Summary
CRISPR‐based directed evolution is an effective breeding biotechnology to improve agronomic traits in plants. However, its gene diversification is still limited using individual single guide RNA. We described here a multiplexed orthogonal base editor (MoBE), and a randomly multiplexed sgRNAs assembly strategy to maximize gene diversification. MoBE could induce efficiently orthogonal ABE (<36.6%), CBE (<36.0%), and A&CBE (<37.6%) on different targets, while the sgRNA assembling strategy randomized base editing events on various targets. With respective 130 and 84 targets from each strand of the 34th exon of rice acetyl‐coenzyme A carboxylase (OsACC), we observed the target‐scaffold combination types up to 27 294 in randomly dual and randomly triple sgRNA libraries. We further performed directed evolution of OsACC using MoBE and randomly dual sgRNA libraries in rice, and obtained single or linked mutations of stronger herbicide resistance. These strategies are useful for in situ directed evolution of functional genes and may accelerate trait improvement in rice.
Keywords: directed evolution, dual base editor, herbicide resistance, multiplexed and orthogonal base editor, randomly multiplexed sgRNAs assembly
Introduction
Directed evolution is a revolutionary biotechnology for improving properties of a given protein or nucleic acids under certain selection methods (Wang et al., 2021). Increasing gene diversification via generating more mutation types is beneficial to directed evolution. Most gene diversification approaches entail error‐prone PCR, DNA synthesis and shuffling in vitro, followed by selection in bacteria or yeast (Morrison et al., 2020; Wang et al., 2021). However, the utility of these ex situ directed evolution approaches is limited when applying to plant functional genes that cannot be selected on plates directly, such as the genes regulating plant architecture, flowering time, yield, quality, and fertility (Engqvist and Rabe, 2019; Zhang and Qi, 2019). Since the CRISPR‐Cas system can create targeted mutagenesis in situ, the CRISPR‐based directed evolution has become a powerful plant breeding biotechnology to improve agronomic traits in plants (Butt et al., 2019; Kuang et al., 2020; Li et al., 2020a; Liu et al., 2020; Wang et al., 2022; Xu et al., 2021b). In particular, the DNA double‐strand break (DSB)‐independent genome editing tools, such as base editors and prime editors, are deemed suitable for performing saturated targeted endogenous mutagenesis in plants via introducing base substitutions (Capdeville et al., 2020; Xu et al., 2021b).
Base editors are fusions of deficient Cas nuclease, such as nCas9, dCas9 and dCas12 with deaminase domains (Anzalone et al., 2020). The cytidine deaminase and adenosine deaminase have been well engineered and several base editors have been generated, including cytosine base editor (CBE), adenine base editor (ABE), C to G base editor (CGBE), A to G and C to T dual base editor (A&CBE), and A to G and C to G/T/A dual base editor (AGBE) (Anzalone et al., 2020; Liang et al., 2022; Wang et al., 2020b). Recently, orthologues or protospacer adjacent motif (PAM) variants of Cas nuclease have been adopted into CBE, ABE, and A&CBE to increase their targeting scope in plants (Cheng et al., 2023; Li et al., 2020a; Tan et al., 2022). The engineered cytidine deaminase and adenosine deaminase were also incorporated into plant base editors to improve their editing activity, purity, and specificity, or expand their editing outcomes (Ren et al., 2021; Tian et al., 2022; Yan et al., 2021; Zeng et al., 2020; Zhu et al., 2020). Some of them have been applied to in situ gene‐directed evolution in mammals or crops; however, most approaches only employed individual single‐guide RNA (sgRNA) and induced base conversions on the targeted 20‐nt region or near once a time (Butt et al., 2019; Kuang et al., 2020; Li et al., 2020a; Liu et al., 2020; Wang et al., 2022; Xu et al., 2021b). We envisioned that gene diversification can be further empowered by assembling randomly multiplexed sgRNAs of various base editors for generating more mutation types simultaneously (Figure 1a).
Figure 1.

Screening adenosine and cytidine deaminases for high base editing activities via sgRNA scaffolds recruiting. (a) Schematic diagram of improving gene diversification in creating CRISPR targeted mutant libraries using randomly multiplexed sgRNAs and various base editors. GOI, gene of interest; POI, protein of interest. (b) Architectures of AD‐fusion, 2A‐AD, CD‐fusion, and 2A‐CD constructs. AD, adenosine deaminase; CD, cytidine deaminase. The grey fragment represents nuclear localization signal (NLS). (c) Frequencies of A to G base editing induced by TadA8e‐fusion control, 2A‐TadA‐TadA7.10, 2A‐TadA8e, and 2A‐TadA9 (n = 3). An untreated protoplast sample served as negative control. Values and error bars indicate mean ± sem of three independent biological replicates. (d) A summary of editing activities induced by TadA‐TadA7.10, TadA8e, and TadA9 in 2A‐AD or AD‐fusion architecture. (e) Frequencies of C to T base editing induced by APOBEC1‐fusion control, 2A‐APOBEC1, 2A‐APOBEC3Bctd, 2A‐CDA1, 2A‐AID and 2A‐evoFERNY (n = 3). An untreated protoplast sample served as negative control. Values and error bars indicate mean ± sem of three independent biological replicates. f, A summary of editing activities induced by APOBEC1, APOBEC3Bctd, CDA1, AID and evoFERNY in 2A‐CD or CD‐fusion architecture. P values in d, f were performed using two‐tailed Student's t‐test: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Here, we engineered a MoBE system that integrates ABE, CBE, and A&CBE modules together via multiple sgRNA scaffold‐recruited cytidine deaminase and/or adenosine deaminase. Moreover, we developed a randomly multiplexed sgRNAs assembly strategy to randomize the occurrence of different base editors on various targets. The targeted mutagenesis potential of MoBE could be largely increased by combining this strategy. We further performed directed evolution of rice acetyl‐coenzyme A carboxylase and acquired single or linked mutations of stronger herbicide resistance. This directed evolution approach provides a useful way to accelerate trait development in rice breeding.
Results
Exploring high activity deaminase recruited by sgRNA scaffolds
Previous works suggested that the base editing properties are different among various deaminases (Gaudelli et al., 2020). To explore deaminases with high editing activity in an RNA aptamer‐recruited architecture, we used esgRNA‐2×boxB and esgRNA‐2×MS2 to recruit adenosine deaminase and cytidine deaminase, respectively, as previously reported (Li et al., 2020b; Figure S1 and Sequence S1). We started with an nCas9 (D10A)‐T2A‐adenosine deaminase (abbreviated 2A‐AD) architecture, and the canonical ABE architecture adenosine deaminase‐nCas9 (D10A) (abbreviated AD‐fusion) served as control (Figure 1b). Three evolved Escherichia coli tRNA adenosine deaminase TadA* variants, including TadA‐TadA7.10 (Komor et al., 2016), TadA8e (Richter et al., 2020) and TadA9 (TadA8e‐V81S/Q153R; Yan et al., 2021), were installed into the 2A‐AD or AD‐fusion architecture (Sequence S2). All the 2A‐AD and AD‐fusion vectors were codon optimized for crop plants, and driven by the maize Ubi‐1 promoter.
To examine their A to G activities, we cloned six sgRNAs (OsCDC48‐T1, OsDEP1‐T1, OsDEP1‐T2, OsNRT1.1B‐T1, OsEV, and OsOD) into pOsU3‐esgRNA‐2×boxB (Table S1). Each sgRNA was co‐transfected into rice protoplasts along with the 2A‐AD or AD‐fusion construct. We evaluated their efficiencies at A2–A12 (counting the distal end to PAM as position 1) of these tested six targets using targeted amplicon deep sequencing. We found that the editing activity of 2A‐Tad8e (average 13.4%) was a little higher compared with TadA8e‐fusion control (12.6%), while that of 2A‐TadA9 (average 19.1%) was improved about 1.5‐fold on these tested target sites (Figure 1c,d; Figure S2). No obvious indel by‐products were observed in 2A‐TadA9 (0%–0.5%), as in the untreated control protoplasts (0%–0.3%) (Figure S2). Thus, TadA9 could be chosen when performing saturated mutagenesis in plants.
Next, we tried to evaluate six cytidine deaminases, including rat APOBEC1, human APOBEC3A, human APOBEC3Bctd, lamprey CDA1, human AID, and an evolved APOBEC1 variant evoFERNY (Thuronyi et al., 2019), using the nCas9 (D10A)‐T2A‐cytidine deaminase‐UGI (abbreviated 2A‐CD) architecture or the canonical CBE architecture (abbreviated CD‐fusion; Figure 1b; Sequence S2). Unfortunately, we were unable to obtain the correct 2A‐APOBEC3A construct from dozens of E. coli clones (Figure S3), likely because of its severe toxicity to bacteria (Liang et al., 2022). Thus, we evaluated the activity of the remaining five 2A‐CD constructs (2A‐APOBEC1, 2A‐APOBEC3Bctd, 2A‐CDA1, 2A‐AID, and 2A‐evoFERNY) in rice protoplasts using six targets (OsCDC48‐T2, OsCDC48‐T3, OsDEP1‐T2, OsPDS‐T1, OsEV, and OsOD) inserted into pOsU3‐esgRNA‐2×MS2 (Table S1). The average C to T base editing activity at C−2‐C15 of these targets ranged from 1.2% to 16.3%. Of these, 2A‐CDA1 was the most efficient with its primary editing window ranging 13 nt (positions C−2‐C11) (Figure 1e,f; Figure S4). Indel frequencies in 2A‐CDA1 group (0%–0.5%) were equivalent to that in the untreated group (0%–0.4%) in rice protoplasts (Figure S4). Therefore, we selected TadA9 and CDA1 for further engineering the architecture of multiplexed orthogonal base editor.
