Summary
Plants experience numerous biotic stresses throughout their lifespan, such as pathogens and pests, which can substantially affect crop production. In response, plants have evolved various metabolites that help them withstand these stresses. Here, we show that two specialized metabolites in the herbaceous perennial Belamcanda chinensis, tectorigenin and its glycoside tectoridin, have diverse defensive effects against phytopathogenic microorganisms and antifeeding effects against insect pest. We further functionally characterized a 7‐O‐uridine diphosphate glycosyltransferase Bc7OUGT, which catalyses a novel reversible glycosylation of tectorigenin and tectoridin. To elucidate the catalytic mechanisms of Bc7OUGT, we solved its crystal structure in complex with UDP and UDP/tectorigenin respectively. Structural analysis revealed the Bc7OUGT possesses a narrow but novel substrate‐binding pocket made up by plentiful aromatic residues. Further structure‐guided mutagenesis of these residues increased both glycosylation and deglycosylation activities. The catalytic reversibility of Bc7OUGT was also successfully applied in an one‐pot aglycon exchange reaction. Our findings demonstrated the promising biopesticide activity of tectorigenin and its glycosides, and the characterization and mechanistic study of Bc7OUGT could facilitate the design of novel reversible UGTs to produce valuable glycosides with health benefits for both plants and humans.
Keywords: Plant chemical defence, Belamcanda chinensis, tectorigenin, tectoridin, glycosylation, isoflavones
Introduction
Plants face numerous biotic and abiotic constraints throughout their lifecycles, including biotic stressors such as pathogens, herbivores, and weeds, which have tremendously negative effects on agricultural crops (Atkinson and Urwin, 2012). These stresses are typically controlled using chemical pesticides and herbicides, but the excessive use of chemical treatments is affecting the environment, agriculture, and poses a threat to human health (Rani et al., 2021). Plants have evolved various metabolites that function as direct defence compounds against biotic stress (Ford et al., 2010; Pretali et al., 2016). For example, pyrethrin and azadirachtin, isolated from Pyrethrum cinerariifolium and Azadirachta indica, respectively, have a long history of traditional and commercial use as pesticides, as they are safe, environmentally degradable, and have low toxicity to mammals (Casida and Quistad, 1995; Chaudhary et al., 2017). Therefore, exploiting plant‐specialized metabolites as biocontrol agents will accelerate the development of eco‐friendly substitutes for hazardous chemical pesticides.
Belamcanda chinensis (blackberry lily) is an herbaceous perennial in the Iridaceae family. A major class of bioactive compounds in B. chinensis is isoflavones, including tectorigenin, irigenin, and their glycosides tectoridin and iridin. Prior research has focused on the biological efficacy of these compounds for human health, for instance, as antioxidants, oestrogen receptor modulators, and potential preventative agents (Ahn et al., 2006; Wu et al., 2011); however, their physiological roles in plants are less well understood. Isoflavones are predominantly found in leguminous plants, where they act as signalling molecules in the production of nitrogen‐fixing root nodules (Eckardt, 2006; Kim, 2022) or as precursors in the production of phytoalexins in other plant–microbe interactions (Ciesielski and Metz, 2020; He and Dixon, 2000). Isoflavones from B. chinensis, a non‐leguminous plant, contain a distinctive 6‐methoxyl group, but few studies have focused on their bioactivity or biosynthesis.
Glycosylation is a process by which a sugar moiety is transferred to an acceptor molecule. The glycosylation of natural plant products significantly affects their bioactivity, generally enhancing their solubility and stability and facilitating storage (Plaza et al., 2014). Direct defence compounds that contain broad‐spectrum toxins are usually cytotoxic and can damage plant tissues. However, due to their glycosylation, these compounds can be safely stored in cells and activated as needed via the removal of sugars to provide protection against pathogens and insect pests (Itkin et al., 2011). A recent comprehensive study in Nicotiana attenuata indicated that the glycosylation of 17‐hydroxygeranyllinalool diterpene glycosides prevents the autotoxicity of this chemical defence compound in vivo (Heiling et al., 2021). Glycosylation is also important for the stability of anthocyanins or terpenoids, contributing to the diversity of pigments, the aroma of flowers, and the taste of fruits (Behr et al., 2020). In metabolic engineering, the glycosylation of compounds such as terpenoid glycosides, glucosinolates, cyanogenic glycosides, and flavonoid glycosides is a necessary step in the production of drug lead compounds, cosmetics, nutrients, and sweeteners (De Bruyn et al., 2015; Huccetogullari et al., 2019; Spahn et al., 2016).
Glycosylation in plants is usually carried out by uridine diphosphate (UDP)‐dependent glycosyltransferases (UGTs) from the glycosyltransferase 1 (GT1) family. The GT1 family is one of the largest groups of natural product‐decorating enzymes in plants, and its gene expansion has greatly contributed to phytochemical diversification during evolution (Yonekura‐Sakakibara and Hanada, 2011). The UGTs accommodate their sugar donors primarily via a conserved 44‐amino acid plant secondary product glycosyltransferase (PSPG) motif (Hughes and Huges, 1994; Ross et al., 2001), and a histidine positioned around the 20th residue was reported to act as a general base to deprotonate the acceptor (Lairson et al., 2008). While some plant UGTs show promiscuous biocatalyst activity with regard to aglycone acceptors, most have strict substrate specificity and regioselectivity (Wen et al., 2021). This specificity might be determined by the correct orientation of the acceptor molecule plus the structure and size of the enzyme, but the residues within the acceptor‐binding pocket are poorly conserved (Li et al., 2007; Modolo et al., 2009b).
Previous phytochemistry studies of B. chinensis indicated that the glycosides of its isoflavones (such as iridin, tectoridin, iristectorin A, and iristectorin B) typically possess regiospecific 7‐O‐glycosylation (Tian et al., 2018). The UGTs that facilitate these modifications remain to be identified, and their catalytic features and mechanisms are currently unclear. In this study, we evaluated the potential defensive effects of tectorigenin and tectoridin towards phytopathogens and herbivores. By sequencing the full‐length transcriptome of B. chinensis, we identified a novel Bc7OUGT that is responsible for the reversible glycosylation of tectorigenin and tectoridin. To explore its catalytic mechanism, we solved the crystal structures of Bc7OUGT in complex with UDP and UDP/tectorigenin. Additional mutagenesis experiments further identified key residues responsible for its catalytic activity. In addition, the reversibility of Bc7OUGT was successfully applied in an one‐pot aglycon exchange reaction. These findings provide a basis for enzyme engineering and a potential versatility of Bc7OUGT in the biosynthesis of bioactive natural products.
Results
Tectorigenin and tectoridin provide biotic resistance against plant pathogens in vitro and in vivo
We isolated tectorigenin and its 7‐O‐glycosylated form tectoridin from the root, stem, leaf, rhizome, flower, and fruit of B. chinensis (Figure S1A). We measured their contents using high‐performance liquid chromatography–tandem mass spectrometry (HPLC–MS) via comparisons to authorized standards (Figure S1B,C). Among the six tested tissues, both compounds were most abundant in the rhizome (162.4 μg/g and 293.4 μg/g, fresh weight), followed by the stem, which is consistent with the previous findings (Figure S1D) (Peng et al., 2009; Wang et al., 2011; Woźniak and Matkowski, 2015).
As tectorigenin and tectoridin are highly abundant specialized metabolites in B. chinensis, we reasoned that they might protect plants against biotic stress, such as bacterial or fungal pathogen attack. We therefore investigated their roles in plant pathogen resistance in vitro. Xanthomonas oryzae pv. oryzicola (Xoc) and X. oryzae pv. oryzae (Xoo) are major phytopathogenic bacteria in rice (Oryza sativa) that cause bacterial leaf streak and bacterial blight respectively (Chu et al., 2022). These bacterial diseases cause serious yield losses, and many high‐yielding rice cultivars are highly susceptible to these pathogens (Wang et al., 2022). We tested the in vitro antibacterial activities of tectorigenin and tectoridin against Xoc strain HGA4 and Xoo strain PXO99 (Figure 1a). Compared with the negative control (DMSO), tectorigenin effectively inhibited the growth of HGA4 and PXO99, with 55.2% and 54.0% reductions in growth, respectively, after 48 h of treatment at a concentration of 200 μm (Figure 1a,b). Treatment with 400 μm tectorigenin led to 50.0% and 59.6% reductions in the growth of HGA4 and PXO99 respectively. By contrast, neither 200 μm nor 400 μm tectoridin affected the growth of either strain in vitro (Figure 1a,b).
Figure 1.
