panel). A Kaplan-Meier (survival) curve for the time to CLL diagnosis from prediagnostic sample collection stratified by BcR stereotyped CLL subsets, indicating that the log-rank test does not find any significant difference (lower panel). (C) Cumulative frequency of clonotypes detected at CLL diagnosis for the prediagnostic sample(s) and diagnostic sample. Cumulative indicates that if >1 clonotype was shared in the prediagnostic sample and diagnostic sample [eg, patient 1 and 2 in panel D], their frequency was summed up. For patients for whom no diagnostic sample was available, the most skewed clonotype in the sample closest to diagnosis was traced instead. (D) The distribution of the BcR IGH gene repertoire of 3 patients with multiple clonotypes that underwent significant shifts over time to diagnosis. Dominant clonotype at CLL diagnosis is shown in red for each sample, with any secondary clonotype at diagnosis shown in blue. All small unrelated clonotypes (frequency <5%) were summed up and shown as background in light green. For more information on all patients with diagnostic material, see supplemental Tables 1 and 3.
Next, we analyzed the IGH VDJ gene rearrangements of the dominant clonotypes of all participants for BcR IG stereotypy. Twenty-five patients were found to carry stereotyped BcR IG up to 17 years prior to CLL diagnosis (Figure 2B; supplemental Table 4). Of these, 10 clonotypes were assigned to minor CLL subsets, and 15 were assigned to major CLL subsets; among the latter, 14 cases belonged to high-risk subsets.12, 15 In most of these cases, a trend for faster disease evolution was observed: high frequency of the dominant clonotype was evident in samples obtained <6 years before diagnosis. High-risk stereotyped clonotypes found longer before diagnosis (as early as 16 years before diagnosis) tended to have a lower dominant clonotype frequency (<20% of IGH gene repertoire). The stereotyped BcR IG matched the clonotype at diagnosis for both patients with diagnostic material. No stereotyped subsets were identified among the dominant clonotypes of the healthy controls.
To investigate the temporal dynamics of the IGH gene repertoire, we sequenced an additional 35 longitudinal samples from 22 patients, of whom 16 had an available diagnostic sample. For 21 of 22 patients, the frequency of the dominant clonotype increased strongly over time or remained stable at high frequency (P < .0001; Figure 2C). For 14 of 16 patients, the CLL BcR IG clonotype was already present at the earliest time point: either as the dominant clonotype (10/14) or in the background (4/14) (Figure 2C; supplemental Table 5). Three patients presented with significant multiclonal dynamics, displaying clonal competition and biclonality (Figure 2D) that were reported previously in LC MBL.16, 17 Interestingly, the repeated samples of patient 2 also captured the emergence of a secondary clonotype in a monoclonal proliferation, which then expanded to become the dominant clonotype at CLL diagnosis. To our knowledge, the dynamics of the emergence of biclonality in a patient with MBL and subsequent progression to CLL have never been captured in such a convincing manner. Furthermore, 2 patients, expressing a stereotyped IGHV3-21 gene (#2) and the highly similar IGHV3-48 gene carried IGLV3-21 light chain rearrangements with a characteristic mutation (R110) in the J-C linker region (Figure 2B), which was reported previously as being critical for cell autonomous signaling capacity in CLL subset #2 and related immunogenetic subgroups, including the IGHV3-48–expressing stereotyped subset #169. This mutation was present in all analyzed samples from both cases, indicating that a particular signaling behavior is already established from early stages in the genesis of a CLL clone.
Our findings extend current knowledge on the evolution of the IGH repertoire prior to CLL diagnosis, highlighting that even high-risk CLL subtypes may display a prolonged indolent preclinical stage. We speculate that somatic genetic aberrations, (auto)stimulation, and epigenetic and/or microenvironmental influences are required for the transformation into overt CLL.18, 19, 20, 21, 22, 23, 24 Because the observed skewing in the IGH gene repertoire often occurs prior to B-cell lymphocytosis, we consider our findings a novel extension of the characterization of MBL. Further studies may prove invaluable in the clinical distinction between “progressing” MBL vs “stable” MBL.6, 25 Notwithstanding the above, we emphasize that early detection is only warranted if it provides clear benefits to patient care. Early therapy, in the absence of symptoms, is not indicated in patients with CLL because it does not confer a survival benefit.26 Therefore, screening through BCR repertoire sequencing for the early detection of MBL/CLL in relatives of patients with CLL or the general population is not indicated.
In conclusion, we detected significant skewing in the IGH gene repertoire at the early steps of the natural history of CLL, supporting its role as an early event in CLL pathogenesis. Notably, our study shows that U-CLL clonotypes and clonotypes belonging to high-risk CLL stereotyped subsets can be present as early as 16 years before CLL diagnosis.
Acknowledgments
The authors thank Michèle van der Klift, Kim Heezen, Francois Kavelaars, Melissa Rijken, Nikos Darzentas, Pia Ostermann, Lotta von Sydow and Anders Dahlin for contributions to the project. They also thank Kostas Stamatopoulos for critical reading of the manuscript and Jorn Assmann and Benjamin Schrijver for stimulating academic discussions. In particular, the authors thank all participants in the initial EPIC study for providing the samples that enabled their research.
The work was supported by TRANSCAN/Dutch Cancer Society grant (179; NOVEL Consortium); Directorate General for Health and Consumer Protection of the European Commission (DG-SANCO); International Agency for Research on Cancer, Danish Cancer Society (Denmark); Ligue Natinale Contre le Cancer; Institut Gustave Roussy; Mutuelle Generale de l'Education Nationale, INSERM (France); German Cancer Aid; German Cancer Research Center; Federal Ministry of Education and Research; German Institute of Human Nutrition Potsdam-Rehbruecke, Nuthetal (Germany); the Hellenic Health Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro (AIRC)Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports; Netherlands Cancer Registry; LK Research Funds; Dutch Prevention Funds; Dutch Zorg Onderzoek Nederland; World Cancer Research Fund; Statistics Netherlands (The Netherlands), grant ERC2009-AdG 232997; Nordforsk; Nordic Centre of Excellence Programme on Food, Nutrition and Health (Norway); German Federal Ministry of Education and Research (BMBF 01EO1303); Centro de Investigación Biomédica en Red: Epidemiologíay Pública (Spain); Spanish Ministry of Economy and Competitiveness -Carlos III Institute of Health cofunded by the European Regional Development Fund - a way to build Europe, grants PI13/00061 (to Granada), PI13/01162 (to EPIC-Murcia, Regional Governments of Andalucıa, Asturias, Basque Country, Murcia, and Navarra), and PI17/01280 and PI14/01219 (to Barcelona); Agència de Gestió d'Ajuts Universitaris i de Recerca; Centres de Recerca de Catalunya (CERCA) Programme/Generalitat de Catalunya for institutional support, grant 2017SGR1085; Swedish Cancer Society; Swedish Research Council and County Councils of Skåne and Vasterbotten (Sweden); Cancer Research UK, grants 14136 (to EPIC-Norfolk) and C570/A11692, C570/A16491, and C8221/A19170 (to EPIC-Oxford); Medical Research Council, grants 1000143 (to EPIC-Norfolk) and MR/M012190/1 (to EPIC-Oxford, United Kingdom); and the Hellenic Foundation for Research and Innovation and the General Secretariat for Research and Technology under grant agreement 336 (Project CLLon).
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
Footnotes
All next-generation sequencing data reported in this article have been deposited in the Gene Expression Omnibus (accession number GSE178208). Access will be become public upon publication of the manuscript.
Data sharing requests should be sent to Anton W. Langerak (a.langerak@erasmusmc.nl).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Kolijn and colleagues provide insights into the earliest phases of emergence of a B-cell clone destined to become monocloncal B-cell lymphocytosis (MBL) and later chronic lymphocytic leukemia (CLL). Through analyses of the B-cell receptor immunoglobulin heavy-chain gene repertoire in serial blood samples collected in a longitudinal study of healthy recruits, they identify clonal skewing up to 16 years before diagnosis of CLL, including in patients later diagnosed with unfavorable-risk disease. While this study extends our insight into the disease's natural history, it has no prognostic impact or implications for clinical assessment.
Authorship
Contribution: P.M.K. and M.S. performed the experiments, P.M.K. analyzed the data, P.M.K., M.S., F.S.H., F.S., M.H., P.J.H., D.C., Y.B., A. Agodo, C.B., A.N., J.D.M., A. Aga., R.C.H.V. and A.W.L. interpreted results; A. Aga. validated CLL subsets; F.S.H., F.S. and M.H. facilitated acquisition of patient material and data; D.C., Y.B., A.B., C.S., M.-D.C., M.S., M.J., M.B.S., G.M., S.G., P.E., C.B. and A.N. collected and managed patient material and data; R.C.H.V. and A.W.L. designed and supervised the study; P.M.K., F.S.H., F.S., R.C.H.V., and A.W.L. wrote the manuscript; and M.H., P.J.H, D.C., Y.B., A. Agudo, M.-J.S., A.B., C.S., M.-D.C., M.S., M.J., M.B.S., G.M., S.G., P.E., C.B., A.N., A. Aga., and J.D.M. critically reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Supplementary Material
REFERENCES
- 1.Hallek M, Cheson BD, Catovsky D, et al. International Workshop on Chronic Lymphocytic Leukemia Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hallek M. Chronic lymphocytic leukemia: 2020 update on diagnosis, risk stratification and treatment. Am J Hematol. 2019;94(11):1266–1287. doi: 10.1002/ajh.25595. [DOI] [PubMed] [Google Scholar]
- 3.Landgren O, Albitar M, Ma W, et al. B-cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360(7):659–667. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strati P, Shanafelt TD. Monoclonal B-cell lymphocytosis and early-stage chronic lymphocytic leukemia: diagnosis, natural history, and risk stratification. Blood. 2015;126(4):454–462. doi: 10.1182/blood-2015-02-585059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henriques A, Rodríguez-Caballero A, Nieto WG, et al. Combined patterns of IGHV repertoire and cytogenetic/molecular alterations in monoclonal B lymphocytosis versus chronic lymphocytic leukemia. PLoS One. 2013;8(7):e67751. doi: 10.1371/journal.pone.0067751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghia P, Caligaris-Cappio F. Monoclonal B-cell lymphocytosis: right track or red herring? Blood. 2012;119(19):4358–4362. doi: 10.1182/blood-2012-01-404681. [DOI] [PubMed] [Google Scholar]
- 7.Slager SL, Lanasa MC, Marti GE, et al. Natural history of monoclonal B-cell lymphocytosis among relatives in CLL families. Blood. 2021;137(15):2046–2056. doi: 10.1182/blood.2020006322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nieto WG, Almeida J, Romero A, et al. Primary Health Care Group of Salamanca for the Study of MBL Increased frequency (12%) of circulating chronic lymphocytic leukemia-like B-cell clones in healthy subjects using a highly sensitive multicolor flow cytometry approach. Blood. 2009;114(1):33–37. doi: 10.1182/blood-2009-01-197368. [DOI] [PubMed] [Google Scholar]
- 9.Rawstron AC, Bennett FL, O'Connor SJ, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359(6):575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 10.Davi F, Langerak AW, de Septenville AL, et al. ERIC, the European Research Initiative on CLL, and the EuroClonality-NGS Working Group Immunoglobulin gene analysis in chronic lymphocytic leukemia in the era of next generation sequencing. Leukemia. 2020;34(10):2545–2551. doi: 10.1038/s41375-020-0923-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parikh SA, Strati P, Tsang M, West CP, Shanafelt TD. Should IGHV status and FISH testing be performed in all CLL patients at diagnosis? A systematic review and meta-analysis. Blood. 2016;127(14):1752–1760. doi: 10.1182/blood-2015-10-620864. [DOI] [PubMed] [Google Scholar]
- 12.Agathangelidis A, Chatzidimitriou A, Gemenetzi K, et al. Higher-order connections between stereotyped subsets: implications for improved patient classification in CLL. Blood. 2021;137(10):1365–1376. doi: 10.1182/blood.2020007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradbury KE, Appleby PN, Key TJ. Fruit, vegetable, and fiber intake in relation to cancer risk: findings from the European Prospective Investigation into Cancer and Nutrition (EPIC) Am J Clin Nutr. 2014;100:394S–398S. doi: 10.3945/ajcn.113.071357. suppl 1. [DOI] [PubMed] [Google Scholar]
- 14.Bystry V, Reigl T, Krejci A, et al. EuroClonality-NGS ARResT/Interrogate: an interactive immunoprofiler for IG/TR NGS data. Bioinformatics. 2017;33(3):435–437. doi: 10.1093/bioinformatics/btw634. [DOI] [PubMed] [Google Scholar]
- 15.Gounari M, Ntoufa S, Apollonio B, et al. Excessive antigen reactivity may underlie the clinical aggressiveness of chronic lymphocytic leukemia stereotyped subset #8. Blood. 2015;125(23):3580–3587. doi: 10.1182/blood-2014-09-603217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agathangelidis A, Galigalidou C, Scarfò L, et al. Infrequent “chronic lymphocytic leukemia-specific” immunoglobulin stereotypes in aged individuals with or without low-count monoclonal B-cell lymphocytosis. Haematologica. 2021;106(4):1178–1181. doi: 10.3324/haematol.2020.247908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanasa MC, Allgood SD, Volkheimer AD, et al. Single-cell analysis reveals oligoclonality among ‘low-count' monoclonal B-cell lymphocytosis. Leukemia. 2010;24(1):133–140. doi: 10.1038/leu.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dühren-von Minden M, Übelhart R, Schneider D, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489(7415):309–312. doi: 10.1038/nature11309. [DOI] [PubMed] [Google Scholar]
- 19.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maity PC, Bilal M, Koning MT, et al. IGLV3-21*01 is an inherited risk factor for CLL through the acquisition of a single-point mutation enabling autonomous BCR signaling. Proc Natl Acad Sci USA. 2020;117(8):4320–4327. doi: 10.1073/pnas.1913810117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roessner PM, Seiffert M. T-cells in chronic lymphocytic leukemia: guardians or drivers of disease? Leukemia. 2020;34(8):2012–2024. doi: 10.1038/s41375-020-0873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerhan JR, Slager SL. Familial predisposition and genetic risk factors for lymphoma. Blood. 2015;126(20):2265–2273. doi: 10.1182/blood-2015-04-537498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YCB, Kipling D, Dunn-Walters DK. Age-related changes in human peripheral blood IGH repertoire following vaccination. Front Immunol. 2012;3:193. doi: 10.3389/fimmu.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart A, Ng JCF, Wallis G, Tsioligka V, Fraternali F, Dunn-Walters DK. Single-cell transcriptomic analyses define distinct peripheral B cell subsets and discrete development pathways. Front Immunol. 2021;12:602539. doi: 10.3389/fimmu.2021.602539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunn-Walters DK. The ageing human B cell repertoire: a failure of selection? Clin Exp Immunol. 2016;183(1):50–56. doi: 10.1111/cei.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131(25):2745–2760. doi: 10.1182/blood-2017-09-806398. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.