Engineering a multiplexed orthogonal base editor
According to the results above, different RNA aptamers in sgRNA scaffolds could efficiently recruit their binding proteins fused with different deaminases. We integrated nCas9 (D10A), MCP‐CDA1‐UGI and TadA9‐N22p modules together spaced by T2A peptide, and the deaminases could be recruited by the RNA aptamers in different sgRNA scaffolds, which would achieve ABE, CBE, and A&CBE tri‐functional editing at multiple sites, generating the MoBE system (Figure 2a; Figure S5a and Sequence S3).
Figure 2.

Engineered multiplexed orthogonal base editor for simultaneous ABE, CBE, and A&CBE base editing on various targets. (a) Architectures of MoBE, esgRNA‐boxB‐MS2, and esgRNA‐MS2‐boxB. (b) Schematic diagram of recruiting adenosine deaminase and cytidine deaminase modules by two different sgRNA scaffolds. (c) Comparison of the A to G and C to T dual base editing frequencies among MoBE and two well developed dual base editors (n = 3). STEME1, a fusion of APOBEC3A, TadA‐TadA7.10, nCas9 (D10A), and UGI. DuBE1, a fusion of TadA8e, nCas9 (D10A), LjCDA1L‐4 and multiple UGIs. An untreated protoplast sample served as negative control. Values and error bars indicate mean ± sem of three independent biological replicates. (d) The allelic outcome of the OsEV site edited by MoBE (n = 1) in rice protoplasts. (e) The concurrent A to G and C to T product purity among edited DNA sequencing reads in rice protoplasts is shown for MoBE, DuBE1, and STEME1. Values and error bars indicate mean ± sem of three independent experiments. (f) Simultaneously ABE, CBE, and A&CBE base editing induced by MoBE. Two sets of sgRNAs were tested (n = 3). One ABE target with esgRNA‐2×boxB, one CBE target with esgRNA‐2×MS2, and one A&CBE target with esgRNA‐boxB‐MS2 were assembled in the same vector.
Next, to verify whether MoBE could perform A to G and C to T dual base editing via sgRNA scaffold recruitment, we re‐engineered two RNA aptamer hairpins boxB‐MS2 and MS2‐boxB chimeras at the 3′ end of esgRNA (Li et al., 2018), generating esgRNA‐boxB‐MS2 and esgRNA‐MS2‐boxB (Figure 2a,b; Figure S1, and Sequence S1). Four rice endogenous targets (OsAAT‐T1, OsCDC48‐T1, OsCDC48‐T2, and OsEV) were used to examine the dual base editing activity (Table S1). The dual base editors STEME1 and DuBE1 were used as the controls (Li et al., 2020a; Xu et al., 2021a). We noticed that MoBE induced higher base editing efficiency on C−3‐C4 and A2–A12 than STEME1, while lower on C5–C16 (Figure 2c; Figure S5b). This result in the A to G and C to T dual base editing efficiency of combing MoBE and esgRNA‐boxB‐MS2 was higher (average 26.4%) than the combinations of MoBE and esgRNA‐MS2‐boxB (average 24.5%), DuBE1 and esgRNA (average 22.4%), and STEME1 and esgRNA (average 21.1%; Figure 2c; Figure S5b). Moreover, the concurrent A to G and C to T product purity of MoBE was 92.2%–99.5% (average 97.0%). In contrast, lower purities were observed when using DuBE1 (70.4%–94.4%; average 83.7%) and STEME1 (0.9%–39.9%; average 12.4%) controls (Figure 2d,e; Figure S5c). Together, these results indicate that base editing activity could be enhanced by using high activity deaminases (TadA9 and CDA1) and sgRNA scaffold engineering (esgRNA‐boxB‐MS2), particularly favourable for concurrent A to G and C to T dual base editing.
To determine whether MoBE could promote tri‐functional ABE, CBE, and A&CBE base editing on various targets, we introduced two groups of targets in front of esgRNA‐2×boxB, esgRNA‐2×MS2, and esgRNA‐boxB‐MS2 under OsU3, OsU6, and TaU6, respectively, and assembled them into a single vector (Figure S6a; Table S1). Amplicon deep sequencing showed that MoBE induced efficient ABE, CBE, and A&CBE editing simultaneously on three different targets on both groups of targets in rice protoplasts (Figure 2f). The efficiency of esgRNA‐2×boxB mediated A to G base editing ranged from 0.4% to 36.6% in A2–A12, and the efficiency of esgRNA‐2×MS2 mediated C to T base editing ranged from 1.1% to 36.0% in C−2‐C8. While esgRNA‐boxB‐MS2 mediated A to G and C to T dual base editing efficiency ranged from 2.0% to 37.6% and from 0.2% to 36.4% in A2–A13 and C1–C13, respectively (Figure 2f). Of these, 88.3%–100% portions of base editing products were concurrent A to G and C to T conversions (Figure S6b). Together, MoBE is not only an efficient A to G and C to T dual base editor, but also a reliable tri‐functional base editing tool to perform ABE, CBE, and A&CBE simultaneously in rice. This merit not only provides a useful tool for improvement of multiple agronomic traits via simultaneous editing of various genes, but also has the potential for in situ directed evolution by increasing gene diversification via mutating the same gene.
MoBE off‐target profiling
We next investigated the sgRNA‐dependent off‐target effects of MoBE‐mediated dual base editing comparing with Cas9 by testing a group of OsCDC48‐T2 mismatched targets with one or two mismatches in rice protoplasts (Table S2). As expected, both MoBE and Cas9 could tolerate most one or two mismatches, especially when these mismatches are located at 5′ region of the spacer (Figure 3a). We noticed that these conclusions were the same as other base editors reported previously (Kim et al., 2019a).
Figure 3.

Off‐target analysis of MoBE. (a) Tolerance of MoBE and Cas9 for mismatched targets (n = 3). The sgRNA scaffolds of esgRNA‐boxB‐MS2 and sgRNA were used for MoBE and Cas9, respectively. Mismatched sgRNAs that differed from the OsCDC48‐T2 site by one to two nucleotides were tested in rice protoplasts. Base editing or indel frequencies were measured using targeted amplicon deep sequencing. Mismatched nucleotides and the PAM sequence are shown in red and bold, respectively. Values and error bars indicate mean ± sem of three independent biological replicates. (b) Schematic of sgRNA‐independent deamination of adenines and cytosines within nSaCas9 (D10A)‐induced R‐loops by MoBE. (c) Frequencies of sgRNA‐independent off‐target base editing induced by MoBE on R‐loop ssDNA (n = 3). Three (OsDEP1‐SaT1, OsNRT1.1B‐SaT1, and OsTAC1‐SaT1) sites targeted by nSaCas9 (D10A) were used for producing the ssDNA regions. Values and error bars indicate mean ± sem of three independent biological replicates. (d) Architecture of MoBE‐HF. Abbreviations: tCDA1EQ, truncated (30–150) lamprey CDA1 variant containing W122E and W139Q. TadA8e‐V106W, TadA8e containing V106W. (e) Comparison of sgRNA‐independent off‐target base editing induced by MoBE and MoBE‐HF on R‐loop ssDNA (n = 3). Values and error bars indicate mean ± sem of three independent biological replicates. (f) A summary of sgRNA‐independent off‐target base editing induced by MoBE and MoBE‐HF on R‐loop ssDNA. P values were performed using two‐tailed Student's t‐test: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Since the deaminase modules of MoBE should present as two forms in cellular, one is complexed with sgRNA scaffold, and the other is a free deaminase module, both may induce sgRNA‐independent off‐target effect (Figure 3b). To evaluate this off‐target effect, we used the nSaCas9 (D10A)‐mediated orthogonal R‐loop assay to detect the off‐target activity of deaminase on exposed ssDNA in plants (Jin et al., 2020). We designed three targets with NNGRRT PAM of rice genes (Table S3), and cloned them into pOsU3‐SasgRNA (Jin et al., 2020). Each SasgRNA (OsDEP1‐SaT1, OsNRT1.1B‐SaT1, and OsTAC1‐SaT1) was co‐transfected into rice protoplasts along with nSaCas9 (D10A) and MoBE. We observed obvious dual base conversions (2.3%–11.3%) on the exposed R‐loop ssDNA (Figure 3c). To reduce this sgRNA‐independent deamination activity, we replaced CDA1 and TadA9 with minimized off‐target activity variants tCDA1EQ and TadA8e‐V106W in MoBE, respectively, generating MoBE‐HF (Figure 3d; Sequence S3). The tCDA1EQ is a truncated (30–150) lamprey CDA1 variant remaining the smallest enzymatic core domain and containing W122E and W139Q amino acid substitutions for improving activity (Li et al., 2022a). The TadA8e‐V106W variant that was proved can reduce both DNA and RNA off‐target editing activities by introducing a V106W amino acid substitution into TadA8e (Richter et al., 2020). We observed a reduced adenine off‐target editing and hardly detectable cytosine off‐target editing on the R‐loop ssDNA using MoBE‐HF, which reduced approximately 1.7‐fold and 22.2‐fold compared to MoBE, respectively (Figure 3e,f). However, for the on‐target editing, MoBE‐HF exhibited a narrower editing window, lower concurrent A to G and C to T conversions products, and about 1.9‐fold and 5.5‐fold reduction on adenine and cytosine editing compared to MoBE, respectively, indicating its limitations when performing saturated targeted endogenous mutagenesis in plants (Figure S7). The trade‐off between on‐target and off‐target editing activities of MoBE could be solved further by deaminase engineering.
Developing a randomly multiplexed sgRNA assembly strategy in plants
High‐throughput methodology and more gene diversification are two limiting factors for directed evolution of proteins (Morrison et al., 2020; Wang et al., 2021). Although there are base editor webtools to simplify sgRNA design, their applications are mainly used for predicting editing outcomes, providing all possible targets in a short region, and other customized purpose (Hwang et al., 2018; Rabinowitz et al., 2020; Xie et al., 2022; Yu et al., 2020). To design saturated 20‐nt targets of plant genes in a simple and high‐throughput manner, we developed PlantBE‐CODE (http://pgec.njau.edu.cn/plantbe‐code), a web application of cover design for directed evolution customized genes based on nCas9 (D10A) and primary base editing windows of different deaminases. For the designated gene, PlantBE‐CODE needs inputing its genomic sequence and its accession number when possible (Figure 4a). PlantBE‐CODE provides recommended targets for saturated targeted mutagenesis, as well as their possible mutations and potential off‐target sites by using different deaminases or their combinations (Figure S8; Note S1).
Figure 4.
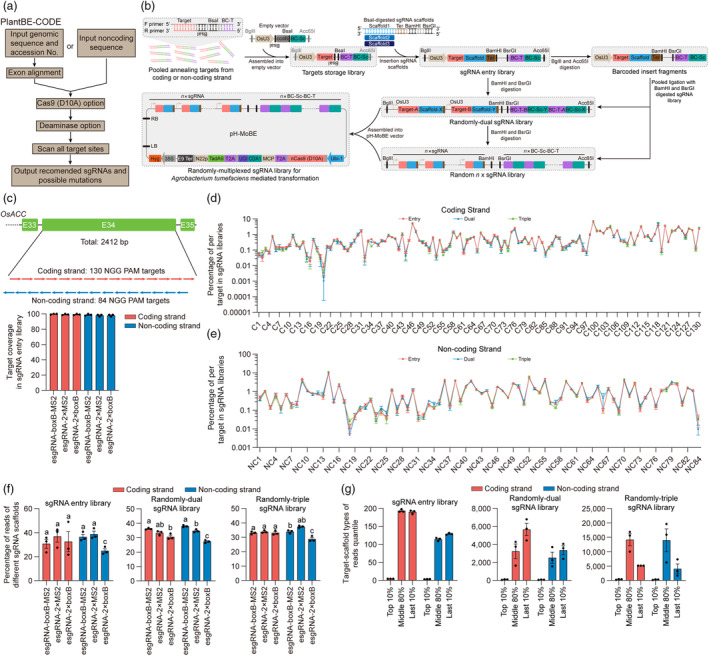
Developing a randomly multiplexed sgRNA assembly strategy in plants. (a) Schematic of the PlantBE‐CODE workflow for design saturated targets of plant genes. PlantBE‐CODE provides recommended targets for directed evolution when using various base editors. (b) Platform for assembling randomly multiplexed sgRNAs. Target oligos with two antitropic BsaI recognition sites and unique barcode of target (BC‐T) were synthesized. The annealed oligos from coding strand or non‐coding strand were pooled together equally and inserted into the empty vector harbouring OsU3 promoter and barcode for sgRNA scaffolds (BC‐Sc), generating targets storage library. To randomize the occurrence of different base editing on various targets, three types of BsaI‐digested sgRNA scaffolds, esgRNA‐boxB‐MS2 (Scaffold1), esgRNA‐2×MS2 (Scaffold2), and esgRNA‐2×boxB (Scaffold3), along with terminator, BamHI, and BsrGI were inserted into corresponding storage library, generating sgRNA entry library. Subsequently, equal amounts of entry libraries from coding strand or non‐coding strand were pooled together and digested by BamHI/BsrGI and BglII/Acc65I to produce linearized backbone with first round sgRNAs and barcoded insert fragments, and then ligated them together, generating randomly dual sgRNA library. Similarly, the former library served as the destination vector for the next insertion round of barcoded fragments, generating a successive randomly multiplexed library that harbouring one more sgRNA than the former round. Target‐A and Target‐B represent any target from one target group of coding strand and non‐coding strand. Scaffold‐X and Scaffold‐Y represent any sgRNA scaffold of esgRNA‐2×boxB, esgRNA‐2×MS2, and esgRNA‐boxB‐MS2. (c) Target coverage in six different sgRNA entry libraries from coding strand and non‐coding strand (n = 3). Percentages of coverage were calculated based on the presence of BC‐T in the deep sequencing reads. (d, e) Percentage of each target in randomly multiplexed sgRNA libraries. The sgRNA entry libraries, dual sgRNA libraries, and triple sgRNA libraries from coding strand (d) and non‐coding strand (e) were analysed (n = 3). (f) The uniformity of three different sgRNA scaffolds in randomly multiplexed sgRNA libraries. Percentages of uniformity were calculated based on the presence of BC‐Sc in the deep sequencing reads (n = 3). (g) The target‐scaffold combination types in randomly multiplexed sgRNA libraries. Reads number of each target‐scaffold combination located in top 10% quantile, middle 80% quantile, and last 10% quantile among total reads were calculated based on the presence of BC‐T and BC‐Sc combinations in the deep sequencing reads (n = 3). Values and error bars indicate mean ± sem of three independent biological replicates in c–g. Multiple comparisons were performed according to one‐way ANOVA and Tukey's multiple comparisons test, P < 0.05.
Multiplexed sgRNA assembly would facilitate gene diversification for in situ directed evolution using MoBE. However, previous multiplexed sgRNA assembly methods rely on a certain tandem order, which limits constructing the sgRNA library in a high‐throughput manner in plants (Čermák et al., 2017; Wang et al., 2015; Xie et al., 2015). Thus, we envisioned that a randomly multiplexed sgRNA assembly strategy may break the trade‐off between high‐throughput sgRNA assembly and gene diversification for in situ directed evolution (Wong et al., 2016). To this end, we developed a platform for assembling randomly multiplexed sgRNAs using a pair of isocaudamers, and the combined sgRNAs can be tracked easily with barcode sequencing (Figure 4b; Sequence S4). In this platform, all the three sgRNA scaffolds (esgRNA‐boxB‐MS2, esgRNA‐2×MS2, and esgRNA‐2×boxB) were used to randomize the occurrence of different base editing on various targets. Then, we designed saturated targets of the 34th Exon, the main region of carboxyltransferase (CT) domain, of the rice acetyl‐coenzyme A carboxylase gene (OsACC) using PlantBE‐CODE. Using parameters of nCas9 (D10A), single TadA9, single CDA1, and combined TadA9 and CDA1, we totally acquired 130 and 84 NGG PAM targets on the coding strand and non‐coding strand, respectively (Figure 4c; Table S4).
All the targets were synthesized, pooled, and assembled into empty vectors. After three corresponding sgRNA scaffolds were inserted into the empty vectors, we acquired six sgRNA entry libraries, with each plasmid harbouring single target. We amplified the target barcode region and analysed target coverage using amplicon deep sequencing (Figure S9a and Code S1). Most of the targets were detected in the sgRNA entry libraries (esgRNA‐boxB‐MS2, esgRNA‐2×MS2, and esgRNA‐2×boxB) of the coding strand (average 99.7%, 99.5%, and 99.7%), and non‐coding strand (average 98.8%, 98.0%, and 98.0%), indicating high target coverage for the entry libraries (Figure 4c; Data S1 and S2). Then, we performed two rounds of isocaudamer digestion and ligation, generating randomly dual and triple sgRNA array of the coding strand and non‐coding strand (Figure 4b). We evaluated the ratios of every target and sgRNA scaffolds in these libraries by barcode sequencing using 100 000–700 000 total reads (Figure S9b, Code S2 and S3). We found that all the targets were covered in three independent experiments, with the target ratio varied, up to 7.5% and 11.7% of the coding strand libraries and non‐coding strand libraries, respectively (Figure 4d,e). Meanwhile, the ratios of different sgRNA scaffolds ranged from 30.6% to 36.7% and from 24.8% to 38.7% on average in the sgRNA libraries of the coding strand and non‐coding strand, respectively, exhibiting an acceptable uniformity of these sgRNA scaffolds (Figure 4f). Moreover, these ratios in dual and triple sgRNA libraries were consistent with the sgRNA entry libraries (Figure 4d–f). Thus, we believe that a carefully equivalent input is the key point to keep target uniform in randomly multiplexed libraries.
We also evaluated the types of target‐scaffold combinations using this barcode sequencing data. We found that the combination types of sgRNA libraries from the coding strand and non‐coding strand reached up to 390 and 249 in the sgRNA entry libraries, 10 656 and 7846 in the dual sgRNA libraries, and 22 538 and 27 294 in the triple sgRNA libraries, respectively, indicating that our randomly multiplexed sgRNA assembly method could markedly increase the types of target‐scaffold combinations (Figure 4g; Data S3–S5). Of them, 36.0%–76.1% combination types were located in the middle 80% quantile of the sequencing reads and became more uniform in the later libraries, exhibiting an acceptable distribution of these target‐scaffold types (Figure 4g). However, theoretical types of target‐scaffold combinations could be up to one hundred thousand and ten million magnitudes for the dual sgRNA libraries and triple sgRNA libraries, respectively. Thus, we recommended that it would be better to assemble randomly multiplexed sgRNAs library less than three rounds. Collectively, as we know, we successfully established and validated an experimental strategy for assembling randomly multiplexed sgRNAs from the same gene for the first time in plant. This strategy would facilitate to generate more gene diversifications for in situ directed evolution in rice.
Directed evolution ACC using randomly dual sgRNA library in rice plants
As a proof‐of‐concept study, we combined MoBE with randomly multiplexed sgRNA library for directed evolution ACC in rice plants (Figure 5a). ACC is an enzyme in fatty acid biosynthesis and the target of herbicides (Powles and Yu, 2010). We assembled the randomly dual sgRNA library of the coding strand and non‐coding strand of OsACC into the binary vector pH‐MoBE (Figure 4b; Figure S10a). Both libraries were transformed into Agrobacterium tumefaciens. We used STEME targeting OsACC‐P1927 as control, which can induce a P1927F mutation tolerating 0.108 mg L−1 haloxyfop selection pressure on plates (Li et al., 2020a). To obtain stronger herbicide resistance mutants, we performed 6 weeks directed selection on plates harbouring herbicide, including 3 weeks selection with 0.108 mg L−1 haloxyfop and another 3 weeks selection with 0.162 mg L−1 haloxyfop (Figure 5a; Figure S10b). Two weeks after callus regeneration step on the medium supplemented with 0.162 mg L−1 haloxyfop, we observed herbicide resistance seedlings were regenerated in the MoBE treatment group, while that in the STEME targeting P1927 control group was not observed, indicating that higher herbicide concentration may inhibit the growth rate of regenerated seedlings harbouring P1927F (Figure 5b; Figure S10c). Finally, we totally obtained 28 herbicide resistance seedlings after 4 weeks regeneration in the MoBE treatment group (Table S5).
Figure 5.

Directed evolution by MoBE with randomly dual sgRNA library generates herbicide‐tolerant mutants. (a) Schematic of the procedure for mutating the OsACC CT domain on medium supplemented with haloxyfop using MoBE with randomly dual sgRNA library. (b) Representative herbicide resistance seedlings regenerated on haloxyfop‐containing medium. STEME targeting P1927 was used as control. Scale bar, 1 cm. (c) Sanger sequencing of the barcode region from herbicide resistance plants. Five representative mutants were showed. Abbreviation: BC, barcode. (d) Genotypes of herbicide resistance seedlings. Five representative mutants were showed. Targeted regions were confirmed according to the barcode sequencing results in c. The genotypes of these mutations with two double peaks were confirmed further by T‐vector cloning. (e) Phenotypes of T1 homozygous rice with linked mutations treated by hydroponic medium containing high haloxyfop levels. The homozygous mutants of T1 generation were treated with 0.54 mg L−1 and 1.62 mg L−1 haloxyfop at two‐to‐three‐leaf stage, which were 5× and 15× of the selection pressure on plates used before (Li et al., 2020a), respectively. Photos were taken at 2 weeks after treatment. The wild‐type seedlings germinate from Japonica rice variety Nipponbare served as control. Scale bar, 5 cm.
To detect the genotype of these herbicide resistance seedlings, we firstly amplified the barcode region of the T‐DNA fragment from their genomic DNA (Figure S10d). We found these seedlings harbouring nine different combinations of target and sgRNA scaffold, three of them from the coding strand, six of them from the noncoding strand, including 13 different targets and all the three sgRNA scaffolds (Figure 5c; Table S5–S7). Then, we amplified the edited region from genomic DNA of these herbicide resistance seedlings based on the barcoded sequencing results. Sanger sequencing indicated that these herbicide resistance seedlings could be divided into three groups according to the carried known mutations: 21 seedlings carried W2125C mutation, three seedlings carried I2139N mutation, and four seedlings carried C2186R mutation (Figure 5d; Table S5). In the W2125C group, two seedlings were linked V1703I‐W2125C heterozygotes, two seedlings were linked R2126T‐G2127A‐E2327K and linked W2125C‐E2327K biallelic mutants, six seedlings were D1970N/W2125C biallelic mutants, and the remaining seedlings were W2125C heterozygotes. In the I2139N group, two seedlings were I2139N/F2207L biallelic mutants and one seedling was I2139N heterozygotes. All the seedlings in C2186R group were C2186R heterozygotes (Figure 5d; Table S5).
In the seedlings containing D1970N, C2186R, and E2327K substitutions, these were the result of canonical C to T or A to G transitions by corresponding RNA aptamer‐recruited deaminase. These canonical base conversions were also observed at amino acids G1702, F2207, and L2328 (causing silent mutations; Figure 5d; Table S5). Moreover, most W2125C substitution and all R2126T, G2127A, and F2207L substitutions were caused by C to G transversion (Figure 5d; Table S5). These were consistent with the editing products of cytidine deaminase‐containing base editors (Koblan et al., 2021; Li et al., 2017). However, esgRNA‐2×boxB recruited TadA9 induced C to T transition was responsible for the observed V1703I and D1970N substitutions (Figure 5c,d and Table S5). Since the former TadA* variants, such as TadA7.10, TadA8e, and TadA8s, were reported to catalyse cytosine deamination (Jeong et al., 2021; Kim et al., 2019b), we speculated that TadA9 variant might inherit this feature. To our surprise, with esgRNA‐boxB‐MS2 recruited TadA9 and CDA1, the G to T and T to A transversions were observed and responsible for three seedlings containing W2125C substitution and all seedlings containing I2139N substitution, respectively (Figure 5c,d; Table S5). Importantly, two different targets (C33 and C120) could be responsible for I2139N substitution, implying that this may not have occasionally occurred during mismatch repair (MMR) and base‐excision repair (BER) when adding herbicide selection pressure. Thus, we speculated that despite the occurrence was non‐canonical, mutations that had no trade‐off on the growth of regenerated seedlings would be enriched after rounds of herbicide selection. Moreover, these non‐canonical base substitutions would further improve gene diversification for in situ directed evolution in plants. Additionally, we also found the off‐target editing on the site of no mismatch of C36 in one C36 contained mutants and one mismatch of NC28 in two NC28 contained mutants (Table S8). W2125C, I2139N, and C2186R were reported herbicide resistance mutations; however, these linked mutations and biallelic mutations have not been reported previously, which provides a case‐by‐case basis for analysing homozygosity or heterozygosity resistance mutations in herbicide resistance research and crop breeding (Powles and Yu, 2010). Together, combining MoBE with randomly multiplexed sgRNA library, we can generate a dozen of functional herbicide resistance single or linked mutations.
To further investigate whether the linked mutation could enhance herbicide resistance, we tested seedlings of T1 generation from V1703I‐W2125C heterozygote (M1) and R2126T‐G2127A‐E2327K/W2125C‐E2327K biallelic mutants (M2). The seedlings were genotyped by Sanger sequencing firstly. We obtained homozygote and heterozygote of linked V1703I‐W2125C mutations, and homozygote of linked W2125C‐E2327K mutations (Figure S11). However, we could not obtain any mutants containing linked R2126T‐G2127A‐E2327K mutations, indicating that this linked mutagenesis may be lethal to rice gamete. Then, we treated the V1703I‐W2125C homozygotes and W2125C‐E2327K homozygotes with 1/2 Murashige‐Skoog hydroponic medium containing 0.54 mg L−1 and 1.62 mg L−1 haloxyfop, which were 5‐fold and 15‐fold higher than the selection pressure on plates initially used (0.108 mg L−1), respectively. The W2125C homozygotes and wild‐type seedlings were used as controls. After 2 weeks, we observed that all the tested homozygotes showed withered phenotype on the whole plant, except for the W2125C‐E2327K homozygote under 0.54 mg L−1 haloxyfop selection pressure (Figure 5e). However, even in the surviving W2125C‐E2327K homozygote, no new leaves had grown. Further investigation and validation of their T2 transgene‐free homologous mutants should be conducted using a wider range of gradient concentrations to evaluate their herbicide resistance capacity. We also noticed that the linked V1703I‐W2125C mutations affect the plant architecture of T1 rice, indicating that it may have fitness cost on rice (Figure 5e). Together, our base editing tool and sgRNA assembly strategy here are useful in carrying out in situ directed evolution of endogenous gene, and in obtaining enhanced trait in rice.
Discussion
Compared to ex situ directed evolution of plant genes, the CRISPR‐based in situ directed evolution methods are more appropriate for evolving functional genes regulating agronomic traits. However, gene diversification methods of in situ directed evolution are still limited when comparing with error‐prone PCR, DNA synthesis, and DNA shuffling. Although prime editors are more powerful in precise editing, they need careful design and they being somewhat labour‐intensive hamper its utility in a high‐throughput manner. We consider that base editors are more practical due to their features of high editing efficiency, easy design, and construction (Anzalone et al., 2020). In the present study, we developed a multiplexed orthogonal base editor (MoBE) tool integrating ABE, CBE, and A&CBE tri‐functional editing together to improve mutagenesis diversification of targeted gene in CRISPR edited mutant library. Our results suggest that high activity and wide base editing window are two key points for choosing deaminase used for in situ directed evolution. The use of highly and equally efficient TadA9 and CDA1 provide an equal chance to act on the same protospacer target simultaneously, thus inducing much higher simultaneous A to G and C to T dual base conversions. However, recently, approaches of genome‐wide sequencing and our R‐loop assay showed their high sgRNA‐independent off‐target editing activity (Li et al., 2022b; Wu et al., 2022). Moreover, we also observed the sgRNA‐dependent off‐target editing on regenerated mutants. We tried to use high‐specificity deaminase variants tCDA1EQ and TadA8e‐V106W to reduce this off‐target editing activity of MoBE. Nevertheless, the adenine off‐target editing of TadA8e‐V106W should be minimized further, while the on‐target editing activity of tCDA1EQ reduced markedly. Therefore, strategies of protein engineering, temporal expression, ribonucleoprotein delivery, and tissue culture optimizing could be further tested in plants to alleviate the off‐target effects of base editors with on‐target editing activity loss, including MoBE (Jin et al., 2020; Li et al., 2022b; Wu et al., 2022).
We developed a randomly multiplexed sgRNA assembly strategy to improve the mutagenesis diversification further. Our results indicated that the initial input library was the key point to keep target uniform in randomly multiplexed libraries. We recommend if the library only contains dozens of targets, assembling each target into empty vector separately will be helpful; but with more than hundreds of targets, equivalently pooled assembly is preferred. We envisage that Cas9 variants, such as Cas9‐NG, SpG, and SpRY, could be engineered into the MoBE to improve the coverage density of targets in the future, which would also improve the mutagenesis diversification remarkably (Nishimasu et al., 2018; Walton et al., 2020). Other multiplexed sgRNA expression strategies, such as tRNA or csy4, could be tried to simplify the expression cassette (Zhang et al., 2019). Moreover, the assembly strategy using one step cloning method rather than isocaudamer could also be developed in the future. When combining MoBE with this randomly multiplexed sgRNA assembly strategy, we can generate linked mutations of stronger herbicide resistance comparing with the former evolved OsACC‐P1927F mutation (Li et al., 2020a). However, the herbicide resistance capacity and potential side effect on other agronomic traits caused by the linked mutations, as well as their separate function, should be further investigated in the future.
Since in situ directed evolution in plant requires a large number of transformants, which is time consuming and labour intensive. We recommend that performing pooled transformation and establishing an effective selection method of the evolved functional genes will facilitate this approach. For those functional genes that can be selected by chemical reagent directly, such as herbicide resistance, or salt stress, we propose to screen the evolved mutants on plates after acquiring the transgenic calli. For those functional genes that can be selected by biotic stress, such as disease resistance, or insect resistance, we propose to screen the evolved mutants after biotic inoculation on regenerated seedlings. For those functional genes without available selection pressure, we propose to identify the mutants using amplicon deep sequencing or T7E1 digestion of pooled amplicons, followed by phenotyping of evolved agronomic traits. Additionally, our directed evolution approaches here may also be suitable for application scenarios in cell lines and animals.
Experimental procedures
Plasmids construction
The adenosine deaminases (TadA8e and TadA9), cytidine deaminases (CDA1, AID, and evoFERNY) were codon‐optimized for cereal plants, and synthesized commercially (GenScript, Nanjing, China). The other deaminases (TadA‐TadA7.10, APOBEC1, and APOBEC3Bctd), nCas9 (D10A), N22p, MCP, and UGI portions of AD‐fusion, 2A‐AD, CD‐fusion, 2A‐CD, MoBE, and MoBE‐HF were amplified from PABEc5, PBEc4, or eAFID‐3 (Li et al., 2020b; Wang et al., 2020a). The DuBE1 modules were amplified from pHUC411‐pDuBE1 (Xu et al., 2021a). The various constructs were cloned into pJIT163 backbone (Li et al., 2020b) by ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The Cas9 were amplified from pHUE411 and assembled into pJIT163 backbone under maize Ubi‐1 promoter (Xing et al., 2014). The nSaCas9 (D10A) used for orthogonal R‐loop assay, as well as sgRNA constructs pOsU3‐sgRNA, pOsU3‐esgRNA, pOsU3‐esgRNA‐2×boxB, pOsU3‐esgRNA‐2×MS2, and pOsU3‐SasgRNA were described previously (Li et al., 2020b). The esgRNA‐boxB‐MS2 and esgRNA‐MS2‐boxB listed in Sequence S1 were synthesized commercially and used to replace the sgRNA scaffold in pOsU3‐sgRNA by One Step Cloning. Annealed oligos were inserted into BsaI (New England BioLabs, Ipswich, MA, USA) digested OsU3‐derived vectors. For triple targets assembling, the target‐sgRNA scaffold fragments were amplified from target installed OsU3‐derived vectors, TaU6 and OsU6 were amplified from pTaU6‐esgRNA (Li et al., 2018) and pYPQ131C (Lowder et al., 2015), respectively. To construct the pH‐MoBE binary vector, the MoBE module was cloned into the pHUE411 backbone without OsU3‐sgRNA expression cassette (Xing et al., 2014). PCR was performed using TransStart FastPfu DNA Polymerase (TransGen Biotech, Beijing, China). All the primer sets used in this work are listed in Table S9 and were synthesized by GenScript.
Protoplast transfection
The Japonica rice variety Nipponbare was used to isolate protoplasts. Protoplast isolation and transformation were performed as described (Li et al., 2020a). Midi extract plasmids used for protoplast transformation were prepared with Wizard Plus Midipreps DNA Purification System (Promega, Madison, WI, USA). Ten micrograms of each of nuclease and target plasmid DNA were introduced into the protoplasts by PEG‐mediated transfection, with a mean transformation efficiency of 30%–45% as measured by haemocytometer. The transfected protoplasts were incubated at 30 °C and 48 h post‐transfection they were collected and genomic DNA extracted for amplicon deep sequencing.
Constructing randomly multiplexed sgRNA libraries
Saturated target oligos were synthesized by GenScript. Fifty micromoles of forward and reverse oligos of each target were annealed individually and pooled together equally according to the targeting strand. For the three empty vectors, the ccdB and barcode for each sgRNA scaffold (BC‐Sc) portions were synthesized commercially (GenScript, Nanjing, China) and assembled into pOsU3‐sgRNA by replacing the native sgRNA scaffold portion. The pooled targets (1 μM each target) were inserted into each BsaI digested empty vector (300 ng) using T4 DNA ligase (New England BioLabs, Ipswich, MA, USA). The ligation products were purified by Monarch PCR & DNA Cleanup Kit (New England BioLabs, Ipswich, MA, USA) and delivered into ElectroMAX DH5‐E Competent Cells (Thermo Fisher Scientific, Waltham, MA, USA) by electroporation. To guarantee the uniformity of each target, three independent biological replicates were performed. Then, the bacterial colony from the same plate was pooled together for plasmid extraction by Wizard Plus Midipreps DNA Purification System (Promega, Madison, WI, USA), and totally 18 BC‐Sc barcoded targets storage libraries were obtained.
Next, the sgRNA scaffolds (esgRNA‐2×boxB, esgRNA‐2×MS2, and esgRNA‐boxB‐MS2) were amplified from corresponding OsU3‐derived vectors with addition of BamHI and BsrGI at 3′ end, and two BsaI sites at both ends. Each BsaI digested sgRNA scaffold (60 ng) was inserted into corresponding BsaI digested BC‐Sc barcoded targets storage libraries (100 ng), and totally 18 sgRNA entry libraries were obtained. The target coverage was analysed by amplifying target barcode region using amplicon deep sequencing. Then, equal amounts of plasmids from three different barcoded sgRNA entry libraries of coding strand or three different barcoded sgRNA entry libraries of non‐coding strand were mixed equally and used for the remaining constructions. The three independent biological replicates were performed individually. For the randomly multiplexed sgRNAs assembly rounds, 200 ng BglII and Acc65I digested target‐scaffold fragments from mixed sgRNA entry libraries were inserted into 400 ng BamHI and BsrGI digested sgRNA library from former round, and the uniformity and target‐scaffold combinations were evaluated by barcode sequencing. All the ligation products here were also purified by Monarch PCR & DNA Cleanup Kit and delivered into ElectroMAX DH5‐E Competent Cells by electroporation. Midi plasmids of sgRNA libraries in every step were prepared with Wizard Plus Midipreps DNA Purification System.
For assembling the randomly dual sgRNAs library with pH‐MoBE, the randomly multiplexed sgRNAs from three independent biological replicates were pooled equally together, and 300 ng target‐scaffold fragments from BglII and Acc65I digested randomly dual sgRNAs library were inserted into 1200 ng BsaI digested pH‐MoBE vector using T4 DNA ligase. The ligation products were purified by Monarch PCR & DNA Cleanup Kit and delivered into E. coli chemical competent cells Trelief 5α (Tsingke Biotech, Beijing, China). Midi plasmids were prepared with Wizard Plus Midipreps DNA Purification System.
Agrobacterium‐mediated transformation of rice callus cells
The pH‐MoBE libraries carrying randomly dual sgRNAs of coding strand and non‐coding strand pooled in equimolar ratios and transformed into A. tumefaciens chemical competent cells EHA105. About 3000 Agrobacterium clones were used to transform 30 000 rice calli. Agrobacterium‐mediated transformation of callus cells of the Japonica rice variety Nipponbare was conducted as reported (Shan et al., 2013). Hygromycin (50 μg/mL) was used to select transgenic calli.
Selection of haloxyfop‐resistant seedlings in the medium
After transformation, the calli were selected on callus induction medium supplemented with hygromycin (50 μg/mL) for 4 weeks. Then, the hygromycin‐resistant calli were transferred to callus induction medium supplemented with 0.108 mg/L haloxyfop for 3 weeks. After 3 weeks selection, the fresh and bright calli were transferred to callus induction medium supplemented with 0.162 mg/L. After another 3 weeks of selection, the small callus pellets differentiated from fresh and bright calli were transferred to regeneration medium supplemented with 0.162 mg/L haloxyfop for regeneration. After 3 weeks regeneration, the regeneration seedlings were transferred to rooting medium supplemented with 0.162 mg/L haloxyfop for 2 weeks.
Selection of haloxyfop‐resistant seedlings of T1 generation
Seeds from T0 rice lines were germinated at 30 °C in the dark for 3 days, then transferred to nutrient soil, and cultivated in growth chamber. The seedlings of T1 generation were genotyped using Sanger sequencing. When growing to two‐to‐three‐leaf‐stage, the identified homozygotes were treated with 1/2 Murashige‐Skoog hydroponic medium (2.15 g/L) containing 0.54 mg/L or 1.62 mg/L haloxyfop. These seedlings were cultured in growth chamber with 30 °C and 16 h light/8 h dark.
DNA extraction
The genomic DNA of protoplasts was extracted with a DNA‐Quick Plant System (Tiangen Biotech, Beijing, China). Genomic DNA of regenerated rice seedlings was extracted with CTAB. The genomic DNA samples were quantified with a NanoDrop One Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Amplicon deep sequencing and data analysis
For the amplicons of protoplasts, two rounds of PCR were performed using barcoded specific primers of the second round (Table S9). To assess the target coverage and sgRNA scaffold ratios in the randomly multiplexed sgRNA libraries, PCR was performed on the extracted plasmids to amplify the barcode combinations region with barcoded primers listed in Table S9. The amplicons were purified with an EasyPure PCR Purification Kit (TransGen Biotech, Beijing, China) and quantified with a NanoDrop One Spectrophotometer. Equal amounts of the PCR products were pooled and sequenced commercially (Beijing Genomics Institute, Shenzhen, China) using the MGI DNBseq‐T7 platform. The protospacer sequences in the reads were examined to identify base substitutions and indels. The amplicon sequencing was repeated three times for each target site, using genomic DNA extracted from three independent protoplast samples. Amplicon reads with a quality score < 30 were filtered out. The distribution of identified alleles around edited targets for each sgRNA were analysed as described previously (Clement et al., 2019). When analysing the details of barcode sequencing, the reads containing N base were abandoned to filter out unreliable sequencing reads.
Off‐target analysis
The sgRNA‐dependent off‐target sites were predicted using the online tool Cas‐OFFinder (Bae et al., 2014). Sites containing up to 3‐nt mismatches were examined. The sgRNA‐independent off‐target sites are listed in Table S9. The orthogonal R‐loop assay was performed and analysed as described previously (Jin et al., 2020).
Statistical analysis
All the statistical data were calculated using GraphPad Prism v.9.0. All numerical values in this work were presented as means ± sem. Pairwise comparison was performed using two‐tailed Student's t‐test. The multiple comparisons were performed according to one‐way ANOVA and Tukey's multiple comparisons test.
Conflicts of interest
The authors have submitted patents application based on the results reported in this paper. The patent does not restrict the research use of the methods in this article.
Author contributions
A.Z., T.S., Y.S., C.L., and J.W. designed the project. A.Z., T.S., Y.S., J.H., Z.H., Z.M., J.He., S.L., and C.L. performed the experiments. A.Z., Z.C., Z.Z., Y.W., and C.L. developed the web application. A.Z., X.L., L.J., X.D., Y.Wang, Y.L., C.L., and J.W. wrote the manuscript. C.L. and J.W. supervised the project.
Code availability
The custom Python scripts to analyse barcode sequencing can be found in Code S1–S3. The web portal server is accessible at http://pgec.njau.edu.cn/plantbe‐code for non‐profit use.
Supporting information
Code S1 The Python script to analyse barcode sequencing reads in sgRNA entry libraries.
Code S2 The Python script to analyse barcode sequencing reads in randomly dual sgRNA libraries.
Code S3 The Python script to analyse barcode sequencing reads in randomly triple sgRNA libraries.
Datasets S1 Datasets of barcode sequencing reads of targets from coding strand in three sgRNA entry libraries.
Datasets S2 Datasets of barcode sequencing reads of targets from non‐coding strand in three sgRNA entry libraries.
Datasets S3 Datasets of barcode sequencing reads of the types of target‐scaffold combinations in sgRNA entry libraries.
Datasets S4 Datasets of barcode sequencing reads of the types of target‐scaffold combinations in randomly dual sgRNA libraries.
Datasets S5 Datasets of barcode sequencing reads of the types of target‐scaffold combinations in randomly triple sgRNA libraries.
Figure S1 The secondary structures of sgRNA scaffolds used in this study.
Figure S2 Activities of AD‐fusion and 2A‐AD constructs in rice protoplasts.
Figure S3 Representative sanger sequencing chromatograms of uncorrected 2A‐APOBEC3A construct.
Figure S4 Activities of CD‐fusion and 2A‐CD constructs in rice protoplasts.
Figure S5 Base editing outcomes generated by MoBE in rice protoplasts.
Figure S6 Schematic of multiple sgRNAs assembly for MoBE in rice protoplasts.
Figure S7 Comparison of on‐target base editing activities between MoBE and MoBE‐HF.
Figure S8 Schematic representation of the PlantBE‐CODE workflow for designing saturated base editing targets.
Figure S9 Schematic of amplified barcode sequencing region on sgRNA libraries.
Figure S10 Selecting haloxyfop resistance calli using pH‐MoBE.
Figure S11 Genotypes of T1 rice.
Note S1 User guide of PlantBE‐CODE.
Sequence S1 DNA sequences of sgRNA, esgRNA, and scRNAs used in this study.
Sequence S2 DNA sequences of the modules composed of 2A‐AD, AD‐fusion, 2A‐CD, and CD‐fusion used in this study.
Sequence S3 DNA sequences of MoBE and MoBE‐HF used in this study.
Sequence S4 DNA sequences of modules for assembling randomly multiplexed sgRNA libraries used in this study.
Table S1 Targets used to test the efficiencies of AD‐fusion, 2A‐AD, CD‐fusion, 2A‐CD, MoBE, and MoBE‐HF in rice protoplasts.
Table S2 Mismatched sequences of OsCDC48‐T2 used to test sgRNA‐dependent off‐target effects of MoBE in rice protoplasts.
Table S3 Target sequences used to test sgRNA‐independent off‐target effects of MoBE and MoBE‐HF in rice protoplasts.
Table S4 Target sequences from the 34th Exon of OsACC for constructing randomly multiplexed libraries.
Table S5 Genotypes of screened herbicide resistance seedlings.
Table S6 Ratios in sgRNA entry libraries of these targets presented in herbicide resistance seedlings.
Table S7 Ratios in sgRNA entry libraries of three different sgRNA scaffolds presented in herbicide resistance seedlings.
Table S8 Potential off‐target sites analysed for herbicide resistance mutants.
Table S9 Primers used in this study.
Acknowledgements
We thank Haiyang Wang (Chinese Academy of Agricultural Sciences) for useful comments and the Bioinformatics Center of Nanjing Agricultural University for supporting high‐performance computing. This work was supported by the Ministry of Agriculture and Rural Affairs of China, the Natural Science Foundation of Jiangsu Province (grant no. BK20212010), the Hainan Yazhou Bay Seed Laboratory (project of B21HJ1004), the Young Elite Scientists Sponsorship Program by CAST (grant no. 2020QNRC001), Jiangsu Agricultural Science and Technology Innovation Fund (grant no. cx(21)1002), Doctor of Entrepreneurship and Innovation in Jiangsu Province (grant no. JSSCBS20210272), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant no. KYCX22_0734). We also acknowledge support from the Collaborative Innovation Center for Modern Crop Production cosponsored by Jiangsu Province and Ministry of Education.
Contributor Information
Chao Li, Email: chaoli@njau.edu.cn.
Jianmin Wan, Email: wanjm@njau.edu.cn, Email: wanjianmin@caas.cn.
Data availability
The authors declare that all data supporting the findings of this study are available in the article, and Supporting Information are available from the corresponding author on request. For sequence data, rice LOC_Os IDs LOC_Os01g55540 (OsAAT), LOC_Os05g22940 (OsACC), LOC_Os03g05730 (OsCDC48), LOC_Os09g26999 (OsDEP1), LOC_Os02g11010 (OsOD, OsEV), LOC_Os10g40600 (OsNRT1.1B), LOC_Os03g08570 (OsPDS), LOC_Os02g07160 (OsHPPD), LOC_Os11g32080 (OsACTG), and LOC_Os10g41310 (OsDL) are available on the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). The deep sequencing data have been deposited with the NCBI BioProject database under accession code numbers PRJNA893550. Plasmids encoding MoBE, pOsU3‐esgRNA‐boxB‐MS2, pOsU3‐EV1, pOsU3‐EV2, pOsU3‐EV3, and pH‐MoBE will be available from Addgene.
References
- Anzalone, A.V. , Koblan, L.W. and Liu, D.R. (2020) Genome editing with CRISPR‐Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844. [DOI] [PubMed] [Google Scholar]
- Bae, S. , Park, J. and Kim, J.S. (2014) Cas‐OFFinder: a fast and versatile algorithm that searches for potential off‐target sites of Cas9 RNA‐guided endonucleases. Bioinformatics, 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt, H. , Eid, A. , Momin, A.A. , Bazin, J. , Crespi, M. , Arold, S.T. and Mahfouz, M.M. (2019) CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 20, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdeville, N. , Schindele, P. and Puchta, H. (2020) Application of CRISPR/Cas‐mediated base editing for directed protein evolution in plants. Sci. China Life Sci. 63, 613–616. [DOI] [PubMed] [Google Scholar]
- Čermák, T. , Curtin, S.J. , Gil‐Humanes, J. , Čegan, R. , Kono, T.J. , Konečná, E. , Belanto, J.J. et al. (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell, 29, 1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Zhang, Y. , Li, G. , Fang, H. , Sretenovic, S. , Fan, A. , Li, J. et al. (2023) CRISPR‐Cas12a base editors confer efficient multiplexed genome editing in rice. Plant Commun. 4, 100601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement, K. , Rees, H. , Canver, M.C. , Gehrke, J.M. , Farouni, R. , Hsu, J.Y. , Cole, M.A. et al. (2019) CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist, M.K.M. and Rabe, K.S. (2019) Applications of protein engineering and directed evolution in plant research. Plant Physiol. 179, 907–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudelli, N.M. , Lam, D.K. , Rees, H.A. , Sola‐Esteves, N.M. , Barrera, L.A. , Born, D.A. , Edwards, A. et al. (2020) Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900. [DOI] [PubMed] [Google Scholar]
- Hwang, G.H. , Park, J. , Lim, K. , Kim, S. , Yu, J. , Yu, E. , Kim, S.T. et al. (2018) Web‐based design and analysis tools for CRISPR base editing. BMC Bioinformatics, 19, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, Y.K. , Lee, S. , Hwang, G.‐H. , Hong, S.‐A. , Park, S.‐E. , Kim, J.‐S. , Woo, J.‐S. et al. (2021) Adenine base editor engineering reduces editing of bystander cytosines. Nat. Biotechnol. 39, 1426–1433. [DOI] [PubMed] [Google Scholar]
- Jin, S. , Fei, H. , Zhu, Z. , Luo, Y. , Liu, J. , Gao, S. , Zhang, F. et al. (2020) Rationally designed APOBEC3B cytosine base editors with improved specificity. Mol. Cell, 79, 728–740. [DOI] [PubMed] [Google Scholar]
- Kim, D. , Kim, D.E. , Lee, G. , Cho, S.I. and Kim, J.S. (2019a) Genome‐wide target specificity of CRISPR RNA‐guided adenine base editors. Nat. Biotechnol. 37, 430–435. [DOI] [PubMed] [Google Scholar]
- Kim, H.S. , Jeong, Y.K. , Hur, J.K. , Kim, J.S. and Bae, S. (2019b) Adenine base editors catalyze cytosine conversions in human cells. Nat. Biotechnol. 37, 1145–1148. [DOI] [PubMed] [Google Scholar]
- Koblan, L.W. , Arbab, M. , Shen, M.W. , Hussmann, J.A. , Anzalone, A.V. , Doman, J.L. , Newby, G.A. et al. (2021) Efficient C•G‐to‐G•C base editors developed using CRISPRi screens, target‐library analysis, and machine learning. Nat. Biotechnol. 39, 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, A.C. , Kim, Y.B. , Packer, M.S. , Zuris, J.A. and Liu, D.R. (2016) Programmable editing of a target base in genomic DNA without double‐stranded DNA cleavage. Nature, 533, 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang, Y. , Li, S. , Ren, B. , Yan, F. , Spetz, C. , Li, X. , Zhou, X. et al. (2020) Base‐editing‐mediated artificial evolution of OsALS1 in planta to develop novel herbicide‐tolerant rice germplasms. Mol. Plant, 13, 565–572. [DOI] [PubMed] [Google Scholar]
- Li, J. , Sun, Y. , Du, J. , Zhao, Y. and Xia, L. (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant, 10, 526–529. [DOI] [PubMed] [Google Scholar]
- Li, C. , Zong, Y. , Wang, Y. , Jin, S. , Zhang, D. , Song, Q. , Zhang, R. et al. (2018) Expanded base editing in rice and wheat using a Cas9‐adenosine deaminase fusion. Genome Biol. 19, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Zhang, R. , Meng, X. , Chen, S. , Zong, Y. , Lu, C. , Qiu, J.L. et al. (2020a) Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38, 875–882. [DOI] [PubMed] [Google Scholar]
- Li, C. , Zong, Y. , Jin, S. , Zhu, H. , Lin, D. , Li, S. , Qiu, J.L. et al. (2020b) SWISS: multiplexed orthogonal genome editing in plants with a Cas9 nickase and engineered CRISPR RNA scaffolds. Genome Biol. 21, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, A. , Mitsunobu, H. , Yoshioka, S. , Suzuki, T. , Kondo, A. and Nishida, K. (2022a) Cytosine base editing systems with minimized off‐target effect and molecular size. Nat. Commun. 13, 4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. , Liu, L. , Sun, W. , Zhou, X. and Zhou, H. (2022b) A large‐scale genome and transcriptome sequencing analysis reveals the mutation landscapes induced by high‐activity adenine base editors in plants. Genome Biol. 23, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Xie, J. , Zhang, Q. , Wang, X. , Gou, S. , Lin, L. , Chen, T. et al. (2022) AGBE: a dual deaminase‐mediated base editor by fusing CGBE with ABE for creating a saturated mutant population with multiple editing patterns. Nucleic Acids Res. 50, 5384–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Qin, R. , Li, J. , Liao, S. , Shan, T. , Xu, R. , Wu, D. et al. (2020) A CRISPR‐Cas9‐mediated domain‐specific base‐editing screen enables functional assessment of ACCase variants in rice. Plant Biotechnol. J. 18, 1845–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder, L.G. , Zhang, D. , Baltes, N.J. , Paul, J.W., 3rd , Tang, X. , Zheng, X. , Voytas, D.F. et al. (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 169, 971–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, M.S. , Podracky, C.J. and Liu, D.R. (2020) The developing toolkit of continuous directed evolution. Nat. Chem. Biol. 16, 610–619. [DOI] [PubMed] [Google Scholar]
- Nishimasu, H. , Shi, X. , Ishiguro, S. , Gao, L. , Hirano, S. , Okazaki, S. , Noda, T. et al. (2018) Engineered CRISPR‐Cas9 nuclease with expanded targeting space. Science, 361, 1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles, S.B. and Yu, Q. (2010) Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. [DOI] [PubMed] [Google Scholar]
- Rabinowitz, R. , Abadi, S. , Almog, S. and Offen, D. (2020) Prediction of synonymous corrections by the BE‐FF computational tool expands the targeting scope of base editing. Nucleic Acids Res. 48, W340–W347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, Q. , Sretenovic, S. , Liu, G. , Zhong, Z. , Wang, J. , Huang, L. , Tang, X. et al. (2021) Improved plant cytosine base editors with high editing activity, purity, and specificity. Plant Biotechnol. J. 19, 2052–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, M.F. , Zhao, K.T. , Eton, E. , Lapinaite, A. , Newby, G.A. , Thuronyi, B.W. , Wilson, C. et al. (2020) Phage‐assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Chen, K. , Liang, Z. , Li, J. , Zhang, Y. , Zhang, K. et al. (2013) Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant, 6, 1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J. , Zeng, D. , Zhao, Y. , Wang, Y. , Liu, T. , Li, S. , Xue, Y. et al. (2022) PhieABEs: a PAM‐less/free high‐efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol. J. 20, 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuronyi, B.W. , Koblan, L.W. , Levy, J.M. , Yeh, W.H. , Zheng, C. , Newby, G.A. , Wilson, C. et al. (2019) Continuous evolution of base editors with expanded target compatibility and improved activity. Nat. Biotechnol. 37, 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y. , Shen, R. , Li, Z. , Yao, Q. , Zhang, X. , Zhong, D. , Tan, X. et al. (2022) Efficient C‐to‐G editing in rice using an optimized base editor. Plant Biotechnol. J. 20, 1238–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton, R.T. , Christie, K.A. , Whittaker, M.N. and Kleinstiver, B.P. (2020) Unconstrained genome targeting with near‐PAMless engineered CRISPR‐Cas9 variants. Science, 368, 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Shen, L. , Fu, Y. , Yan, C. and Wang, K. (2015) A simple CRISPR/Cas9 system for multiplex genome editing in rice. J. Genet. Genomics, 42, 703–706. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Zong, Y. , Lin, Q. , Zhang, H. , Chai, Z. , Zhang, D. , Chen, K. et al. (2020a) Precise, predictable multi‐nucleotide deletions in rice and wheat using APOBEC‐Cas9. Nat. Biotechnol. 38, 1460–1465. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Zhou, L. , Tao, R. , Liu, N. , Long, J. , Qin, F. , Tang, W. et al. (2020b) sgBE: a structure‐guided design of sgRNA architecture specifies base editing window and enables simultaneous conversion of cytosine and adenosine. Genome Biol. 21, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Xue, P. , Cao, M. , Yu, T. , Lane, S.T. and Zhao, H. (2021) Directed evolution: methodologies and applications. Chem. Rev. 121, 12384–12444. [DOI] [PubMed] [Google Scholar]
- Wang, H. , He, Y. , Wang, Y. , Li, Z. , Hao, J. , Song, Y. , Wang, M. et al. (2022) Base editing‐mediated targeted evolution of ACCase for herbicide‐resistant rice mutants. J. Integr. Plant Biol. 64, 2029–2032. [DOI] [PubMed] [Google Scholar]
- Wong, A.S.L. , Choi, G.C.G. , Cui, C.H. , Pregernig, G. , Milani, P. , Adam, M. , Perli, S.D. et al. (2016) Multiplexed barcoded CRISPR‐Cas9 screening enabled by CombiGEM. Proc. Natl Acad. Sci. USA, 113, 2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Ren, Q. , Zhong, Z. , Liu, G. , Han, Y. , Bao, Y. , Liu, L. et al. (2022) Genome‐wide analyses of PAM‐relaxed Cas9 genome editors reveal substantial off‐target effects by ABE8e in rice. Plant Biotechnol. J. 20, 1670–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, K. , Minkenberg, B. and Yang, Y. (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA‐processing system. Proc. Natl Acad. Sci. USA, 112, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X. , Li, F. , Tan, X. , Zeng, D. , Liu, W. , Zeng, W. , Zhu, Q. et al. (2022) BEtarget: a versatile web‐based tool to design guide RNAs for base editing in plants. Comput. Struct. Biotechnol. J. 20, 4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. , Kong, F. , Qin, R. , Li, J. , Liu, X. and Wei, P. (2021a) Development of an efficient plant dual cytosine and adenine editor. J. Integr. Plant Biol. 63, 1600–1605. [DOI] [PubMed] [Google Scholar]
- Xu, R. , Liu, X. , Li, J. , Qin, R. and Wei, P. (2021b) Identification of herbicide resistance OsACC1 mutations via in planta prime‐editing‐library screening in rice. Nat. Plants, 7, 888–892. [DOI] [PubMed] [Google Scholar]
- Yan, D. , Ren, B. , Liu, L. , Yan, F. , Li, S. , Wang, G. , Sun, W. et al. (2021) High‐efficiency and multiplex adenine base editing in plants using new TadA variants. Mol. Plant, 14, 722–731. [DOI] [PubMed] [Google Scholar]
- Yu, H. , Wu, Z. , Chen, X. , Ji, Q. and Tao, S. (2020) CRISPR‐CBEI: a designing and analyzing tool kit for cytosine base editor‐mediated gene inactivation. mSystems 5, e00350‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, D. , Liu, T. , Tan, J. , Zhang, Y. , Zheng, Z. , Wang, B. , Zhou, D. et al. (2020) PhieCBEs: plant high‐efficiency cytidine base editors with expanded target range. Mol. Plant, 13, 1666–1669. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. and Qi, Y. (2019) CRISPR enables directed evolution in plants. Genome Biol. 20, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Malzahn, A.A. , Sretenovic, S. and Qi, Y. (2019) The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants, 5, 778–794. [DOI] [PubMed] [Google Scholar]
- Zhu, H. , Li, C. and Gao, C. (2020) Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Code S1 The Python script to analyse barcode sequencing reads in sgRNA entry libraries.
Code S2 The Python script to analyse barcode sequencing reads in randomly dual sgRNA libraries.
Code S3 The Python script to analyse barcode sequencing reads in randomly triple sgRNA libraries.
Datasets S1 Datasets of barcode sequencing reads of targets from coding strand in three sgRNA entry libraries.
Datasets S2 Datasets of barcode sequencing reads of targets from non‐coding strand in three sgRNA entry libraries.
Datasets S3 Datasets of barcode sequencing reads of the types of target‐scaffold combinations in sgRNA entry libraries.
Datasets S4 Datasets of barcode sequencing reads of the types of target‐scaffold combinations in randomly dual sgRNA libraries.
Datasets S5 Datasets of barcode sequencing reads of the types of target‐scaffold combinations in randomly triple sgRNA libraries.
Figure S1 The secondary structures of sgRNA scaffolds used in this study.
Figure S2 Activities of AD‐fusion and 2A‐AD constructs in rice protoplasts.
Figure S3 Representative sanger sequencing chromatograms of uncorrected 2A‐APOBEC3A construct.
Figure S4 Activities of CD‐fusion and 2A‐CD constructs in rice protoplasts.
Figure S5 Base editing outcomes generated by MoBE in rice protoplasts.
Figure S6 Schematic of multiple sgRNAs assembly for MoBE in rice protoplasts.
Figure S7 Comparison of on‐target base editing activities between MoBE and MoBE‐HF.
Figure S8 Schematic representation of the PlantBE‐CODE workflow for designing saturated base editing targets.
Figure S9 Schematic of amplified barcode sequencing region on sgRNA libraries.
Figure S10 Selecting haloxyfop resistance calli using pH‐MoBE.
Figure S11 Genotypes of T1 rice.
Note S1 User guide of PlantBE‐CODE.
Sequence S1 DNA sequences of sgRNA, esgRNA, and scRNAs used in this study.
Sequence S2 DNA sequences of the modules composed of 2A‐AD, AD‐fusion, 2A‐CD, and CD‐fusion used in this study.
Sequence S3 DNA sequences of MoBE and MoBE‐HF used in this study.
Sequence S4 DNA sequences of modules for assembling randomly multiplexed sgRNA libraries used in this study.
Table S1 Targets used to test the efficiencies of AD‐fusion, 2A‐AD, CD‐fusion, 2A‐CD, MoBE, and MoBE‐HF in rice protoplasts.
Table S2 Mismatched sequences of OsCDC48‐T2 used to test sgRNA‐dependent off‐target effects of MoBE in rice protoplasts.
Table S3 Target sequences used to test sgRNA‐independent off‐target effects of MoBE and MoBE‐HF in rice protoplasts.
Table S4 Target sequences from the 34th Exon of OsACC for constructing randomly multiplexed libraries.
Table S5 Genotypes of screened herbicide resistance seedlings.
Table S6 Ratios in sgRNA entry libraries of these targets presented in herbicide resistance seedlings.
Table S7 Ratios in sgRNA entry libraries of three different sgRNA scaffolds presented in herbicide resistance seedlings.
Table S8 Potential off‐target sites analysed for herbicide resistance mutants.
Table S9 Primers used in this study.
Data Availability Statement
The authors declare that all data supporting the findings of this study are available in the article, and Supporting Information are available from the corresponding author on request. For sequence data, rice LOC_Os IDs LOC_Os01g55540 (OsAAT), LOC_Os05g22940 (OsACC), LOC_Os03g05730 (OsCDC48), LOC_Os09g26999 (OsDEP1), LOC_Os02g11010 (OsOD, OsEV), LOC_Os10g40600 (OsNRT1.1B), LOC_Os03g08570 (OsPDS), LOC_Os02g07160 (OsHPPD), LOC_Os11g32080 (OsACTG), and LOC_Os10g41310 (OsDL) are available on the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). The deep sequencing data have been deposited with the NCBI BioProject database under accession code numbers PRJNA893550. Plasmids encoding MoBE, pOsU3‐esgRNA‐boxB‐MS2, pOsU3‐EV1, pOsU3‐EV2, pOsU3‐EV3, and pH‐MoBE will be available from Addgene.