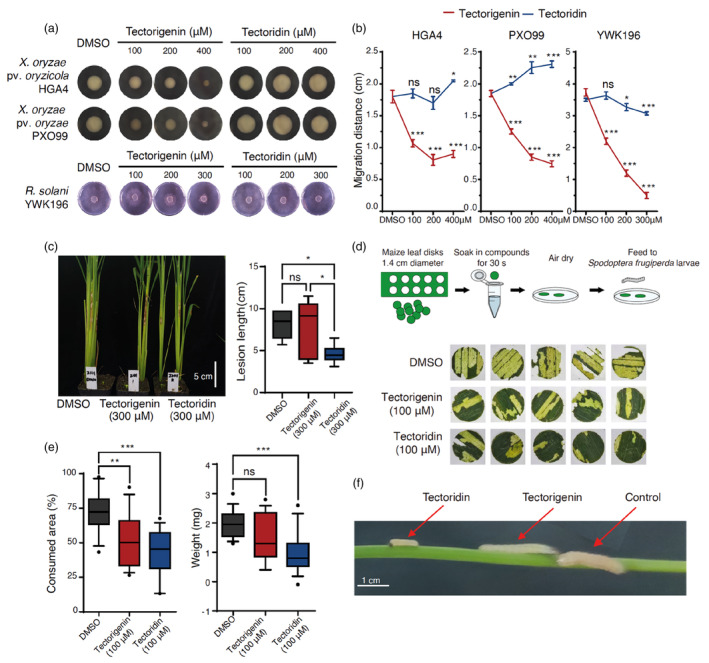
Tectorigenin and tectoridin show phytopathogen resistance and antifeeding activity. (a) Xanthomonas oryzae pv. oryzicola (Xoc) strain HGA4, X. oryzae pv. oryzae (Xoo) strain PXO99, and Rhizoctonia solani strain YWK196 were grown for 48 h on medium containing DMSO, tectorigenin, or tectoridin at the indicated concentrations. (b) Migration distances of the HGA4, PXO99, and YWK196 on medium containing tectorigenin and tectoridin after 48 h of growth (N = 3). Asterisks indicate significant differences between the DMSO and treatment groups (ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.001). (c) Lesion growth on rice leaf sheaths pretreated with DMSO control or 300 μm of either compound for 48 h after inoculation with R. solani strain YWK196 (N = 8; ns, not significant; *P < 0.05). The middle line represents the median. Lower and upper hinges correspond to the first (Q1) and third (Q3) quartiles. The lower whisker extends to Q1 − 1.5 × IOR (where IOR is the interquartile range) and the upper whisker extends to Q3 + 1.5 × IOR. Outliers are shown as solid data points. (d) Antifeeding activity experiment with third‐instar Spodoptera frugiperda larvae. Representative consumed leaf discs after 48 h of feeding are shown. (e) Left, percentage of consumed leaf area after 48 h of feeding by the larvae (N = 15; ns, not significant; **P < 0.01, ***P < 0.001). Right, larval weight after 72 h of feeding on food premixed with 100 μm of each test compound (N = 15; final concentration = 0.4 μmol/g; ns, not significant; **P < 0.01, ***P < 0.001). (f) Representative S. frugiperda larvae fed with food that was premixed with the tested compounds.
The plant pathogenic fungus Rhizoctonia solani causes economically important diseases in many agricultural and horticultural crops worldwide, including rice sheath blight (Méndez‐Chávez et al., 2022). We therefore tested the antifungal activities of tectorigenin and tectoridin against R. solani strain YWK196 in vitro (Figure 1a). Tectorigenin effectively inhibited the growth of R. solani, resulting in 40.5%, 67.6%, and 86.5% reductions in growth relative to the control (DMSO) when applied at 100, 200, and 300 μm respectively (Figure 1a,b). By contrast, tectoridin had a relatively weak effect, reducing R. solani growth by 12.4% when applied at concentrations up to 300 μm (Figure 1a,b).
To determine whether tectorigenin and/or tectoridin enhances the resistance of rice to R. solani in vivo, we carried out inoculation experiments with R. solani strain YWK196 in rice leaf sheaths in vivo (Figure 1c). Compared with the solvent control (DMSO), rice leaf sheaths pretreated with 300 μm tectoridin exhibited milder disease symptoms, with a significantly shorter lesion length of 4.5 ± 1.0 cm compared to 7.1 ± 2.3 cm for the solvent control. By contrast, tectorigenin treatment at the same concentration (300 μm) did not inhibit lesion progression, resulting in a similar lesion length to the control treatment (Figure 1c). Thus, even though tectoridin did not show a significant antifungal effect in vitro, it might trigger plant immune responses and improve rice resistance in vivo.
Tectorigenin and tectoridin show antifeeding activity against Spodoptera frugiperda larvae
Spodoptera frugiperda is an economically important herbivore pest whose larval stage affects a wide variety of crops, including maize (Zea mays), sorghum (Sorghum bicolor), rice, and soybean (Glycine max) (Lemus de la Cruz et al., 2022). Identifying and developing new biopesticides is highly desirable (Fang et al., 2019). Here, we evaluated the antifeeding activity of tectorigenin and tectoridin against S. frugiperda larvae. We fed third‐instar larvae of S. frugiperda with maize leaf discs presoaked with the tested compounds (100 μm) and measured the consumed area after 48 h of feeding (Figure 1d). A significantly smaller leaf area was consumed from leaf discs treated with tectorigenin (52.3%) or tectoridin (43.1%) compared to the DMSO control (72.1%), revealing that both tectorigenin and tectoridin have remarkable feeding deterrent activities (Figure 1d,e). When third‐instar larvae were fed with food premixed with tectorigenin or tectoridin for 72 h (final concentration: 0.4 μmol/g), the larval mass exhibited a 23.2% and 47.7% decrease compared with the control group respectively (Figure 1e,f). The diminished larval weights were in line with the antifeedant activity, as less consumption results in a lower larval body mass. These results reveal that both tectorigenin and tectoridin have significant antifeeding activity against S. frugiperda larvae.
Full‐length transcriptome sequencing reveals candidate 7‐O ‐glycosyltransferase genes in B. chinensis
To identify the glycosylation transferases involved in tectorigenin decoration, we sequence mixed samples of root, rhizome, leaf, flower, and fruit tissues of B. chinensis using PacBio HiFi full‐length transcriptome sequencing. A total of 16 500 transcripts were assembled, and 46 putative UGT genes were identified using the UGT superfamily signature motif PF00201. Their amino acid lengths were predicted to range from 290 to 615. Phylogenetic analysis of the 46 UGTs with other functionally characterized plant UGTs suggested that the putative B. chinensis UGTs cluster into the previously identified A, C, D, E, F, G, K, L, O, and R subgroups (Figure 2a; marked in red). Notably, 10 putative B. chinensis UGTs were phylogenetically closer to the monocot‐specific genes within the D and E subgroups (Figure 2a, marked in blue), which were reported to catalyse the 7‐O‐glycosylation of flavonoids (Figure 2b). We thus considered these UGTs to be candidate enzymes for the glycosylation of tectorigenin into tectoridin. Among these, Bch3/13402.p1 and Bch3/12632.p1 have the same protein sequence and showed the most similarity to UGT706C1 and UGT706D1, which were previously identified as flavonoid 7‐O‐glycosyltransferases in rice (Peng et al., 2017).
Figure 2.
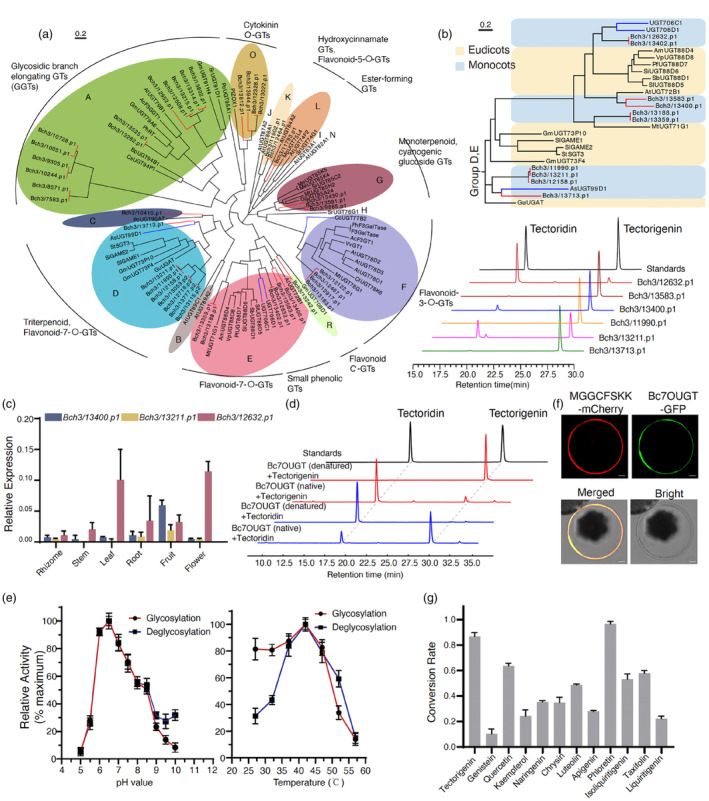
Functional characterization and catalytic features of Bc7OUGT. (a) Phylogenetic tree of the 46 putative B. chinensis UGTs and other functionally characterized plant UGTs. The tree was adapted from Louveau and Osbourn (2019). (b) Upper panel, UGT phylogenetic subgroups D and E. Lower panel, HPLC analysis of the glycosylation products with tectorigenin using candidate UGTs. (c) Relative expression of Bch3/13400.p1, Bch3/13211.p1, and Bch3/12632.p1 in six organs of B. chinensis. (d) HPLC analysis of the products of bidirectional reactions with tectorigenin or tectoridin using purified Bc7OUGT (denatured Bc7OUGT was used as a negative control). (e) Effects of various temperatures and pH levels on the bidirectional catalytic activity of Bc7OUGT. (f) Subcellular localization of Bc7OUGT in Arabidopsis protoplasts; MGGCFSKK‐mCherry is a membrane localization marker. Scale bars = 10 μm. (g) Conversion rates of Bc7OUGT acting on various flavone substrates.
Bc7OUGT catalyses the reversible glycosylation of tectorigenin and tectoridin
Six of the candidate genes were successfully amplified, cloned into the pET28a (+) vector, and expressed in Escherichia coli strain C43 (DE3) (Figure 2b). In the presence of UDP‐Glu and tectorigenin, reactions with Bch3/11990.p1, Bch3/13583.p1, and Bch/13713.p1 did not yield any new products, whereas reactions with Bch3/13400.p1, Bch3/13211.p1, and Bch3/12632.p1 resulted in a peak with the same retention time as tectoridin (19.6 min; Figure 2b). The identity of the product was further confirmed using HPLC–MS/MS with a comparison to the authorized standard of tectoridin. Among these, Bch3/12632.p1 had the highest conversion rate, while Bch3/13211.p1 generated a by‐product with a retention time of 20.2 min. We then measured the transcription level of Bch3/13400.p1, Bch3/13211.p1, and Bch3/12632.p1 in six B. chinensis organs. The expression of these UGTs are very low in rhizome, only Bch3/12632.p1 showed the highest transcript levels in the leaf and flower (Figure 2c). These results identify Bch3/12632.p1 as a functional regiospecific 7‐O‐UGT of tectorigenin in B. chinensis, which we refer to hereafter as Bc7OUGT.
We purified the recombinant proteins using Ni‐NTA affinity chromatography for functional characterization (Figure 2d). In the presence of tectorigenin and UDP‐Glu, Bc7OUGT exhibited a K m of 4.13 ± 0.82 μm and a V max of 1040 ± 72 nmol/min/μg protein. The purified protein also catalysed a reverse reaction that removed the sugar moieties from tectoridin (Figure 2d); in the presence of tectoridin and UDP, Bc7OUGT exhibited a K m of 2.95 ± 0.96 μm and a V max of 1042 ± 329 nmol/min/μg. We then tested whether variations in reaction conditions, such as pH or temperature, would affect the balance of this bidirectional reaction. The maximum catalytic activities in both directions occurred at pH 6.5 and 42 °C (Figure 2e). The equilibrium constant K eq = 0.93 (pH 6.5, 42 °C) indicates that glycosylation is a reversible reaction, with the formation and removal of glycosides occurring simultaneously.
We next explored the subcellular localization of Bc7OUGT in planta. Sequence prediction using TargetP (https://services.healthtech.dtu.dk/services/TargetP‐2.0) suggested that Bc7OUGT might be localized to the chloroplast. We fused Bc7OUGT to the N terminus of GFP and transiently expressed it in Arabidopsis mesophyll protoplasts. The eight N‐terminal amino acids (MGGCFSKK) of tomato (Solanum lycopersicum) LeCPK1 were fused to the N terminus of mCherry to generate a membrane localization marker (Rutschmann et al., 2002; Vermeer et al., 2004). Bc7OUGT appeared to completely colocalize with the marker (Figure 2f), indicating that it localizes exclusively to the plasma membrane. We therefore reasoned that Bc7OUGT might glycosylate not only tectorigenin but also other substrates. To explore the catalytic promiscuity of Bc7OUGT, we used a library containing flavones and isoflavones for substrate screening. Bc7OUGT glycosylated genistein, quercetin, kaempferol, naringenin, chrysin, luteolin, apigenin, phloretin, and isoliquiritigenin, producing multiple glycosylated products, indicating that Bc7OUGT is highly promiscuous towards flavonoid and isoflavone substrates (Figure 2g). We also tested its activity with other small molecules, such as gallic acid and 4‐O‐methyl gallic acid, but no glycosylated products were detected.
The structural basis for the glycosylation mechanism of Bc7OUGT
To explore the catalytic mechanisms of Bc7OUGT, we determined its crystal structures in complex with UDP‐Glu or with UDP and tectorigenin in space‐group P 2 21 21 at 2.12 Å and 2.50 Å resolution respectively. We used the Alphafold2 server to generate the initial model for structural determination using the molecular replacement method. One Bc7OUGT molecule is present in the asymmetric unit. Data collection and refinement statistics are presented in Table S1. The N‐terminal domain (NTD; residues 1–233) and the C‐terminal domain (CTD; residues 253–467) of Bc7OUGT both include a core region of parallel β strands surrounded by α helices (β/α/β) (Figure 3a). The NTD of Bc7OUGT comprises a typical Rossmann fold, with a central seven‐stranded twisted parallel β‐sheet surrounded by eight helices. The CTD comprises a twisted β‐sheet, with six strands flanked by eight helices. The C‐terminal helix containing residues 451–467 crosses from the CTD and completes the NTD fold by forming an α‐helix. The NTD and CTD are connected by a flexible loop containing residues 234–252. These two structural domains are tightly bound together, with a cleft in the middle providing space for the sugar donor and acceptor to bind. According to the DALI server, the structure of Bc7OUGT is highly similar to the structures of UGT74AC2 (PDB: 7BV3), UGT71G1 (PDB: 2ACV), UGT85H2 (PDB: 2PQ6), and UGT78G1 (PDB:3HBJ), with root mean square deviations (RMSDs) ranging from 0.97 to 2.04 Å, but Bc7OUGT shares only 28.1%–37.4% sequence identity with these proteins (Figure S2, Table S2). Among these, UGT74AC2 and UGT78G1 can use both flavonoids and isoflavonoids as substrates, resulting in 3‐O‐ or 7‐O‐glycosides, but only UGT78G1 is reported to catalyse reversible reactions of isoflavonoids (Li et al., 2021; Modolo et al., 2009b).
Figure 3.
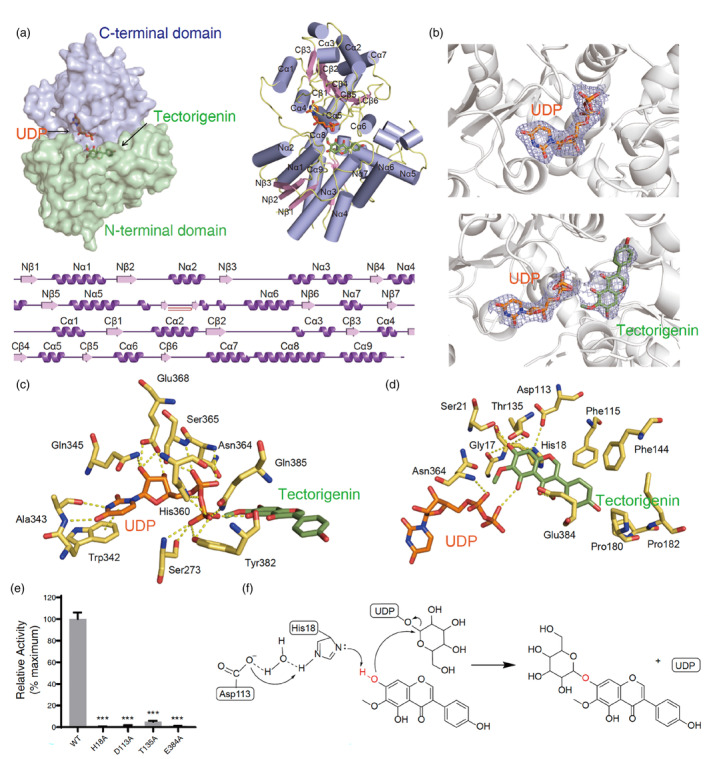
Structural basis for the glycosylation mechanism of Bc7OUGT. (a) Surface (left), cartoon (right), and secondary structure (bottom) representation of the structure of Bc7OUGT. The N‐terminal domain (NTD) and C‐terminal domain (CTD) are shown in green and purple respectively. The α helices are shown in dark purple and the β strands are shown in pink. (b) The electron density of Bc7OUGT with UDP (upper) or with UDP and tectorigenin (lower). The 2F o –F c map is contoured at 1σ. The structure of Bc7OUGT is represented as cartoon in grey colour, UDP is shown as a coloured stick diagram, and atoms are coloured by element (carbon: orange; nitrogen: dark blue; oxygen: red; phosphorus: dark orange), tectorigenin is also shown as a coloured stick diagram, and atoms are coloured by element (carbon: green; oxygen: red). (c) Interactions between UDP and the residues of Bc7OUGT. Hydrogen bonds are represented by yellow dashed lines. Atoms are shown in stick models and are coloured by element. (d) Interactions between tectorigenin and Bc7OUGT residues. Hydrogen bonds are represented by yellow dashed lines. (e) Relative glycosylation activities of wild‐type Bc7OUGT and its mutants. Significance was tested by a two‐tailed unpaired t‐test method (N = 3, error bars, mean ± SD) with asterisks indicating P‐value (***P < 0.001). (f) Proposed glycosylation mechanism of Bc7OUGT.
In the complex structures of Bc7OUGT with UDP‐Glu or with UDP and tectorigenin, we observed the electron densities for UDP or UDP and tectorigenin (Figure 3b). UDP interacts with Bc7OUGT mainly through the conserved residues of the PSPG motif (Trp342‐Gln385), which is involved in the recognition and binding of the sugar donor (Figure 3c). The uracil ring of the UDP molecule forms parallel π‐stacking interactions with the indole ring of Trp342 and forms hydrogen bonds with Ala343. The ribose ring of the UDP molecule forms hydrogen bonds with Gln345 and Glu368. The α‐phosphate group interacts with Asn364 and Ser365. The β‐phosphate forms hydrogen bonds with Ser273, His360, Tyr382, and Gln385. All the above‐mentioned residues are conserved except Tyr382, which was Phe in the other UGTs examined. Tyr382 contains a hydrophilic hydroxyl group that forms hydrogen bonds with the β‐phosphate group of UDP, resulting in a stronger interaction than would occur with Phe.
Several hydrogen bonds also exist between Bc7OUGT and the sugar acceptor tectorigenin. Glu384 forms a hydrogen bond with the 4‐carbonyl group of the tectorigenin, while the hydrogen group of Gly17, His18, Ser21, and Thr135 forms hydrogen bonds with both the 7‐hydroxyl groups (Figure 3d). These interactions promote the correct orientation of tectorigenin in the binding pocket and multiple hydrogen bonds for the 7‐hydroxyl groups enhance the specificity of substrate recognition. Consistently, our mutagenesis experiments showed that His18Ala, Asp113Ala, Thr135Ala, and Glu384Ala substitutions led to a nearly complete loss of glycosylation activity (Figure 3e), indicating that these residues play important roles in the glycosylation process.
The SN2‐like displacement mechanism, which is found in a variety of plant UGTs, involves a catalytic triad formed by the highly conserved Asp‐His‐sugar acceptor (Wang, 2009). Based on the structure of the Bc7OUGT and tectorigenin complex, the His18 forms hydrogen bonds with 7‐hydroxyl group and Asp113 (Figure 3d). In our mutagenesis experiments, the His18Ala and Asp113Ala substitution completely abolished the enzyme activity of Bc7OUGT, suggesting that His18 and Asp113 indeed play a key role in 7‐hydroxyl glycosylation (Figure 3e). We therefore propose that, while Asp113 helps balance the charge from His18, and His18 residue detaches a proton from the 7‐hydroxyl group of the tectorigenin substrate, facilitating nucleophiles to attack the anomeric carbon of UDP‐Glu specifically at the 7‐hydroxyl group, resulting in the formation of the 7‐O‐glycoside product, tectoridin (Figure 3f).
Substrate recognition of Bc7OUGT
The conserved residues of Bc7OUGT were analysed by the Consurf Web server (Ashkenazy et al., 2016). The C‐terminal domain, which comprised the sugar donor‐binding pocket showed a very conserved pattern, while the N‐terminal domain, especially the residues around the substrate‐binding pocket exhibited a novel and unique substrate recognition pattern for Bc7OUGT (Figure 4a). By a B factor analysis of the Bc7OUGT structure, we found His44‐Gly48, Pro72‐His83, and Ser305‐Arg316, three loop regions around the substrate‐binding pocket that are highly flexible (Figure 4b). The flexibility of these three loops could function as ‘lids’ of the substrate pocket and participate in substrate recognition. The electrostatic potential surface map shows that the substrate‐binding pocket of Bc7OUGT is quite small and hydrophobic. Compared to that of UGT74AN2 complexed with UDP and digitoxigenin (Huang et al., 2022), the substrate‐binding pocket of Bc7OUGT is in a narrow channel shape, with the entrance diameter only 6.8 Å, much smaller than that in UGT74AN2 (15.8 Å) (Figure 4c).
Figure 4.
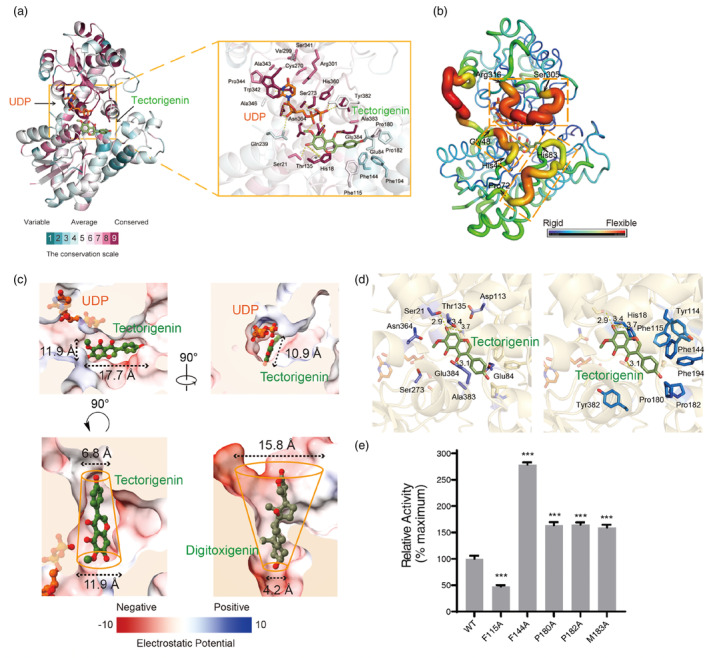
Catalyse mechanism of the substrate recognition of Bc7OUGT. (a) The three‐dimensional structure diagram and the substrate‐binding pocket diagram of conserved sequence analysis of Bc7OUGT. (b) The three‐dimensional structure diagram of B factor analysis of Bc7OUGT. Three loop regions of high B factor are marked by yellow dashed boxes. (c) Upper panel—the electrostatic potential surface map of the substrate‐binding pocket of Bc7OUGT. Lower panel, the comparison of the substrate‐binding pockets between Bc7OUGT and UGT74AN2 (PDB: 7W1B). The top and bottom diameters are labelled. (d) The acceptor‐binding pocket of Bc7OUGT. Aromatic or hydrophobic residues are represented by blue sticks, other residues are represented by purple sticks. (e) Relative glycosylation activities of wild‐type Bc7OUGT and its mutants. Significance was tested by a two‐tailed unpaired t‐test method (N = 3, error bars, mean ± SD) with asterisks indicating P‐value (***P < 0.001).
We also found the pocket is mainly formed by hydrophobic and negatively charged residues, and the hydrophobic residues are mainly composed of aromatic rings of sidechains, such as Phe115, Phe144, Pro180, Pro182, and Met183 (Figure 4d), leading to a more rigid of pocket entrances. To confirm their roles in the catalytic process, we substituted each of the above‐mentioned residues with Ala. Four of the five substitutions—Phe144Ala, Pro180Ala, Pro182Ala, and Met183Ala—increased the glycosylation activity of Bc7OUGT (Figure 4e). We propose that substituting these residues with Ala, which has a smaller sidechain, makes the substrate‐binding pocket more spacious, promoting its flexibility and possibly increasing the speeds of entrance and departure of the substrate or its glycosides. Therefore, the residual composition together with the narrow channel shape lead to the specificity of substrate recognition.
Mechanism and application of deglycosylation activity of Bc7OUGT
Bc7OUGT is one of the few plant UGTs that has been reported to catalyse a reversible glycosylation reaction. However, very limited information on the deglycosylation mechanism is available. It has been suggested that glucosidases require two catalytic carboxylates, Glu191 and Glu406, to facilitate the hydrolysis of glycosidic bonds (Modolo et al., 2009a). Moreover, a mutagenesis study on the Medicago glycosyltransferase UGT78G1 confirmed that Glu192 and Asp376, two acidic residues that are close to the sugar moiety of the glucoside, play key roles in its deglycosylation activity (Modolo et al., 2009b). In this regard, we screened amino acids around the substrate‐binding pocket of Bc7OUGT and found two acidic residues Asp113 and Glu384 (Figure 5a). Consistently, mutating either of these residues to Ala in Bc7OUGT led to reduced deglycosylation activity (Figure 5b). Consistent with its glycosylation activity, the mutation of His18Ala completely abolished the deglycosylation activities, suggesting that the His residue acts as a general base in the reactions of both directions. Moreover, four of the five substitutions of the hydrophobic residues in the substrate‐binding pocket (Phe144Ala, Pro180Ala, Pro182Ala, and Met183Ala) exhibit equal or increased deglycosylation activities, implying that the possible expansion of the binding pocket would equivalently improve its bidirectional activities. Further structural and biochemical studies are needed to understand the detailed mechanism for the reverse reaction.
Figure 5.
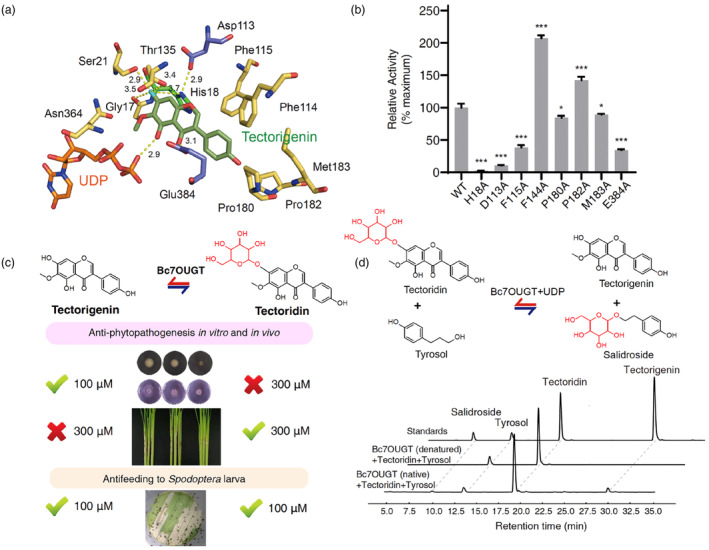
Mechanism and application of deglycosylation activity of Bc7OUGT. (a) Diagram of acceptor‐binding pockets of Bc7OUGT, two acidic residues Glu384 and Asp113 of Bc7OUGT are represented by purple sticks. (b) Relative deglycosylation activities of wild‐type Bc7OUGT and its mutants. Significance was tested by a two‐tailed unpaired t‐test method (N = 3, error bars, mean ± S.D.) with asterisks indicating P‐value (***P < 0.001; *P < 0.05). (c) Summary of the biological activities of tectorigenin and tectoridin against various biotic stresses. ‘√’ indicates inhibitory activity at shown concentration. ‘×’ indicates no or very low inhibitory activity at shown concentration. (d) Bc7OUGT incubated with tectoridin and tyrosol transferred the sugar moiety from tectoridin to tyrosol, producing salidroside in vitro; HPLC analysis of reaction products of tectoridin and tyrosol with Bc7OUGT (denatured as negative control).
Based on our activity tests with tectorigenin and tectoridin, we found they show diverse defensive effects against various biotic stresses (Figure 5c). As a central coordinator, the use of reversible UGTs could be a highly cost‐effective solution to the problem of stress in plants. Besides, these proteins could be also employed to transfer sugars from glycoside scaffolds to other aglycons, expanding the glycosylation diversity with limited but valuable UDP‐Glu in planta. In a preliminary experiment, when we incubated Bc7OUGT with both tectoridin and tyrosol, the sugar moiety was successfully transferred from tectoridin to tyrosol in vitro, producing salidroside, a compound with potential pharmacological uses (Figure 5d). This finding indicates that UGTs not only play regulatory roles in various stress responses, but they also participate in complex regulatory networks in planta. The identification of reversible UGTs may therefore provide enzymatic tools for producing valuable glycosides in microbial factories using metabolic engineering.
Discussion
Plants produce a wide variety of secondary metabolites to adapt to the changing environment. Previous research has mainly focused on the pharmaceutical performance of these metabolites, leaving their detailed effects in plant defence and their potential roles as pesticides largely overlooked. Here, we showed that two specialized metabolites from B. chinensis, tectorigenin and tectoridin, are involved in chemical defence against various biotic stresses, such as resistance to phytopathogens, antifeeding activity against larval pests. These findings provide a foundation for their comprehensive use as biopesticides. In addition, the isoflavonoid compound tectorigenin degrades in the environment due to heat and oxidation (Niamnuy et al., 2012), meaning it will not build up in natural ecosystems and could be a more environmentally friendly alternative to the chemical pesticides currently on the market.
We confirm the results with several repeats that tectorigenin showed a significant growth inhibition against R. solani at 300 μm in vitro, it failed to inhibit the lesion expanding in vivo, and the phenotype is opposite for tectoridin (Figure 5c). Physically, R. solani is a nectrophic fungal pathogen inoculated to the inner face of the plant leaf sheaths, whereas the chemicals were sprayed directly to the outer face of leaf sheaths. On the one hand, tectorigenin may not be a systemic fungicide which is hard to permeate the leaf sheath to direct inhibit the R. solani hypha growth. The other hand, tectorigenin may not trigger the rice immune response or trigger the responses independent of the resistance to R. solani. Previous studies had revealed that flavonoid regulator genes of AtMYB12 and OsF3H could negatively resist to some pathogens or pest, but positively resist to others (Ding et al., 2021; Wu et al., 2022). Ideally, more pathogen types can be tested here.
Glycosylation significantly affects the bioactivities of natural plant products. We found that tectorigenin and tectoridin, which differ by one glucose decoration at the 7‐hydroxyl group, provide plants with variable resistance against several biotic stresses, especially phytopathogens, in vitro and in vivo. This is an example of a highly efficient adaptation of plants to various biotic stresses via the regulation of just one reversible reaction. The large number of UGT genes in the plant genome provides many possibilities for the decoration of various metabolites, leading to numerous functional diversifications throughout the plant's lifespan. While preparing this article, we learned that two research groups characterized seven UGTs from Iris tectorum (UGT73CD1) and B. chinensis (BcUGT1‐2, BcUGT4‐7), all of which can glycosylate tectorigenin into tectoridin (Huang et al., 2021; Zhang et al., 2021). Sequence alignment showed that our candidate genes are different from these genes and are novel putative UGT genes.
Our solved Bc7OUGT structure and the mutagenesis experiments in this study revealed the molecular basis for the glycosylation mechanism of this enzyme. The Asp113‐His18‐acceptor catalytic triad, which is conserved in most plant UGTs, formed at the 7‐hydroxyl group of tectorigenin. We also found that plentiful aromatic residues form a novel substrate recognition pattern, play important roles in its catalytic activities of both directions (Figure 4). Mutating these residues to Ala might cause the displacement of the flavone‐conjugated plate of tectorigenin, eventually affecting its deprotonation at the His18 residue. These results could provide a basis for enzyme engineering to produce certain types of compounds in the future.
Although many bacterial GT1 enzymes exhibit reversible glycosylation and deglycosylation activities (Zhang et al., 2006), plant UGTs that can remove sugar moieties from glucosides seem to be rare. To the best of our knowledge, only five plant UGTs have been reported to catalyse the reverse reaction from flavonoid or isoflavonoid glycosides, including one PcOGT from Pyrus communis (Gutmann et al., 2014), MiCGT from Mangifera indica (Chen et al., 2015), UGT73AE1 and UGT71E5 from Carthamus tinctorius (Xie et al., 2014, 2017), and UGT78G1 from Medicago truncatula (Modolo et al., 2009b). Analysis of the crystal structure of UGT78G1 identified a Glu192 residue that plays a key role in its reverse reaction (Modolo et al., 2009b). Interestingly, a single Val200Glu mutation in UGT85H2, corresponding to Glu192 of UGT78G1, conferred a gain of deglycosylation function, suggesting that this residue in UGT78G1 might play a key role in the reverse reaction (Modolo et al., 2009a). However, this is the only known example of a reverse reaction in plant UGTs, and this mechanism was not applicable in Bc7OUGT. Given that the reversibility of plant UGTs is important for the dynamic regulation of natural product properties, a better understanding of the structure/activity relationship of plant UGTs will provide insights into the mechanisms involved and enable the rational design of new enzymes.
In this study, we examined the roles of tectorigenin and its glycosides as promising biopesticides. We also functionally characterized Bc7OUGT, which catalyses the reversible glycosylation of tectorigenin to tectoridin. Bc7OUGT might play an important role in regulating plant chemical defences against various biotic stresses. Our structural determination of Bc7OUGT and mutagenesis analysis also provided insights into the mechanism underlying both glycosylation and deglycosylation reactions. The catalytic reversibility was also modelled in an one‐pot aglycon exchange reaction. These findings could facilitate the design of novel reversible UGTs to produce valuable glycosides with health benefits for both plants and humans.
Materials and methods
Plant materials, chemicals, and reagents
Belamcanda chinensis was grown for 1 year under natural day/night conditions in the greenhouse located in Wuhan University, Wuhan, China (30°31′48″ N, 114°21′20″ E). The fresh and healthy root, stem, leaf, rhizome, flower, and fruit tissues were collected in September 2021. Upon harvest, plant materials were washed clean and quickly frozen in liquid nitrogen and stored at −80 °C until extraction of chemical constituents and RNA. Tectorigenin and tectoridin standards were purchased from Chengdu Herb Substance Biotechnology Co, Ltd. (Chengdu, China). Crystallization screening kits were purchased from Hampton Research (Laguna Niguel, CA). All solvents in this study were in analytical reagent grade and purchased from Sangon Biotech Co., Ltd. (Shanghai, China).
Plant isoflavonoids extraction and quantitative analysis
B. chinensis root, stem, leaf, rhizome, flower, and fruit samples were ground to a fine powder in liquid nitrogen using a mortar and pestle. To each 100 mg of plant material, we added 1 mL of 75% methanol and sonicated the samples in an ultrasonic bath for 30 min at room temperature. After centrifuging at 14 000 g for 10 min, the supernatant was then filtered using a 0.22‐μm syringe filter (Jinlong Biotech Co., Ltd., Beijing, China). The standard compounds were accurately weighed using an analytical balance, dissolved in 75% methanol, and uniformly formulated to a concentration of 1 mm for quantitative analysis.
LC–MS analysis
The samples (10 μL) were fractionated using an Acclaim C18 column (4.6 × 250 mm, 5 μm; Thermo Scientific, MA, USA) in a Dionex UltiMate 3000 HPLC system (Thermo Scientific) with a flow rate of 0.5 mL/min. Solvent A was Milli‐Q water containing 1‰ (v/v) formic acid, while solvent B was acetonitrile containing 1‰ (v/v) formic acid. The elution programme commenced at 90% (v/v) solvent A and was ramped from 10% to 20% (v/v) solvent B over 10 min, from 20% to 50% (v/v) solvent B over 14 min, and from 50% to 100% (v/v) solvent B over 6 min, then remained isocratic at 100% (v/v) solvent B for 4 min. Thereafter, it was ramped from 100% to 5% (v/v) solvent B over 0.1 min, and then remained isocratic at 95% (v/v) solvent A for 3.9 min. The total run time was 38 min. Chromatograms were acquired at 254 nm, while photodiode array spectra were recorded from 180 to 400 nm. The HPLC and LC–MS chromatograms followed consistent methods. The mass spectrometry condition and software are consistent with that we used previously (Cheng et al., 2022). Flavonoids identification was achieved using exact masses, retention times, and CID spectra of available authentic standards or previously published data. The contents of different organs were determined by the peak area from liquid chromatography data and the calibration curve was drawn using the standard substance.
The antibacterial and antifungal effects of the compounds in vitro and in vivo
Xanthomonas oryzae pv. oryzicola (Xoc) strain HGA4 and Xanthomonas oryzae pv. oryzae (Xoo) strain PXO99 were cultured at 28 °C on potato sucrose agar (PSA; 0.7% infusion from potatoes, 2% sucrose, and 2% agar, w/v) medium. The R. solani AG1‐IA strain YWK196 was cultured on potato dextrose agar (PDA; 20% potato, 2% dextrose, and 2% agar, w/v) at 28 °C.
The in vitro antibacterial effects of tectorigenin and tectoridin against the Xoc, Xoo, and YWK19 were determined by agar microdilution as previously described (Baker et al., 1991). The Xoc and Xoo was inoculated in 5 mL of potato sucrose broth medium (0.7% infusion from potatoes and 2% sucrose, w/v) for 17 h and then adjusted to OD620 = 0.05. Tectorigenin and tectoridin were added to the medium at the desired final concentration (100 μm, 200 μm, or 400 μm) using a 200 mm stock solution in DMSO. A total quantity of 2 mL medium with the tested compounds was added to each well of 6‐well plates (CostarTM 3516, Corning Inc., Corning, NY, USA). DMSO was used as a negative control. For each treatment, 2 μL of Xoc and Xoo were added to the centre of 6‐well plates, which were then cultured at 28 °C for 48 h. The colony migration area was then determined. The R. solani AG1‐IA strain YWK196 was activated on potato dextrose agar medium at 28 °C, and a mycelial plug cut from the edge of a 3‐day‐old colony was transferred onto potato dextrose agar modified with the compound at the indicated concentration (100 μm, 200 μm, or 400 μm). After incubated for 48 h at 28 °C, the diameter of the colony was measured. Three replicates were performed for each experiment.
The Oryza sativa cv. Zhonghua 11 was used in the inoculation experiment of rice sheath blight disease. The methods were followed as previously described with modifications (Park et al., 2008). The rice seedlings were grown in the greenhouse at 22 °C and 60% relative humidity with a 16 : 8 h (light: dark) cycle for about 10 weeks (before the heading stage). The R. solani AG1‐IA strain YWK196 was activated on potato dextrose agar medium at 28 °C. Short sterilized strips were placed on potato dextrose broth medium. The activated mycelium was inoculated in a petri dish, cultured at 28 °C for 2–3 days, and short strips with mycelium were selected for inoculation. Eight plants with pretreated compounds in the sheath were inoculated with R. solani strain YWK196, and the lesion length was measured at 5 days post‐inoculation. Independent experiments were repeated twice, and significant analysis was performed by student's t‐test.
Antifeeding bioassays with Spodoptera frugiperda
The leaf disc assay of target compounds (tectorigenin and tectoridin) and negative controls (DMSO) against Spodoptera frugiperda were tested indoors according to the literature (Boiça Júnior et al., 2015). Tested compounds were dissolved in DMSO and diluted to 100 μm with distilled water. Leaves of young maize were cut into 1.4 cm diameter discs, soaked in compounds for 30 s, dried naturally, wrapped in wet absorbent cotton to moisturize the lower edge of the leaves, and placed in petri dishes (2.0 cm in diameter). Treatment with distilled water containing 0.01% (v/v) DMSO was used as the negative control. Fifteen pre‐starved (for approximately 6 h) third‐instar larvae of well‐developed S. frugiperda raised in the laboratory were placed in each petri dish containing pretreated leaf disks. Petri dishes were then incubated at 26 °C, 85% relative humidity, and a photoperiod of 16 : 8 h (light: dark). Consumed leaf areas (%) were determined 48 h after feeding to the larvae. The diet‐incorporation bioassay of tectorigenin and tectoridin followed a method recommended before (Boiça Júnior et al., 2015). Tested compounds were dissolved in DMSO and mixed with feed by hand thoroughly. The final concentration was 0.4 μmol/g and DMSO was used as a negative control. A total quantity of 10 mg of premixed feed was placed in each of the 24‐well plates. Fifteen pre‐weighed and pre‐starved (for approximately 6 h) third‐instar larvae of well‐developed S. frugiperda raised in the laboratory were placed in each well. Feeding was refilled every 24 h for a total of two times. Weight differences were measured 24 h after the last feeding.
RNA extraction, real‐time PCR, RNA sequencing, and bioinformatics analysis
Total RNA was extracted from plant materials using the FastPure Plant Total RNA Isolation Kit (Vazyme Biotech Co., Ltd., Nanjing, China) according to the manufacturer's instructions. One microgram RNA was reverse‐transcribed into cDNA with SuperScript III (ThermoFisher) followed by quantitative PCR assay with SYBR Green Master Mix using CFX96 Real‐Time System 690 (Bio‐Rad). Primers used in this study are listed in Table S3.
RNAs from different tissues were mixed for library construction and PacBio Iso‐seq were performed by Novogene Biotechnology Co., Ltd (Beijing, China).
The open reading frames (ORFs) were identified using TransDecoder software (version 5.5.0, https://github.com/TransDecoder/TransDecoder) to obtain putative genes, coding sequences (CDS), and protein sequences. For gene annotation, the DIAMOND program (version 2.0.4) (Buchfink et al., 2021) was conducted between the predicted proteins of B. chinensis and Swiss‐Prot protein database (https://www.uniprot.org) with an E‐value threshold of 1e −5.
UDP‐glycosyltransferase genes were identified using the Hidden Markov Model (HMM) (Eddy, 2004) in the PFAM database. Subsequently, multiple sequence alignments of these proteins were performed by the MAFFT tool (version 7.475) (Katoh et al., 2002). Finally, a phylogenetic tree was constructed by the FastTree tool (version 2.1.11) (Price et al., 2009) using the maximum‐likelihood (ML) method. The bootstrap process was replicated 1000 times.
Prokaryotic expression and purification of recombinant protein
The gene of Bc7OUGT with optimized codons was cloned into the pET28a (+) vector with an N‐terminal 8 × His tag, an N‐terminal SUMO tag, and a TEV Protease cleavage site. The resulting recombinant plasmid was transformed into E. coli C43 (DE3) for heterologous expression. A single colony was then grown in 10 mL LB media containing 50 μg/mL kanamycin at 37 °C and 220 rpm overnight. The cultures were then added into 1 L of LB with 50 μg/mL kanamycin and shaken at 37 °C and 220 rpm until OD600 reached 0.8. The cultures were induced with 0.15 mm isopropyl‐β‐D‐1‐thiogalactopyranoside (IPTG) and further grown for 15 h at 20 °C. Subsequently, the cells were harvested by centrifugation at 4000 g and 10 °C for 10 min. Then, the resulting cell pellets were resuspended in 50 mL buffer A (20 mm Tris–HCl, pH 8.0, and 300 mm NaCl), and disrupted using a high‐pressure homogenizer at 800 bar. Cellular debris was removed by centrifugation at 16 000 g and 4 °C for 50 min. The supernatant was loaded onto a Ni‐NTA HisTrap FF column (Cytiva) equilibrated with buffer B (20 mm Tris–HCl, pH 8.0, 300 mm NaCl, and 10 mm imidazole). Then, the column was washed with 20 CV (column volume) of buffer C (20 mm Tris–HCl pH 8.0, 300 mm NaCl, and 30 mm imidazole) and eluted with 2 CV of buffer D (20 mm Tris–HCl pH 8.0, 300 mm NaCl, and 300 mm imidazole). The eluted protein was further purified by passing it through a HiLoad 16/60 SuperdexTM 200 pg column (Cytiva) and exchanged into buffer A. The target protein was pooled and incubated with TEV Protease at 4 °C for 12 h to remove the SUMO tag and the 8 × His tag. The digestion mixture was reloaded onto the Ni‐NTA affinity column, and the column was washed with 3 CV of buffer B. The flow through, containing the tag‐free protein, was collected and further purified using a SuperdexTM 200 Increase 10/300 GL column (Cytiva) and exchanged into buffer A. The peak fractions containing Bc7OUGT were collected and checked by 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE). The protein concentrations were determined using the BCA Protein Assay Kit (Solarbio Science), and the purified protein was stored at −80 °C until use.
Enzymatic assays
The reaction was carried out according to methods reported previously (Peng et al., 2017). In brief, the enzyme assays were performed in a final volume of 100 μL of 150 mm PB (pH 7.5) containing 1.5 mm UDP‐Glu, 40 μg of purified protein, and various concentrations of substrate. The purified recombinant protein was denatured in boiling water for 10 min as a negative control. The reaction was incubated at 42 °C for 4 h. Then, an equal volume of methanol was added to the reaction mixture to quench it. After filtering through a 0.22‐μm syringe filter, assay samples were analysed using the HPLC system. Substrate conversion rates were calculated based on substrate loss. The kinetic parameters were measured at substrate concentrations from 1 to 200 μm and a fixed UDP‐Glu concentration of 1.5 mm. All reactions were performed in triplicate. The products were confirmed by LC–MS. The peak area of the product after each reaction was determined using the standard curve obtained from the standard compound. The K m and V max values were evaluated by fitting the initial velocity‐versus‐substrate concentration to the Michaelis–Menten kinetic equation using GraphPad Prism 8 software.
Optimization of the enzymatic reaction
The reaction mixture was kept at a constant pH of 6.5 at 27, 32, 37, 42, 47, 52, and 57 °C to identify the optimal temperature for the enzymatic reaction. Three buffer systems at a constant temperature of 42 °C were utilized to investigate the influence of pH on enzyme activity: Na2HPO4‐CiH3 buffer (30 mm and pH 2.0–6.0), NaH2PO4‐Na2HPO4 buffer (30 mm, pH 6.5–7.5), and glycine‐NaOH buffer (30 mm, pH 8.0–11.0). All reactions were performed in triplicate. Each experiment began with the addition of enzymes and a 4 h incubation period. The reaction mixture was processed in the same way as the enzymatic tests described above.
Subcellular localization in Arabidopsis protoplast
The lower epidermis of 2‐week‐old leaves from Arabidopsis thaliana ectype Col‐0 was peeled with tape and then soaked in 10 mL enzyme solution (0.4 M mannitol, 20 mm KCl, 20 mm MES, and pH 5.7) containing 0.1 g cellulase R10, 0.02 g R10, 10 mm CaCl2 macerozyme, 0.5 mm b‐mercaptoethanol, and 0.1% bovine serum albumin for 2 h with slow shaking. The solution was then filtered through 75 mm nylon mesh. Protoplasts were collected by 100 g centrifugation for 5 min and then washed twice with W5 buffer (154 mm NaCl, 125 mm CaCl2, 5 mm KCl, 2 mm MES, and pH 5.7). After 40 min incubation on ice in 10 mL W5 buffer, protoplasts were collected and resuspended with 200 mL MMG buffer (0.4 M mannitol, 15 mm MgCl2, 4 mm MES, and pH 5.7). For each transformation, 10 mg plasmid and 220 mL PEG buffer (40% [w/v] PEG4000, 100 mm CaCl2, 200 mm mannitol) were added into 200 mL protoplast and mixed well slowly. After 20 min incubation at room temperature, the reaction was stopped by adding 800 mL W5 buffer. Pellet was collected and resuspended with 2mL W5 buffer and incubated on a 6‐well plate.
Crystallization of Bc7OUGT
The purified Bc7OUGT (17 mg/mL) was incubated with 2 mm UDP‐Glu at 4 °C in advance for 1 h before crystallization screening. Crystal optimization of the initial hits was further performed. Flaky crystals of Bc7OUGT complexed with UDP‐Glu were obtained at 20 °C within 3 days using the sitting‐drop vapour‐diffusion method by mixing 0.4 μL of protein solution with 0.2 μL of the reservoir solution (0.17 M Lithium sulfate monohydrate, 0.085 M Tris hydrochloride pH8.5, and 25.5% w/v PEG 4000). The crystals of Bc7OUGT complexed with UDP and tectorigenin were obtained by soaking the crystals into 100 mm UDP and 100 mm tectorigenin dissolved in the mother liquor for 10 min. All crystals were cryoprotected with 10% (v/v) glycerol with mother liquid and flash‐frozen in liquid nitrogen before data collection.
Data collection and structure determination of Bc7OUGT
The crystallographic data sets were collected on the beamlines 10 U2 and 18 U1 at the Shanghai Synchrotron Radiation Facility, China (SSRF). Collected data sets were processed with XDS and scaled and merged with Aimless of CCP4 suite. The structure of Bc7OUGT in complex with UDP‐Glu was solved by molecular replacement using Phaser (Bunkóczi et al., 2013) from the CCP4 suite with the model predicted by AlphaFold2 as the initial search model. The structure of Bc7OUGT in complex with UDP and tectorigenin was determined by molecular replacement using the structure of Bc7OUGT in complex with UDP‐Glu as the search model. REFMAC 5 and PHENIX were used for refinement. Restriction files of the ligands were generated by AceDrug using SMILES as input. (Long et al., 2017). All the electron density maps shown in the figures were generated using the CCP4 FFT function. PyMol is used to visualize the models and create the figures.
Conflict of interest statement
The authors declare no competing interests.
Author contributions
WC, XF, ZG, YY, ZX, YB, KR, and JL performed the experiments. WC, FC, XC, WM, ZC, ZD, ZZ, and LL analysed the data. ZZ and LL conceived the project, designed the experiments, and prepared the article. All authors read and approved the final article.
Supporting information
Figure S1 Tectorigenin and its glycosides tectoridin are most abundant in the rhizome of B. chinensis.
Figure S2 The superimposition of Bc7OUGT to UGT74AC2, UGT71G1, UGT85H2 and UGT78G1.
Table S1 Summary of data collection and refinement statistics for Bc7OUGT.
Table S2 Four plant glycosyltransferases with the most similar structure to the Bc7OUGT.
Table S3 Primers used in this work.
Acknowledgements
This work was supported by grants from the National Key Research and Development Program (2022YFA0912100), Natural Science Foundation of China (32100413) to L. Lu., and the National Key R&D Program of China (2021YFC2100600) to Z. Zhang. The numerical calculations have been done on the supercomputing system in the Supercomputing Center of Wuhan University. The X‐ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF). We thank all the staff at the SSRF for their assistance with data collection.
Contributor Information
Zhengyu Zhang, Email: zhengyu.zhang@whu.edu.cn.
Li Lu, Email: luliwhu@whu.edu.cn.
Data availability statement
Pacbio HiFi Iso‐seq raw reads have been submitted to NCBI as a BioProject under accession PRJNA930589. Sequence information from this article can be found in the GenBank/National Center for Biotechnology Information databases under the following accession numbers: Bch3/12632.p1 (OR260275), Bch3/13400.p1 (OR260277), and Bch3/13211.p1 (OR260279). The coordinates and structure factors for the structures of Bc7OUGT, in complex with UDP‐Glu or with UDP and tectorigenin, have been deposited in the PDB (www.rcsb.org) with PDB ID 8INP and 8ITA.
References
- Ahn, K.S. , Noh, E.J. , Cha, K.‐H. , Kim, Y.S. , Lim, S.S. , Shin, K.H. and Jung, S.H. (2006) Inhibitory effects of Irigenin from the rhizomes of Belamcanda chinensis on nitric oxide and prostaglandin E2 production in murine macrophage RAW 264.7 cells. Life Sci. 78, 2336–2342. [DOI] [PubMed] [Google Scholar]
- Ashkenazy, H. , Abadi, S. , Martz, E. , Chay, O. , Mayrose, I. , Pupko, T. and Ben‐Tal, N. (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson, N.J. and Urwin, P.E. (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. [DOI] [PubMed] [Google Scholar]
- Baker, C.N. , Stocker, S.A. , Culver, D.H. and Thornsberry, C. (1991) Comparison of the E Test to agar dilution, broth microdilution, and agar diffusion susceptibility testing techniques by using a special challenge set of bacteria. J. Clin. Microbiol. 29, 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr, M. , Neutelings, G. , El Jaziri, M. and Baucher, M. (2020) You want it sweeter: how glycosylation affects plant response to oxidative stress. Front. Plant Sci. 11, 571399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiça Júnior, A.L. , Souza, B.H.S.D. , Costa, E.N. , Ribeiro, Z.A. and Stout, M.J. (2015) Factors influencing expression of antixenosis in soybean to Anticarsia gemmatalis and Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Econ. Entomol. 108, 317–325. [DOI] [PubMed] [Google Scholar]
- Buchfink, B. , Reuter, K. and Drost, H.‐G. (2021) Sensitive protein alignments at tree‐of‐life scale using DIAMOND. Nat. Methods, 18, 366–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkóczi, G. , Echols, N. , McCoy, A.J. , Oeffner, R.D. , Adams, P.D. and Read, R.J. (2013) Phaser. MRage: automated molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 69, 2276–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casida, J.E. and Quistad, G.B. (1995) Pyrethrum flowers: production, chemistry, toxicology, and uses. In International Symposium on Pyrethrum Flowers: Production, Chemistry, Toxicology and Uses, Honolulu, Hawaii (USA), 1992, pp. 345– 350. Oxford University Press. [Google Scholar]
- Chaudhary, S. , Kanwar, R.K. , Sehgal, A. , Cahill, D.M. , Barrow, C.J. , Sehgal, R. and Kanwar, J.R. (2017) Progress on Azadirachta indica based biopesticides in replacing synthetic toxic pesticides. Front. Plant Sci. 8, 610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D. , Chen, R. , Wang, R. , Li, J. , Xie, K. , Bian, C. , Sun, L. et al. (2015) Probing the catalytic promiscuity of a regio‐and stereospecific C‐glycosyltransferase from Mangifera indica . Angewandte Chemie, 127, 12869–12873. [DOI] [PubMed] [Google Scholar]
- Cheng, W. , Yao, Y. , Wang, Q. , Chang, X. , Shi, Z. , Fang, X. , Chen, F. et al. (2022) Characterization of benzylisoquinoline alkaloid methyltransferases in Liriodendron Chinese provides insights into the phylogenic basis of angiosperm alkaloid diversity. Plant J. 112, 535–548. [DOI] [PubMed] [Google Scholar]
- Chu, Q.‐R. , He, Y.‐H. , Tang, C. , Zhang, Z.‐J. , Luo, X.‐F. , Zhang, B.‐Q. , Zhou, Y. et al. (2022) Design, synthesis, and antimicrobial activity of quindoline derivatives inspired by the cryptolepine alkaloid. J. Agric. Food Chem. 70, 2851–2863. [DOI] [PubMed] [Google Scholar]
- Ciesielski, P. and Metz, P. (2020) Asymmetric one‐pot transformation of isoflavones to pterocarpans and its application in phytoalexin synthesis. Nat. Commun. 11, 3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyn, F. , Van Brempt, M. , Maertens, J. , Van Bellegem, W. , Duchi, D. and De Mey, M. (2015) Metabolic engineering of Escherichia coli into a versatile glycosylation platform: production of bio‐active quercetin glycosides. Microb. Cell Fact. 14, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, X. , Zhang, H. , Li, M. , Yin, Z. , Chu, Z. , Zhao, X. , Li, Y. et al. (2021) AtMYB12‐expressing transgenic tobacco increases resistance to several phytopathogens and aphids. Front. Agron. 3, 694333. [Google Scholar]
- Eckardt, N.A. (2006) The role of flavonoids in root nodule development and auxin transport in Medicago truncatula . Plant Cell, 18, 1539–1540. [Google Scholar]
- Eddy, S.R. (2004) What is a hidden Markov model? Nat. Biotechnol. 22, 1315–1316. [DOI] [PubMed] [Google Scholar]
- Fang, W. , Peng, Y. , Muir, D. , Lin, J. and Zhang, X. (2019) A critical review of synthetic chemicals in surface waters of the US, the EU and China. Environ. Int. 131, 104994. [DOI] [PubMed] [Google Scholar]
- Ford, K.A. , Casida, J.E. , Chandran, D. , Gulevich, A.G. , Okrent, R.A. , Durkin, K.A. , Sarpong, R. et al. (2010) Neonicotinoid insecticides induce salicylate‐associated plant defense responses. Proc. Natl. Acad. Sci. 107, 17527–17532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann, A. , Krump, C. , Bungaruang, L. and Nidetzky, B. (2014) A two‐step O‐to C‐glycosidic bond rearrangement using complementary glycosyltransferase activities. Chem. Commun. 50, 5465–5468. [DOI] [PubMed] [Google Scholar]
- He, X.‐Z. and Dixon, R.A. (2000) Genetic manipulation of isoflavone 7‐O‐methyltransferase enhances biosynthesis of 4′‐O‐methylated isoflavonoid phytoalexins and disease resistance in alfalfa. Plant Cell, 12, 1689–1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiling, S. , Llorca, L.C. , Li, J. , Gase, K. , Schmidt, A. , Schäfer, M. , Schneider, B. et al. (2021) Specific decorations of 17‐hydroxygeranyllinalool diterpene glycosides solve the autotoxicity problem of chemical defense in Nicotiana attenuata. Plant Cell, 33, 1748–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J. , Li, J. , Yue, J. , Huang, Z. , Zhang, L. , Yao, W. , Guan, R. et al. (2021) Functional characterization of a novel glycosyltransferase (UGT73CD1) from iris tectorum maxim. for the substrate promiscuity. Mol. Biotechnol. 63, 1030–1039. [DOI] [PubMed] [Google Scholar]
- Huang, W. , He, Y. , Jiang, R. , Deng, Z. and Long, F. (2022) Functional and structural dissection of a plant steroid 3‐O‐glycosyltransferase facilitated the engineering enhancement of sugar donor promiscuity. ACS Catalysis, 12, 2927–2937. [Google Scholar]
- Huccetogullari, D. , Luo, Z.W. and Lee, S.Y. (2019) Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell Fact. 18, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J. and Hughes, M.A. (1994) Multiple secondary plant product UDP‐glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons. DNA Seq. 5, 41–49. [DOI] [PubMed] [Google Scholar]
- Itkin, M. , Rogachev, I. , Alkan, N. , Rosenberg, T. , Malitsky, S. , Masini, L. , Meir, S. et al. (2011) GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell, 23, 4507–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K. , Misawa, K. , Kuma, K.i. and Miyata, T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, I.‐S. (2022) Current perspectives on the beneficial effects of soybean isoflavones and their metabolites on plants. Food Sci. Biotechnol. 31, 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairson, L. , Henrissat, B. , Davies, G. and Withers, S. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555. [DOI] [PubMed] [Google Scholar]
- Lemus de la Cruz, A.S. , Barrera‐Cortés, J. , Lina‐García, L.P. , Ramos‐Valdivia, A.C. and Santillán, R. (2022) Nanoemulsified formulation of Cedrela odorata essential oil and its larvicidal effect against Spodoptera frugiperda (J.E. Smith). Molecules, 27, 2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. , Modolo, L.V. , Escamilla‐Trevino, L.L. , Achnine, L. , Dixon, R.A. and Wang, X. (2007) Crystal structure of Medicago truncatula UGT85H2–insights into the structural basis of a multifunctional (iso) flavonoid glycosyltransferase. J. Mol. Biol. 370, 951–963. [DOI] [PubMed] [Google Scholar]
- Li, J. , Qu, G. , Shang, N. , Chen, P. , Men, Y. , Liu, W. , Mei, Z. et al. (2021) Near‐perfect control of the regioselective glucosylation enabled by rational design of glycosyltransferases. Green Synth. Catal. 2, 45–53. [Google Scholar]
- Long, F. , Nicholls, R.A. , Emsley, P. , Gražulis, S. , Merkys, A. , Vaitkus, A. and Murshudov, G.N. (2017) AceDRG: a stereochemical description generator for ligands. Acta Crystallogr. Section D: Struct. Biol. 73, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louveau, T. and Osbourn, A. (2019) The sweet side of plant‐specialized metabolism. Cold Spring Harb. Perspect. Biol. 11, a034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez‐Chávez, M. , Ledesma‐Escobar, C.A. , Hidalgo‐Morales, M. , Rodríguez‐Jimenes, G.D.C. and Robles‐Olvera, V.J. (2022) Antifungal activity screening of fractions from Annona cherimola Mill. leaf extract against Fusarium oxysporum . Arch. Microbiol. 204, 330. [DOI] [PubMed] [Google Scholar]
- Modolo, L.V. , Escamilla‐Treviño, L.L. , Dixon, R.A. and Wang, X. (2009a) Single amino acid mutations of Medicago glycosyltransferase UGT85H2 enhance activity and impart reversibility. FEBS Lett. 583, 2131–2135. [DOI] [PubMed] [Google Scholar]
- Modolo, L.V. , Li, L. , Pan, H. , Blount, J.W. , Dixon, R.A. and Wang, X. (2009b) Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso) flavonoids. J. Mol. Biol. 392, 1292–1302. [DOI] [PubMed] [Google Scholar]
- Niamnuy, C. , Nachaisin, M. , Poomsa‐ad, N. and Devahastin, S. (2012) Kinetic modelling of drying and conversion/degradation of isoflavones during infrared drying of soybean. Food Chem. 133, 946–952. [Google Scholar]
- Park, D.‐S. , Sayler, R.J. , Hong, Y.‐G. , Nam, M.‐H. and Yang, Y. (2008) A method for inoculation and evaluation of rice sheath blight disease. Plant Dis. 92, 25–29. [DOI] [PubMed] [Google Scholar]
- Peng, C. , Liang, Y. , Wang, X. , Xie, H. , Zhang, T. and Ito, Y. (2009) Preparative isolation and purification of flavonoids from the Chinese medicinal herb Belamcanda by high‐speed countercurrent chromatography. J. Liq. Chromatogr. Relat. Technol. 32, 2451–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, M. , Shahzad, R. , Gul, A. , Subthain, H. , Shen, S. , Lei, L. , Zheng, Z. et al. (2017) Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV‐tolerance. Nat. Commun. 8, 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza, M. , Pozzo, T. , Liu, J. , Gulshan Ara, K.Z. , Turner, C. and Nordberg Karlsson, E. (2014) Substituent effects on in vitro antioxidizing properties, stability, and solubility in flavonoids. J. Agric. Food Chem. 62, 3321–3333. [DOI] [PubMed] [Google Scholar]
- Pretali, L. , Bernardo, L. , Butterfield, T.S. , Trevisan, M. and Lucini, L. (2016) Botanical and biological pesticides elicit a similar Induced Systemic Response in tomato (Solanum lycopersicum) secondary metabolism. Phytochemistry, 130, 56–63. [DOI] [PubMed] [Google Scholar]
- Price, M.N. , Dehal, P.S. and Arkin, A.P. (2009) FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani, L. , Thapa, K. , Kanojia, N. , Sharma, N. , Singh, S. , Grewal, A.S. , Srivastav, A.L. et al. (2021) An extensive review on the consequences of chemical pesticides on human health and environment. J. Clean. Prod. 283, 124657. [Google Scholar]
- Ross, J. , Li, Y. , Lim, E.‐K. and Bowles, D.J. (2001) Higher plant glycosyltransferases. Genome Biol. 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutschmann, F. , Stalder, U. , Piotrowski, M. , Oecking, C. and Schaller, A. (2002) LeCPK1, a calcium‐dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 129, 156–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, P.N. , Hansen, A.H. , Hansen, H.G. , Arnsdorf, J. , Kildegaard, H.F. and Lewis, N.E. (2016) A Markov chain model for N‐linked protein glycosylation–towards a low‐parameter tool for model‐driven glycoengineering. Metab. Eng. 33, 52–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, M. , Zhang, X. , Zhu, Y. , Xie, G. and Qin, M. (2018) Global transcriptome analyses reveal differentially expressed genes of six organs and putative genes involved in (Iso) flavonoid biosynthesis in Belamcanda chinensis . Front. Plant Sci. 9, 1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer, J.E. , Van Munster, E.B. , Vischer, N.O. and Gadella, T.W., Jr. (2004) Probing plasma membrane microdomains in cowpea protoplasts using lipidated GFP‐fusion proteins and multimode FRET microscopy. J. Microsc. 214, 190–200. [DOI] [PubMed] [Google Scholar]
- Wang, X. (2009) Structure, mechanism and engineering of plant natural product glycosyltransferases. FEBS Lett. 583, 3303–3309. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Liang, Y. , Peng, C. , Xie, H. , Pan, M. , Zhang, T. and Ito, Y. (2011) Preparative isolation and purification of chemical constituents of belamcanda by MPLC, HSCCC, and prep‐HPLC. J. Liq. Chromatogr. Relat. Technol. 34, 241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Liu, H. , Yin, Z. , Li, Y. , Lu, C. , Wang, Q. and Ding, X. (2022) A novel guanine elicitor stimulates immunity in arabidopsis and rice by ethylene and jasmonic acid signaling pathways. Front. Plant Sci. 13, 841228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Z. , Zhang, Z.‐M. , Zhong, L. , Fan, J. , Li, M. , Ma, Y. , Zhou, Y. et al. (2021) Directed evolution of a plant glycosyltransferase for chemo‐ and regioselective glycosylation of pharmaceutically significant flavonoids. ACS Catalysis, 11, 14781–14790. [Google Scholar]
- