Abstract
Synaptic plasticity plays a crucial role in memory formation by regulating the communication between neurons. Although actin polymerization has been linked to synaptic plasticity and dendritic spine stability, the causal link between actin polymerization and memory encoding has not been identified yet. It is not clear whether actin polymerization and structural changes in dendritic spines are a driver or a consequence of learning and memory. Using an extrinsically disordered form of the protein kinase LIMK1, which rapidly and precisely acts on ADF/cofilin, a direct modifier of actin, we induced long-term enlargement of dendritic spines and enhancement of synaptic transmission in the hippocampus on command. The activation of extrinsically disordered LIMK1 in vivo improved memory encoding and slowed cognitive decline in aged mice exhibiting reduced cofilin phosphorylation. The engineered memory by an extrinsically disordered LIMK1 supports a direct causal link between actin-mediated synaptic transmission and memory.
Chemogenetic control of LIMK1 activity provides evidence for direct causality between actin-mediated spine plasticity and memory.
INTRODUCTION
The ability of dendritic spines to change their structure has been proposed to be involved in the long-term potentiation (LTP) of glutamatergic synapses (1–5), a cellular correlate of learning, which is thought to be essential for creating long-term memories (6–8). Actin polymerization plays a crucial role in the structural plasticity of dendritic spines by allowing their cytoskeletons to remodel (9). During LTP, the influx of Ca2+ and subsequent activation of Ca2+/calmodulin-dependent protein kinase II (CaMKII) lead to the activation of various downstream proteins compartmentalized within dendritic spines (5, 10). These proteins downstream of CaMK include Rho guanine exchange factors and Rho guanosine triphosphatases, which may activate many proteins, including Rho kinase (ROCK) and p21-activated kinase (PAK). Both PAK and ROCK activate LIM (Lin-11/Isl-1/Mec-3) domain–containing protein kinase 1 (LIMK1) (11), resulting in the inactivation of actin-depolymerizing factor (ADF)/cofilin, leading to actin polymerization and subsequent enlargement and stabilization of dendritic spines (Fig. 1A) (5). However, PAK and ROCK are multifunctional kinases having numerous substrates that may act in an antagonist manner (12). Thus, there is still much to learn about the molecular mechanisms and timing of the core LTP signaling in dendritic spines and how they contribute to the formation of memories. In addition, the role of actin polymerization in modulating synaptic plasticity and its potential impact on memory formation are poorly understood and need further investigation. This is mainly due to the lack of genetically encoded technologies enabling the precise and rapid control of actin polymerization in vivo.
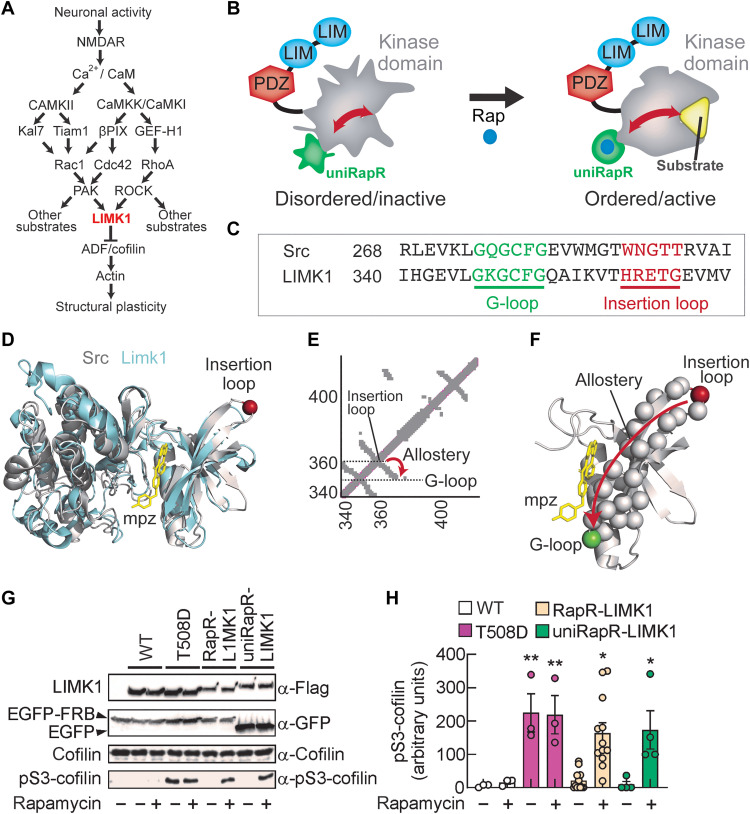
Fig. 1. Engineering extrinsically disordered LIMK1.
(A) Schematic of the canonical signaling cascade leading to structural long-term potentiation (LTP). The arrows indicate the activation, and T-shaped bar indicates inhibition. (B) The chemogenetic LIMK1 is generated by inserting the unimolecular rapamycin regulatable domain (uniRapR) into the kinase domain. This enables the order/disorder transition in only the kinase domain without affecting other regulatory domains PDZ and LIM domains. (C) Sequence alignment of Src and LIMK1 shows the presence of G-loop in green and insertion loop in red. (D) The crystal structure of LIMK1 (PDB ID: 5L6W) and Src (PDB ID: 1Y57) is shown in cyan and gray, respectively. The adenosine 5′-triphosphate (ATP) analog mpz is shown in yellow. The insertion loop is shown with a red sphere. (E and F) The pairwise contact map. The insertion loop and G-loop are connected by an antiparallel β strand. (G) Cells expressing LIMK1 constructs are used to immunoprecipitate the protein via Flag tag antibody. The rapamycin-dependent activity of LIMK1 analogs is shown by monitoring the pS3-cofilin. (H) Summary graphs of pS3-cofilin in enzymatic activity assay performed with wild-type (WT), T508D, RapR, and uniRapR analogs in the presence of vehicle or rapamycin. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.001; statistics by one-way analysis of variance (ANOVA) with the Dunnett’s post hoc test comparisons.
Investigating the proteins involved in learning and memory by knock-in/out or knockdown strategies can lead to molecular compensations, introducing confounding factors in data interpretation. A notable example is the effect of inhibitory peptides on protein kinase M-ζ (PKM-ζ) that hamper the late-phase LTP, learning, and memory (12). This finding was later challenged by a study investigating PKM-ζ knockout mice that did not exhibit impaired learning and memory (13). Similar conflicting data have been reported for other proteins, such as Src family kinases (14) and AP-1 proteins (15). These examples highlight the necessity of acute and specific protein perturbation methods, such as molecular chemogenetics and optogenetics, which allow to establish causal links between proteins and phenotypes (16, 17). Moreover, interfering with proteins downstream in the molecular cascades can provide more precise information and insight into compensatory mechanisms, as they are often more directly involved in cellular responses, making them a more relevant target for studying the role of a signaling pathway. For example, activation of an engineered photo-activatable Rac1 in the mouse motor cortex shrinks the dendritic spines that are potentiated upon motor learning (18), while the chemogenetic activation of the Rac1 downstream protein PAK1 leads to the enlargement of dendritic spines in hippocampal circuits (19). However, both Rac1 and PAK1 are relatively distant upstream of actin, as the manipulation of these proteins may influence many other downstream proteins. Therefore, previously unutilized strategies are required to make a direct molecular link between actin depolymerization–induced spine stabilization and memory.
To directly and acutely manipulate actin polymerization in the mouse brain, we sought to engineer a controllable LIMK1 with the following rationales. First, an actin modifier ADF/cofilin is the only known substrate of LIMK1 in the actin polymerization pathway (20); thus, direct perturbation of LIMK1 renders specific control of cofilin (Fig. 1A). Second, we have previously engineered controllable kinases using an extrinsic disorder approach (21, 22); thus, the application of extrinsic disorder to LIMK1 would be straightforward as most kinase domain structures are evolutionarily conserved. Third, LIMK1-deficient mice have abnormal spine morphology and impaired learning and memory (23), which are further exaggerated in double-knockout mice lacking both Limk1 and Limk2 genes (24). Last, aging-related cognitive and memory encoding decline has been proposed to be associated with decreased dendritic spine volumes (25), and we hypothesize that induction of direct actin polymerization in aging mice can reverse this process. The extrinsically disordered LIMK1 reported here allowed controllable phosphorylation of cofilin in living neurons. Its activation triggered spine enlargement bypassing the activation of canonical pathways involved in LTP, enhanced glutamatergic synaptic transmission, and rescued memory deficits in aged mice.
RESULTS
A designed extrinsically disordered LIMK1 promotes a direct and inducible control of cofilin phosphorylation in living cells
To generate a controllable LIMK1, we have used the extrinsic disorder (21). In this approach, a rapamycin-regulatable engineered protein domain (uniRapR) or light-oxygen-voltage-sensing domain 2 (LOV2) is used to transform a protein domain of interest into a ligand or light switch (21). These domains are inserted into the target protein domains’ surface exposed tight loops connecting two interacting structural units (helices or strands), which are also evolutionarily nonconserved. This insertion site is also selected to be allosterically coupled to the active site of the host domain. To control LIMK1 activity, we inserted the uniRapR domain into the kinase domain without perturbing other domains, including LIM and PDZ domains. Because uniRapR is designed to be disordered in the absence of rapamycin, its insertion into the kinase domain causes a structural perturbation in the absence of rapamycin (Fig. 1B). The delivery of rapamycin (or its non-immunosuppressive analogs) induces the disorder to order transition in uniRapR domain and thereby in the host domain. This conformational transition should activate LIMK1. Notably, rapamycin is blood-brain permeable; consequently, this system should provide exogenous control of LIMK1 in the brain in vivo.
To identify the uniRapR insertion site, we first performed sequence and structural alignment between LIMK1 and Src, a tyrosine kinase that has been previously engineered using the same approach (22). On the basis of the sequence (Fig. 1C) and structural (Fig. 1D) alignment analyses, we identified the residues 346 to 351 (GKGCFG, highly conserved across mice, rats, and humans; fig. S1) as the G-loop of LIMK1 and the residues 359 to 363 (HRETG) as the insertion loop (Fig. 1, C and D). On the basis of the contact map analyses (21), the insertion loop should be allosterically coupled to the G-loop (Fig. 1E), so the perturbation at the loop should be transmitted to the G-loop through the β strand connecting the insertion loop to the G-loop (Fig. 1F). Hence, we hypothesize that uniRapR domain inserted at an allosteric site provides extrinsic disorder for LIMK1.
We inserted the uniRapR domain into the identified insertion loop by replacing it with E360. We also generated the previous version of uniRapR approach, called RapR, a dual-chain LIMK1 switch by inserting insertable FKBP12 (iFKBP) domain into the insertion loop. As previously described, iFKBP requires the coexpression of FKBP12-rapamycin-binding (FRB) domain to turn on kinase in the presence of rapamycin (19, 22, 26, 27). In both uniRapR and RapR approaches, we included Gly-Pro-Gly (GPG) flexible linkers (fig. S2). In our designs, we avoided interfering with the VAIK motif (MVMK sequence) that contributes to forming the hydrophobic cage for adenosine 5′-triphosphate (ATP) indispensable for kinase catalytic activity (28). Both PAK and ROCK activate LIMK1 by phosphorylating T508 located in the activation loop (5). Consequently, LIMK1 T508D or T508EE mutants (in which threonine in position 508 is replaced by either aspartic acid or two glutamic acids, respectively), mimicking phosphorylation on T508, represent the constitutively active mutants maintaining the catalytic kinase activity independently of PAK1- and ROCK1-mediated LIMK1 phosphorylation (29, 30). We included the phosphomimetic T508D mutation in both RapR and uniRapR constructs for a direct comparison of the activity in the presence of rapamycin and the activity of the constitutively active T508D mutant.
To investigate whether our engineered LIMK1 constructs promoted a direct and inducible control of cofilin phosphorylation at residue S3 (pS3-cofilin) (20), we assessed the in vitro enzymatic activity of LIMK1 immunoprecipitated from transfected human embryonic kidney (HEK) 293T cells on purified cofilin in the presence of either rapamycin or vehicle. Wild-type (WT) LIMK1 did not phosphorylate cofilin in the absence or presence of rapamycin, indicating that rapamycin had no effects on LIMK1 activity (normalized pS3-cofilin levels: 5.50 ± 2.75%, n = 3, versus 15.80 ± 5.71%, n = 3, in the presence of vehicle or 500 nM rapamycin, respectively, P = 0.999; Fig. 1, G and H). The levels of pS3-cofilin were substantially increased in the presence of purified LIMK1 T508D mutant (normalized pS3-cofilin levels: 224.56 ± 57.57%, n = 3, P = 0.007 compared to LIMK1 WT treated with vehicle, n = 3). The presence of rapamycin did not affect cofilin phosphorylation (normalized pS3-cofilin levels: 218.94 ± 57.23%, n = 3, P = 0.009 compared to LIMK1 WT treated with vehicle, n = 3; P = 0.999 compared to LIMK1 T508D mutant treated with vehicle, n = 3; Fig. 1, G and H). As expected, the inactive T508A LIMK1 mutant was unable to phosphorylate cofilin either in the absence or presence of rapamycin (fig. S3). To test RapR-LIMK1 function, we coexpressed LIMK1-iFKBP with enhanced green fluorescent protein (EGFP)–FRB. RapR-LIMK1 phosphorylated cofilin in the presence of rapamycin, whereas its activity was significantly lower with vehicle (normalized pS3-cofilin levels: 163.43 ± 61.70%, n = 12, P = 0.016 in the presence of 500 nM rapamycin versus 20.98 ± 16.73% in the presence of vehicle, n = 12, P = 0.999 compared to LIMK1 WT treated with vehicle; Fig. 1, G and H). Rapamycin did not trigger cofilin phosphorylation in cells coexpressed with RapR-LIMK1 T508A mutant and EGFP-FRB, indicating that phosphomimetic mutation at T508 is still needed for activation (fig. S3). Without FRB coexpression, rapamycin did not activate RapR-LIMK1 T508D (fig. S3). In contrast, rapamycin alone was sufficient to activate uniRapR-LIMK1 (normalized pS3-cofilin levels: 173.19 ± 66.34%, n = 4, in the presence of 500 nM rapamycin versus 9.12 ± 10.53%, n = 4, in the presence of vehicle, P = 0.035; P = 0.999 uniRapR-LIMK1 versus LIMK1 WT, both treated with vehicle; Fig. 1, G and H). The uniRapR-LIMK1 T508A mutant was unable to phosphorylate cofilin even in the presence of rapamycin (fig. S3). These results suggest that the replacement of E360 with iFKBP or uniRapR flanked by GPG flexible linkers enables a robust extrinsically disordered LIMK1.
Next, we tested whether the perturbation we introduced in LIMK1 to generate RapR-LIMK1 and uniRapR-LIMK1 affected the ability of LIMK1 to form complexes with its binding partners. As LIMK1 interacts with tropomyosin receptor kinase B (TrkB) (31), we investigated whether RapR-LIMK1 and uniRapR-LIMK1 also interact with it. We performed coimmunoprecipitation experiments in HEK293T cells cotransfected with TrkB-EGFP and either RapR-LIMK1-Flag or uniRapR-LIMK1-Flag. TrkB formed a complex with LIMK1-Flag WT (amount of immunoprecipitated TrkB signal normalized to the cell lysate amount of TrkB: 88.49 ± 9.44% for LIMK1 WT, n = 3; fig. S4). TrkB-EGFP did not form a complex with mCherry-Flag (fig. S4A). A comparable level of TrkB was detected in cells cotransfected with RapR-LIMK1-Flag or uniRapR-LIMK1-Flag, indicating that neither RapR nor uniRapR affected the ability of LIMK1 to associate with TrkB (94.29 ± 11.08% for RapR-LIMK1 treated with vehicle, n = 3, P = 0.997 compared with LIMK1 WT treated with vehicle, n = 3; 95.84 ± 6.99% for uniRapR-LIMK1 treated with vehicle, n = 3, P = 0.992 compared with vehicle-treated LIMK1 WT; fig. S4). Moreover, rapamycin did not affect the complex formation, suggesting that LIMK1-TrkB interaction is independent of LIMK1 activity (normalized binding to TrkB: 100.07 ± 18.41% for LIMK1 WT treated with 500 nM rapamycin, n = 3, P = 0.946 compared with vehicle-treated LIMK1 WT; 98.62 ± 14.66% for RapR-LIMK1 treated with 500 nM rapamycin, n = 3, P = 0.968 compared with vehicle-treated LIMK1 WT; 103.68 ± 12.40% for uniRapR-LIMK1 treated with 500 nM rapamycin, n = 3, P = 0.862 compared with vehicle-treated LIMK1 WT; fig. S4). We performed other coimmunoprecipitation experiments to assess the interaction between engineered LIMK1 and protein phosphatase slingshot homolog 1 (SSH1). This was particularly important due to prior evidence indicating SSH1’s ability to interact with the kinase domain of LIMK1 (32), which we had modified by incorporating iFKBP or uniRapR domains. We found that both LIMK1 and uniRapR-LIMK1 coimmunoprecipitate with SSH1 in the presence or absence of rapamycin (fig. S4C), indicating that the perturbation we introduced in our constructs did not affect the ability of LIMK1 to form complexes with its binding partners.
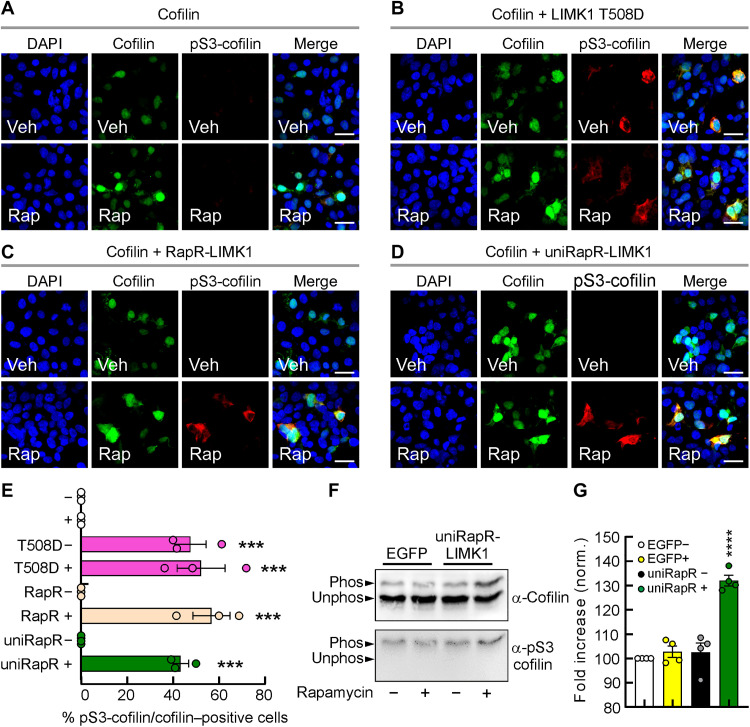
Next, we tested the ability of LIMK1 constructs to phosphorylate cofilin in living cells. We performed immunostaining of pS3-cofilin in HEK293T cells transfected with cofilin and a series of LIMK1 constructs. In cells transfected with cofilin alone (cofilin-EGFP), pS3-cofilin was not detected, even following 1-hour treatment with 500 nM rapamycin (Fig. 2A), whereas a robust signal was observed in cells cotransfected with the phosphomimetic LIMK1 T508D mutant and cofilin (Fig. 2B). The cofilin phosphorylation was unaffected by 1-hour application of 500 nM rapamycin (Fig. 2B), indicating that, also in living cells, rapamycin per se had no effects on LIMK1 activity. In contrast, cells coexpressing cofilin, RapR-LIMK1 and FRB showed pS3-cofilin immunofluorescence only after the addition of rapamycin (1 hour, 500 nM) but not the vehicle (Fig. 2C). Similarly, rapamycin application induced cofilin phosphorylation in cells transfected with cofilin and uniRapR-LIMK1 (Fig. 2D). The pS3-cofilin immunofluorescence signals were similar in cells transfected with T508D mutant with or without rapamycin treatment (52.25 ± 10.43% versus 47.63 ± 6.81%, in cells treated with rapamycin and vehicle, respectively, P = 0.788; Fig. 2E), RapR-LIMK1 and FRB in the presence of rapamycin (56.76 ± 8.03, P = 0.548 compared with T508D mutant treated with vehicle; Fig. 2E) and uniRapR-LIMK1 in the presence of rapamycin (43.40 ± 3.34%, P = 0.788 compared with T508D mutant treated with vehicle; Fig. 2E). These data demonstrate the ability of engineered LIMK1 constructs to control cofilin phosphorylation in living cells.
Fig. 2. Inducible cofilin phosphorylation in living cells.
(A to D) Representative images of pS3-cofilin immunofluorescence (red fluorescence) in HEK293T cells coexpressing cofilin (EGFP, enhanced green fluorescent protein) and LIMK1 analogs and treated with either vehicle (Veh) or 500 nM rapamycin (Rap). (E) The percentage of pS3-cofilin/cofilin–positive HEK293T cells transfected with LIMK1 analogs and treated with Veh (−) or Rap (+). The experiment was repeated three times [statistics by one-way analysis of variance (ANOVA) with the Dunnett’s post hoc test comparisons]. Scale bars, 10 μm. Data are expressed as mean ± SEM. ***P < 0.001. (F) HEK293T cells expressing EGFP or unimolecular rapamycin regulatable domain (uniRapR)–LIMK1 were treated with or without rapamycin for 3 hours. Phosphorylated (Phos) from unphosphorylated (Unphos) cofilin was separated using Phos-tag SDS-PAGE and blotted with antibodies against pS3-cofilin and total cofilin, quantified in (G). Data are expressed as mean ± SEM. ****P < 0.0001.
To quantify the cofilin phosphorylation induced by engineered LIMK1 activation, we use the Phos-tag SDS–polyacrylamide gel electrophoresis (SDS-PAGE) approach. We found that the stoichiometry of S3 phosphorylation in cells expressing the engineered LIMK1 after 3 hours of activation was about 30% higher (P < 0.001) than those of vehicle-treated cells expressing the engineered LIMK1 or cells expressing EGFP (0.028 ± 2.36%, 0.250 ± 3.87%, and 32.0 ± 2.16% of cofilin were phosphorylated at S3 in EGFP cells + rapamycin, in uniRapR-LIMK1 cells + vehicle or rapamycin, compared with EGFP cells + vehicle, n = 4 each condition; Fig. 2, F and G). On the basis of the Phos-tag SDS-PAGE experiments, we found that (i) endogenous cofilin was phosphorylated in cells transfected with EGFP and treated with vehicle and (ii) the increase of endogenous pS3-cofilin induced by activation of the engineered LIMK1 was much lower than the expected level of saturation.
Because cofilin phosphorylation is associated with actin polymerization, we also investigated this phenomenon in living cells. To evaluate the effect of uniRapR-LIMK1 activation on actin polymerization, we expressed uniRapR-LIMK1 and EGFP-actin in COS-7 cells. Transfected and untransfected cells showed similar cellular morphologies, indicating that transgene expression did not alter the cellular phenotype. Analyses of EGFP-actin distribution provided that there is a substantial actin localization to the plasma membrane upon the addition of rapamycin. Before rapamycin, the actin is in the cytosol, mainly in the perinuclear cytosolic region. Before rapamycin, the nuclear region with a lower fluorescence intensity can be observed, as EGFP-actin is predominantly localized to the cytosol (fig. S5A). Upon addition of rapamycin, the appearance of the plasma membrane and actin aggregates presumably reflecting actin polymerization (33), as well as the disappearance of the low-fluorescence nuclear region, shows that actin polymerization emerges at the plasma membrane, region 1 (fig. S5, A and B). Also, the accumulated actin in the perinuclear region is substantially reduced and distributed to the plasma membrane (region 2) upon rapamycin addition (fig. S5, A and B). On the basis of the quantification of actin dynamics from 18 cells, the normalized actin polymerization upon activation of uniRapR-LIMK1 with rapamycin, compared to the vehicle, is approximately more than fivefold (fig. S5C). These results suggest that uniRapR-LIMK1 activation with rapamycin induces rapid actin polymerization in living cells.
The controlled activation of engineered LIMK1 induces dendritic spine enlargement
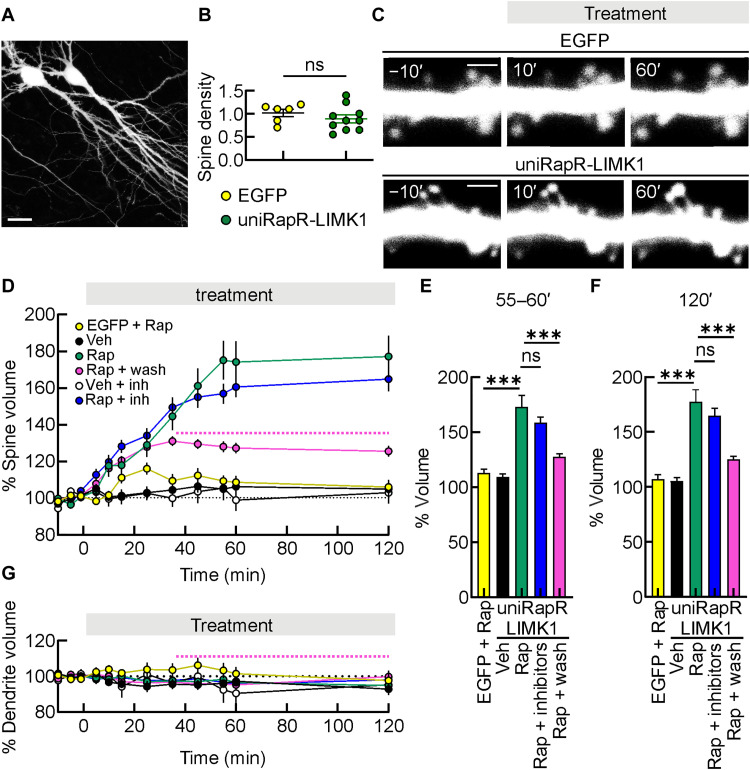
LIMK1 regulates the architecture of actin cytoskeleton in subcellular compartments of most cells, including dendritic spines of neurons, thereby playing a critical role in structural LTP (5, 23, 34–36). This prompted us to test the ability of engineered LIMK1 to promote inducible dendritic spine enlargement. We expressed either EGFP alone or uniRapR-LIMK1 together with freely soluble EGFP in organotypic hippocampal slice cultures using ballistic transfection, and 48 hours later, we imaged dendritic spines on primary or secondary dendrites from the distal part of the main apical dendrite of CA1 pyramidal neurons using two-photon microscopy (Fig. 3A). As controls, we monitored (i) EGFP-transfected neurons treated with rapamycin and (ii) neurons transfected with uniRapR-LIMK1 and EGFP treated with vehicle. Dendritic spine density, measured 48 hours after the transfection, was similar for each experimental condition (1.02 ± 0.08 spines/μm versus 0.89 ± 0.08 spines/μm, in neurons transfected with EGFP, n = 6, and uniRapR-LIMK1 and EGFP, n = 10, respectively, P = 0.337; Fig. 3B), indicating that expression of uniRapR-LIMK1 did not affect the number of spines per dendrite length. Then, we monitored the volume of dendritic spines 5 to 120 min after vehicle or rapamycin addition. In neurons transfected with EGFP, 120-min rapamycin application did not significantly affect the volume of dendritic spines (108.73 ± 2.77%, P = 0.102 compared with 5-min pre-application and 106.07 ± 3.79%, P = 0.362 compared with 5-min pre-application, n = 98 spines recorded in eight different organotypic hippocampal slices, n = 98/8, volume increment after 55 to 60 min and 120 min of rapamycin addition, respectively; Fig. 3, C to F). Similarly, the volume and number of dendritic spines of neurons transfected with uniRapR-LIMK1 and treated with vehicle remained stable throughout recordings (106.35 ± 2.30%, P = 0.141 compared with 5-min pre-application and 104.97 ± 3.22%, P = 0.306 compared with 5-min pre-application, n = 93/7, volume increment after 55 to 60 min and 120 min of vehicle addition, respectively; Fig. 3, E and F). However, uniRapR-LIMK1 activation with rapamycin led to an enlargement of dendritic spines (173.13 ± 10.33%, P < 0.001 and 177.17 ± 11.22%, P < 0.001, n = 121/10, volume increment after 55 to 60 min and 120 min of rapamycin addition, respectively; Fig. 3, C to F). Our results indicate that uniRapR-LIMK1 activation induced a long-term dendritic spine enlargement starting 10 min after rapamycin application and remained potentiated after 120 min. UniRapR-LIMK1–mediated spine enlargement was also caused by short rapamycin application [i.e., 30 min followed by washout with artificial cerebrospinal fluid (ACSF)], although these enlargements were lower than those obtained with longer rapamycin application (127.6 ± 2.73% and 125.6 ± 2.95%, n = 188/10, volume increment after 25 to 30 and 90 min of the rapamycin washout with ACSF, respectively, P < 0.001 compared with the same time points without rapamycin washout; Fig. 3, E and F).
Fig. 3. Enlargement of dendritic spines induced by engineered LIMK1 activation in hippocampal organotypic slice cultures.
(A) A representative two-photon image of hippocampal CA1 pyramidal neurons in organotypic slice cultures. Scale bar, 25 μm. (B) Spine density in neurons transfected with unimolecular rapamycin regulatable domain (uniRapR)–LIMK1 and enhanced green fluorescent protein (EGFP). ns, not significant; unpaired Student’s t test. (C) A series of two-photon images in a dendrite of an EGFP-transfected neuron (top) and in a dendrite of a neuron transfected with uniRapR-LIMK1 + EGFP (bottom), both before and after the application of Rap. Scale bars, 2 μm. (D) Mean time courses of spine volume obtained in EGFP-transfected neurons treated with Rap (yellow circles, EGFP + Rap), uniRapR-LIMK1 + EGFP-transfected neurons treated with Veh (black circles), Rap (green circles), Rap for 30 min before washout with artificial cerebrospinal fluid (ACSF) (pink curve, the dashed line indicates the washout), Veh in slices pretreated with a combination of KN93, IPA3, and GSK429286A (white circles, Veh + inh), and Rap in slices pretreated with a combination of KN93, IPA3, and GSK429286A (blue circles, Rap + inhibitors). (E and F) Bar graphs show the percentage of dendritic spine increments after 55 to 60 min (E) or 120 min (F) of Rap application. (G) Mean time courses of dendrite volume obtained in conditions shown in (D). Data are expressed as mean ± SEM. ***P < 0.001. Statistics by one-way analysis of variance (ANOVA) with the Dunnett’s post hoc test comparisons.
Chemogenetic activation of LIMK1 in the presence of a combination of upstream key synaptic protein blockers (i.e., 10 μM KN93, 30 μM IPA3, and 10 μM GSK429286A, for the inhibition of CaMKII, PAK and ROCK, respectively) still triggered spine enlargement (158.72 ± 5.18%, P < 0.001, and 164.83 ± 6.65%, P < 0.001, n = 98/4, volume increment after 55 to 60 min and 120 min of rapamycin addition, respectively; Fig. 3, E and F), indicating that LIMK1 activation is sufficient for structural changes in dendritic spines. Combined inhibition of CaMKII, PAK, and ROCK did not significantly affect the volume of dendritic spines throughout the recordings in neurons on which engineered LIMK1 was not activated (102.19 ± 6.13%, P = 0.626 compared with 5-min pre-application and 102.92 ± 5.92%, P = 0.543 compared with 5-min pre-application, n = 98/8, volume increment after 55 to 60 min and 120 min of vehicle addition, respectively; Fig. 3, E and F). We also monitored the time course of pS3-cofilin during the chemogenetic activation of LIMK1. We found that cofilin phosphorylation after 30 to 120 min of rapamycin application (30 min: 35.79 ± 7.83, 60 min: 31.45 ± 3.82, 120 min: 21.33 ± 8.92) was significantly higher (P < 0.001) than those observed in neurons transfected with uniRapR-LIMK1 and treated with vehicle or controls but consistently decreased from 30 to 120 min (fig. S6). All these results suggest that chemogenetic activation of uniRapR-LIMK1 with rapamycin is sufficient to induce rapid and relatively transient cofilin phosphorylation in second-to-minute time scales and persistent dendritic enlargement in the minute-to-hour time scales.
The effects of rapamycin in neurons transfected with uniRapR-LIMK1 were specific on dendritic spines because the volume of dendritic shafts was not modified throughout the recordings (101.46 ± 3.75% and 97.74 ± 5.45%, n = 8, in neurons transfected with EGFP after 60 and 120 min of rapamycin addition, respectively; 97.33 ± 2.38% and 92.82 ± 3.29%, n = 7, in neurons transfected with uniRapR-LIMK1 after 60 and 120 min of vehicle addition, respectively; 96.55 ± 1.33% and 94.96 ± 2.55%, n = 10, in neurons transfected with uniRapR-LIMK1 after 60 and 120 min of rapamycin addition, respectively; 95.26 ± 1.75% and 99.89 ± 2.18%, n = 7, in neurons transfected with uniRapR-LIMK1 at 60 and 120 min in slices treated for 30 min with rapamycin and washed with ACSF, respectively; 90.50 ± 8.32% and 95.29 ± 3.66%, n = 4, in neurons transfected with uniRapR-LIMK1 and treated with CaMKII, PAK and ROCK inhibitors after 60 and 120 min of vehicle addition, respectively; 95.67 ± 3.29% and 98.10 ± 2.97%, n = 4, in neurons transfected with uniRapR-LIMK1 and treated with CaMKII, PAK, and ROCK inhibitors after 60 and 120 min of vehicle addition, respectively; Fig. 3G).
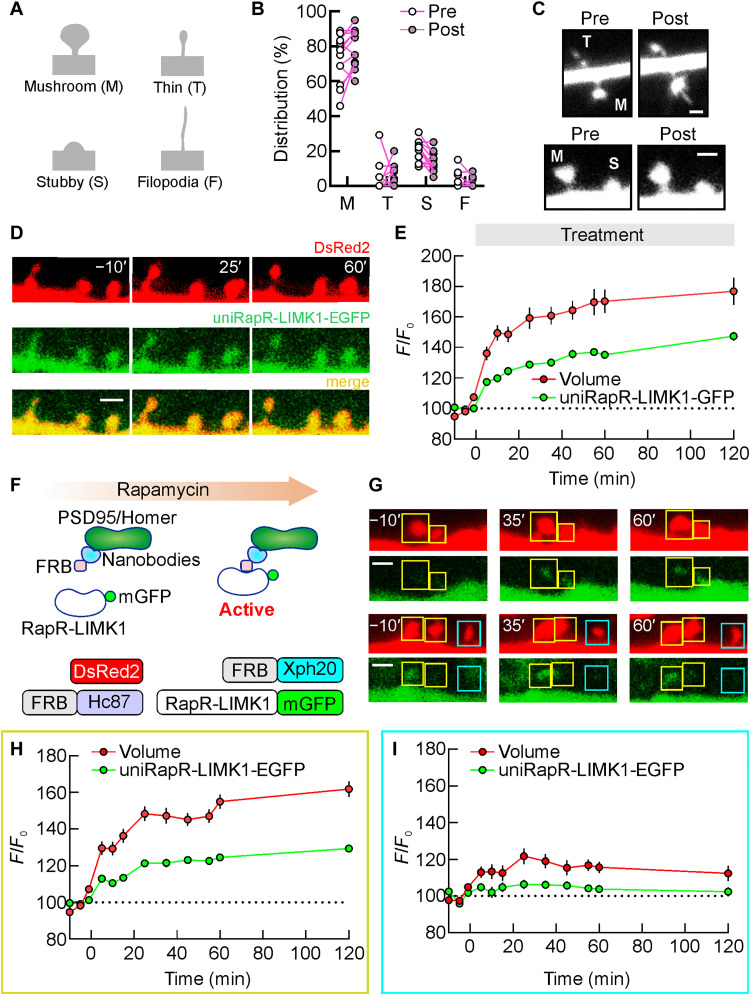
We also evaluated the effects of uniRapR-LIMK1 activation on different spine subtypes. Spine type analysis showed that the vast majority of spines in our preparations are mushroom (72.53 ± 3.93%, n = 12 dendrites) followed by stubby (20.68 ± 1.83%, n = 12 dendrites), and there is only a small percentage of thin (4.18 ± 2.49%, n = 12 dendrites) and filopodia (2.60 ± 1.37%, n = 12 dendrites) (Fig. 4, A and B). After 120 min of uniRapR-LIMK1 activation, we did not observe any notable changes in the distribution of spine types (mushroom: 79.19 ± 3.17%; stubby: 14.75 ± 1.85%; thin: 4.64 ± 1.83%; filopodia: 1.42 ± 0.80%) (Fig. 4B). For each spine subtype, we found a notable enlargement upon uniRapR-LIMK1 activation (mushroom: 52.08 ± 6.86%, n = 235/12; stubby: 43.16 ± 12.74%, n = 65/12; thin: 275.30 ± 107.60%, n = 13/12; filopodia: 48.03 ± 42.31%, n = 4/12; table S1). These findings indicate that uniRapR-LIMK1 activation can induce structural changes in different spine subtypes (Fig. 4C), which may affect synaptic function.
Fig. 4. Engineered LIMK1-mediated enlargement of different spine subtypes.
(A) Classification of dendritic spine types (mushroom, thin, stubby, and filopodia) based on morphological criteria. (B) A comparison of the spine type distribution in neurons expressing unimolecular rapamycin regulatable domain (uniRapR)–LIMK1 before and after rapamycin application. (C) Representative spine subtypes before and after activation by rapamycin. (D) A series of two-photon images in a dendrite of a transfected hippocampal CA1 pyramidal neuron before and after rapamycin application [green, enhanced green fluorescent protein (EGFP)–tagged uniRapR-LIMK1; red, DsRed2]. Spine volume (DsRed2, red) and amount of uniRapR-LIMK1 protein in the spine (EGFP, green) were quantified by measuring the total fluorescence intensity (F) relative to the averaged baseline fluorescence intensity (F0). (E) Spine volume and uniRapR-LIMK1 protein amount (mean ± SEM) were monitored for 120 min after rapamycin application. (F) Schematic of the strategy to accumulate engineered LIMK1 in the spines (top). The constructs used to accumulate engineered LIMK1 in the spines included nanobodies against Homer1 (Hc87) and PSD95 (Xph20) fused with FKBP12-rapamycin-binding (FRB) and RapR-LIMK1-EGFP (bottom). DsRed2 was used as a volume marker. (G) A series of two-photon images in a dendrite of CA1 pyramidal neuron transfected with (DsRed2, FRB-Xc87, FRB-Xph20, and RapR-LIMK1-EGFP) before and after rap application. Spines with rapamycin-mediated RapR-LIMK1-EGFP accumulation are denoted by yellow squares, while spines without accumulation are denoted by cerulean squares. Spine volume was monitored for 120 min after rapamycin application and quantified in spines with (H) or without (I) RapR-LIMK1 accumulation. Data are expressed as mean ± SEM. Scale bars, 1.5 μm.
To build a relationship between the localization of uniRapR-LIMK1 and the structural dynamics of spines, we performed additional two-photon live imaging experiments. We coexpressed uniRapR-LIMK1-EGFP along with a volumetric marker DsRed2 in CA1 neurons. We found that uniRapR-LIMK1 did not follow the volumetric change, indicating that the localization of uniRapR-LIMK1 remained mostly unchanged after rapamycin application (Fig. 4, D and E), probably because LIMK1 is localized and anchored to the plasma membrane via palmitoyl motif (36).
To enhance the accumulation of engineered LIMK1 in the spines, we transfected CA1 neurons of organotypic slices with nanobodies targeting Homer1 (Hc87) and PSD95 (Xph20) (37, 38) fused with FRB, along with RapR-LIMK1-EGFP and DsRed2. The activation of RapR-LIMK1-EGFP required a rapamycin-mediated interaction with FRB within the spines (fig. S3). Upon rapamycin application, the interaction between RapR-LIMK1-EGFP and FRB nanobodies promoted the localization of RapR-LIMK1-EGFP into the spines (Fig. 4F). Our two-photon live imaging experiments showed that spines exhibiting a rapamycin-mediated RapR-LIMK1-EGFP signal (>20%) had a significantly larger volume, as evaluated with the DsRed2 signal, compared to those with a low RapR-LIMK1-EGFP signal (154.94 ± 3.84%, n = 70/10 versus 115.66 ± 3.01%, n = 32/10, P < 0.001 and 161.84 ± 4.32%, n = 70/10 versus 112.32 ± 4.16%, n = 32/10, P < 0.001, after 60 and 120 min of rapamycin addition, respectively; Fig. 4, G to I). On the basis of these results, we conclude that the induced spine enlargement is due to the action of engineered LIMK1 within the spines.
Postsynaptic activation of engineered LIMK1 potentiates glutamatergic synaptic responses
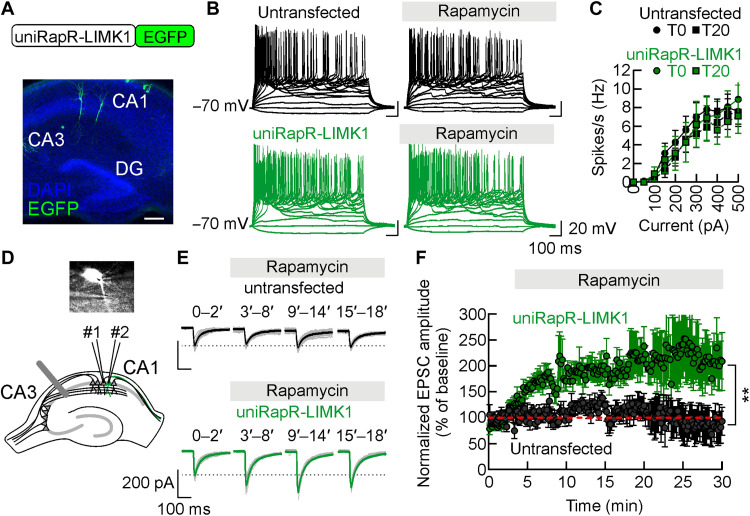
Looking for a functional correlate of the observed changes in structural plasticity induced by uniRapR-LIMK1 activation, we investigated the neuronal activity of transfected CA1 pyramidal neurons in organotypic hippocampal slice cultures using whole-cell patch-clamp recordings (Fig. 5A). Active and passive membrane properties were indistinguishable between untransfected and uniRapR-LIMK1–transfected neurons, including the number of action potentials upon current injection (50 to 500 pA, 50-pA steps from a holding potential of −70 mV). Specifically, (i) firing rate following 500-pA current injection was 7.60 ± 1.36 Hz (n = 10) in untransfected neurons and 8.90 ± 1.83 Hz (n = 10) in uniRapR-LIMK1–transfected neurons (P = 0.555; Fig. 5, B and C); (ii) resting membrane potential (untransfected: −61.92 ± 1.37 mV, n = 13; uniRapR-LIMK1: −64.71 ± 1.46 mV, n = 14; P = 0.434; fig. S7A); and (iii) input resistance (untransfected: 330.8 ± 26.7 megohm, n = 13; uniRapR-LIMK1: 292.7 ± 29.2 megohm, n = 14; P = 0.875; fig. S7B). These findings indicate that uniRapR-LIMK1 did not, per se, affect the electrical properties of neurons. In both untransfected and uniRapR-LIMK1–transfected neurons, no significant changes in excitability and membrane properties were observed upon application of rapamycin: (i) firing rate following 500-pA current injection, 7.50 ± 1.53 Hz, n = 10, in untransfected neurons and 7.10 ± 1.86 Hz, n = 10, in uniRapR-LIMK1–transfected neurons, P = 0.863; (ii) resting membrane potential, untransfected: −61.31 ± 0.55 mV, n = 13; uniRapR-LIMK1: −62.79 ± 1.17 mV, n = 14; P = 0.773; (iii) input resistance, untransfected: 327.9 ± 34.4 megohm, n = 13; uniRapR-LIMK1: 280.2 ± 47.6 megohm, n = 14; P = 0.783 (Fig. 5, B and C, and fig. S7). In voltage-clamp recordings, both inward and outward peak currents during 800-ms pulses delivered from −80 to +10 mV were not significantly different in untransfected and neighboring transfected neurons (inward peak current observed at −30 mV was 4.80 ± 0.53 nA and 3.98 ± 0.49 nA, P = 0.251; outward peak current observed at 10 mV was 0.95 ± 0.10 nA and 0.99 ± 0.11 nA, P = 0.829, in untransfected, n = 11, and uniRapR-LIMK1–transfected neurons, n = 13, respectively; fig. S7). Membrane capacitance of hippocampal CA1 pyramidal neurons was also unaffected by uniRapR-LIMK1 expression (116.9 ± 3.1 pF in untransfected neurons, n = 13, versus 112.9 ± 3.0 pF in uniRapR-LIMK1 neurons, n = 14, P = 0.357; fig. S7). Together, these data indicate that the expression and activation of uniRapR-LIMK1 did not influence neuronal excitability.
Fig. 5. Engineered LIMK1 boosts glutamatergic synaptic transmission.
(A) The construct used to study the unimolecular rapamycin regulatable domain (uniRapR)–LIMK1 activity in neurons (top). Immunofluorescent image of an organotypic hippocampal slice with few CA1 pyramidal neurons expressing uniRapR-LIMK1–enhanced green fluorescent protein (EGFP) (bottom). DG, Dentate gyrus. Scale bar, 250 μm. (B) Example traces showing action-potential generation in response to current pulses applied with 50-pA step sizes in untransfected (black traces) and uniRapR-LIMK1–transfected (green traces) neurons. (C) Frequency of action-potentials plotted against current pulse values. (D) Overlay of DIC and uniRapR-LIMK1-EGFP fluorescence images (gray scale) in hippocampal organotypic slice culture. A stimulating electrode was placed in the stratum radiatum to activate Schaffer collaterals. Dual patch-clamp whole-cell recordings were performed in two neighboring, one transfected and one untransfected, CA1 pyramidal neurons. (E) Representative traces of excitatory postsynaptic current (EPSC) recorded in untransfected and uniRapR-LIMK1–transfected neurons. In black and green, sample traces show an average of baseline recordings (0 to 2 min) and recordings during rapamycin application (3 to 8, 9 to 14, and 15 to 18 min) from untransfected and uniRapR-LIMK1–transfected neurons, respectively. (F) The time course of normalized EPSC amplitudes before and after rapamycin application. Data are expressed as mean ± SEM. **P < 0.001; statistics by two-tailed Student’s t test.
Last, we studied the effects of uniRapR-LIMK1 activation on glutamatergic synaptic transmission by dual patch-clamp recordings from transfected CA1 pyramidal neurons and adjacent untransfected neurons (Fig. 5D). We first compared the AMPA receptor–mediated excitatory postsynaptic currents (EPSCs) measured in CA1 pyramidal neurons 48 hours after transfection. Neurons expressing uniRapR-LIMK1 showed EPSC amplitudes similar to those only expressing EGFP (162.78 ± 54.37 versus 189.78 ± 37.65 pA in untransfected and uniRapR-LIMK1 expressing neurons, respectively, n = 7, P = 0.657), indicating that uniRapR-LIMK1 expression did not affect AMPA receptor–mediated transmission. Then, we measured the EPSC amplitude after rapamycin addition. In untransfected neurons, rapamycin did not significantly affect the EPSC amplitude that was 106.70 ± 13.39 (P = 0.934) and 85.70 ± 25.73% (P = 0.687) of pre-application values (n = 7) after 13 to 16 min and 25 to 28 min of rapamycin application, respectively (Fig. 5, E and F). In contrast, uniRapR-LIMK1 activation by rapamycin significantly increased the EPSC amplitude (189.07 ± 16.22%, P = 0.015 compared with pre-application and 206.94 ± 40.36%, P = 0.008 compared with pre-application, n = 7, after 13 to 16 min and 25 to 28 min of rapamycin application, respectively; Fig. 5, E and F), indicating that acute and selective LIMK1 activation enhanced glutamatergic synaptic transmission. Our data suggest that uniRapR-LIMK1 activation boosts structural and functional plasticity at glutamatergic synapses.
LIMK1 activation improves memory in aged mice
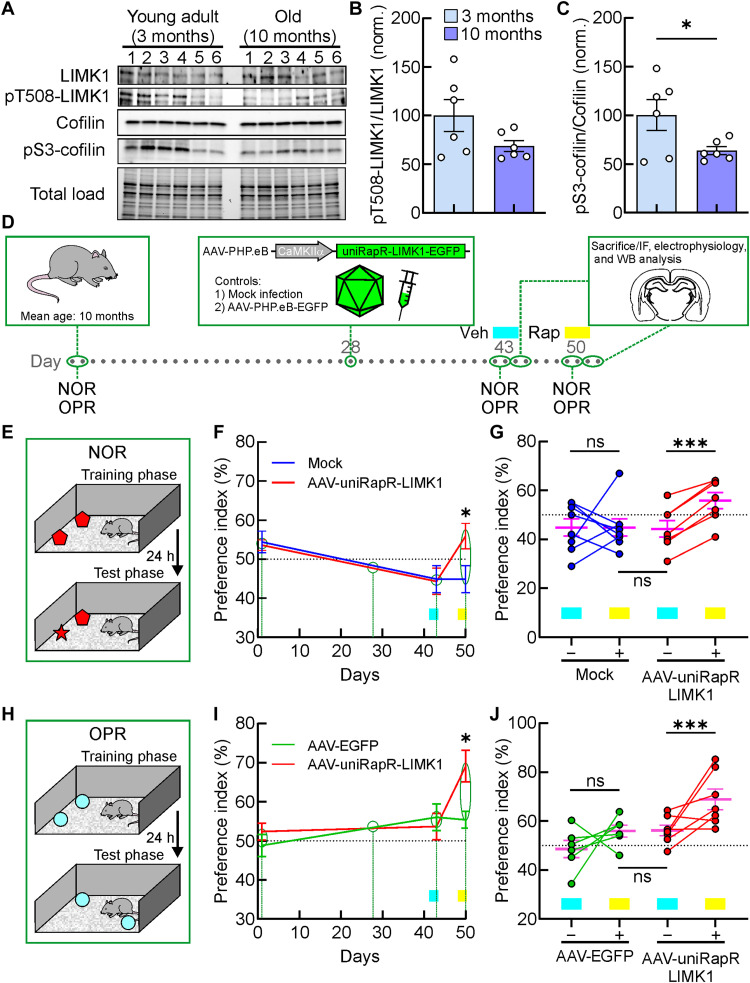
As a next step, we sought to determine the effect of our chemogenetic strategy in vivo to support a direct causal link between LIMK1-mediated actin polymerization and memory. Considering that aging can affect cognitive functions, leading to memory impairment and hippocampal dysfunctions (39), also involving alterations in dendritic spine number and morphology (25), we wondered whether the selective LIMK1 activation in vivo could counteract age-induced cognitive decline. Thus, we used the C57BL/6 mouse model, showing a spontaneous, age-related cognitive decline (39) and hippocampal dysfunctions (40) starting from about 6 months of age. We hypothesized that the progressive worsening of cognitive performance in these mice could be related to different levels of hippocampal cofilin phosphorylation. Thus, first of all, we assessed whether aging influences both LIMK1 and cofilin phosphorylation in the hippocampus of C57BL/6 mice by Western blot analysis. In 10-month-old C57BL/6 mice, we found lower levels of pS3-cofilin compared with younger mice (3 months of age), along with a trend decrease of pT508-LIMK1 in aged mice (normalized pT508-LIMK1: 100.0 ± 20.3%, n = 6, versus 70.2 ± 26.3%, n = 6, in 3 and 10 months of age C57BL/6 mice, respectively, P = 0.348; normalized pS3-cofilin: 100.2 ± 17.4%, n = 6, versus 63.7 ± 4.5%, n = 6, in 3- and 10 month-old C57BL/6 mice, respectively, P = 0.049; Fig. 6, A to C). Mice virally expressing an inactive version of cofilin into the hippocampus reduced both losses of neuronal connections and memory deficits (41).
Fig. 6. Chemogenetic activation of LIMK1 improves the hippocampal-dependent memory in vivo.
(A to C) The protein levels of LIMK1, pT508-LIMK1, cofilin, and pS3-cofilin in the hippocampus of young adult and old mice. *P < 0.05; Statistics by two-tailed Student’s t test. (D) The experimental procedure for the timing of viral injection. AAV-CaMKIIα-uniRapR-LIMK1-EGFP was injected into behaviorially tested mice before a new round of the tests, after which they were subsequently sacrified. IF, immunofluorescence; WB, Western blotting. (E) The positions of the objects during the training and the test phases. (F) Preference index (PI) in Novel Object Recognition (NOR) experiments of mock (blue line) and AAV-uniRapR-LIMK1 (red line) animals plotted with days of the experimental procedures shown in (D). Cyan and yellow rectangles indicate the timing of intranasal treatment with vehicle (Veh) or rapamycin (Rap), respectively. *P < 0.05. Statistics by two-way repeated-measures analysis of variance (ANOVA) with the Bonferroni’s post hoc test. (G) Analyses of PI in NOR experiments of each animal before (−) and after (+) intranasal Rap treatment (mock: blue circles; AAV-uniRapR-LIMK1: red circles; cumulative mean values are shown in magenta). AAV-uniRapR-LIMK1–infected mice intranasally treated with Rap explored the new object related to the familiar one significantly more, compared to the mock-infected animals intranasally treated with Rap. ***P < 0.001; statistics by paired Student’s t test. (H) A schematic for the position of the objects in object place recognition (OPR) experiments. (I) PI in OPR experiments plotted with days of the experimental procedures shown in (D). Cyan and yellow rectangles indicate the timing of intranasal treatment with Veh or Rap, respectively. *P < 0.05; statistics by two-way repeated-measures ANOVA with the Bonferroni’s post hoc test. (J) Preference index in OPR experiments of each animal before (−) and after (+) intranasal Rap injection (AAV-EGFP: green circles; AAV-uniRapR-LIMK1: red circles; cumulative mean values: magenta). ***P < 0.001; statistics by paired Student’s t test. Data in (B), (C), (F), (G), (I), and (J) are expressed as mean ± SEM. ns, not significant.
To engineer the learning and memory capacity of aged mice, we delivered our extrinsically disordered LIMK1 construct in adeno-associated virus (AAV) into the mouse brain (Fig. 6D). We first tested the neurobehavioral phenotype of WT male old C57BL/6 mice by means of the Novel Object Recognition (NOR) task (Fig. 6, D and E). In a group of old mice (mean age 10.20 ± 0.38, n = 15), we found that the preference index (PI) for the novel object was 54.00 ± 1.64%. We randomly divided animals into two groups maintaining similar PI and age parameters (group #1, PI: 54.38 ± 2.82%, mean age 10.25 ± 0.56%, n = 8; group #2, PI: 53.57 ± 1.38%, mean age 10.14 ± 0.60%, n = 7, P = 0.811; Fig. 6F). After 29 days, we randomly selected the two groups for mock and AAV infections (Fig. 6F). AAV-PHP.eB carrying uniRapR-LIMK1-EGFP were delivered by retro-orbital injection C57BL/6 mice (n = 7) at 3 × 1011 vector genomes (vg)/virus (9 × 1011 vg total). For mock-infected mice, we followed the same procedure for AAV-infected mice without using viral particles. The treatments were well tolerated, and no changes in body weight or drinking water consumption were observed. Two weeks later, both groups of animals were intranasally treated with the vehicle before the training and the test (Fig. 6F). Two weeks after infection, PI was similar for the two groups: 44.88 ± 3.46%, n = 8 versus 44.29 ± 3.39%, n = 7 for group #1–mock and group #2–AAV-uniRapR-LIMK1, respectively (P = 0.906; Fig. 6F). One week later, both groups of animals received intranasal administration of rapamycin before the training and the test (Fig. 6F). We used a dose of rapamycin solution (0.1 μg/μl) in 10 μl (1 μg per mouse; 0.05 mg/kg per mouse). The rapamycin dose was chosen from a dose-response pilot study confirming the rapamycin distribution in mouse brains (42). After rapamycin treatment, AAV-infected mice showed a higher preference for the novel object compared to mock-infected animals for the novel object (55.86 ± 3.46%, n = 7, versus 44.88 ± 3.46%, n = 8, for AAV-uniRapR-LIMK1–infected and mock-infected animals, respectively, P = 0.040; Fig. 6, F and G).
We also used WT male old C57BL/6 mice to evaluate the hippocampus-dependent learning and memory performances with the object place recognition (OPR) task (Fig. 6H). Two groups of randomly selected animals were tested for OPR, showing a similar PI (group #3, PI: 48.82 ± 2.85%, n = 12; group #4, PI: 52.39 ± 2.16%, n = 13, P = 0.987; Fig. 6I). After 29 days, group #3 was selected for retro-orbital AAV infections with AAV-PHP.eB carrying EGFP alone as an additional control over the group received mock infections, and group #4 was the AAV-uniRapR-LIMK1 group. Two weeks later, both groups of animals were intranasally treated with the vehicle before the training and the test. The PI was similar for the two groups: 56.06 ± 3.41%, n = 12 versus 53.66 ± 3.39%, n = 13 for group #3–AAV-EGFP and group #4–AAV-uniRapR-LIMK1, respectively (P > 0.999; Fig. 6I). At this point, half of the animals of the two groups were sacrificed for immunohistochemistry, electrophysiology, or molecular experiments. One week later, both groups of the remaining animals received intranasal administration of rapamycin before the training and the test. Also, in the OPR task, we found that animals infected with AAV-uniRapR-LIMK1 and treated with rapamycin showed a significantly higher PI compared to controls (69.12 ± 4.04%, n = 7, versus 55.37 ± 2.18%, n = 6, for AAV-uniRapR-LIMK1– and AAV-EGFP–infected animals, respectively, P = 0.045; Fig. 6, I and H). We excluded the possibility that the behavioral effects were due to the intranasal injection of the rapamycin per se, as the same dose of rapamycin used in mock- and AVV-EGFP–infected mice did not affect the PI in NOR and OPR tests (Fig. 6, E to G). Moreover, no changes were observed in the exploration levels during the test phase (NOR: 14.99 ± 2.01 s, n = 8 versus 18.79 ± 3.42 s, n = 7 for group #1–mock and group #2–AAV-uniRapR-LIMK1, respectively, P = 0.307; OPR: 30.48 ± 7.42 s, n = 6 versus 21.82 ± 4.50 s, n = 7 for group #3–AAV-EGFP and group #4–AAV-uniRapR-LIMK1, respectively, P = 0.412; fig. S8), suggesting that engineered LIMK1 activation did not change the level of exploration for the familiar objects but specifically modulated the preference for the novel object during memory tests.
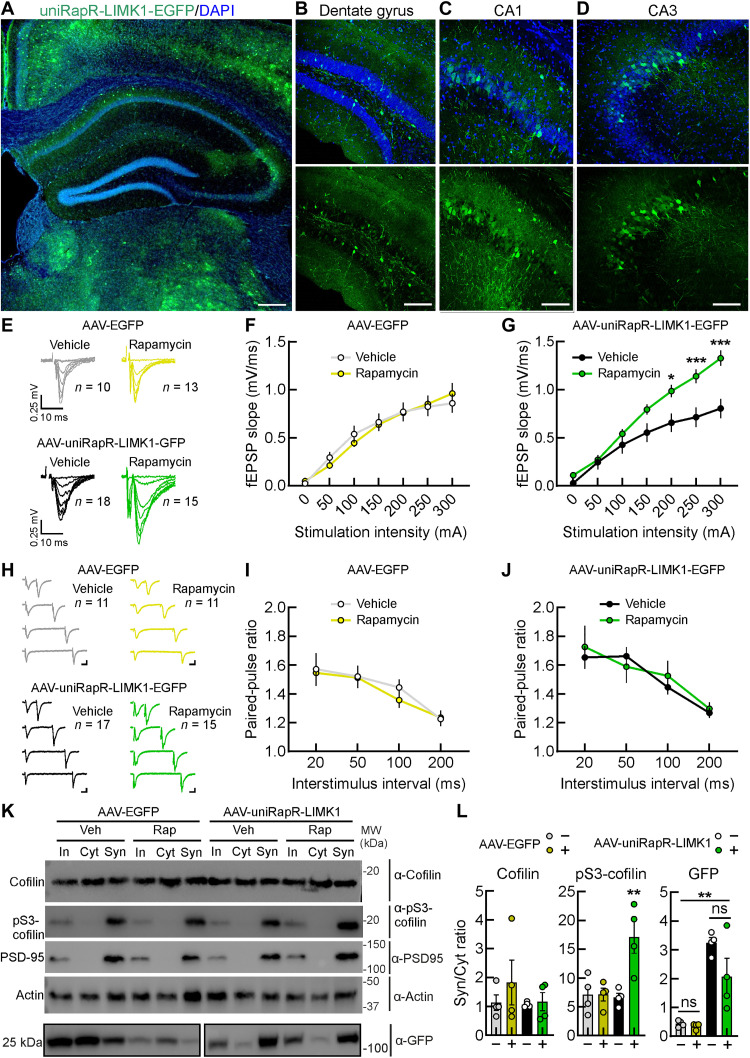
UniRapR-LIMK1-EGFP was packaged in AAV-PHP.eB, and its expression was driven by CaMKIIα promoter, allowing in excitatory neurons throughout the brain, including the hippocampus and dendritic spines (Fig. 7 and fig. S9). In coronal brain slices obtained from the AAV-EGFP– and AAV-uniRapR-LIMK1–infected mice, we found a similar and relatively sparse % of EGFP-positive neurons (AAV-EGFP: 18.47 ± 4.17% from five hippocampal slices and AAV-uniRapR-LIMK1: 17.07 ± 3.87% from five hippocampal slices; Fig. 7, A to D, and fig. S9, A and B). However, the EGFP-positive neurons from the AAV-uniRapR-LIMK1–infected mice showed a significant increase in the fluorescent intensity for LIMK1 (44.47 ± 3.28, n = 48/3 versus 120.70 ± 8.20, n = 50/3 in EGFP fluorescent neurons from AAV-EGFP– and AAV-uniRapR-LIMK1–infected mice, respectively, P < 0.0001; fig. S9, C and D). Considering that it has been demonstrated that a small percentage of neurons (roughly 15%) could be sufficient to encode memories (43), the infection rate of our AAV should be enough to manipulate memory in vivo.
Fig. 7. Enhanced synaptic transmission upon in vivo chemogenetic activation of LIMK1.
(A) A coronal brain slice of a mouse (10 months old) infected with AAV-uniRapR-LIMK1-EGFP and examined 3 weeks after infection using confocal microscopy. Insets show a zoomed-in view of the hippocampus: Dentate gyrus (B), CA1 (C), and CA3 (D). Scale bars, 300 μm (A) and 100 μm (insets). (E) Representative traces of CA1 field excitatory postsynaptic potentials (fEPSPs) evoked at different stimulation intensities in slices obtained from AAV-EGFP– and AAV-uniRapR-LIMK1-EGFP–infected mice injected intranasally with vehicle or rapamycin. (F) Input-output curves constructed from the fEPSP slope recorded with increasing stimulation intensities in hippocampal slices of AAV-EGFP–infected mice intranasally treated with vehicle (n = 10 from five mice) and rapamycin (n = 13 from five mice). (G) Average input-output slopes measured between 1 and 300 μA resulted in a significant increase in input-output slope in AAV-uniRapR-LIMK1-EGFP–infected mice and intranasal treated with rapamycin relative to those injected with vehicle. *P < 0.05; ***P < 0.001. (H) Representative traces of paired CA1 fEPSPs evoked with different interstimulus intervals (50, 100, 150, and 200 ms) in slices obtained from AAV-EGFP– and AAV-uniRapR-LIMK1-EGFP–infected mice intranasally injected with vehicle or rapamycin. Scale bars, 0.25 mV and 10 ms. Quantitative analysis of fEPSP slope paired-pulse ratio in AAV-EGFP (I)– and AAV-uniRapR-LIMK1-EGFP (J)–infected mice intranasally injected with vehicle or rapamycin. (K and L) Western blot analyses of total lysate [input (In)], cytosolic (Cyt), and synaptosomal (Syn) fractions of hippocampi from AAV-EGFP– and AAV-uniRapR-LIMK1-EGFP–infected mice following intranasal administration of either vehicle or rapamycin. **P < 0.01. Statistics by one-way analysis of variance (ANOVA) with the Dunnett’s post hoc test comparisons. Data in (F), (G), (I), (J), and (L) are expressed as mean ± SEM. Statistics in (F), (G), (I), and (J) by two-way repeated-measures ANOVA with the Bonferroni’s post hoc test. MW, molecular weight.
To evaluate the synaptic impact of the hippocampus-dependent learning and memory improvements observed, we assessed the basal synaptic transmission from Schaffer collateral–CA1 synapses from acute hippocampal slices of AAV-EGFP– and AAV-uniRapR-LIMK1–infected mice that were intranasally treated with the vehicle or rapamycin. To generate input/output curves, we measured field excitatory postsynaptic potentials (fEPSPs) in response to increasing stimulus intensities. We found that slices from AAV-uniRapR-LIMK1–infected mice intranasally treated with rapamycin displayed a substantial increase in synaptic responses (Fig. 7, E and G). We excluded that these effects were due to rapamycin per se (Fig. 7, E and F) or transgene expression (slices from AAV-uniRapR-LIMK1–infected mice intranasally treated with vehicle; Fig. 7, E and G). In the same Schaffer collateral–CA1 synapses, we studied the paired-pulse ratio of fEPSPs in response to two consecutive stimulations with different interstimulus intervals (50, 100, 150, and 200 ms) to evaluate presynaptic function. We did not find any differences in paired-pulse ratios among the groups (Fig. 7, H to J), suggesting no major effects of chemogenetic LIMK1 activation on presynaptic mechanisms, e.g., modification of the glutamate release probability. These electrophysiological studies suggest a prevalent role in chemogenetic LIMK1 activation on postsynaptic sites, i.e., dendritic spines.
Last, in the hippocampal extracts of infected mice treated with either vehicle or rapamycin, we isolated the synaptosomes and cytosols to evaluate the levels of cofilin and pS3-cofilin. The elevated levels of PSD95 observed in synaptosomes merely indicate the quality of the preparation (Fig. 7K).
Intranasal rapamycin administration significantly increased the amount of pS3-cofilin in synaptosomes of the hippocampal extracts of AAV-uniRapR-LIMK1–infected mice (P = 0.0079, P = 0.0078, and P = 0.0056 for AAV-uniRapR-LIMK1-EGFP + Rap versus AAV-EGFP + Veh, AAV-EGFP + Rap, and AAV-uniRapR-LIMK1-EGFP + Veh, respectively). In contrast, the total cofilin amount was unaffected (Fig. 7, K to L). This effect could result from a positive feedback cycle in which the activation of uniRapR-LIMK1 promotes pS3-cofilin in some neurons, which has the consequence of facilitating the activation of neuronal circuits and promoting more generalized cofilin phosphorylation. We also observed an enrichment of uniRapR-LIMK1-EGFP, but not EGFP alone, in the synaptosomes (Fig. 7, K to L). The presence of rapamycin or vehicle did not change either the expression or the localization of uniRapR-LIMK1, but rapamycin increased the uniRapR-LIMK1–mediated cofilin phosphorylation in synaptosomes, a mechanism required for increasing spine morphogenesis through actin polymerization. All these results show that cofilin is phosphorylated by rapamycin-activated uniRapR-LIMK1 in synaptosomes in vivo.
DISCUSSION
Genetically encoded engineered proteins that can be spatiotemporally controlled by safe, inert molecules offer an unprecedented opportunity for both understanding the physiological role of a specific protein and for next-generation therapeutic applications. The technology used here enables protein activation in vivo with rapamycin or its non-immunosuppressive analogs, which was shown to have life-spanning effects along with some manageable side effects (42, 44, 45). In this study, our genetically encoded chemically activatable LIMK1 is successfully used to control actin dynamics through cofilin phosphorylation both in vitro, ex vivo, and in vivo. The selective activation of the engineered LIMK1 was sufficient to induce long-term enlargement of dendritic spines, boost glutamatergic synaptic transmission at CA3-CA1 synapses in the hippocampus, and improve memory in aged mice.
Several strategies for spatiotemporal regulation of cofilin in living cells have been developed (46, 47). Ghosh et al. (47) used purified and chemically modified cofilin that can be locally photoactivated, but it requires the microinjection of the purified protein into the cells, which negatively affects cell health, hindering in vivo applications. Stone and colleagues (46) designed a genetically encoded optically controllable cofilin by using LOV2 domain, but this approach cannot inactivate cofilin, hindering the investigation of the spine enlargement in neurons. Because even small amounts of either active or inactive cofilin can influence actin dynamics, we decided to develop an approach to control its direct inactivator LIMK1 (5, 48). Our engineered LIMK1 has similar expression levels compared to LIMK1 WT, and engineering did not affect its interaction with its native binders TrkB. Potentially, it should not affect other interactors that bind to LIM and PDZ domains, as the extrinsic disorder is engineered only to affect the kinase domain. Also related to the kinase domain interactors, we demonstrated that engineered LIMK1 did not lose the ability to interact with SSH1 (fig. S4C). We found that selective chemical activation of LIMK1 controlled phosphorylation of cofilin at S3 in vitro (Fig. 1H) and in living cells (Fig. 2). Notably, the background level of pS3-cofilin in the presence of inactivated LIMK1 analogs was almost zero, as expected from an ideal engineered on/off protein switch, enabling the precise control of LIMK1 activation over time (Figs. 1H and 2).
In addition to LIMK1, another important cofilin regulatory protein is LIMK2 (49, 50). However, despite its expression in the brain, LIMK2 cannot compensate for LIMK1 loss, as demonstrated by spine abnormalities and cognitive impairment in LIMK1 knockout mice and LIMK1 mutations in humans (3, 23, 36, 51). The spine-specific role of LIMK1, compared with LIMK2, is related to the presence of a palmitoyl motif responsible for localizing and anchoring LIMK1 in spines (36). Thus, the LIMK1-cofilin axis may play an essential role in regulating actin filament dynamics in dendritic spines (3, 23, 36).
Our findings provide insights into the role of LIMK1 and actin polymerization in glutamatergic synaptic physiology. We found that acute and selective activation of LIMK1 is sufficient to persistently increase the size of dendritic spines in CA1 pyramidal neurons (Fig. 3). Previous studies have demonstrated the importance of LIMK1 in dendritic spine plasticity, as LIMK1 knockout mice showed altered hippocampal LTP (23). Similarly, knocking down LIMK1 in organotypic brain slices by short hairpin RNA caused a substantial reduction in spine enlargement and impaired persistent retention of cofilin (3). An exacerbated LIMK1 activation in mouse models genetically overexpressing neuregulin1 showed dendritic spine impairment (52). Similarly, pharmacological inhibition of LIMK1 provides dendritic spine resilience against β amyloid protein (53). We found that the overexpression of uniRapR-LIMK1 did not, per se, affect either spine density (Fig. 3B), active and passive electrophysiological membrane properties (Fig. 5, B and C, and fig. S7), and basal synaptic transmission (Fig. 7, E to J). However, activation of uniRapR-LIMK1 alone was sufficient to induce dendritic spine enlargement also in neurons treated with CaMKII, PAK, and ROCK inhibitors (Fig. 3, D to F). LIMK1 can be phosphorylated by either PAK or ROCK family kinases (5, 54–57). Acute activation of PAK1 is also sufficient to trigger long-term dendritic spine enlargement (19), and PAK inhibition, but not ROCK, reduced LIMK1 phosphorylation in hippocampal neurons, suggesting that PAK is predominant for LIMK1 activation in spines (36). Once phosphorylated, LIMK1 promotes a rapid polymerization of actin by inhibiting the cofilin that accumulates at the bottom of the spine head in the early phase of LTP, enlarging the spine structure (3, 9, 36, 58, 59). Intriguingly, we also found that acute and selective activation of LIMK1 is sufficient to boost the AMPA receptor–mediated EPSCs in CA1 pyramidal neurons of organotypic hippocampal slice cultures in a similar time course (Fig. 5). In a typical induction protocol, multiple pathways such as CaMKII, PKCα, and PKA are all activated in parallel, and it is a challenge to dissect the contribution of each axis into the spine stability or instability (60–62). LIMK1 is downstream of the canonical molecular mechanism involved in structural LTP (Fig. 1) (5); thus, its selective activation has been expectedly associated with an increase in spine volume only. We found that activation of LIMK1 is also sufficient to potentiate EPSCs and to increase the input-output curves (Figs. 5 and 7), suggesting possible unrevealed roles for LIMK1 in glutamatergic synaptic transmission (Fig. 8). The increased pS3-cofilin levels resulting from chemogenetic LIMK1 activation might lead to an elevated presence of F-actin. This could, in turn, potentially create a conducive environment for glutamate receptors to indirectly associate with scaffolding proteins via binding sites (63).
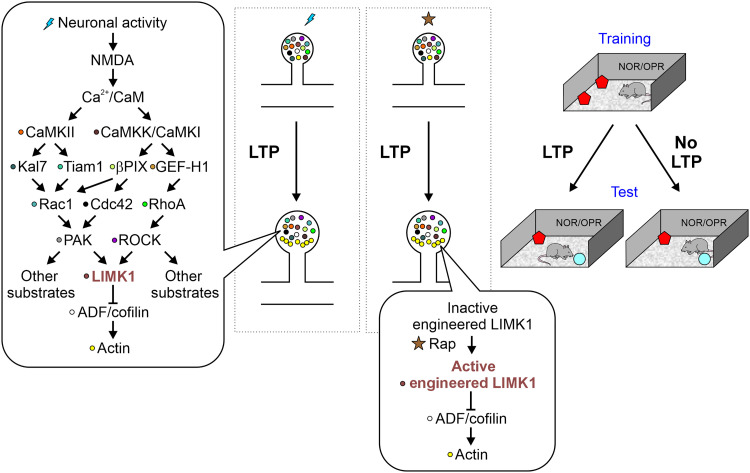
Fig. 8. A signaling mechanism during plasticity of dendritic spines.
The dendritic spine signaling during structural long-term potentiation (LTP) underlying the convergence of protein signaling on LIMK1 phosphorylation. The activation of LIMK1 promotes cofilin phosphorylation suppressing its function and promoting actin polymerization. Because pS3-cofilin/actin-depolymerizing factor (ADF) is tightly controlled as an almost exclusive substrate for LIMK1 in spines, chemogenetic activation of LIMK1 causes the inactivation of endogenous cofilin and eventually leads to spine enlargements bypassing the canonical plasticity signaling. Alterations of the signaling pathway at any level can potentially interfere with LTP and actin polymerization. The activation of LIMK1 has a strong correlation with memory formation.
LIMK1 has additional roles in synaptic physiology, playing a part in the late phase of LTP and in long-term memory (64). LIMK1 phosphorylates and regulates the transcription factor cAMP-responsive element-binding protein (CREB) (64, 65), an extensively studied protein with a well-documented role in neuronal plasticity and long-term memory formation (66). Notably, overexpression of CREB in a sparsely distributed subpopulation (~15%) of neurons in the lateral amygdala promoted the recruitment of these neurons into the fear memory trace (43).
LIMK1 is also important for the brain-derived neurotrophic factor (BDNF)–mediated signal transduction leading to the axonal outgrowth of neurons (29, 31). Nonetheless, the LIMK1-cofilin-actin axis has not been sufficiently investigated to develop strategies to convert the intracellular signaling pathway into synaptic plasticity, a crucial phenomenon for learning and memory. Our work reveals that controlled cofilin inhibition obtained by using engineered LIMK1 positively affects on learning and memory ability in vivo (Fig. 6). A more in-depth understanding of LIMK’s role in synaptic plasticity can pave the way to identify the therapeutic strategies for disorders associated with altered memory formation and cognitive processing. Notably, stimuli promoting synaptic plasticity have been suggested to determine functional recovery from neurological disorders (67). Our approach is based on a safe and well-tolerated clinically approved drug such as rapamycin that (i) crosses the blood-brain barrier (68), (ii) activates the engineered proteins (19, 21, 69–71), and (iii) has well-known beneficial effects on cognition (42, 45, 68), allowing a potential synergic beneficial effect for future translational applications.
In conclusion, here, we shed further light on the role of LIMK1-cofilin–mediated actin polymerization in dendritic spine stability, glutamatergic synaptic function, and memory using an engineered LIMK1. This technology will also be a case study for developing genetically encoded therapeutic strategies in the future.
MATERIALS AND METHODS
Ethics and animal use statement
All animal procedures were approved by the Ethics Committee of the Università Cattolica del Sacro Cuore, Sapienza University of Rome, Istituto Italiano di Tecnologia and Kyoto University. Procedures were fully compliant with Italian (Ministry of Health guidelines, Legislative Decree No. 116/1992), Japanese (Med Kyo 22042) and European Union (Directive No. 86/609/EEC) legislation on animal research. The methods were carried out in strict accordance with the approved guidelines.
Animals
Rats from the Sprague-Dawley strain (both males and females) were used for hippocampal slice culture. C57BL/6 male mice were used for infection, behavioral, immunohistochemistry, electrophysiology, and Western blotting experiments.
Reagents
KN-93, IPA3, GSK429286A, and TTX were purchased from Tocris Bioscience; Phos-tag acrylamide was purchased from Wako. Rapamycin was purchased from LC Laboratories. Cofilin monoclonal antibody (#5175), phosphorylated S3 (pS3) cofilin monoclonal antibody (#3313), and phosphorylated T508 LIMK1 polyclonal antibody (#3841) were purchased from Cell Signaling Technology. LIMK1 monoclonal antibody (#611748) was purchased from BD Biosciences. EGFP polyclonal antibody (#A11122) and 4′,6-diamidino-2-phenylindole (DAPI, #D1306) were purchased from Thermo Fisher Scientific. Flag-M2 monoclonal antibody (#F1804), Flag-M2-agarose beads (#A2220), and Flag peptide (#F3290) were purchased from Sigma-Aldrich. Myc antibody (clone 4A6, #05-724) was purchased from Sigma-Aldrich. Anti-PSD95 (#ab18258) and anti-actin (ab8227) antibodies were purchased from Abcam.
Molecular cloning and protein structures
Point mutations on LIMK1 cDNA (corresponding to T508A and T508D) were introduced with a QuikChange II system (Agilent Technologies). Insertions of iFKBP, EGFP, and uniRapR sequences into LIMK1 or nanobodies were obtained through the Gibson Assembly Reaction (Gibson Cloning MasterMix, New England Biolabs, M5520A). All restriction enzymes were purchased from New England Biolabs. The crystal structure of the kinase domain was obtained from the Protein data bank (PDB ID: 5L6W; PDB ID: 1Y57). Protein structures were predicted using AlphaFold2 as previously described (72). Sequence data were inspected using Sanger Sequencing and Fragment Analysis Software SeqScape of Applied Biosystems (Thermo Fisher Scientific) and assisted by SnapGene software (GSL Biotech).
Cell cultures, DNA transfections, and treatments
HEK293T and COS-7 cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) supplemented with 10% (v/v) fetal bovine serum (Sigma-Aldrich) and incubated at 37°C temperature and 5% CO2 conditions. For the immunoprecipitation and LIMK1 activity assay experiments, dissociated cells were plated on 100-mm cell culture dishes (Corning). DNA plasmid vectors were transfected at 60 to 90% cell confluency with (pH 7.3) polyethylenimine HCl max solution (2 mg/ml; PEI max, Polysciences). Each cell culture dish containing 3 × 106 cells received a DNA-PEI max mixture consisting of 10 μg total DNA along with 20 μg of PEI max in Opti-MEM Reduced Serum Media (Thermo Fisher Scientific). For immunofluorescence experiments, cells were plated at a density of 105 cells on 20-mm coverslips precoated with poly-l-lysine (0.1 mg/ml; Sigma-Aldrich). After 48 hours of transfection, cells were treated with 500 nM rapamycin or 99% ethanol as a vehicle (final concentration, 0.005%) for 30 or 60 min before lysis or fixation and two-photon fluorescence microscopy experiments.
Immunoprecipitation from HEK293T cells
Following the procedures previously described in (70), we lysed ~3 × 106 HEK293T cells in 1 ml of ice-cold lysis buffer [150 mM NaCl, 50 mM tris-HCl (pH 7.4), and 2 mM EDTA] containing 1% Triton X-100, 0.1% SDS, 10% glycerol, 1 × cOmplete Ultra tablets protease inhibitor cocktail (Roche), 1 mM sodium orthovanadate (Sigma-Aldrich), 11 mM β-glycerolphosphate, and 10 mM sodium fluoride (Sigma-Aldrich) with or without 500 nM rapamycin. Lysates were collected in a centrifuge tube and spun down at 14,000g at 4°C. We used 100 μl of the supernatant of each cell culture dish as input and 900 μl for immunoprecipitation with 10 μl of the anti-Flag antibody beads for 2 to 4 hours at 4°C. We washed the beads with 1 ml of lysis buffer three times. Bound proteins were eluted with SDS-PAGE sample buffer or Flag-peptide. Input was diluted with SDS-PGE sample buffer, and immunoprecipitated proteins were boiled for 5 min and subjected to Western blotting.
In vitro LIMK1 activity assay
Flag-tagged LIMK1 WT, T508D, T508A, RapR-LIMK1, and uniRapR-LIMK1 were purified from HEK293T cell using Flag-M2-agarose. After elution with Flag peptide, the concentration of LIMK1 was quantified by Western blotting using purified LIMK1 protein as a standard (73). The kinase assay was performed in a buffer containing 25 mM Hepes (pH 7.5), 5 mM MgCl2, 5 mM MnCl2, 0.5 mM EGTA, 2.5 mM dithiothreitol, bovine serum albumin (BSA) (0.1 mg/ml), and 50 mM ATP with or without 500 nM rapamycin. The kinase reaction was performed for 30 min at 30°C and 4 μg of cofilin (Cytoskeleton). The reaction was stopped by adding SDS-PAGE sample buffer.
Coimmunoprecipitation
Coimmunoprecipitation experiments were performed similarly to the protocol described by Soosairajah and colleagues (32). For the coimmunoprecipitation of TrkB and LIMK1, cells were cotransfected with TrkB-EGFP along with mCherry or Flag-tagged LIMK1 WT, Flag-tagged RapR-LIMK1 with EGFP-FRB, or Flag-tagged uniRapR-LIMK1 constructs. For the coimmunoprecipitation of SSH1 and LIMK1, we first cloned SSH1 cDNA with a myc tag at the N terminus. Then, we coexpressed myc-SSH1 alone, with LIMK1-Flag, or with uniRapR-LIMK1-Flag in cells. Cells were lysed with lysis buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, protease inhibitor cocktail, 10 mM NaF, 1 mM Na3VO4, and 10 mM NaP2O7]. The mouse Myc antibody was attached to Pierce Protein G Agarose beads for 2 to 3 hours at 4°C in the presence of BSA. The cleared lysate and the bead-antibody complex were mixed for 2 to 3 hours at 4°C. The samples were washed with the same lysis buffer three times, denatured, and prepared for Western blot. The immunoprecipitated samples (IP) and cleared lysates (input) were blotted with Flag or Myc antibodies.
Western blotting
The supernatant was quantified for protein content (DC Protein Assay; Bio-Rad). Equal amounts of protein were diluted in Laemmli buffer, boiled, and resolved by SDS-PAGE. The primary antibodies were incubated for 1 hour or overnight at 4°C and revealed with horseradish peroxidase–conjugated secondary antibodies (Cell Signaling Technology). Expression was evaluated and documented by using UVItec Cambridge Alliance.
Phos-tag SDS-PAGE
Experiments were performed as described previously (74). Acrylamide gels (12.5%) were mixed with 50 μM MnCl2 and 50 μM Phos-tag acrylamide (AAL-107, Wako). The gels containing samples separated by electrophoresis were washed with 20 mM EDTA to remove Mn2+ before Western blotting.
Immunofluorescence analysis
HEK293T cells cotransfected with cofilin-EGFP and Flag-tagged LIMK1 constructs were fixed with 4% paraformaldehyde for 20 min at room temperature (RT; 22° to 25°C), permeated with 0.1% Triton X-100 for 15 min before being blocked in 0.3% BSA for 20 min. Samples were incubated for 3 hours with anti–pS3-cofilin primary antibody diluted 1:200 in 0.3% BSA in phosphate-buffered saline (PBS), washed twice in PBS, and incubated with goat anti-rabbit secondary antibody (Alexa Fluor 546, Thermo Fisher Scientific) diluted 1:1000 in PBS at RT for 90 min, light-protected. Cell nuclei were counterstained with DAPI (Thermo Fisher Scientific; 1:1000 in PBS) for 10 min at RT, light-protected. Samples were coverslipped with an antifade medium (ProLong Gold; Thermo Fisher Scientific). Images were taken by a confocal laser scanning microscope (Nikon Ti-E, Confocal Head A1 MP) with a 40× objective lens. Analysis of cofilin phosphorylation was performed by counting pS3-cofilin–labeled cells by using ImageJ software (version 1.51s) for each condition. Data are expressed as the percentage of the pS3-cofilin–positive cells on the number of cells expressing EGFP signal.
Organotypic hippocampal slice cultures
Organotypic hippocampal slice (350 μm) cultures were prepared from postnatal day 4 to 7 rats through a McIllwain tissue chopper and placed on semi-porous membranes (Millipore) as previously described (70, 73, 75, 76). Plasmids were biolistically transfected into slices at 6 to 8 days in culture (DIV) by using Gene-Gun (Bio-Rad). Patch-clamp, two-photon laser-scanning imaging, and immunofluorescence experiments were performed 2 to 4 days later. Slices were treated with 2 μM rapamycin or 99% ethanol as a vehicle (final concentration, 0.005%).
Two-photon laser-scanning imaging
Organotypic hippocampal slice cultures were transferred to a recording chamber and superfused with ACSF containing 119 mM NaCl, 2.5 mM KCl, 4 mM CaCl2, 4 mM MgCl2, 1 mM NaH2PO4, 26 mM NaHCO3, 11 mM d-glucose, 1 mM tetrodotoxin (Tocris), and 50 mM picrotoxin (Tocris), gassed with 95% O2/5% CO2. All two-photon imaging experiments were carried out at RT. Two-photon imaging was performed at 8 to 12 DIV in primary or secondary dendrites from the distal part of the main apical dendrite of CA1 pyramidal neurons using an Alcor 920 pulsed laser (80-MHz pulse frequency, 920-nm output wavelength; Spark Lasers) or a Spirit laser (80-MHz pulse frequency, 920-nm output wavelength; Spectra Physiscs). Two-photon imaging was performed at 0.45 to 2.5 Hz, 8-μs dwell time, and at ~9-mW average power. Rapamycin or vehicle was applied through bath perfusion. Three-dimensional reconstructions of dendritic morphology were generated by the summation of EGFP and DsRed2 fluorescence values separated by 0.5 μm. The spine-head volumes were estimated from the total fluorescent intensity. Dendritic spine subtypes were characterized as previously described (70, 77, 78). Spines with a head-to-neck ratio less than 1 were labeled as thin spines, those with a ratio around 1 were categorized as stubby spines, and those with a ratio exceeding 1 were designated as mushroom spines.
Electrophysiology
Hippocampal subfields and electrode positions were identified with the aid of 4× and 40× water-immersion objectives on an upright microscope equipped with differential interference contrast optics under infrared illumination (BX51WI; Olympus) and video observation (BTE-B050-U CMOS camera; Mightex). Neighboring pairs of pyramidal cells were recorded simultaneously in CA1 as described previously (75, 76). Organotypic hippocampal slices were incubated in ACSF gassed with 95% O2/5% CO2. Action potentials and resting membrane potential were recorded in whole-cell, current-clamp mode. EPSCs were recorded in voltage-clamp configuration, with CA1 pyramidal neurons held at −70 mV. Schaffer collateral fibers were stimulated with a bipolar tungsten electrode (FHC). All experiments were performed at RT. Whole-cell recording pipettes (3 to 4 megohm) were filled with a solution containing 145 mM k-gluconate, 2 mM MgCl2, 0.1 mM EGTA, 2 mM Na2ATP, 0.2 mM NaGTP, and 10 mM Hepes pH adjusted to 7.2 with KOH for excitability experiments, and 135 mM CsMeSO3, 8 mM NaCl, 10 mM Hepes, 0.25 mM EGTA, 2 mM Mg2ATP, 0.3 mM Na3GTP, 0.1 mM spermine, 7 mM phosphocreatine, and 5 mM QX-314, pH 7.25 to 7.30 (osmolarity 300) for synaptic transmission experiment. Data were collected with a MultiClamp 700B amplifier (Molecular Devices), digitized at 10 kHz using the Digidata 1440A data acquisition system (Molecular Devices), and analyzed offline using pClamp 11 software (Molecular Devices). Rapamycin or vehicle was applied through a gravity-regulated perfusion system (flow rate, 0.3 to 0.5 ml/min). We monitored the access resistance and membrane capacity before and at the end of the experiments to ensure the recording stability and the health of the studied cells.
Immunofluorescence in organotypic hippocampal slices
Organotypic hippocampal slices were biolistically transfected with DsRed2 and empty vector or with DsRed2 and uniRapR-LIMK1-Flag. After 48 to 72 hours, organotypic hippocampal slices were treated with 2.5 μM rapamycin or 99% ethanol as a vehicle for 30, 60, 120 min and then fixed with paraformaldehyde 4% for 30 min at RT. Slices were washed and permeabilized with 0.3% Triton X-100 in PBS for 15 min and then were incubated with a blocking buffer containing PBS and 10% normal goat serum (NGS) for 2 hours. Slices were incubated with primary antibodies rabbit anti-pS3-cofilin (77G2, Cell Signaling) diluted 1:100 in PBS containing 10% NGS and 0.1% Triton X-100. The next day, slices were incubated with secondary antibody Alexa Fluor 488 donkey anti-rabbit 1:500 (Thermo Fisher Scientific; #A-21206, RRID: AB_2535792) diluted in 3% NGS in PBS for 2 hours at RT, and nuclei were counterstained with DAPI (1:1000; D1306, Invitrogen) for 15 min. Last, slices were coverslipped with ProLong Gold antifade reagent (Invitrogen). Confocal stacks made of images (1024 × 1024 pixels) were acquired at ×100 magnification with a confocal laser scanning system (Nikon Ti-E, Confocal Head A1 MP) and an oil-immersion objective.
AAV vector production, characterization, and intravenous (retro-orbital) injection
UniRapR-LIMK1 was cloned in pAAV-CWB-EGFP plasmid (AAV vector with CaMKIIa promoter and EGFP, Addgene, #61462). Plasmids were amplified and purified using a NucleoBond Xtra Maxi endotoxin-free plasmid purification kit (Macherey-Nagel, #740424.50), following the manufacturer’s instructions. AAV vectors serotype PHP.eB (AAV-PHP.eB) were produced by InnovaVector (Italy) by triple transfection of HEK293T cells using pAAV-CWB-EGFP, pUCmini-iCAP-PHP.eB, and pHelper plasmid DNA. After viral purification, physical titers were determined by averaging the titer achieved by dot blot analysis and by polymerase chain reaction quantification using TaqMan (Applied Biosystems). The titer of AAV PHP.eB-CaMKIIa-uniRapR-LIMK1-EGFP produced resulted in 3.1 × 1012 genome copies/ml. An AAV-PHB.eB-CMW-EGFP was used as a control (2.6 × 1012 genome copies/ml). Purified viruses are intravenously delivered via retro-orbital injection to mice deeply anesthetized with ketamine (70 mg/kg) and medetomidine (0.5 mg/kg). AAVs were retro-orbitally injected in anesthetized mice using an insulin syringe with permanently attached needles. Each animal received 2 × 1011 viral genome of AAV-PHP.eB-uniRapR-LIMK1-EGFP or AAV-PHP.eB-EGFP in sterile PBS. Mice were intranasally treated with PBS (vehicle) or rapamycin solution (0.1 μg/μl) in 10 μl (1 μg per mouse; 0.05 mg/kg per mouse).
Animal behavior
Behavioral analyses were performed in C57BL76 mice from 9 a.m. to 4 p.m., and data were analyzed using the behavioral tracking system ANY-MazeTM (Stoelting Co.) by an experimenter blind to the treatments. NOR and OPR were performed as previously described with slight modifications (79). Briefly, both tests consisted of a habituation phase, a training phase, and a test phase, separated from each other by 24 hours. During the habituation phase, animals were allowed to explore the testing arena (45 cm by 45 cm) to familiarize with the arena for a total time of 5 min. In the training phase, animals were placed within the testing arena and allowed to explore two identical objects placed symmetrically in the center of the arena for 5 min. The main difference between the NOR and OPR training phases was the presence of a visual cue on the wall inside the testing arena for the OPR. During the test phase, for the NOR test, one of the objects was replaced with a novel one, while for the OPR test, one of the objects was moved to a different location within the arena. For both tests, animals were allowed to explore the objects for 5 min. The exploration time (time the animal snout was directed at the object at a distance of <2 cm) recorded during the test phase was then expressed as the PI, which is the percentage of time spent exploring the novel object compared to the total object exploration. The object position and identity were alternated between animals. The objects and the arena were cleaned with 70% ethanol solution before a new animal placed in the arena.
Acute slices for field recordings
Field recordings were performed on coronal slices containing the hippocampus as previously described (80, 81). Briefly, animals were anesthetized by isoflurane inhalation (Esteve) and decapitated. The brains were rapidly removed and placed in ice-cold, sucrose-based cutting solution containing the following 72 mM tris-HCl, 18 mM TRIZMA base, 1.2 mM NaH2PO4, 30 mM NaHCO3, 2.5 mM KCl, 25 mM glucose, 20 mM Hepes, 10 mM MgSO4, 3 mM sodium pyruvate, 5 mM ascorbic acid, 0.5 mM CaCl2, and 20 mM sucrose previously saturated with carbogen (5% CO2 and 95% O2). Coronal slices (400 μm) were obtained by cutting the brain in the same solution with a vibratome (VT1200S; Leica Microsystems) and immediately transferred to an incubation chamber held at 32°C and filled with ACSF containing the following 124 mM NaCl, 3.2 mM KCl, 1.2 mM NaH2PO4, 1 mM MgCl2, 2 mM CaCl2, 26 mM NaHCO3, and 10 mM glucose (pH 7.4). During incubations, the chambers were continuously bubbled with 95% O2/5% CO2. Last, slices were equilibrated at RT for at least 45 min. Slices were maintained at 25°C, and the recordings were performed in the same ACSF solution used for recovery. Data were collected with a MultiClamp 700B amplifier (Molecular Devices), digitized at 10 kHz using the Digidata 1440A data acquisition system (Molecular Devices), and analyzed offline using pClamp 11 software (Molecular Devices).
Synaptosome preparation
For the preparation of synaptosomes, all procedures were conducted at a temperature of 4°C or on ice. The buffers used in all steps were supplemented with protease and phosphatase inhibitors from Thermo Fisher Scientific, as previously described (82). The hippocampi from infected mice were homogenized in Syn-PER Synaptic Protein Extraction Reagent (Thermo Fisher Scientific, #87793) at a ratio of approximately 1 ml per 100 mg of tissue while maintaining a cold environment. After homogenization, the samples were centrifuged at 1200g for 10 min at 4°C. A portion of the resulting supernatant was saved as the total homogenate (input). Next, this homogenate was centrifuged again at 15,000g for 20 min at 4°C, yielding a supernatant that represents the cytosolic fraction. The obtained synaptosome pellets were then resuspended in Syn-Per at a ratio of approximately 0.1 ml per 100 mg tissue. Last, the samples were subjected to resolution using SDS-PAGE.
Immunohistochemistry
Mice were deeply anesthetized with ketamine (70 mg/kg) and medetomidine (1 mg/kg) and were transcardially perfused with PBS (0.1 M, pH 7.4) followed by 4% paraformaldehyde. Brains were collected, postfixed overnight at 4°C in paraformaldehyde, and then transferred to a solution of 30% sucrose in 0.1 M PBS. Coronal brain sections (40 μm thick) were then obtained using a vibratome (VT1000S, Leica Microsystems). Sections were incubated with a blocking solution containing PBS with 1% BSA, 10% NGS (Sigma-Aldrich), and 0.5% Triton X-100 for 1 hour at RT. Then, slices were treated with rabbit anti-LIMK1 monoclonal antibody diluted 1:100 in PBS containing 1% BSA, 5% NGS, and 0.3% Triton X-100 for 48 hours at 4°C. Then, slices were incubated with Alexa Fluor 546 donkey anti-mouse (Thermo Fisher Scientific; #A10036) secondary antibody diluted 1:500 in PBS for 90 min at RT. Cell nuclei were counterstained for 10 min with DAPI (0,5 mg/ml; Thermo Fisher Scientific), and the sections were mounted on glass slides and cover-slipped with ProLong Gold antifade reagent (Invitrogen). Confocal stacks made of images (1024 × 1024 pixels) were acquired at ×20 magnification with a confocal laser scanning system (Nikon Ti-E, Confocal Head A1 MP).
Statistical analysis
The statistical tests used [i.e., Student’s t test, one-way analysis of variance (ANOVA) with the Dunnett’s post hoc test comparisons, and two-way repeated-measures ANOVA with the Bonferroni’s post hoc test] are indicated in the corresponding figure legends for each experiment. All statistical tests were two-tailed, and the level of significance was set at 0.05. Results are shown as mean ± SEM.
Acknowledgments
Funding: This work was supported by the Alzheimer’s Association Research Grant (AARG-21-847593 to C.R.) from the Alzheimer’s Association, Italian Ministry of Health (GR-2021-12373657) and Italian Ministry of University and Research (PRIN: 20223S7JKZ). C.R. research was also supported by the Italian Ministry of Health (GR-2016-02363982), Japan Society for the Promotion of Science (JSPS) short-term fellow, and intramural funds (D1 and D3.2). T.S. was supported by MEXT, JP21H02595, JP21H05692, and the Takeda Foundation. Y.H. was supported by Grant-in-Aid for Scientific Research 18H04733, 18H05434 from the MEXT, Japan; Human Frontier Science Program (RGP0020/2019); Japan Science and Technology Corporation (CREST JPMJCR20E4); and Ministry of Education, Culture, Sports, Science, and Technology (JP22K21353). The research in O.D.’s group was supported by the European Research Council Starting Grant (E-CTRL, 101078163), Swedish Research Council (2021-02848), and The Strategic Research Area Neuroscience (StratNeuro). We would like to acknowledge the contribution of the Core Facility G-STeP “Electrophysiology”, Fondazione Policlinico Universitario “A. Gemelli” IRCCS and the financial support of “Ricerca Corrente 2023” from Fondazione Policlinico Universitario “A. Gemelli” IRCCS to C.G..
Author contributions: C.R. and C.G. conceived and coordinated the study. C.R., T.S., and O.D. engineered the chemogenetic LIMK1. C.R. carried out and analyzed all research not otherwise attributed. O.D. carried out image analyses of actin polymerization, biochemistry experiments of SSH1, and structural analyses. P.R. cloned AAV PHP.eB-CaMKIIa-uniRapR-LIMK1-EGFP and performed the sequencing of the plasmids. F.Pac. and M.B. performed and analyzed the immunofluorescent experiments. M.B. and A.S. contributed to the live-neuronal two-photon imaging experiments. R.S. contributed to Western blotting experiments and performed Phos-Tag SDS-PAGE experiments. T.F. and Y.H. provided the resources and supervised the live-neuronal two-photon imaging studies. F.Pas. contributed to the patch-clamp experiments and performed Western blotting experiments from synaptosomes. A.T. and E.B. performed Western blotting experiments from hippocampi homogenates. P.R., S.M., and M.R. infected mice and performed behavioral experiments. C.R., O.D., and C.G. wrote the manuscript with input from all authors. All authors approved the final version of the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, C.R. (cristian.ripoli@unicatt.it).
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Table S1
REFERENCES AND NOTES
- 1.Y. Hayashi, A. K. Majewska, Dendritic spine geometry: Functional implication and regulation. Neuron 46, 529–532 (2005). [DOI] [PubMed] [Google Scholar]
- 2.R. Yasuda, Biophysics of biochemical signaling in dendritic spines: Implications in synaptic plasticity. Biophys. J. 113, 2152–2159 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.M. Bosch, J. Castro, T. Saneyoshi, H. Matsuno, M. Sur, Y. Hayashi, Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron 82, 444–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.M. Matsuzaki, G. C. R. Ellis-Davies, T. Nemoto, Y. Miyashita, M. Iino, H. Kasai, Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat. Neurosci. 4, 1086–1092 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.T. Saneyoshi, Y. Hayashi, The Ca2+ and Rho GTPase signaling pathways underlying activity-dependent actin remodeling at dendritic spines. Cytoskeleton 69, 545–554 (2012). [DOI] [PubMed] [Google Scholar]
- 6.M. A. Lynch, Long-term potentiation and memory. Physiol. Rev. 84, 87–136 (2004). [DOI] [PubMed] [Google Scholar]
- 7.R. G. M. Morris, E. Anderson, G. S. Lynch, M. Baudry, Selective impairment of learning and blockade of long-term potentiation by an N-methyl-D-aspartate receptor antagonist, AP5. Nature 319, 774–776 (1986). [DOI] [PubMed] [Google Scholar]
- 8.S. Nabavi, R. Fox, C. D. Proulx, J. Y. Lin, R. Y. Tsien, R. Malinow, Engineering a memory with LTD and LTP. Nature 511, 348–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.J. Noguchi, T. Hayama, S. Watanabe, H. Ucar, S. Yagishita, N. Takahashi, H. Kasai, State-dependent diffusion of actin-depolymerizing factor/cofilin underlies the enlargement and shrinkage of dendritic spines. Sci. Rep. 6, 32897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Y. Nakahata, R. Yasuda, Plasticity of spine structure: Local signaling, translation and cytoskeletal reorganization. Front. Synaptic Neurosci. 10, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. K. Chetty, B. H. Ha, T. J. Boggon, Rho family GTPase signaling through type II p21-activated kinases. Cell. Mol. Life Sci. 79, 598 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.R. Shema, T. C. Sacktor, Y. Dudai, Rapid erasure of long-term memory associations in the cortex by an inhibitor of PKMζ. Science 317, 951–953 (2007). [DOI] [PubMed] [Google Scholar]
- 13.A. M. Lee, B. R. Kanter, D. Wang, J. P. Lim, M. E. Zou, C. Qiu, T. McMahon, J. Dadgar, S. C. Fischbach-Weiss, R. O. Messing, Prkcz null mice show normal learning and memory. Nature 493, 416–419 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.P. H. Chu, D. Tsygankov, M. E. Berginski, O. Dagliyan, S. M. Gomez, T. C. Elston, A. V. Karginov, K. M. Hahn, Engineered kinase activation reveals unique morphodynamic phenotypes and associated trafficking for Src family isoforms. Proc. Natl. Acad. Sci. U.S.A. 111, 12420–12425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.E. L. Yap, N. L. Pettit, C. P. Davis, M. A. Nagy, D. A. Harmin, E. Golden, O. Dagliyan, C. Lin, S. Rudolph, N. Sharma, E. C. Griffith, C. D. Harvey, M. E. Greenberg, Bidirectional perisomatic inhibitory plasticity of a Fos neuronal network. Nature 590, 115–121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O. Dagliyan, K. M. Hahn, Controlling protein conformation with light. Curr. Opin. Struct. Biol. 57, 17–22 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D. Tischer, O. D. Weiner, Illuminating cell signalling with optogenetic tools. Nat. Rev. Mol. Cell Biol. 15, 551–558 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A. Hayashi-Takagi, S. Yagishita, M. Nakamura, F. Shirai, Y. I. Wu, A. L. Loshbaugh, B. Kuhlman, K. M. Hahn, H. Kasai, Labelling and optical erasure of synaptic memory traces in the motor cortex. Nature 525, 333–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O. Dagliyan, A. V. Karginov, S. Yagishita, M. E. Gale, H. Wang, C. DerMardirossian, C. M. Wells, N. V. Dokholyan, H. Kasai, K. M. Hahn, Engineering Pak1 Allosteric Switches. ACS Synth. Biol. 6, 1257–1262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.S. Hamill, H. J. Lou, B. E. Turk, T. J. Boggon, Structural basis for noncanonical substrate recognition of cofilin/ADF proteins by LIM kinases. Mol. Cell 62, 397–408 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O. Dagliyan, M. Tarnawski, P.-H. Chu, D. Shirvanyants, I. Schlichting, N. V. Dokholyan, K. M. Hahn, Engineering extrinsic disorder to control protein activity in living cells. Science 354, 1441–1444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O. Dagliyan, D. Shirvanyants, A. V. Karginov, F. Ding, L. Fee, S. N. Chandrasekaran, C. M. Freisinger, G. A. Smolen, A. Huttenlocher, K. M. Hahn, N. V. Dokholyan, Rational design of a ligand-controlled protein conformational switch. Proc. Natl. Acad. Sci. U.S.A. 110, 6800–6804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Y. Meng, Y. Zhang, V. Tregoubov, C. Janus, L. Cruz, M. Jackson, W. Y. Lu, J. F. MacDonald, J. Y. Wang, D. L. Falls, Z. Jia, Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 35, 121–133 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Y. Meng, H. Takahashi, J. Meng, Y. Zhang, G. Lu, S. Asrar, T. Nakamura, Z. Jia, Regulation of ADF/cofilin phosphorylation and synaptic function by LIM-kinase. Neuropharmacology 47, 746–754 (2004). [DOI] [PubMed] [Google Scholar]
- 25.C. K. Walker, J. H. Herskowitz, Dendritic spines: Mediators of cognitive resilience in aging and Alzheimer’s disease. Neuroscientist 27, 487–505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. V. Karginov, F. Ding, P. Kota, N. V. Dokholyan, K. M. Hahn, Engineered allosteric activation of kinases in living cells. Nat. Biotechnol. 28, 743–747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.A. V. Karginov, K. M. Hahn, Allosteric activation of kinases: Design and application of RapR kinases. Curr. Protoc. Cell Biol. 53, 14.13.1–14.13.16 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D. Chatterjee, F. Preuss, V. Dederer, S. Knapp, S. Mathea, Structural aspects of LIMK regulation and pharmacology. Cell 11, 142 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.A. Saito, K. Miyajima, J. Akatsuka, H. Kondo, T. Mashiko, T. Kiuchi, K. Ohashi, K. Mizuno, CaMKIIβ-mediated LIM-kinase activation plays a crucial role in BDNF-induced neuritogenesis. Genes Cells 18, 533–543 (2013). [DOI] [PubMed] [Google Scholar]
- 30.A. Jayo, M. Parsons, J. C. Adams, A novel Rho-dependent pathway that drives interaction of fascin-1 with p-Lin-11/Isl-1/Mec-3 kinase (LIMK) 1/2 to promote fascin-1/actin binding and filopodia stability. BMC Biol. 10, 72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Q. Dong, Y.-S. Ji, C. Cai, Z.-Y. Chen, LIM Kinase 1 (LIMK1) interacts with Tropomyosin-related kinase B (TrkB) and mediates Brain-derived Neurotrophic Factor (BDNF)-induced axonal elongation. J. Biol. Chem. 287, 41720–41731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. Soosairajah, S. Maiti, O. Wiggan, P. Sarmiere, N. Moussi, B. Sarcevic, R. Sampath, J. R. Bamburg, O. Bernard, Interplay between components of a novel LIM kinase-slingshot phosphatase complex regulates cofilin. EMBO J. 24, 473–486 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.A. Nagasaki, S. T. Kijima, T. Yumoto, M. Imaizumi, A. Yamagishi, H. Kim, C. Nakamura, T. Q. P. Uyeda, The position of the GFP tag on actin affects the filament formation in mammalian cells. Cell Struct. Funct. 42, 131–140 (2017). [DOI] [PubMed] [Google Scholar]
- 34.N. Yang, O. Higuchi, K. Mizuno, Cytoplasmic localization of LIM-kinase 1 is directed by a short sequence within the PDZ domain. Exp. Cell Res. 241, 242–252 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Y. Fukazawa, Y. Saitoh, F. Ozawa, Y. Ohta, K. Mizuno, K. Inokuchi, Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron 38, 447–460 (2003). [DOI] [PubMed] [Google Scholar]
- 36.J. George, C. Soares, A. Montersino, J.-C. Beique, G. M. Thomas, Palmitoylation of LIM Kinase-1 ensures Spine-Specific actin polymerization and morphological plasticity. eLife 4, e06327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.C. Rimbault, K. Maruthi, C. Breillat, C. Genuer, S. Crespillo, V. Puente-Muñoz, I. Chamma, I. Gauthereau, S. Antoine, C. Thibaut, F. W. J. Tai, B. Dartigues, D. Grillo-Bosch, S. Claverol, C. Poujol, D. Choquet, C. D. Mackereth, M. Sainlos, Engineering selective competitors for the discrimination of highly conserved protein-protein interaction modules. Nat. Commun. 10, 4521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J. X. Dong, Y. Lee, M. Kirmiz, S. Palacio, C. Dumitras, C. M. Moreno, R. Sando, L. F. Santana, T. C. Südhof, B. Gong, K. D. Murray, J. S. Trimmer, A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons. eLife 8, e48750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. O. Hendrickx, S. de Moudt, E. Calus, P. P. de Deyn, D. van Dam, G. R. Y. de Meyer, Age-related cognitive decline in spatial learning and memory of C57BL/6J mice. Behav. Brain Res. 418, 113649 (2022). [DOI] [PubMed] [Google Scholar]
- 40.D. Beckmann, M. Feldmann, O. Shchyglo, D. Manahan-Vaughan, Hippocampal synaptic plasticity, spatial memory, and neurotransmitter receptor expression are profoundly altered by gradual loss of hearing ability. Cereb. Cortex 30, 4581–4596 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R. Havekes, A. J. Park, J. C. Tudor, V. G. Luczak, R. T. Hansen, S. L. Ferri, V. M. Bruinenberg, S. G. Poplawski, J. P. Day, S. J. Aton, K. Radwańska, P. Meerlo, M. D. Houslay, G. S. Baillie, T. Abel, Sleep deprivation causes memory deficits by negatively impacting neuronal connectivity in hippocampal area CA1. eLife 5, e13424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. Tramutola, C. Lanzillotta, E. Barone, A. Arena, I. Zuliani, L. Mosca, C. Blarzino, D. A. Butterfield, M. Perluigi, F. Di Domenico, Intranasal rapamycin ameliorates Alzheimer-like cognitive decline in a mouse model of Down syndrome. Transl. Neurodegener. 7, 28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.J. H. Han, S. A. Kushner, A. P. Yiu, H.-L. L. Hsiang, T. Buch, A. Waisman, B. Bontempi, R. L. Neve, P. W. Frankland, S. A. Josselyn, Selective erasure of a fear memory. Science 323, 1492–1496 (2009). [DOI] [PubMed] [Google Scholar]
- 44.R. Strong, R. A. Miller, M. Bogue, E. Fernandez, M. A. Javors, S. Libert, P. A. Marinez, M. P. Murphy, N. Musi, J. F. Nelson, M. Petrascheck, P. Reifsnyder, A. Richardson, A. B. Salmon, F. Macchiarini, D. E. Harrison, Rapamycin-mediated mouse lifespan extension: Late-life dosage regimes with sex-specific effects. Aging Cell 19, e13269 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R. Selvarani, S. Mohammed, A. Richardson, Effect of rapamycin on aging and age-related diseases—Past and future. GeroScience 43, 1135–1158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O. J. Stone, N. Pankow, B. Liu, V. P. Sharma, R. J. Eddy, H. Wang, A. T. Putz, F. D. Teets, B. Kuhlman, J. S. Condeelis, K. M. Hahn, Optogenetic control of cofilin and αTAT in living cells using Z-lock. Nat. Chem. Biol. 15, 1183–1190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M. Ghosh, X. Song, G. Mouneimne, M. Sidani, D. S. Lawrence, J. S. Condeelis, Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743–746 (2004). [DOI] [PubMed] [Google Scholar]
- 48.R. W. Scott, M. F. Olson, LIM kinases: Function, regulation and association with human disease. J. Mol. Med. 85, 555–568 (2007). [DOI] [PubMed] [Google Scholar]
- 49.T. Sumi, K. Matsumoto, Y. Takai, T. Nakamura, Cofilin phosphorylation and actin cytoskeletal dynamics regulated by Rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol. 147, 1519–1532 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.T. Sumi, K. Matsumoto, T. Nakamura, Specific activation of LIM kinase 2 via phosphorylation of threonine 505, by ROCK, a Rho-dependent protein kinase. J. Biol. Chem. 276, 670–676 (2001). [DOI] [PubMed] [Google Scholar]
- 51.J. M. Frangiskakis, A. K. Ewart, C. A. Morris, C. B. Mervis, J. Bertrand, B. F. Robinson, B. P. Klein, G. J. Ensing, L. A. Everett, E. D. Green, C. Pröschel, N. J. Gutowski, M. Noble, D. L. Atkinson, S. J. Odelberg, M. T. Keating, LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell 86, 59–69 (1996). [DOI] [PubMed] [Google Scholar]
- 52.P. Chen, H. Jing, M. Xiong, Q. Zhang, D. Lin, D. Ren, S. Wang, D. Yin, Y. Chen, T. Zhou, B. Li, E. Fei, B.-X. Pan, Spine impairment in mice high-expressing neuregulin 1 due to LIMK1 activation. Cell Death Dis. 12, 403 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.B. W. Henderson, K. M. Greathouse, R. Ramdas, C. K. Walker, T. C. Rao, S. V. Bach, K. A. Curtis, J. J. Day, A. L. Mattheyses, J. H. Herskowitz, Pharmacologic inhibition of LIMK1 provides dendritic spine resilience against β-amyloid. Sci. Signal. 12, eaaw9318 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D. C. Edwards, L. C. Sanders, G. M. Bokoch, G. N. Gill, Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1, 253–259 (1999). [DOI] [PubMed] [Google Scholar]
- 55.C. Dan, A. Kelly, O. Bernard, A. Minden, Cytoskeletal changes regulated by the PAK4 serine/threonine kinase are mediated by LIM kinase 1 and cofilin. J. Biol. Chem. 276, 32115–32121 (2001). [DOI] [PubMed] [Google Scholar]
- 56.M. Maekawa, T. Ishizaki, S. Boku, N. Watanabe, A. Fujita, A. Iwamatsu, T. Obinata, K. Ohashi, K. Mizuno, S. Narumiya, Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 285, 895–898 (1999). [DOI] [PubMed] [Google Scholar]
- 57.K. Ohashi, K. Nagata, M. Maekawa, T. Ishizaki, S. Narumiya, K. Mizuno, Rho-associated kinase ROCK activates LIM-kinase 1 by phosphorylation at threonine 508 within the activation loop. J. Biol. Chem. 275, 3577–3582 (2000). [DOI] [PubMed] [Google Scholar]
- 58.K. I. Okamoto, T. Nagai, A. Miyawaki, Y. Hayashi, Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 7, 1104–1112 (2004). [DOI] [PubMed] [Google Scholar]
- 59.A. Goto, A. Bota, K. Miya, J. Wang, S. Tsukamoto, X. Jiang, D. Hirai, M. Murayama, T. Matsuda, T. J. McHugh, T. Nagai, Y. Hayashi, Stepwise synaptic plasticity events drive the early phase of memory consolidation. Science 374, 857–863 (2021). [DOI] [PubMed] [Google Scholar]
- 60.J. Boehm, M. G. Kang, R. C. Johnson, J. Esteban, R. L. Huganir, R. Malinow, Synaptic incorporation of AMPA receptors during LTP is controlled by a PKC phosphorylation site on GluR1. Neuron 51, 213–225 (2006). [DOI] [PubMed] [Google Scholar]
- 61.N. Otmakhov, L. Khibnik, N. Otmakhova, S. Carpenter, S. Riahi, B. Asrican, J. Lisman, Forskolin-induced LTP in the CA1 hippocampal region is NMDA receptor dependent. J. Neurophysiol. 91, 1955–1962 (2004). [DOI] [PubMed] [Google Scholar]
- 62.N. Otmakhov, J. H. Tao-Cheng, S. Carpenter, B. Asrican, A. Dosemeci, T. S. Reese, J. Lisman, Persistent accumulation of calcium/calmodulin-dependent protein kinase II in dendritic spines after induction of NMDA receptor-dependent chemical long-term potentiation. J. Neurosci. 24, 9324–9331 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.K. Futai, Y. Hayashi, Dynamism of postsynaptic proteins as the mechanism of synaptic plasticity, in Excitatory-Inhibitory Balance (Springer US, 2003), pp. 45–58. [Google Scholar]
- 64.Z. Todorovski, S. Asrar, J. Liu, N. M. N. Saw, K. Joshi, M. A. Cortez, O. C. Snead, W. Xie, Z. Jia, LIMK1 regulates long-term memory and synaptic plasticity via the Transcriptional Factor CREB. Mol. Cell. Biol. 35, 1316–1328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.E. J. Yang, J.-H. Yoon, D. S. Min, K. C. Chung, LIM kinase 1 activates cAMP-responsive element-binding protein during the neuronal differentiation of immortalized hippocampal progenitor cells. J. Biol. Chem. 279, 8903–8910 (2004). [DOI] [PubMed] [Google Scholar]
- 66.A. J. Silva, J. H. Kogan, P. W. Frankland, S. Kida, CREB and memory. Annu. Rev. Neurosci. 21, 127–148 (1998). [DOI] [PubMed] [Google Scholar]
- 67.M. S. Bassi, E. Iezzi, L. Gilio, D. Centonze, F. Buttari, Synaptic plasticity shapes brain connectivity: Implications for network topology. Int. J. Mol. Sci. 20, 6193 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.A. Caccamo, S. Majumder, A. Richardson, R. Strong, S. Oddo, Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-β, and Tau. J. Biol. Chem. 285, 13107–13120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O. Dagliyan, N. V. Dokholyan, K. M. Hahn, Engineering proteins for allosteric control by light or ligands. Nat. Protoc. 14, 1863–1883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.P. Renna, C. Ripoli, O. Dagliyan, F. Pastore, M. Rinaudo, A. Re, F. Paciello, C. Grassi, Engineering a switchable single-chain TEV protease to control protein maturation in living neurons. Bioeng. Transl. Med. 7, e10292 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.F. Pastore, M. Battistoni, R. Sollazzo, P. Renna, F. Paciello, D. D. Li Puma, E. Barone, O. Dagliyan, C. Ripoli, C. Grassi, A bioengineering strategy to control ADAM10 activity in living cells. Int. J. Mol. Sci. 24, 917 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov, M. Steinegger, ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. Saneyoshi, H. Matsuno, A. Suzuki, H. Murakoshi, N. G. Hedrick, E. Agnello, R. O’Connell, M. M. Stratton, R. Yasuda, Y. Hayashi, Reciprocal activation within a kinase-effector complex underlying persistence of structural LTP. Neuron 102, 1199–1210.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.T. Hosokawa, D. Mitsushima, R. Kaneko, Y. Hayashi, Stoichiometry and phosphoisotypes of hippocampal AMPA-type glutamate receptor phosphorylation. Neuron 85, 60–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.E. Barone, A. Tramutola, F. Triani, S. Calcagnini, F. di Domenico, C. Ripoli, S. Gaetani, C. Grassi, D. A. Butterfield, T. Cassano, M. Perluigi, Biliverdin reductase-a mediates the beneficial effects of intranasal insulin in Alzheimer disease. Mol. Neurobiol. 56, 2922–2943 (2019). [DOI] [PubMed] [Google Scholar]
- 76.W. Gulisano, M. Melone, C. Ripoli, M. R. Tropea, D. D. Li Puma, S. Giunta, S. Cocco, D. Marcotulli, N. Origlia, A. Palmeri, O. Arancio, F. Conti, C. Grassi, D. Puzzo, Neuromodulatory action of picomolar extracellular Aβ42 oligomers on presynaptic and postsynaptic mechanisms underlying synaptic function and memory. J. Neurosci. 39, 5986–6000 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.E. N. Falk, K. J. Norman, Y. Garkun, M. P. Demars, S. Im, G. Taccheri, J. Short, K. Caro, S. E. McCraney, C. Cho, M. R. Smith, H.-M. Lin, H. Koike, J. Bateh, P. Maccario, L. Waltrip, M. Janis, H. Morishita, Nicotinic regulation of local and long-range input balance drives top-down attentional circuit maturation. Sci. Adv. 7, eabe1527 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.S. W. Weyer, M. Zagrebelsky, U. Herrmann, M. Hick, L. Ganss, J. Gobbert, M. Gruber, C. Altmann, M. Korte, T. Deller, U. C. Müller, Comparative analysis of single and combined APP/APLP knockouts reveals reduced spine density in APP-KO mice that is prevented by APPsα expression. Acta Neuropathol. Commun. 2, 36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.M. Rinaudo, F. Natale, F. la Greca, M. Spinelli, A. Farsetti, F. Paciello, S. Fusco, C. Grassi, Hippocampal estrogen signaling mediates sex differences in retroactive interference. Biomedicine 10, 1387 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.C. Sánchez-Castillo, M. I. Cuartero, A. Fernández-Rodrigo, V. Briz, S. López-García, R. Jiménez-Sánchez, J. A. López, M. Graupera, J. A. Esteban, Functional specialization of different PI3K isoforms for the control of neuronal architecture, synaptic plasticity, and cognition. Sci. Adv. 8, eabq8109 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.S. Fusco, C. Ripoli, M. V. Podda, S. C. Ranieri, L. Leone, G. Toietta, M. W. McBurney, G. Schütz, A. Riccio, C. Grassi, T. Galeotti, G. Pani, A role for neuronal cAMP responsive-element binding (CREB)-1 in brain responses to calorie restriction. Proc. Natl. Acad. Sci. U.S.A. 109, 621–626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.H. E. Speed, M. Kouser, Z. Xuan, J. M. Reimers, C. F. Ochoa, N. Gupta, S. Liu, C. M. Powell, Autism-associated insertion mutation (InsG) of Shank3 Exon 21 causes impaired synaptic transmission and behavioral deficits. J. Neurosci. 35, 9648–9665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Table S1