Abstract
Mammalian centromeres direct faithful genetic inheritance and are typically characterized by regions of highly repetitive and rapidly evolving DNA. We focused on a mouse species, Mus pahari, that we found has evolved to house centromere-specifying centromere protein-A (CENP-A) nucleosomes at the nexus of a satellite repeat that we identified and termed π-satellite (π-sat), a small number of recruitment sites for CENP-B, and short stretches of perfect telomere repeats. One M. pahari chromosome, however, houses a radically divergent centromere harboring ~6 mega–base pairs of a homogenized π-sat–related repeat, π-satB, that contains >20,000 functional CENP-B boxes. There, CENP-B abundance promotes accumulation of microtubule-binding components of the kinetochore and a microtubule-destabilizing kinesin of the inner centromere. We propose that the balance of pro- and anti-microtubule binding by the new centromere is what permits it to segregate during cell division with high fidelity alongside the older ones whose sequence creates a markedly different molecular composition.
Chromatin and kinetochore alterations arise in response to evolutionarily rapid changes to underlying repetitive centromere DNA.
INTRODUCTION
Centromeres are the loci that coordinate chromosome segregation during cell division (1). They do so by assembling a proteinaceous structure, the kinetochore, at cell division that attaches to spindle microtubules, housing the chromatin that regulates microtubule attachment to ensure error-free segregation, and serving as the final site of sister chromatid cohesion. In many species, including mammals, the site for all of these functions is epigenetically specified by the presence of nucleosomes harboring the histone H3 variant, CENP-A.
Despite generally shared and essential functional roles, there is marked diversity in the DNA sequences and molecular composition of centromeres between different eukaryotic species. Many, but certainly not all, eukaryotic centromeres have highly repetitive DNA sequences. In some species, the repeats are common across each of the chromosomes [e.g., many mammalian species, including humans (2–4)], whereas in others, the repeats are divergent between different chromosomes (e.g., in the fruit fly) (5). In some eukaryotes, including primates, a high degree of homogenization between DNA repeats is interpreted as evidence of active evolution, while not supplying any direct information about the timing of the emergence of a particular sequence (4, 6–8). In some species, such as Mus musculus, the centromere repeats are especially highly repetitive and homogeneous (9) to the point that assembling beyond portions of the centromere (10) is difficult to imagine accomplishing even when using current long-read sequencing methodologies that have succeeded in the full genome assemblies of human (and those of other species) centromeres (4).
Centromere formation can influence evolution by allowing some centromeres to be preferentially inherited during female meiosis by biasing segregation outcomes in a process called “centromere drive” or, more generally, “meiotic drive” (11, 12). A prime example from plants is in monkeyflowers, where a driving centromere element can bias transmission during reproduction but comes at the cost of reducing pollen production (13). Centromeres that direct biased segregation to the egg are referred to as “stronger” centromeres. Among other factors, expanding the region of DNA housing CENP-A nucleosomes can strengthen the centromere (9). Female meiotic drive is thought to be the major driver of rapid evolution of centromeric DNA (14).
One powerful model system for assessing the molecular basis for female meiotic drive is the mouse (15). Prior work has demonstrated that major differences in the abundance of repetitive centromere DNA between inbred laboratory strains or species lead to differences in which chromosomes are more likely to be inherited through meiosis (15). While centromere DNA sequence and architecture differ between mouse species, in each of the reported cases, centromere DNA differences between chromosomes within a strain or species are thought to be negligible (e.g., every Mus spretus chromosome has a nearly identical repeat at each centromere at a similar abundance) (9, 16, 17). Of course, centromeres are present on separate chromosomes, implying that DNA sequence–based differences between centromeres are homogenized across the genome through some undefined selective pressure to do so. More precisely, individual chromosomes are physically unlinked and subject to the independent accrual of new mutations. Nonallelic homologous repair processes can homogenize centromeres from different chromosomes, erasing signals of chromosome-level centromere divergence (18, 19). Such mechanisms have likely been particularly active on acrocentric mouse chromosome, where centromeres colocalize at the nuclear periphery during meiosis onset, before the completion of double-stranded DNA (dsDNA) break repair (20). Nonetheless, the rapid evolution of centromeric DNA suggests that genomes with heterologous centromere composition are potentially pervasive, even if only transiently manifest in mouse genomes.
Thus, evolutionary intermediates must have existed before homogenization, and the molecular consequences remain unclear of having divergent centromeric DNA within a single mouse strain and/or species. In other eukaryotes, there are examples of different centromeres within a species, but it is unclear how they relate to the mouse model for strengthening through modulation of DNA repeat number or sequence due to major differences at the centromere. For instance, plant neocentromeres, like a famous example in maize (21–23), can function not through an actual centromere/kinetochore but by directing independent movements through tethering a specialized motor protein to the spindle. In mammals, evolutionarily young centromeres have been found on up to half of the centromeres of individual equine species (24). Furthermore, many are present (albeit on smaller numbers of chromosomes) in several other vertebrate systems (7, 25–27). In all these documented cases, the young centromeres consist of nonrepetitive DNA. Given the recent successful studies using the mouse model system to reveal the role of centromere strength in centromere evolution (9, 17, 28), advances in mice on isolating and studying new radical changes in repetitive centromere DNA are likely to have important implications for advancing models of centromere evolution in diverse eukaryotic species.
CENP-B is the only known sequence-specific DNA binding protein found at many eukaryotic centromeres, including at the centromeres in diverse mammalian species. It recognizes a conserved 17-mer sequence termed the CENP-B box, in which nine positions are essential for CENP-B binding (29, 30). The CENP-B box is found within the sequences of the centromere repeat monomers (i.e., within the 171-bp α-satellite repeat in Homo sapiens and within the 120-bp repeat in the minor satellite in M. musculus) (31, 32). In another mouse species, Mus caroli, the functional centromere appears assembled on an array consisting of a 79-bp repeat that harbors a functional CENP-B box (table S1) (33). While not essential for centromere function [indeed, CENP-B boxes are absent on the Y chromosome in humans and mice (34)], CENP-B can buffer against other molecular insults and is a prime candidate to play a role in modulating centromere strength (28, 34, 35). CENP-B serves to support the pericentromeric enrichment of constitutive heterochromatin [i.e., chromatin enriched with nucleosomes marked with histone H3 lysine 9 trimethylation (H3K9me3)] that, in turn, enhances the recruitment of inner centromere components involved in sister chromatid cohesion and the process of mitotic error correction (28, 36–38). CENP-B, likewise, enhances kinetochore formation through its ability to bind an essential centromere protein, CENP-C (34). Removal of CENP-B enhances functional differences in female meiosis between diverged strains of M. musculus that have approximately 10-fold differences in minor satellite abundance relative to one another (28). Thus, there is a strong support for the notion that CENP-B can play a key role in modulating centromere strength.
Here, we find that CENP-B is dispensable for CENP-A nucleosome positioning on minor satellite DNA, suggesting that its roles are likely limited to strengthening the centromere by other proposed means that rely on the amount of CENP-B at the centromere. We then identify a single chromosome in the mouse species Mus pahari that has a massive expansion of a newly evolved repeat array that houses >20,000 functional CENP-B boxes: ~100-fold more than on the other M. pahari centromeres. Using a comprehensive set of short- and long-read sequencing–based methodologies, we define this centromere and the more typical centromeres in M. pahari. The latter accumulate kinetochore-forming CENP-A chromatin at a subset of repeats that harbor a relatively small number (hundreds) of CENP-B boxes, as well as up to 68,000 telomere repeats. Together, our sequencing efforts predict a difference in the molecular composition of the two types of centromeres within a single organismal genome. We test this notion and determine how the opposing recruitment of microtubule-binding and microtubule-destabilizing factors coexist in the same mouse species.
RESULTS
Positioning of CENP-A nucleosomes on the minor satellite is independent of CENP-B
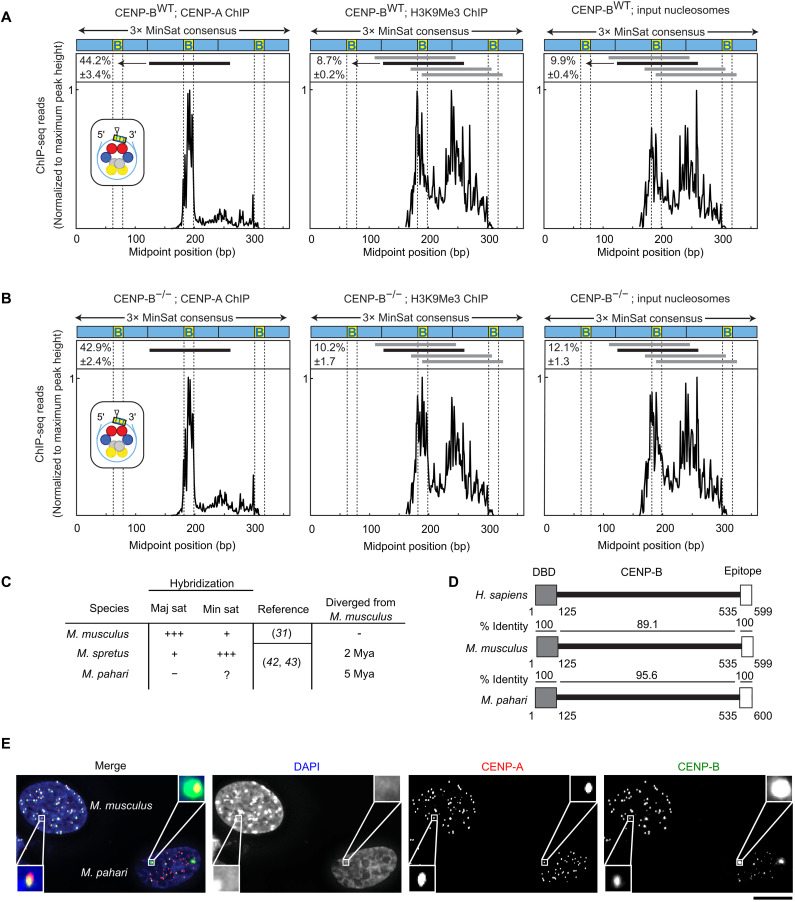
Earlier studies have revealed in mouse and human that centromere DNA sequence plays an important role in specific positioning of CENP-A nucleosome within the monomer repeats (9, 39, 40). In M. musculus, CENP-A nucleosomes are assembled on a single predominant site within the minor satellite repeat, with their centers (also known as the “nucleosomal dyad” position) within the CENP-B box, with flanking CENP-B boxes 120 bp on either side (9). For conventional nucleosomes, this nucleosome position is only one of several used in minor satellite repeats (9). In humans, where the spacing between CENP-B box position is 171 bp in α-satellite DNA, CENP-A also has a specific favored position (relative to the more heterogeneous positioning of conventional nucleosomes harboring histone H3), but it is located between CENP-B boxes (40). Furthermore, recent analysis of single-molecule chromatin fiber sequencing data suggests that the CENP-B occupancy may phase nucleosomes on α-satellite DNA (41). In considering M. musculus centromeres, we considered two possibilities. First, minor satellite DNA could directly affect the positioning of CENP-A nucleosomes independently of CENP-B. Second, alternatively, the CENP-B protein could affect CENP-A nucleosome positioning upon binding to the CENP-B box and through its direct and indirect interactions with the CENP-A nucleosomes. To distinguish between these possibilities, we enriched for nucleosomes containing either CENP-A or H3K9me3 via chromatin immunoprecipitation (ChIP) from chromatin isolated from wild-type (WT; C57BL/6J) or CENP-B−/− (C57BL/6J) mice. We found that positioning on the minor satellite of CENP-A nucleosomes, H3K9me3 nucleosomes, and the total pool of nucleosomes (input to the native ChIP) was essentially unchanged in the absence of CENP-B protein (Fig. 1, A and B). Thus, our data support the notion that minor satellite DNA sequence is uniquely responsible for positioning of CENP-A nucleosomes, independently of the presence of CENP-B protein. Our results suggest that CENP-B protein, the CENP-B box, and centromere satellite sequences are important for us to consider in contributing to centromere drive.
Fig. 1. CENP-B occupancy on centromere DNA does not affect CENP-A nucleosome phasing but does vary widely between and within mouse species.
(A) Midpoint position of CENP-A ChIP H3K9Me3 or input reads (size 100 to 160 bp) from WT M. musculus along the trimer minor satellite consensus sequence. Vertical lines indicate the 17-bp CENP-B box. The major CENP-A nucleosome position (identified in the CENP-A ChIP samples) is indicated by a horizontal black line above the respective midpoint values and schematized (inset) for CENP-A ChIP with a triangle representing the dyad position. The same nucleosome position is indicated in the H3K9Me3 and input samples. Numbers to the left of the positions indicate the percentage of reads (means ± SEM; n = 3 independent experiments) where the midpoint spans the 10 bp at the 3′ end of the CENP-B box (yellow, labeled B). Horizontal gray lines indicate other major nucleosome positions in the H3K9Me3 and input samples. (B) Midpoint position of CENP-A ChIP H3K9Me3 or input reads (size 100 to 160 bp) from CENP-B KO M. musculus along the trimer minor satellite consensus sequence. (C) Centromere satellites from M. musculus, M. spretus, and M. pahari. (D) CENP-B is highly conserved in mouse species, with 100% identical sequences in both the DNA binding domain and the epitope targeted by the CENP-B antibody used in our study. (E) Immunofluorescence of CENP-A and CENP-B from lung fibroblast cells derived from M. musculus [with their nuclei identified by strong 4′,6-diamidino-2-phenylindole (DAPI)–staining pericentromeres] or M. pahari. Scale bar, 10 μm.
Rapid centromere DNA repeat evolution affects the amount of CENP-B at centromeres
We next considered the conservation of CENP-B boxes in other Mus species. Early hybridization studies indicate that closely related house mouse species are undergoing evolutionarily rapid changes in centromere DNA sequence (Fig. 1C) (31, 42, 43). This divergence can include the number of CENP-B boxes and/or the sequence of the repeat itself (44). One way to alter CENP-B box number is to vary the abundance of homogeneous centromere repeats. For instance, in M. spretus, the minor satellite is the most abundant centromere satellite, and the major satellite is much less abundant (31), the opposite of what is found in M. musculus. Changes also include apparent drastic alterations in DNA sequence, as in M. pahari where the major satellite is undetectable (42, 43).
Two initial observations suggested that investigating the centromere diversity in M. pahari could yield insights into the mechanism governing centromere strength. First, the M. pahari genome encodes the CENP-B protein, which is almost identical to its counterpart in M. musculus and 100% identical in its DNA binding domain (Fig. 1D). Such high species–level protein conservation is highly unlikely to persist over evolutionary time in the absence of purifying selection to retain CENP-B function. Thus, we anticipated that M. pahari centromeres would contain repeats—minor satellite DNA or other divergent repetitive centromere DNA—that harbor functional CENP-B box sequences capable of CENP-B binding. Second, we found that while most M. pahari centromeres have low (relative to those from M. musculus cells co-seeded for immunofluorescence measurements) yet detectable levels of CENP-B, a pair of very strong foci of CENP-B are present (Fig. 1E). We concluded that the pair of foci likely represents a single pair of homologous chromosomes. Thus, our initial observations suggested that in M. pahari, major changes exist in centromere DNA both relative to M. musculus and between different M. pahari chromosomes, and that affects the CENP-B abundance at the centromeres.
π-sat, a divergent centromere satellite, is identified
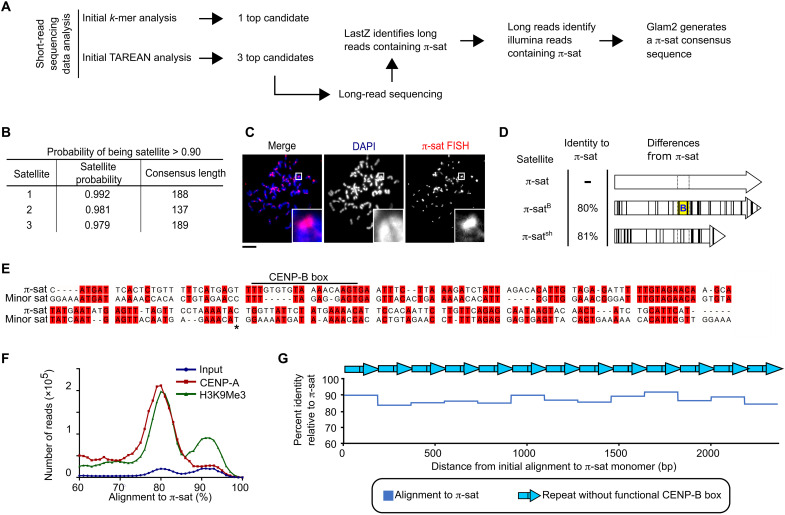
Since no centromere satellite has been identified in M. pahari, we used several strategies to identify candidate centromere repeats (Fig. 2A). The first strategy was a k-mer–based approach using an existing short-read sequencing dataset (45). This yielded a top hit with a repeat unit length of 189 bp (fig. S1). The second strategy was an analysis of total nucleosomal DNA and CENP-A nucleosome–enriched (native ChIP) short-read data with the computational pipeline TAREAN (46) coupled to downstream analysis of native Oxford Nanopore Technologies (ONT) long-read sequencing we performed of the M. pahari genome (Fig. 2A; see Materials and Methods for details of the strategy we used). This produced a total of three sequences with a high likelihood of satellite DNA (Fig. 2B and Materials and Methods). Of the three sequences, the 189-bp satellite, which we term π-satellite (π-sat), is nearly identical to the top hit identified by the k-mer strategy (fig. S1). Consistent with our hypothesis that is a centromere repeat, π-sat hybridizes to a single locus on each chromosome in a chromosome spread of mitotic M. pahari cells (Fig. 2C). However, the π-sat sequence lacks an intact CENP-B box (Fig. 2D). The two remaining repeats we identified are related to π-sat: One (π-satsh) is ~50-bp shorter, whereas the other (π-satB) contains an intact CENP-B box (Fig. 2D).
Fig. 2. Identification of the most abundant form of centromere repeats in M. pahari: π-sat.
(A) Two approaches to identify M. pahari centromeric repeats. (B) Satellite sequences derived from TAREAN analysis on input sequencing data. Satellite probability was calculated as described in Materials and Methods. (C) Representative image of M. pahari centromeric DNA labeled with FISH probe using consensus sequence derived from k-mer approach. Insets: 7.9× magnification. Scale bar, 10 μm. (D) Schematized representation of the three satellites identified by TAREAN analysis. Differences between each satellite compared to π-sat are marked with black lines. The location corresponding to the CENP-B box position is indicated with dotted lines, and the functional CENP-B box from π-satB is indicated with yellow background and a blue “B.” (E) Alignment of the π-sat consensus sequence to minor sat consensus sequence. A dimer of π-sat was aligned to a trimer of the minor satellite, and the first monomer of π-sat is shown. The end of the first monomer of the minor satellite is marked with an asterisk. (F) Histograms show distribution of reads from input, CENP-A ChIP, or H3K9Me3 ChIP aligning to π-sat. (G) Representative example of a π-sat containing ONT long read that was divided into monomers. The percent identity of each monomer to π-sat is plotted.
We noted that none of the three π-sats we identified were closely related to the major satellite from M. musculus, explaining why early hybridization studies failed to identify the major satellite in M. pahari (42, 43). The minor satellite similarly has only small regions of identity with π-sat, and the region of π-sat aligning to the CENP-B box has several substitutions (Fig. 2E). Alignment of enriched sequences from CENP-A (functional centromere) and H3K9me3 (enriched in pericentromeric heterochromatin) native ChIP with π-sat yielded strong peaks of high sequence identity (Fig. 2F). Furthermore, we noted that many of the long reads that align to π-sat consisted of homogeneous stretches where π-sat contained no intervening sequences, including any π-satsh or π-satB (Fig. 2G). Together with the fluorescence in situ hybridization (FISH) and native ChIP data, these experiments suggest that most or all M. pahari centromeres harbor long and uninterrupted stretches of π-sat repeats that lack functional CENP-B boxes.
A chromosome pair harbors highly homogenized π-satB
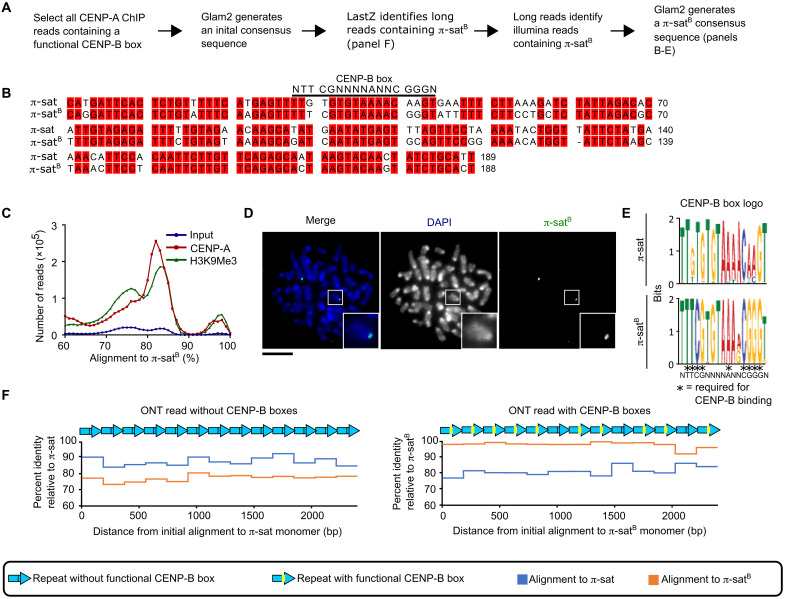
To gain an understanding of the centromere sequences that harbor functional CENP-B boxes, we used another strategy (Fig. 3A), starting with native ONT long reads that harbor functional CENP-B box sequences. This approach yielded a refined centromere consensus sequence (Fig. 3B) that corresponded to what we had initially identified as π-satB (Fig. 2). CENP-A and H3K9me3 native ChIP reads contained many sequences that align well to the π-satB consensus sequence (Fig. 3C). Peaks around 83 to 86% sequence identity likely correspond to alignments with general π-sat, while a peak around 94 to 96% sequence identity likely represents π-satB sequences. We designed a FISH probe using the π-satB consensus sequence and found that it hybridized to a pair of mitotic chromosomes in M. pahari cells (Fig. 3D). Furthermore, in interphase M. pahari cells, the π-satB probe colocalized with a probe specific to the CENP-B box (fig. S2). This supported our prior conclusion that the two nuclear puncta with high amounts of CENP-B (Fig. 1E) correspond to a single pair of homologous chromosomes. Alignment of sequences found on long reads containing either π-sat or π-satB showed that π-satB has near invariance at the CENP-B box positions that are required for CENP-B binding, including at the positions that diverge from the π-sat consensus (Fig. 3E). Most ONT long reads containing centromere repeats were homogeneous stretches that align more closely to π-sat and were devoid of CENP-B boxes. On the other hand, a smaller proportion contain centromere repeats that, while also comprising homogeneous stretches, contain many functional CENP-B boxes and align more closely to π-satB (Fig. 3F). Our findings indicate that a homologous pair of chromosomes that bind high levels of CENP-B harbors a large and highly homogeneous derivative of the satellite present on the other chromosomes.
Fig. 3. π-satB is highly homogeneous, restricted to a single pair of chromosomes, and present in long, contiguous blocks that lack generic π-sat.
(A) Approach to identify CENP-B box containing satellite. (B) Alignment of π-sat and π-satB. (C) Histograms show distribution of reads from input, CENP-A ChIP, or H3K9Me3 ChIP aligning to π-satB. (D) Representative image of M. pahari centromeric DNA labeled with FISH probe using π-satB consensus sequence. Insets: 2.5× magnification. Scale bar, 10 μm. (E) Logo representation of the CENP-B box consensus of π-sat and π-satB. (F) Plots of the percent identity of satellites along a portion of representative ONT reads with (right) and without (left) CENP-B boxes to the π-sat and π-satB consensus sequences.
Sequences of M. pahari centromeres are assembled with high accuracy
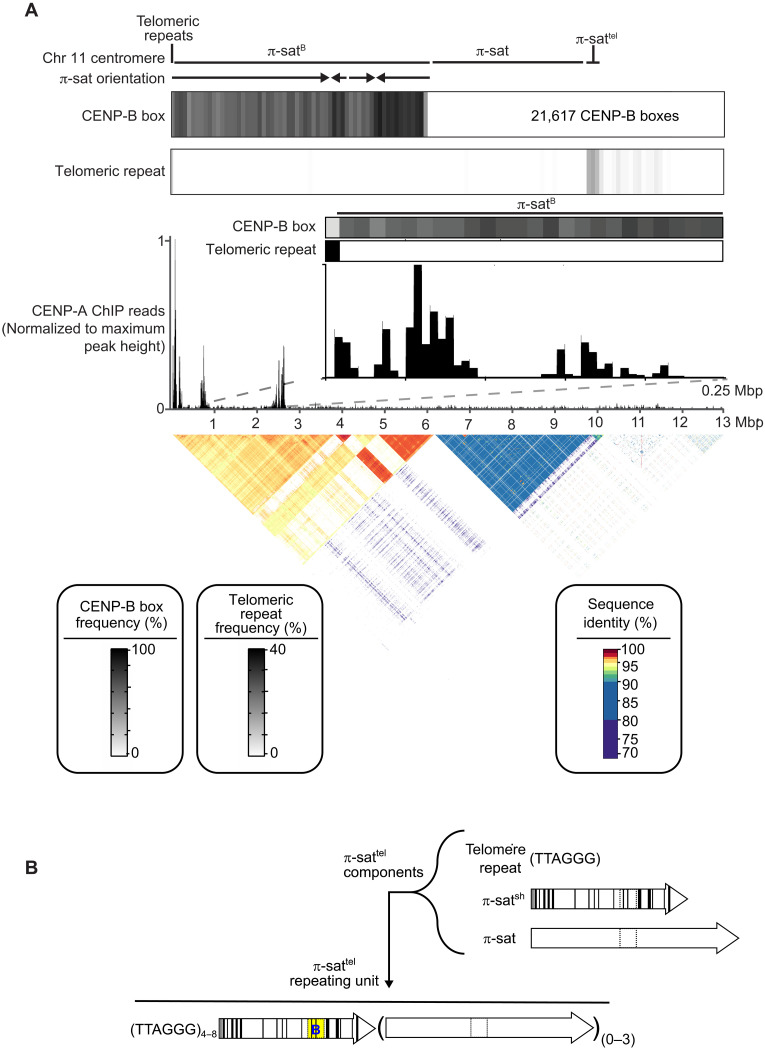
To identify the chromosome with high amounts of CENP-B, as well as to more broadly understand centromere structure in M. pahari, we set out to generate centromere sequence assemblies from several M. pahari chromosomes. While murine centromeres have long been assumed to be relatively intractable to sequence assembly due to high repeat homogeneity and apparent lack of higher-order repeat patterns (e.g., this is true of the best-known murine centromere repeat for centromere function in cell division, minor satellite from M. musculus), we were encouraged by two aspects. The first was the success of Pacific Biosciences high-fidelity (PacBio HiFi) long-read sequencing in assembling human centromeres with high accuracy (2–4). The second was our finding that π-sat is not as homogeneous as the minor satellite (Fig. 2G). Our initial focus for sequence assembly was of the chromosome containing a large array of π-satB (Fig. 4A). Therefore, we generated a 22-fold coverage of PacBio HiFi data from the M. pahari genome and assembled it with the whole-genome assembler hifiasm (47). This generated a whole-sequence assembly that was 4.54 giga–base pairs in length, consistent with its diploid nature, and containing a contiguous assembly from the telomere through the first 13 mega–base pairs (Mbp) of the chromosome containing arrays of π-satB. Aligning this contig to an adjacent contig was sufficient to extend to complex sequence that matches chromosome 11 from the initial genome build of M. pahari (45). This chromosome is telocentric, with no intervening sequence between perfect telomere repeats and centromeric repeats (Fig. 4A).
Fig. 4. Genomic assembly reveals the identity and nature of the centromere harboring π-satB.
(A) The composition of the centromere of chromosome 11. The assembly consists of, in order, 8 kb of telomeric repeats, 6 Mbp of π-satB, 3.6 Mb of π-sat, and 400 kb of π-sattel, followed by other repetitive elements. The total number of CENP-B boxes (21,617) on this centromere is denoted. The fraction of π-sat repeats containing a functional CENP-B box (NTTCGNNNNANNCGGGN) and the frequency of telomeric repeats (TTAGGG) are shown. CENP-A ChIP-seq reads were aligned to the chromosome 11 centromere assembly. A pairwise sequence identity heatmap indicates that the centromere consists of 6 Mbp of highly homogeneous π-satB. (B) Schematized representation of the three types of repeats that make up repeating units of π-sattel. The repeating unit of π-sattel consists of a variable number of telomere repeats, a single unit of π-satsh, and from zero to three repeats of π-sat. Functional CENP-B boxes are typically found on π-satsh. An example of a single unit of π-sattel is shown. While the overall makeup of π-sattel contains the listed components, the number of telomere repeats and the units of π-sat can vary in different centromeres and even within a single centromere.
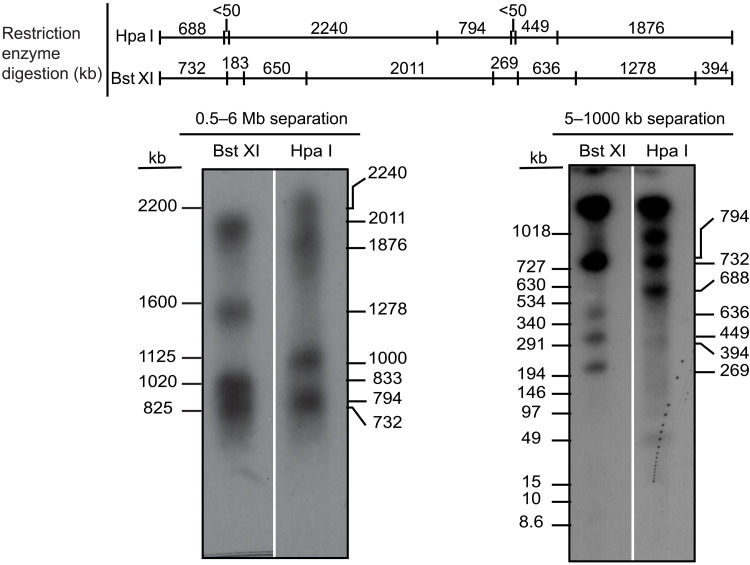
The first centromere repeats consist of a ~6-Mbp block of contiguous π-satB. The first 3.7 Mbp of this π-satB array includes monomers in exclusive head-to-tail orientation. The directionality of the head-to-tail repeats switches three times over the next 2.3 Mbp. In total, this 6-Mbp block houses 21,617 functional CENP-B boxes (Fig. 4A), explaining the massive enrichment of CENP-B on this chromosome (Fig. 1E). A ~4-Mbp contiguous stretch of π-sat lies distal to the π-satB arrays, followed by a shorter stretch of π-sat variant, π-sattel (Fig. 4A). π-sattel is a more complex composite repeat monomer composed of elements built from π-sat, π-satsh, and 2 to 16 telomere repeats (Fig. 4B). CENP-A association is not uniform across the chromosome 11 centromere, with enrichment localized to three sites: a site of enrichment adjacent to the telomere [0 to 250 kilo–base pairs (kbp)] and two more regions marked by peaks at ~750 kbp and 2.5 Mbp from the telomere, respectively (Fig. 4A). CENP-A peaks are only observed on π-satB but not on π-sat or π-sattel (Fig. 4A). Southern blots of M. pahari DNA digested with two restriction enzymes, Bst XI and Hpa I, and probed with π-satB almost perfectly match the pattern predicted by our assembly (Fig. 5). Two predicted bands (183 and 650 kb) for BxtXI digestion were not detected, but one at 833 kb was (Fig. 5). This minor difference is likely due to a sequence polymorphism between the animal used to generate the assembly versus the one used to harvest DNA for the blot. Thus, despite the high degree of sequence identity between repeat monomers and the lack of other unique sequences for a >6-Mbp span of π-satB, our approach with PacBio HiFi long-read sequencing downstream assembly strategy is extremely faithful.
Fig. 5. Restriction digest analysis confirms chromosome 11 assembly.
Schematic of predicted restriction digest sites of chromosome 11 with Bst XI and Hpa I. Pulsed-field gel Southern blot of Mus pahari DNA confirms the structure and organization of the chromosome 11 centromeric higher-order repeat (HOR) array. For each gel, left corresponds to ethidium bromide (EtBr) staining and right corresponds to 32P-labeled chromosome 11 π-satB–specific probe. The left gel was run at conditions to separate DNA from 0.6 to 5 Mb, and the right gel was run at conditions to separate DNA from 5 to 1000 kb.
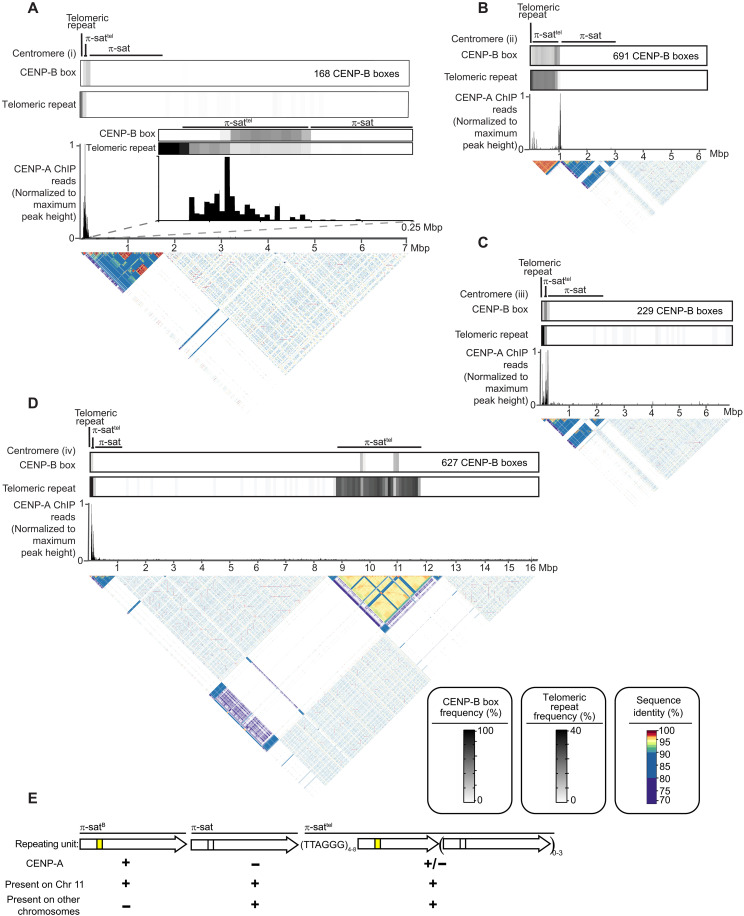
We successfully assembled seven other M. pahari centromeres (Fig. 6, A to D, and fig. S4). Note that all seven have unmapped regions between the centromere and the rest of the chromosome that preclude assignment to a particular M. pahari chromosome, so we have numbered them centromeres (i) to (vii). They vary in size and precise arrangement, are commonly telocentric, and house π-sattel between the telomere and a long stretch of π-sat (Fig. 6, A to D, and fig. S4). None contain π-satB (Fig. 6, A to D, and fig. S4). CENP-A peaks are almost entirely restricted to π-sattel, as are functional CENP-B boxes (Fig. 6, A to D, and fig. S4). The functional CENP-B boxes are almost exclusively confined to π-sattel repeats and vary in their sequence from those found on chromosome 11 in π-satB and are much less abundant (Fig. 6, A to D, and figs. S3 and S4). Most of the π-sat repeats harbor nonfunctional CENP-B boxes that do not match the consensus required for CENP-B binding (Fig. 6, A to D, and fig. S4). Thus, other assembled centromeres harbor 27 to 143 times fewer total functional CENP-B boxes than chromosome 11. For all centromeres that we assembled, the major site of CENP-A enrichment spans 100 to 300 kbp (Fig. 6, A to D, and fig. S4). As far as the role of the different specific forms of π-sat, general π-sat is the most abundant and represent a candidate pericentromeric satellite (analogous to major satellite DNA in M. musculus), while both π-sattel and π-satB are primary sites for kinetochore forming chromatin containing CENP-A nucleosomes (Fig. 6E). Compared to chromosome 11, the other centromeres contain π-sat wherein monomer units are less similar to one another (Fig. 3F). Thus, it appears that the highly homogeneous chromosome 11 centromere is evolutionarily more active. In total, our long-read analysis define the general sequence features of M. pahari centromeres, including the evolutionary young centromere on chromosome 11.
Fig. 6. Evolutionarily older Mus pahari centromeres harbor CENP-A nucleosomes near CENP-B boxes and π-sattel.
(A to D) The composition of a representative M. pahari centromere. Each of the assembly consists of, in order, an array of telomeric repeats, an array of π-sattel, and an array of π-sat followed by various repetitive elements. The fraction of π-sat repeats containing a functional CENP-B box (NTTCGNNNNANNCGGGN) and the frequency of telomeric repeats (TTAGGG) are shown. CENP-A ChIP-seq reads were aligned to the assembly revealing that CENP-A is primarily present on π-sattel. A pairwise sequence identity heatmap indicates the degree of homogeneity in centromeric DNA. (E) The types of repeating units found at M. pahari centromeres.
Chromosomes with markedly different abundance of centromere factors co-exist
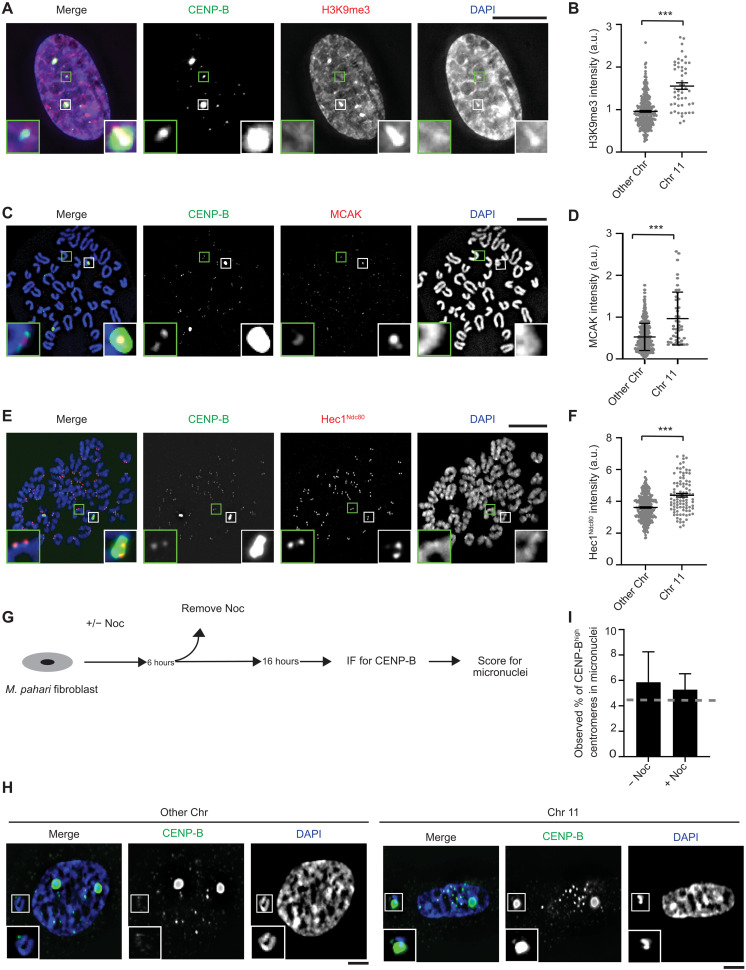
Chromosome 11 has a markedly different centromere repeat that leads to massive differences in CENP-B abundance (Fig. 4A). To test whether or not the large difference of CENP-B leads to higher levels of H3K9me3 accumulation, we performed quantitative immunofluorescence on interphase cells (Fig. 7, A and B). Chromosome 11 has 1.6-fold higher H3K9me3 relative to that measured at the centromeres of other chromosomes that have low yet detectable CENP-B levels (Fig. 7, A and B).
Fig. 7. Chromosome 11 harbors levels of both pro- and anti-microtubule–binding proteins that are higher than the other Mus pahari centromeres.
(A) Immunofluorescence of H3K9Me3 from lung fibroblast cells derived from M. pahari. Insets: 4.0× magnification. Scale bar, 10 μm. (B) Quantification corresponding to (A). The mean ratio (± SEM) is shown. n = 314 for the centromeres with low abundance of CENP-B and n = 50 for the centromeres with high abundance of CENP-B, pooled from two independent experiments (***P < 0.0001). a.u., arbitrary units. (C) Immunofluorescence of MCAK from lung fibroblast cells derived from M. pahari. Insets: 6.5× magnification. Scale bar, 10 μm. (D) Quantification corresponding to (C). The mean ratio (± SEM) is shown. n = 389 for the centromeres with low abundance of CENP-B and n = 45 for the centromeres with high abundance of CENP-B, pooled from two independent experiments (***P < 0.0001). (E) Immunofluorescence of Hec1Ndc80 from lung fibroblast cells derived from M. pahari. Insets: 5.1× magnification. Scale bar, 10 μm. (F) Quantification corresponding to (E). The mean ratio (± SEM) is shown. n = 324 for the centromeres with low abundance of CENP-B and n = 94 for the centromeres with high abundance of CENP-B, pooled from three independent experiments (***P < 0.0001). (G) Schematic for measuring micronuclei containing chromosome 11 or other chromosomes. (H) Immunofluorescence of micronuclei with low and high abundance of CENP-B centromeres from lung fibroblast cells derived from M. pahari. Insets: 1.8× magnification. Scale bars, 10 μm. (I) Quantification corresponding to (H). Welch’s t test showed no significant difference between the actual micronuclei frequency and the expected frequency if there is no bias. A gray line represents the expected frequency given no bias, n = 133 (−Noc) and n = 419 (+Noc), pooled from four independent experiments, (P = 0.8374).
Differences in centromere repeats between different mouse strains and species also have downstream molecular consequences that direct changes in the abundance of factors involved in microtubule attachment (i.e., microtubule-binding proteins of the kinetochore, such as Hec1Ndc80) or in microtubule destabilization (i.e., the kinesin, MCAK, that uses its motor activity to disassemble kinetochore microtubules) (9, 16, 17). The M. pahari genome harbors chromosomes with divergent centromere architectures that must undergo mitosis in unison, and therefore, it presents a unique opportunity for investigating the regulation of microtubule dynamics at the kinetochore. One likely scenario we considered is that the molecular changes yield a similar balance of microtubule couplers (e.g., Hec1Ndc80) and destabilizers (e.g., MCAK) so that their ratio is similar enough to each align and segregate on the mitotic spindle with similar fidelity. Current models suggest that CENP-B recruits MCAK, which is thought to be via its role in enriching H3K9me3 chromatin. Per our expectation, we observed an approximately 1.8-fold enrichment of MCAK on chromosome 11 relative to the other M. pahari chromosomes during mitosis (Fig. 7, C and D). Thus, the heterochromatin pathway governing centromere strength leads to greater accumulation of a primary microtubule destabilizer on chromosome 11. We hypothesized that the kinetochore pathway stimulated by CENP-B would likely be affected as well. To measure this, we detected the kinetochore microtubule coupler Hec1Ndc80 and found that it is also recruited on 1.2-fold higher levels on chromosome 11 than on other chromosomes (Fig. 7, E and F).
Since there are both increased levels of MCAK and Hec1Ndc80 on chromosome 11, we predict that this chromosome will properly segregate at rates comparable to the other M. pahari chromosomes. In unperturbed cells, chromosome segregation errors lead to a small percentage (1.5 ± 0.14% in our experiment) of cells having micronuclei. This is increased to 4.2 ± 0.91% in our experiment by transient incubation with the microtubule poison, nocodazole. In both cases, chromosome 11 missegregation to micronuclei (Fig. 7, G to I) is near the expected value if there is no bias simply based on chromosome number (Fig. 7I, dashed gray line). Note that the slightly higher than expected value is explained by a likely undercount of the other chromosomes that are present in micronuclei since their levels of CENP-B, which is used to identify missegregated chromosomes, are lower than on chromosome 11. Together, our cell-based measurements support the notion that that despite chromosome 11 having different centromere DNA, its segregation fidelity is the same relative to the remaining centromeres.
DISCUSSION
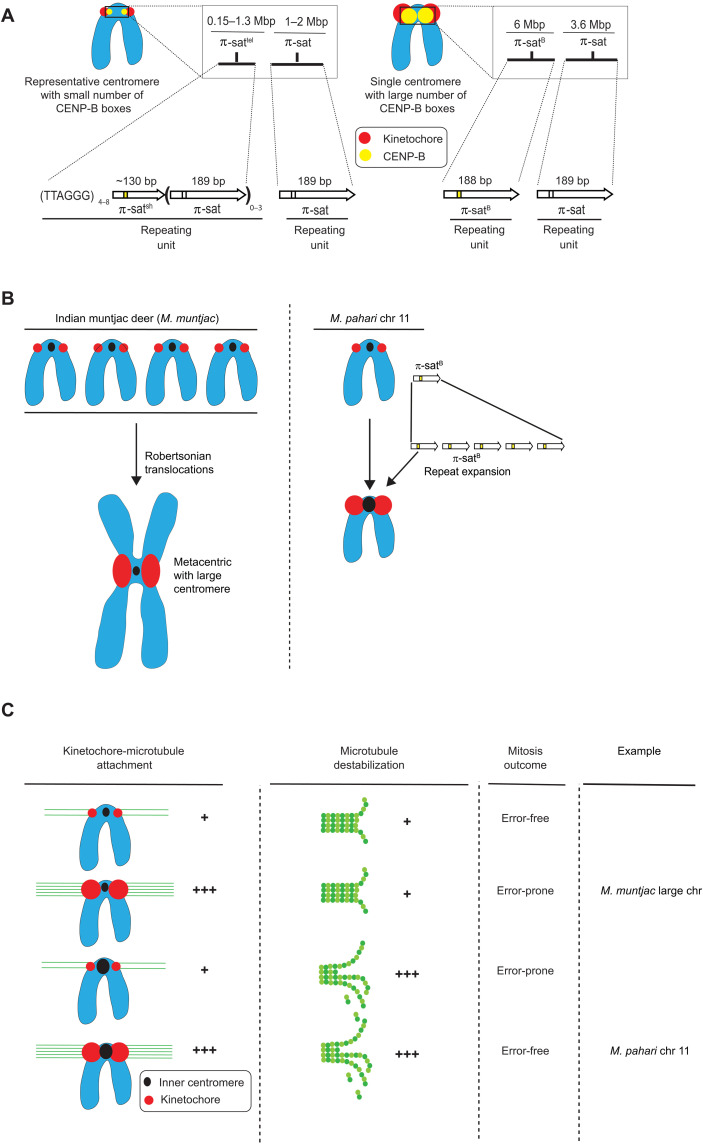
For rapid centromere evolution to occur, a new innovation within a species would have to initiate on a single chromosome. Some innovations will strengthen centromeres and spread to other chromosomes, eventually becoming the dominant form within a species. We have identified and characterized a new repeat, π-satB, in M. pahari that exists as a homogeneous 6-Mbp array that confers centromere function on chromosome 11. The chromosome 11 centromere is an outlier compared to centromeres in other species. In M. musculus, all centromeres have similar numbers of CENP-B boxes, and even in H. sapiens where centromeric sequence diverge, the range of CENP-B box numbers varies only ~10-fold between chromosomes (2, 4). On the other hand, chromosome 11 has 27 to 143 times more CENP-B boxes than presumably evolutionarily older centromeres in M. pahari that we sequenced and assembled. The chromosome 11 centromere directly recruits high levels of CENP-B that, in turn, generates a larger kinetochore (Figs. 7, E and F, and 8A).
Fig. 8. Divergent centromere DNA, molecular composition, and implications for mitotic chromosome segregation in M. pahari.
(A) Cartoon drawing summarizing the different types of M. pahari centromeres. Most M. pahari centromeres contain a low density of functional CENP-B boxes. Furthermore, these centromeres have two kinds of π-sat. First, the CENP-A–containing region is a stretch of repeating units of π-sat that is short (~130 bp) or long (189 bp) and interspersed with telomeric repeats. This is adjacent to a longer stretch of repeating units of 189-bp π-sat. The second type of M. pahari centromere has a high density of CENP-B boxes and is only found on chromosome 11. This centromere consists of 6 Mbp of homogeneous π-satB. The higher homogeneity of this centromeric DNA suggests that it is evolutionarily more recent relative to the other M. pahari centromeres. (B) A summary of the distinct mechanisms by which Indian munjtac and chromosome 11 from M. pahari centromeres likely became large repetitive arrays observed in the present day. (C) Model to understand different possible outcomes of centromere innovations during mitosis. The typical centromere has relatively low numbers of kinetochore attachments and relatively low amounts of microtubule destabilizer. These two factors balance each other, allowing normal segregation during mitosis. If either pro- or anti-microtubule binding factors are increased in the absence of the other, there will be an imbalance resulting in incorrect segregation during mitosis. Chromosome 11 has higher levels of microtubule destabilizer and more microtubule attachments, but because both factors are increased together, the chromosomes can still undergo error-free mitosis. Large Indian muntjac centromeres, on the other hand, have too strong pro-microtubule binding forces, compromising chromosome segregation (50).
Our finding that chromosome 11 in M. pahari has counterbalanced pro- and anti-microtubule binding behavior (Fig. 8) should be considered in light of recent observations in the Indian muntjac deer species, Muntiacus muntjac. In M. muntjac, very large centromeres arose not due to repeat expansion but rather by extensive rounds of Robertsonian translocations of acrocentric chromosomes present in related deer (Fig. 8B) (48, 49). The large M. muntjac centromeres lead to inappropriately strong connections to the spindle that lead to chromosome segregation errors (50) (Fig. 8C). For present-day M. pahari, we conclude that there is no imbalance of forces at the chromosome 11 centromere or chromosome segregation errors. In the past, though, we envision that compensatory kinetochore would have evolved to resolve any imbalance and restore faithful chromosome segregation. Invasion of stronger centromere sequences into other chromosomes is likely to lead to imbalances during female meiosis that would favor the biased segregation of the new centromeres into the egg. Such a model would put female meiosis as the driver of the rapid expansion of new, stronger centromere sequences through an entire genome. Testing this model with M. pahari will require the identification (and/or isolation) of strains or closely related species where interspecies crosses produce viable animals with functional oocytes (note that M. pahari does not productively mate with M. musculus or M. spretus). Our study also opens up the prospect that other experimentally tractable model systems exist where centromere innovation similarly initiates from one specific chromosome.
To understand the arrangement of M. pahari centromeres, including the location of CENP-A nucleosomes, we started with information from short-read sequencing. In the final analysis, however, clarity on the situation would never have been achieved without using long-read sequencing that yielded complete centromere assemblies. Our approach was modeled after the recent success in human centromere assemblies that has been a centerpiece accomplishment of the telomere-to-telomere consortium (2–4). Our work exemplifies how these approaches can be successfully used to identify centromere repeats in a nontraditional model system (such as M. pahari, which has had only modest genomic resources) for understanding mammalian chromosome evolution. Furthermore, it succeeded in assembling centromeres harboring several megabases of repetitive DNA that are even more homogeneous in sequence than are human centromeres. For the older, more numerous M. pahari centromeres, our experiments revealed an association between CENP-A accumulation and repeats containing short spans of perfect telomere sequences as well as CENP-B boxes (Fig. 6). This suggests that for most centromeres in this species, the genetic contribution to centromere identity is particularly high. On the other hand, within the 6 Mbp of the most homogeneous repeat, π-satB on chromosome 11, there is no strong sequence correlation with the specific peaks of CENP-A enrichment since the sequences are almost identical at sites of either high or low CENP-A enrichment (Fig. 4A). On chromosome 11, the highest peak of CENP-A enrichment is also adjacent to the telomere repeats at the natural telomere (Fig. 4). The lack of DNA sequence differences within the chromosome 11 centromere would suggest a strong epigenetic feedback that organizes the functional centromere at discrete sites within a large “sea” of homogenized DNA repeats. Similar observations have been made in M. musculus where large stretches of homogeneous centromeric DNA contain different kinds of chromatin at discrete locations despite no apparent sequence differences (51). On a technical note, our findings indicate that current sequencing methodologies and sequence assembly approaches can tackle some of the longest stretches of the most homogenized centromere sequences known in biology. Thus, massive stretches of similarly repetitive regions in other species (i.e., major satellite in M. musculus) should now be feasibly assembled using these methodologies.
As mentioned above in the context of the recent genome assemblies of human centromeres (2, 4, 8), local homogeneity of sequence is interpreted as evidence of a region that is more actively evolving but not necessarily recently emerged. We have found that the level of heterogeneity within and across M. pahari centromeres is perhaps more reminiscent of some primate centromeres than what had been assumed in mouse species because of the strong attention given to M. musculus where there is much higher homogeneity within and across centromeres. Future work to identify centromere sequence heterogeneity within and between Mus species should help reveal the origins of π-sat and π-sat variants that we describe in this study.
Rapid centromere evolution is thought to be tied to karyotypic changes that separate closely related species (52–56). On one hand, the position (i.e., telocentric versus metacentric), size, and sequence of centromere DNA are malleable since closely related species harbor notable changes between these attributes (53, 56). On the other hand, centromere repeats are generally homogenized within a species (54), supporting the concept that there is a positive functional consequence of having similar centromere function (i.e., recruitment of similar amounts of centromere proteins) across centromeres within a single species. The example in this study of M. pahari suggests that radical functional change to one of these attributes (centromere repeat) can be tolerated through counterbalancing pro-and anti-attachment of the centromere to spindle microtubules during cell division. We propose that the selective force to counterbalance functional centromere strength properties within a species shapes the nature and magnitude of innovations that would have the chance to “take hold” in a population during the evolution of centromeres.
MATERIALS AND METHODS
Experimental model and subject details
Mice
Mouse strains were purchased from the Jackson Laboratory (C57BL/6J no. 000664 and PAHARI/EiJ no. 002655). The CENP-B WT and knock-out (KO) mouse lines were generated as described previously (28). M. pahari used for ChIP were male, and the age was 6 months. CENP-B WT/KO mice were female, and the age was 3.5 months. All animal experiments were approved by the Institutional Animal Care and Use Committee (no. 804882) and were consistent with the National Institutes of Health guidelines.
Cell lines
Primary lung fibroblasts were isolated from M. musculus or M. pahari as described previously (57). Cells were immortalized by transfection of SV40 large T antigen (58), a gift from B. Johnson (Upenn), using the TransIT-X2 Dynamic Delivery System (Mirus). T-antigen integration was confirmed by polymerase chain reaction (PCR) (5′-GGAATCTTTGCAGCTAATGGACCTTC-3′ and 5′-CCTCCAAAGTCAGGTTGATGAGCA-3′ primers yield a 246-bp product).
Cell culture
The immortalized mouse primary fibroblasts were cultured in Dulbecco’s modified Eagle’s medium (DMEM/F-12) supplemented with 10% fetal bovine serum (FBS, Sigma) and 1% penicillin-streptomycin (Gibco) at 37°C in a humidified atmosphere with 5% CO2.
Primary MEFs cultures
Mouse embryonic fibroblast (MEF) lines were isolated from a pregnant female embryonic day 12.5 (E12.5) to E13.5 embryos from M. pahari (PAHARI/EiJ). MEFs were cultured in MEF media composed of DMEM supplemented with 10% FBS (Lonza), Primocin (100 μg/ml; Invivogen), and 1× GlutaMAX (Thermo Fisher Scientific/GIBCO) at 37°C in a humidified atmosphere with 5% CO2.
Method details
MNase-digested chromatin and native ChIP
ChIP was performed as described previously (9). Briefly, nuclei were isolated from flash-frozen mouse livers. Livers were homogenized in 4-ml ice-cold buffer I [0.32 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM tris-Cl (pH 7.5), 5 mM MgCl2, 0.1 mM EGTA, 0.5 mM dithiothreitol (DTT), 0.1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM leupeptin/pepstatin, and 1 mM aprotinin] per gram of tissue by dounce homogenization. Homogenate was filtered through a 100-μm cell strainer (Falcon) and centrifuged at 6000g for 10 min at 4°C. The pellet was resuspended in the same volume of buffer I. An equivalent volume of ice-cold buffer I supplemented with 0.2% IGEPAL was added, and samples were incubated on ice for 10 min. Four milliliters of nuclei was layered on top of 8 ml of ice-cold buffer III [1.2 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 15 mM tris (pH 7.5), 0.5 mM DTT, 0.1 mM PMSF, 1 mM leupeptin/pepstatin, and 1 mM aprotinin] and centrifuged at 10,000g for 20 min at 4°C with no brake applied. Pelleted nuclei were resuspended in buffer A [0.34 M sucrose, 15 mM Hepes (pH 7.4), 15 mM NaCl, 60 mM KCl, 4 mM MgCl2, 1 mM DTT, 0.1 mM PMSF, 1 mM leupeptin/pepstatin, and 1 mM aprotinin], flash-frozen in liquid nitrogen, and stored at −80°C. Nuclei were digested with MNase (Affymetrix) using chromatin (0.05 to 0.15 U/μg) in buffer A supplemented with 3 mM CaCl2 for 10 min at 37°C. The reaction was quenched with 10 mM EGTA on ice for 5 min and an equal volume of 2× Post-MNase buffer [40 mM tris (pH 8.0), 220 mM NaCl, 4 mM EDTA, 2% Triton X-100, 0.5 mM DTT, 0.5 mM PMSF, 1 mM leupeptin/pepstatin, and 1 mM aprotinin] was added before centrifugation at 18,800g for 15 min at 4°C. The supernatant containing the MNase-digested chromatin was precleared with 100 μl of 50% Protein G Sepharose bead (GE Healthcare) slurry in 1× Post-MNase buffer for ~2 hours at 4°C with rotation. Beads were blocked in NET buffer [150 mM NaCl, 50 mM tris (pH 7.5), 1 mM EDTA, 0.1% IGEPAL, 0.25% gelatin, and 0.03% NaN3]. Precleared supernatant was divided so that an estimated 250 μg of chromatin was used for ChIP 10 μg H3K9me3 antibody (Abcam, catalog no. ab8898, RRID:AB_306848) or 10 μg anti-mouse specific CENP-A antibody (custom-made and affinity purified by Covance) and 12.5 μg was saved as input. The custom polyclonal antibody was raised against a peptide corresponding to mouse CENP-A amino acids 6-30. Briefly, a New Zealand White rabbit was immunized the peptide in phosphate-buffered saline (PBS) and Freund’s adjuvant. ChIP samples were rotated at 4°C for 2 hours. Immunocomplexes were recovered by the addition of 100 μl of 50% NET-blocked protein G Sepharose bead slurry followed by overnight rotation at 4°C. The beads were washed three times with wash buffer 1 [150 mM NaCl, 20 mM tris-HCl (pH 8.0), 2 mM EDTA, 0.1% SDS, and 1% Triton X-100], once with high-salt wash buffer [500 mM NaCl, 20 mM tris-HCl (pH 8.0), 2 mM EDTA, 0.1% SDS, and 1% Triton X-100], and the chromatin was eluted 2× each with 200 μl of elution buffer [50 mM NaHCO3, 0.32 mM sucrose, 50 mM tris (pH 8.0), 1 mM EDTA, and 1% SDS] at 65°C for 10 min at 1500 rpm. The input sample was adjusted to a final volume of 400 μl with elution buffer. To each 400-μl input and ChIP sample, 16.8 μl of 5 M NaCl and 1 μl of ribonuclease (RNase) A (10 mg/ml) were added. After 1 hour at 37°C, 4 μl of 0.5 M EDTA and 12 μl of Proteinase K (2.5 mg/ml, Roche) were added, and samples were incubated for another 2 hours at 42°C. The resulting Proteinase K–treated samples were subjected to a phenol-chloroform extraction followed by purification of DNA with a QiaQuick PCR Purification column (Qiagen) in preparation for high-throughput sequencing.
High-throughput sequencing
Purified, unamplified input or ChIP DNA (see the “MNase-digested chromatin and native ChIP” section) was quantified using an Agilent 2100 Bioanalyzer high-sensitivity kit. DNA libraries were prepared for multiplexed sequencing according to Illumina recommendations as previously described (40) with minor modifications using New England BioLabs (NEB) enzymes. Briefly, 5 ng of input or ChIP DNA was end-repaired and A-tailed. Illumina TruSeq adaptors were ligated, libraries were size-selected to exclude polynucleosomes, and adapter-modified DNA fragments were enriched by PCR using KAPA polymerase. Libraries were assessed by Bioanalyzer, and the degree of nucleosome digestion for each experiment was assessed to avoid any potentially overdigested samples. Libraries were submitted for 150-bp, paired-end Illumina sequencing on a NextSeq 500 instrument.
Paired-end sequencing analysis
Paired-end sequencing analysis was performed as described previously (9). Briefly, paired-end reads were converted to a name-sorted SAM file using picard-tools and samtools (59) then joined in MATLAB using the “localalign” function to determine the overlapping region between the paired-end reads [requiring ≥95% overlap identity; (40)], and adapter sequences were removed if present. For analysis of minor and major satellite DNA, we used a custom tandem repeat analysis as described (40) with the following modifications. Joined reads were aligned to a trimerized mouse minor satellite consensus (GenBank: X14464.1) (60) or dimerized π-sat consensus or to the reverse complement of those tandem consensus sequences. Those joined reads aligning with ≥80% identity were chosen for further analysis. To calculate the percentage of total reads, the number of joined reads aligning to the consensus sequence in either the forward or reverse complement orientation (without double-counting any joined read) was divided by the total number of joined reads. ChIP fold enrichment was calculated as the fraction of reads mapping to the minor satellite from the ChIP divided by the fraction of reads mapping to the minor satellite in the input. Alignment of satellites was visualized with Matlab scripts Code3_plotting_fixIncrement_1sizeClass_JDM20170206_allPlots or 2020-04-29-INP-consensus-align-hist-line. Logos were generated via Glam2 with the command (glam2 -2 -a 190 -b 220 n pahari_input_all_to_2nd_pisat_read.CENPBbox.10reads.fa -o 2nd_pisat_region_CBBox_10) (61). Sequence alignments were generated using CLC Sequence Viewer.
TAREAN
Putative satellite sequences were identified with TAREAN (46) from Illumina input sequencing data (500,000 paired-end reads). Quality filtered and interlaced input fasta files were prepared from fastq files as recommended. TAREAN was run with the following parameters: cluster merging performed, no custom repeat database, cluster size threshold 0.0, no automatic filtering of abundant repeats, and similarity search options: Illumina reads and read length of 100 nt or more.
ONT long-read sequencing of the M. pahari genome
To generate ONT long-read sequencing data from the M. pahari genome, we first extracted high–molecular weight DNA from ~2.5 million M. pahari liver nuclei by resuspending them in 1 ml of Puregene Cell Lysis Solution (catalog no. 158113) in a 2-ml microfuge tube. Then, we added 6 μl of RNase A solution (catalog no. 158153) and incubated the mixture at 37°C for 40 min. We let the mixture cool to room temperature before adding 333 μl of Puregene Protein Precipitation Solution (catalog no. 158123), vortexing for 20 s, and then placing the tube on ice for 10 min. We spun the tube containing the mixture at maximum speed in a 4°C microfuge for 3 min. Then, we split the supernatant into two separate 1.5-ml tubes with 700 μl in each. We added 750 μl of isopropanol to each tube, inverted 50 times to mix, and then spun the tubes at maximum speed in a 4°C microfuge for 1 min. We discarded the supernatant and then added 666 μl of 70% ethanol to one of the tubes. We vortexed the single tube for 1 s and then transferred all of the ethanol solution plus the pellet into the second tube. We vortexed the second tube for 1 s and then spun it at maximum speed in a 4°C microfuge for 1 min. We washed the pelleted DNA with 666 μl of 70% ethanol two more times (pouring off the supernatant, adding new 70% ethanol, briefly vortexing, and then spinning at maximum speed in a 4°C microfuge for 1 min). After the second wash, we removed as much ethanol as possible from the tube and let it air-dry for 25 min, until all traces of ethanol were gone. We then added 110 μl of Qiagen’s DNA Hydration Solution (catalog no. 158133) to the DNA pellet and stored it at 4°C for 2 days. Once the DNA was fully resuspended, we prepared the DNA for ONT long-read sequencing using the ONT ligation sequencing kit (catalog no. SQK-LSK109), following the manufacturer’s instructions. The library was loaded onto a primed FLO-MIN106 R9.4.1 flow cell for sequencing on the GridION. All ONT data were basecalled with Guppy 3.6.0 with the HAC model.
PACBio HiFi sequencing of the M. pahari genome
DNA extraction, library preparation, quality control, and sequencing were performed by the Genome Technologies Scientific Service at the Jackson Laboratory. Approximately 60 μg of high–molecular weight DNA was isolated from spleen tissue of a single M. pahari (PAHARI/EiJ) male using the Monarch HMW DNA (NEB) according to the manufacturer’s protocols with an agitation speed of 2000 rpm. DNA concentration and quality were assessed using the Nanodrop 2000 spectrophotometer (Thermo Scientific; 434 ng/μl), the Qubit 3.0 dsDNA BR Assay (Thermo Scientific; 406 ng/μl), and the Genomic DNA ScreenTape Analysis Assay (Agilent Technologies). DNA quality was assessed to be high (260/280 = 1.83, 260/230 = 2.29) and suitable for input for PacBio HiFi library construction. A PacBio HiFi library was constructed using the SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences) according to the manufacturer’s protocols. Briefly, the protocol entails shearing DNA using the g-TUBE (Covaris), ligating PacBio–specific barcoded adapters, and size selection on the Blue Pippin (Sage Science). The quality and concentration of the library were assessed using the Femto Pulse Genomic DNA 165 kb Kit (Agilent Technologies) and Qubit dsDNA HS Assay (Thermo Fisher), respectively, according to the manufacturers’ instructions. The resultant library was sequenced on two SMRT cells on the Sequel II platform (Pacific Biosciences) using a 30-hour movie time. The two SMRT cells yielded 71.25 and 93.94 Gb of unique sequence data, respectively, with an average read length of 13.9 kb.
Assembly of the M. pahari genome
We assembled the M. pahari genome using PacBio HiFi data and the whole-genome assembler, hifiasm [v0.16.1; (47)] using standard parameters. The assembled contigs were not scaffolded into the entire chromosomes.
Alignment of CENP-A ChIP-seq and bulk nucleosomal data to the M. pahari genome assembly
To identify the location of centromeric chromatin, we took advantage of the M. pahari CENP-A ChIP sequencing (ChIP-seq) and bulk nucleosomal (input) data that we had generated. We first assessed the reads for quality using FastQC (v0.11.9) (https://github.com/s-andrews/FastQC), trimmed them with Sickle (v1.33) (https://github.com/najoshi/sickle) to remove low-quality 5′ and 3′ end bases, and trimmed them with Cutadapt (62) to remove adapters. We aligned the processed CENP-A ChIP-seq reads to the whole-genome M. pahari assembly using BWA (v0.7.17) with the following parameters: bwa mem -t {threads} -k 50 -c 1000000 {path_to_index} {path_to_read1.fastq} {path_to_read2.fastq}. We filtered the resulting SAM file to remove partial and supplementary alignments (retaining only primary alignments) with SAMtools flag-F4 before normalizing the data to the input data using DeepTools (v3.4.3) and the following command: bamCompare -b {path_to_CENP-A.bam} -b2 {path_to_input.bam} --operation ratio --binSize 5000 --minMappingQuality 60 -p 20 -o {out.bw}.
Identifying CENP-B boxes and telomere repeats within the M. pahari sequence assembly
To identify the location of CENP-B boxes within the M. pahari genome assembly, we used a custom python script (findKmers.py) to detect the location of the following sequences within the assembly: 5’-TTCGNNNNANNCGGG-3′ (the 17-bp CENP-B box) and 5′-CCCGNNTNNNNCGAA-3′ (the reverse complement of the 17-bp CENP-B box) or 5′-TTAGGG-3′ (telomere repeat) and 5’-CCCTAA-3′ (reverse complement of the telomere repeat). We ran the script with the following command: ./findKmers.py --kmers {CENP-B_box_sequences} --fasta {genome_assembly.fasta} --out {out.bed}/. We visualized the resulting BED file on the UCSC genome browser with the M. pahari reference genome assembly.
Metaphase chromosome spreads of MEFs, FISH, and image capture
FISH images of metaphase spreads of M. pahari cells were obtained using two different protocols. To obtain FISH images of π-sat, MEFs were cultured in MEF media to ~80% confluency at 37°C in a humidified atmosphere with 5% CO2. Cells were subsequently serum starved on MEF media without FBS and exposed to Colcemid (0.02 μg/ml; Thermo Fisher Scientific/GIBCO) for 12 hours to synchronize and arrest cells in metaphase. MEFs were subsequently shaken off and resuspended in hypotonic solution (56 mM KCl) for 60 min. The harvested cells were then gradually fixed in 3:1 methanol:glacial acetic acid under constant agitation. Cells were pelleted by centrifugation, and the fixative was decanted off and refixed for a total of three to four times. Following the final fixation round, cells were suspended in 1 to 2 ml of fixative and dropped onto slides from a height of ~1 m. Slides were allowed to air dry for approximately 10 min and then stored at −20°C until hybridization. Commercially synthesized oligos corresponding to the M. pahari sequence was PCR amplified and fluorescently labeled via nick translation. The genomic DNA sequence of putative M. pahari centromere sequence, π-sat, is AAAACATGTATGTTTCTTCCTGCTCTATTAGACGCATTGTAAAGATATCTGTAGAACAAGCATAGGAATATGAGTGCACTTCTTGAAACACATGGTATTCTAAGAATAATTTCCTCCATGGCAGTTCAGAGCACTAAGTACAACTATGTGCACTCATGATTCACTCTGTTTTTCGTGAGTTTTGCATGT and the primers used were as follows: forward: 5’-AACATGTATGTTTCTTCCTGCTCT-3′, reverse: 5’-TGTACTTAGTGCTCTGAACTGCC-3′.
Briefly, 250 to 1000 ng of PCR-amplified DNA was combined with nick translation buffer [200 mM tris (pH 7.5), 500 mM MgCl2, 5 mM DTT, and bovine serum albumin (500 mg/ml)], 0.2 mM deoxynucleotide triphosphate, 0.2 mM fluorescent nucleotides, 1 U deoxyribonuclease (Promega), and 1 U DNA Pol I (Thermo Fisher Scientific). One of three fluorescent nucleotides was used for each satellite probe set: Fluorescein-12–deoxyuridine triphosphate (dUTP) (Thermo Fisher Scientific), ChromaTide Texas Red-12-dUTP (Thermo Fisher Scientific/Invitrogen), and Alexa Fluor 647–aha-dUTP (Thermo Fisher Scientific/Invitrogen). The reaction mixture was incubated at 14.5°C for 90 min and then terminated by the addition of 10 mM EDTA. Probes ranged from 50 to 200 bp in size, as assessed by gel electrophoresis. Probes were used in FISH reactions on MEF metaphase cell spreads. Probes were denatured in hybridization buffer [50% formamide, 10% dextran sulfate, 2× saline-sodium citrate (SSC), and mouse Cot-1 DNA] at 72°C for 10 min and then allowed to reanneal at 37°C until slides were ready for hybridization. Slides were dehydrated in a sequential ethanol series (70, 90, and 100%; each 5 min) and dried at 42°C. Slides were then denatured in 70% formamide/2× SSC at 72°C for 3 min and immediately quenched in ice-cold 70% ethanol for 5 min. Slides were subjected to a second ethanol dehydration series (90 and 100%; each 5 min) and air-dried. The probe hybridization solution was then applied to the denatured slide. The hybridized region was then cover-slipped and sealed with rubber cement. Hybridization reactions were allowed to occur overnight in a humidified chamber at 37°C. After gently removing the rubber cement and soaking off coverslips, slides were washed two times in 50% formamide/2× SSC followed by an additional two washes in 2× SSC for 5 min at room temperature. Slides were counterstained in 4′,6-diamidino-2-phenylindole (DAPI, 80 ng/ml; Thermo Fisher Scientific/Invitrogen) for 10 min and air-dried at room temperature. Last, slides were mounted with ProLong Gold AntiFade (Thermo Fisher Scientific/Invitrogen) and stored at −20°C until imaging. FISH reactions were imaged at 63× magnification on a Leica DM6B upright fluorescent microscope equipped with fluorescent filters (Leica model numbers: 11504203, 11504207, and 11504164), light-emitting diode illumination, and a cooled monochrome Leica DFC7000 GT 2.8 megapixel digital camera. Images were captured using LAS X (version 3.7) at a resolution of 1920 × 1440 pixels.
FISH of π-satB and the π-satB CENP-B box was performed as described earlier (63) with some modifications. For FISH on metaphase spreads, M. pahari lung fibroblast cells were treated with 50 μM S-trityl-L-cysteine (STLC) (Sigma-Aldrich) for 2 to 4 hours to arrest cells during mitosis. Mitotic cells were blown off using a transfer pipette and swollen in a hypotonic buffer consisting of a 75 mM KCl for 15 min. Cells (3 × 104) were cytospun in an EZ Single Cytofunnel in a Shandon Cytospin 4 onto an ethanol-washed positively charged glass slide and allowed to adhere for 1 min before permeabilizing with KCM buffer for 15 min. For interphase FISH, cells were seeded on a positively charged glass slide before permeabilizing with KCM buffer for 15 min. Slides were washed three times in KCM for 5 min at room temperature. Slides were fixed in 4% formaldehyde in PBS before washing three times in dH2O for 1 min each. Slides were incubated with RNase A (5 μg/ml) in 2× SSC at 37°C for 5 min. Cells were subjected to an ethanol series to dehydrate the cells and then denatured in 70% formamide/2× SSC at 77°C for 2.5 min. Cells were dehydrated with an ethanol series.
Biotinylated π-satB DNA probe was generated by PCR using the template sequence TTTGAATCTAGATTTGTTTAGCTTAGAATACCATGTTTTCCGGAACTGCACTCATATTGATCTGCTTTTACTACAGAAATCTCTACAAAGCGTCTAATAGAGCAGGAAGAAAAATACCCGTTTTACACGAAAAACTCTTGAAATACAGAGTGAATCCTGAGTGCAGATACTTGTACTTAGTGCTCTGAACAAGAATTGAGGAATGTAAAGGATCCTAT, and the primers used were as follows: forward: 5’-GTTTAGCTTAGAATACCATGTTT-3′ and reverse: 5’-TTCCTCAATTCTTGTTCAGAG-3′ with Biotin-11 dUTP (Thermo Fisher Scientific; AM8450), purified with a G-50 spin column (Illustra) and ethanol-precipitated with salmon sperm DNA and Cot-1 DNA. Precipitated π-satB was suspended in 50% formamide/10% dextran sulfate in 2× SSC and denatured at 77°C for 5 to 10 min before being placed at 37°C for at least 20 min. One hundred nanograms of DNA probe was incubated with the cells on a glass slide at 37°C overnight in a dark, humidified chamber. The CENP-B box probe was ordered from PNABio with a Cy3 fluorophore conjugated to the sequence TTTCGTGTAAAACGGGT. PNA probe was prepared as described previously (https://pnabio.com/pdf/FISH_protocol_PNABio.pdf). PNA probe (50 μM) was resuspended in formamide, heated to 55°C for 5 min, and stored in aliquots at −80°C. After thawing, the probe was diluted 1:100 in 10 mM tris-HCl (pH 7.2), 70% formamide, 10 mM maleic acid, 15 mM NaCl, and 0.5% blocking reagent (Roche 11096176001). The probe was denatured at 77°C for 5 to 10 min before being placed at 37°C for at least 20 min. Ten microliters of probe was incubated with the cells on a glass slide at 37°C overnight in a dark, humidified chamber. The next day, slides were washed two times with 50% formamide in 2× SSC for 5 min at 45°C. Next, slides were washed two times with 0.1× SSC for 5 min at 45°C. Slides were blocked with 2.5% milk in 4× SSC with 0.1 Tween-20 for 10 min. For the π-satB FISH, cells were incubated with NeutrAvidin–fluorescein isothiocyanate (FITC, Thermo Fisher Scientific; 31006) diluted to 25 μg/ml in 2.5% milk with 4× SSC and 0.1% Tween-20 for 1 hour at 37°C in a dark, humidified chamber. Cells were washed three times with 4× SSC and 0.1% Tween 20 at 45°C, DAPI-stained, and mounted on a glass coverslip with Vectashield (Vector Labs).
Pulsed-field gel electrophoresis and Southern blot
Pahari mouse genomic DNA was prepared in agarose plugs and digested with Bst XI and Hpa I enzymes according to the manufacturer’s recommendation. The digested DNA was separated with the CHEF Mapper system (Bio-Rad; Run conditions for the 5- to 1000-kbp range: 0.5× Tris-borate-EDTA, 1% pulse-field certified agarose, 14°C, auto program, 16 hours run; Run conditions for the 500- to 6000-kbp range: 1× tris-acetate-EDTA, 1% pulse-field certified agarose, 14°C, 2 V/cm, 106° included angle, 5 to 40 min field switching with linear ramp, 92 hours run), transferred to a membrane (Amersham Hybond-N+), and blot-hybridized with a 30-bp probe specific to the M. pahari centromeres (5′-TTCGTGTAAAACGGGTATTTTTCTTCCTGC-3′). To label the probe, 5′ and 3′ adapters below primers were added. The probe was labeled with 32P by PCR-amplifying a synthetic DNA template (5′-TTTGTGGAAGTGGACATTTCTTCGTGTAAAACGGGTATTTTTCTTCCTGCTAAAAATAGACAGAAGCATT-3′) with the following primers: forward: 5′-TTTGTGGAAGTGGACATTTC-3′ and reverse: 5′-AATGCTTCTGTCTATTTTTA-3′. The blot was incubated for 2 hours at 65°C for prehybridization in Church’s buffer (0.5 M Na-phosphate buffer containing 7% SDS and 100 μg/ml of unlabeled salmon sperm carrier DNA). The labeled probe was heat-denatured in a boiling water bath for 5 min and snap-cooled on ice. The probe was added to the hybridization Church’s buffer and allowed to hybridize for 48 hours at 65°C. The blot was washed twice in 2× SSC [300 mM NaCl and 30 mM sodium citrate (pH 7.0)], 0.05% SDS for 10 min at room temperature, and four times in 2× SSC, 0.05% SDS for 5 min each at 60°C. The blot was exposed to x-ray film for 1 to 16 hours at −80°C.
Immunofluorescence and microscopy for immortalized mouse lung fibroblast cells
For a co-seed experiment involving CENP-A and CENP-B immunofluorescence, M. pahari and M. musculus immortalized lung fibroblast cells were coplated in 1:1 ratio. For experiments involving H3K9Me3 immunofluorescence, mouse lung fibroblast cells were fixed in 4% formaldehyde for 10 min at room temperature and quenched with 100 mM tris (pH 7.5) for 5 min, followed by permeabilization with 0.5% Triton X-100 for 5 min at room temperature. All coverslips were then blocked in PBS supplemented with 2% FBS, 2% bovine serum albumin, and 0.1% Tween before antibody incubation. The following primary antibodies were used: mouse monoclonal antibody anti-mouse CENP-B (1:200, Santa Cruz Biotechnology, catalog no. sc-376283, RRID:AB_10988421), rabbit polyclonal antibody (pAb) anti-mouse CENP-A (1:500, 0.535 μg/ml; custom-made by Covance and affinity-purified in-house), rabbit pAb anti-human H3K9Me3 (1:500, Abcam, catalog no. ab8898, RRID:AB_306848), rabbit pAb anti-human MCAK [a gift from D. Compton (Dartmouth)], and rabbit pAb anti-mouse Hec1Ndc80 antibody (64). Secondary antibodies conjugated to fluorophores were used: FITC goat anti-mouse (1:200, Jackson ImmunoResearch Labs, catalog no. 115-095-146, RRID:AB_2338599) and Cy3 goat anti-rabbit (1:200, Jackson ImmunoResearch Labs, catalog no. 111-165-144, RRID:AB_2338006). Samples were stained with DAPI before mounting with VectaShield medium (Vector Laboratories). For metaphase chromosome spread, cells were treated with 50 μM STLC for 4 hours to arrest the cells during mitosis. Mitotic cells were blown off using a transfer pipette and swollen in a hypotonic buffer consisting of a 1:1:1 ratio of 75 mM KCl, 0.8% Na citrate, 3 mM CaCl2, and 1.5 mM MgCl2 for 15 min at room temperature. Cells (5 × 104) were cytospun onto an ethanol-washed Superfrost Plus glass slide at 1500 rpm for 5 min and allowed to adhere for 2 min before fixing with 4% formaldehyde. Cells were permeabilized with 0.5% Triton X-100 for 15 min at room temperature followed by immunostaining. Images were captured at room temperature on an inverted fluorescence microscope (DFC9000 GT; Leica) equipped with a charge-coupled device camera (ORCA AG; Hamamatsu Photonics) and a 100×, 1.4 numerical aperture oil immersion objective. Images were collected as 0.2-μm z sections using identical acquisition conditions, and z series were deconvolved using LAS-X software (Leica). The fluorescence intensity was measured from deconvolved and maximum-projected images by ImageJ using 8 × 8 for H3K9Me3, 1.3 × 1.3 for MCAK, and 2.4 × 2.4 pixel box for Hec1Ndc80 using CENP-B as a reference channel. The local background intensity was subtracted from the measured fluorescence intensity. A minimum of 300 centromeres with low abundance of CENP-B and a minimum of 40 centromeres with high abundance of CENP-B were counted from at least two independent experiments. The mean ratio ± SEM is reported. For micronuclei experiment, M. pahari cells were arrested with nocodazole for 6 hours and then released for 16 hours. Cells were fixed with 4% formaldehyde for 10 min at room temperature and immunofluorescence was performed as described above.
Acknowledgments
We thank our UPenn colleagues G. Birchak and K. McCannell for discussion, and L. Chmátal for isolating the M. pahari primary lung fibroblast cells. We also thank B. Sullivan (Duke) for sharing the protocol for mouse CENP-A antibody production, B. Johnson (UPenn) for providing the SV40 large T antigen plasmid, and D. Compton (Dartmouth) for providing the human MCAK antibody.
Funding: This work was supported by NIH grants GM130302 (B.E.B.), GM108360 (J.M.D.-M.), K99 GM147352 (G.A.L.), GM133415 (B.L.D.), F31 CA268727 (U.P.A.), and a Basser Center for BRCA Early Career Award (N.P.).
Author contributions: C.W.G., N.P., J.D.M., U.P.A., M.A.Li., M.A.La., G.A.L., B.L.D., and B.E.B. designed experiments. C.W.G., N.P., J.D.M., U.P.A., M.A.Li., and G.A.L. performed experiments and analyzed data. V.L. supervised Southern blot analysis. J.M., P.L., and M.A.La. provided animal reagents. C.W.G., N.P., and B.E.B. wrote the paper. All authors edited the manuscript. B.E.B. directed the research.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All sequencing data are available on GenBank (centromere sequence assemblies) and SRA (raw sequencing files from Illumina, PACBio HiFi, and ONT) (accession no. PRJNA966193). All codes used in the study are available on Dryad (doi:10.5061/dryad.tqjq2bw51). All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Reasonable requests for materials not available from commercial sources used in this study can be provided by B.E.B. pending standard material transfer agreement procedures mandated by the University of Pennsylvania.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Table S1
REFERENCES AND NOTES
- 1.K. Kixmoeller, P. K. Allu, B. E. Black, The centromere comes into focus: From CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol. 10, 200051 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.N. Altemose, G. A. Logsdon, A. V. Bzikadze, P. Sidhwani, S. A. Langley, G. V. Caldas, S. J. Hoyt, L. Uralsky, F. D. Ryabov, C. J. Shew, M. E. G. Sauria, M. Borchers, A. Gershman, A. Mikheenko, V. A. Shepelev, T. Dvorkina, O. Kunyavskaya, M. R. Vollger, A. Rhie, A. M. McCartney, M. Asri, R. Lorig-Roach, K. Shafin, J. K. Lucas, S. Aganezov, D. Olson, L. G. de Lima, T. Potapova, G. A. Hartley, M. Haukness, P. Kerpedjiev, F. Gusev, K. Tigyi, S. Brooks, A. Young, S. Nurk, S. Koren, S. R. Salama, B. Paten, E. I. Rogaev, A. Streets, G. H. Karpen, A. F. Dernburg, B. A. Sullivan, A. F. Straight, T. J. Wheeler, J. L. Gerton, E. E. Eichler, A. M. Phillippy, W. Timp, Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.G. A. Logsdon, M. R. Vollger, P. Hsieh, Y. Mao, M. A. Liskovykh, S. Koren, S. Nurk, L. Mercuri, P. C. Dishuck, A. Rhie, L. G. de Lima, T. Dvorkina, D. Porubsky, W. T. Harvey, A. Mikheenko, A. V. Bzikadze, M. Kremitzki, T. A. Graves-Lindsay, C. Jain, K. Hoekzema, S. C. Murali, K. M. Munson, C. Baker, M. Sorensen, A. M. Lewis, U. Surti, J. L. Gerton, V. Larionov, M. Ventura, K. H. Miga, A. M. Phillippy, E. E. Eichler, The structure, function and evolution of a complete human chromosome 8. Nature 593, 101–107 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.K. H. Miga, S. Koren, A. Rhie, M. R. Vollger, A. Gershman, A. Bzikadze, S. Brooks, E. Howe, D. Porubsky, G. A. Logsdon, V. A. Schneider, T. Potapova, J. Wood, W. Chow, J. Armstrong, J. Fredrickson, E. Pak, K. Tigyi, M. Kremitzki, C. Markovic, V. Maduro, A. Dutra, G. G. Bouffard, A. M. Chang, N. F. Hansen, A. B. Wilfert, F. Thibaud-Nissen, A. D. Schmitt, J.-M. Belton, S. Selvaraj, M. Y. Dennis, D. C. Soto, R. Sahasrabudhe, G. Kaya, J. Quick, N. J. Loman, N. Holmes, M. Loose, U. Surti, R. A. Risques, T. A. Graves Lindsay, R. Fulton, I. Hall, B. Paten, K. Howe, W. Timp, A. Young, J. C. Mullikin, P. A. Pevzner, J. L. Gerton, B. A. Sullivan, E. E. Eichler, A. M. Phillippy, Telomere-to-telomere assembly of a complete human X chromosome. Nature 585, 79–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.C.-H. Chang, A. Chavan, J. Palladino, X. Wei, N. M. C. Martins, B. Santinello, C.-C. Chen, J. Erceg, B. J. Beliveau, C.-T. Wu, A. M. Larracuente, B. G. Mellone, Islands of retroelements are major components of Drosophila centromeres. PLoS Biol. 17, e3000241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C. Alkan, M. Ventura, N. Archidiacono, M. Rocchi, S. C. Sahinalp, E. E. Eichler, Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput. Biol. 3, 1807–1818 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Rocchi, R. Stanyon, N. Archidiacono, Evolutionary new centromeres in primates. Prog. Mol. Subcell. Biol. 48, 103–152 (2009). [DOI] [PubMed] [Google Scholar]
- 8.G. A. Logsdon, A. N. Rozanski, F. Ryabov, T. Potapova, V. A. Shepelev, Y. Mao, M. Rautiainen, S. Koren, S. Nurk, D. Porubsky, J. K. Lucas, K. Hoekzema, K. M. Munson, J. L. Gerton, A. M. Phillippy, I. A. Alexandrov, E. E. Eichler, The variation and evolution of complete human centromeres. bioRxiv 542849 [Preprint]. 30 May 2023. 10.1101/2023.05.30.542849. [DOI]
- 9.A. Iwata-Otsubo, J. M. Dawicki-McKenna, T. Akera, S. J. Falk, L. Chmátal, K. Yang, B. A. Sullivan, R. M. Schultz, M. A. Lampson, B. E. Black, Expanded satellite repeats amplify a discrete CENP-A nucleosome assembly site on chromosomes that drive in female meiosis. Curr. Biol. 27, 2365–2373.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. Packiaraj, J. Thakur, DNA satellite and chromatin organization at house mouse centromeres and pericentromeres. bioRxiv 549612 [Preprint]. 18 July 2023. 10.1101/2023.07.18.549612. [DOI] [PMC free article] [PubMed]
- 11.M. Dumont, D. Fachinetti, DNA sequences in centromere formation AND function. Prog. Mol. Subcell. Biol. 56, 305–336 (2017). [DOI] [PubMed] [Google Scholar]
- 12.M. A. Lampson, B. E. Black, cellular and molecular mechanisms of centromere drive. Cold Spring Harb. Symp. Quant. Biol. 82, 249–257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.L. Fishman, A. Saunders, Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322, 1559–1562 (2008). [DOI] [PubMed] [Google Scholar]
- 14.S. Henikoff, K. Ahmad, H. S. Malik, The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102 (2001). [DOI] [PubMed] [Google Scholar]
- 15.D. Dudka, M. A. Lampson, Centromere drive: Model systems and experimental progress. Chromosome Res. 30, 187–203 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.T. Akera, L. Chmátal, E. Trimm, K. Yang, C. Aonbangkhen, D. M. Chenoweth, C. Janke, R. M. Schultz, M. A. Lampson, Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358, 668–672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.T. Akera, E. Trimm, M. A. Lampson, Molecular strategies of meiotic cheating by selfish centromeres. Cell 178, 1132–1144.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.E. V. Linardopoulou, E. M. Williams, Y. Fan, C. Friedman, J. M. Young, B. J. Trask, Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature 437, 94–100 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Nurk, S. Koren, A. Rhie, M. Rautiainen, A. V. Bzikadze, A. Mikheenko, M. R. Vollger, N. Altemose, L. Uralsky, A. Gershman, S. Aganezov, S. J. Hoyt, M. Diekhans, G. A. Logsdon, M. Alonge, S. E. Antonarakis, M. Borchers, G. G. Bouffard, S. Y. Brooks, G. V. Caldas, N.-C. Chen, H. Cheng, C.-S. Chin, W. Chow, L. G. de Lima, P. C. Dishuck, R. Durbin, T. Dvorkina, I. T. Fiddes, G. Formenti, R. S. Fulton, A. Fungtammasan, E. Garrison, P. G. S. Grady, T. A. Graves-Lindsay, I. M. Hall, N. F. Hansen, G. A. Hartley, M. Haukness, K. Howe, M. W. Hunkapiller, C. Jain, M. Jain, E. D. Jarvis, P. Kerpedjiev, M. Kirsche, M. Kolmogorov, J. Korlach, M. Kremitzki, H. Li, V. V. Maduro, T. Marschall, A. M. McCartney, J. McDaniel, D. E. Miller, J. C. Mullikin, E. W. Myers, N. D. Olson, B. Paten, P. Peluso, P. A. Pevzner, D. Porubsky, T. Potapova, E. I. Rogaev, J. A. Rosenfeld, S. L. Salzberg, V. A. Schneider, F. J. Sedlazeck, K. Shafin, C. J. Shew, A. Shumate, Y. Sims, A. F. A. Smit, D. C. Soto, I. Sović, J. M. Storer, A. Streets, B. A. Sullivan, F. Thibaud-Nissen, J. Torrance, J. Wagner, B. P. Walenz, A. Wenger, J. M. D. Wood, C. Xiao, S. M. Yan, A. C. Young, S. Zarate, U. Surti, R. C. McCoy, M. Y. Dennis, I. A. Alexandrov, J. L. Gerton, R. J. O’Neill, W. Timp, J. M. Zook, M. C. Schatz, E. E. Eichler, K. H. Miga, A. M. Phillippy, The complete sequence of a human genome. Science 376, 44–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.M. Zetka, D. Paouneskou, V. Jantsch, The nuclear envelope, a meiotic jack-of-all-trades. Curr. Opin. Cell Biol. 64, 34–42 (2020). [DOI] [PubMed] [Google Scholar]
- 21.R. K. Dawe, E. G. Lowry, J. I. Gent, M. C. Stitzer, K. W. Swentowsky, D. M. Higgins, J. Ross-Ibarra, J. G. Wallace, L. B. Kanizay, M. Alabady, W. Qiu, K.-F. Tseng, N. Wang, Z. Gao, J. A. Birchler, A. E. Harkess, A. L. Hodges, E. N. Hiatt, A kinesin-14 motor activates neocentromeres to promote meiotic drive in maize. Cell 173, 839–850.e18 (2018). [DOI] [PubMed] [Google Scholar]
- 22.T. A. K. Yamakake, “Cytological studies of maize [Zea mays L.] and teosinte [Zea mexicana Schrader Kuntze] in relation to their origin and evolution,” thesis, Massachusetts Agricultural Experiment Station (1976). [Google Scholar]
- 23.M. M. Rhoades, H. Vilkomerson, On the anaphase movement of chromosomes. Proc. Natl. Acad. Sci. U.S.A. 28, 433–436 (1942). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C. M. Wade, E. Giulotto, S. Sigurdsson, M. Zoli, S. Gnerre, F. Imsland, T. L. Lear, D. L. Adelson, E. Bailey, R. R. Bellone, H. Blöcker, O. Distl, R. C. Edgar, M. Garber, T. Leeb, E. Mauceli, J. N. MacLeod, M. C. T. Penedo, J. M. Raison, T. Sharpe, J. Vogel, L. Andersson, D. F. Antczak, T. Biagi, M. M. Binns, B. P. Chowdhary, S. J. Coleman, G. D. Valle, S. Fryc, G. Guérin, T. Hasegawa, E. W. Hill, J. Jurka, A. Kiialainen, G. Lindgren, J. Liu, E. Magnani, J. R. Mickelson, J. Murray, S. G. Nergadze, R. Onofrio, S. Pedroni, M. F. Piras, T. Raudsepp, M. Rocchi, K. H. Røed, O. A. Ryder, S. Searle, L. Skow, J. E. Swinburne, A. C. Syvänen, T. Tozaki, S. J. Valberg, M. Vaudin, J. R. White, M. C. Zody; Broad Institute Genome Sequencing Platform; Broad Institute Whole Genome Assembly Team, E. S. Lander, K. Lindblad-Toh, Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326, 865–867 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D. P. Locke, L. W. Hillier, W. C. Warren, K. C. Worley, L. V. Nazareth, D. M. Muzny, S.-P. Yang, Z. Wang, A. T. Chinwalla, P. Minx, M. Mitreva, L. Cook, K. D. Delehaunty, C. Fronick, H. Schmidt, L. A. Fulton, R. S. Fulton, J. O. Nelson, V. Magrini, C. Pohl, T. A. Graves, C. Markovic, A. Cree, H. H. Dinh, J. Hume, C. L. Kovar, G. R. Fowler, G. Lunter, S. Meader, A. Heger, C. P. Ponting, T. Marques-Bonet, C. Alkan, L. Chen, Z. Cheng, J. M. Kidd, E. E. Eichler, S. White, S. Searle, A. J. Vilella, Y. Chen, P. Flicek, J. Ma, B. Raney, B. Suh, R. Burhans, J. Herrero, D. Haussler, R. Faria, O. Fernando, F. Darré, D. Farré, E. Gazave, M. Oliva, A. Navarro, R. Roberto, O. Capozzi, N. Archidiacono, G. D. Valle, S. Purgato, M. Rocchi, M. K. Konkel, J. A. Walker, B. Ullmer, M. A. Batzer, A. F. A. Smit, R. Hubley, C. Casola, D. R. Schrider, M. W. Hahn, V. Quesada, X. S. Puente, G. R. Ordoñez, C. López-Otín, T. Vinar, B. Brejova, A. Ratan, R. S. Harris, W. Miller, C. Kosiol, H. A. Lawson, V. Taliwal, A. L. Martins, A. Siepel, A. Roychoudhury, X. Ma, J. Degenhardt, C. D. Bustamante, R. N. Gutenkunst, T. Mailund, J. Y. Dutheil, A. Hobolth, M. H. Schierup, O. A. Ryder, Y. Yoshinaga, P. J. de Jong, G. M. Weinstock, J. Rogers, E. R. Mardis, R. A. Gibbs, R. K. Wilson, Comparative and demographic analysis of orang-utan genomes. Nature 469, 529–533 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.S. G. Nergadze, F. M. Piras, R. Gamba, M. Corbo, F. Cerutti, J. G. W. McCarter, E. Cappelletti, F. Gozzo, R. M. Harman, D. F. Antczak, D. Miller, M. Scharfe, G. Pavesi, E. Raimondi, K. F. Sullivan, E. Giulotto, Birth, evolution, and transmission of satellite-free mammalian centromeric domains. Genome Res. 28, 789–799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.W.-H. Shang, T. Hori, A. Toyoda, J. Kato, K. Popendorf, Y. Sakakibara, A. Fujiyama, T. Fukagawa, Chickens possess centromeres with both extended tandem repeats and short non-tandem-repetitive sequences. Genome Res. 20, 1219–1228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.T. Kumon, J. Ma, R. B. Akins, D. Stefanik, C. E. Nordgren, J. Kim, M. T. Levine, M. A. Lampson, Parallel pathways for recruiting effector proteins determine centromere drive and suppression. Cell 184, 4904–4918.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.H. Masumoto, H. Masukata, Y. Muro, N. Nozaki, T. Okazaki, A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Y. Tanaka, O. Nureki, H. Kurumizaka, S. Fukai, S. Kawaguchi, M. Ikuta, J. Iwahara, T. Okazaki, S. Yokoyama, Crystal structure of the CENP-B protein-DNA complex: The DNA-binding domains of CENP-B induce kinks in the CENP-B box DNA. EMBO J. 20, 6612–6618 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D. F. Pietras, K. L. Bennett, L. D. Siracusa, M. Woodworth-Gutai, V. M. Chapman, K. W. Gross, C. Kane-Haas, N. D. Hastie, Construction of a small Mus musculus repetitive DNA library: Identification of a new satellite sequence in Mus musculus. Nucleic Acids Res. 11, 6965–6983 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.H. F. Willard, J. S. Waye, Chromosome-specific subsets of human alpha satellite DNA: Analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J. Mol. Evol. 25, 207–214 (1987). [DOI] [PubMed] [Google Scholar]
- 33.D. Kipling, A. R. Mitchell, H. Masumoto, H. E. Wilson, L. Nicol, H. J. Cooke, CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli. Mol. Cell. Biol. 15, 4009–4020 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D. Fachinetti, J. S. Han, M. A. McMahon, P. Ly, A. Abdullah, A. J. Wong, D. W. Cleveland, DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell. 33, 314–327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. Dumont, R. Gamba, P. Gestraud, S. Klaasen, J. T. Worrall, S. G. De Vries, V. Boudreau, C. Salinas-Luypaert, P. S. Maddox, S. M. Lens, G. J. Kops, S. E. McClelland, K. H. Miga, D. Fachinetti, Human chromosome-specific aneuploidy is influenced by DNA-dependent centromeric features. EMBO J. 39, e102924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.H. Nakagawa, J.-K. Lee, J. Hurwitz, R. C. Allshire, J.-I. Nakayama, S. I. S. Grewal, K. Tanaka, Y. Murakami, Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 16, 1766–1778 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.T. Okada, J. Ohzeki, M. Nakano, K. Yoda, W. R. Brinkley, V. Larionov, H. Masumoto, CENP-B controls centromere formation depending on the chromatin context. Cell 131, 1287–1300 (2007). [DOI] [PubMed] [Google Scholar]
- 38.K. Otake, J.-I. Ohzeki, N. Shono, K. Kugou, K. Okazaki, T. Nagase, H. Yamakawa, N. Kouprina, V. Larionov, H. Kimura, W. C. Earnshaw, H. Masumoto, CENP-B creates alternative epigenetic chromatin states permissive for CENP-A or heterochromatin assembly. J. Cell Sci. 133, jcs243303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C. W. Gambogi, B. E. Black, The nucleosomes that mark centromere location on chromosomes old and new. Essays Biochem. 63, 15–27 (2019). [DOI] [PubMed] [Google Scholar]
- 40.D. Hasson, T. Panchenko, K. J. Salimian, M. U. Salman, N. Sekulic, A. Alonso, P. E. Warburton, B. E. Black, The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat. Struct. Mol. Biol. 20, 687–695 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D. Dubocanin, A. E. Sedeno Cortes, G. A. Hartley, J. Ranchalis, A. Agarwal, G. A. Logsdon, K. M. Munson, T. Real, B. J. Mallory, E. E. Eichler, R. J. O’Neill, A. B. Stergachis, Conservation of chromatin organization within human and primate centromeres. bioRxiv 537689 [Preprint]. 20 April 2023. 10.1101/2023.04.20.537689. [DOI]
- 42.B. Dod, E. Mottez, E. Desmarais, F. Bonhomme, G. Roizés, Concerted evolution of light satellite DNA in genus Mus implies amplification and homogenization of large blocks of repeats. Mol. Biol. Evol. 6, 478–491 (1989). [DOI] [PubMed] [Google Scholar]
- 43.Y. Nishioka, Genome comparison in the genus Mus: A study with B1, MIF (mouse interspersed fragment), centromeric, and Y-chromosomal repetitive sequences. Cytogenet. Cell Genet. 50, 195–200 (2004). [DOI] [PubMed] [Google Scholar]
- 44.U. P. Arora, C. Charlebois, R. A. Lawal, B. L. Dumont, Population and subspecies diversity at mouse centromere satellites. BMC Genomics 22, 279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D. Thybert, M. Roller, F. C. P. Navarro, I. Fiddes, I. Streeter, C. Feig, D. Martin-Galvez, M. Kolmogorov, V. Janoušek, W. Akanni, B. Aken, S. Aldridge, V. Chakrapani, W. Chow, L. Clarke, C. Cummins, A. Doran, M. Dunn, L. Goodstadt, K. Howe, M. Howell, A.-A. Josselin, R. C. Karn, C. M. Laukaitis, L. Jingtao, F. Martin, M. Muffato, S. Nachtweide, M. A. Quail, C. Sisu, M. Stanke, K. Stefflova, C. Van Oosterhout, F. Veyrunes, B. Ward, F. Yang, G. Yazdanifar, A. Zadissa, D. J. Adams, A. Brazma, M. Gerstein, B. Paten, S. Pham, T. M. Keane, D. T. Odom, P. Flicek, Repeat associated mechanisms of genome evolution and function revealed by the Mus caroli and Mus pahari genomes. Genome Res. 28, 448–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.P. Novák, L. Ávila Robledillo, A. Koblížková, I. Vrbová, P. Neumann, J. Macas, TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 45, e111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.H. Cheng, E. D. Jarvis, O. Fedrigo, K.-P. Koepfli, L. Urban, N. J. Gemmell, H. Li, Haplotype-resolved assembly of diploid genomes without parental data. Nat. Biotechnol. 40, 1332–1335 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.B. R. Brinkley, M. M. Valdivia, A. Tousson, S. L. Brenner, Compound kinetochores of the Indian muntjac. Evolution by linear fusion of unit kinetochores. Chromosoma 91, 1–11 (1984). [DOI] [PubMed] [Google Scholar]
- 49.R. P. Zinkowski, J. Meyne, B. R. Brinkley, The centromere-kinetochore complex: A repeat subunit model. J. Cell Biol. 113, 1091–1110 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D. Drpic, A. C. Almeida, P. Aguiar, F. Renda, J. Damas, H. A. Lewin, D. M. Larkin, A. Khodjakov, H. Maiato, Chromosome segregation is biased by kinetochore size. Curr. Biol. 28, 1344–1356.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.A. H. F. M. Peters, S. Kubicek, K. Mechtler, R. J. O’Sullivan, A. A. H. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. A. Martens, T. Jenuwein, Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 12, 1577–1589 (2003). [DOI] [PubMed] [Google Scholar]
- 52.O. J. Marshall, A. C. Chueh, L. H. Wong, K. H. A. Choo, Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 82, 261–282 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.G. Montefalcone, S. Tempesta, M. Rocchi, N. Archidiacono, Centromere repositioning. Genome Res. 9, 1184–1188 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.F. Pardo-Manuel de Villena, C. Sapienza, Female meiosis drives karyotypic evolution in mammals. Genetics 159, 1179–1189 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. Rocchi, N. Archidiacono, W. Schempp, O. Capozzi, R. Stanyon, Centromere repositioning in mammals. Heredity 108, 59–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.M. Ventura, N. Archidiacono, M. Rocchi, Centromere emergence in evolution. Genome Res. 11, 595–599 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.A. Seluanov, A. Vaidya, V. Gorbunova, Establishing primary adult fibroblast cultures from rodents. J. Vis. Exp. 2033, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Y. Yu, J. C. Alwine, Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J. Virol. 76, 3731–3738 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.H. Li, B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis, R. Durbin, The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.A. K. Wong, J. B. Rattner, Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 16, 11645–11661 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.M. C. Frith, N. F. W. Saunders, B. Kobe, T. L. Bailey, Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput. Biol. 4, e1000071 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. Martin, Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 17, 10–12 (2011). [Google Scholar]
- 63.W. Bickmore, Fluorescence in situ hybridization analysis of chromosome and chromatin structure. Methods Enzymol. 304, 650–662 (1999). [DOI] [PubMed] [Google Scholar]
- 64.E. Diaz-Rodríguez, R. Sotillo, J.-M. Schvartzman, R. Benezra, Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc. Natl. Acad. Sci. U.S.A. 105, 16719–16724 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Table S1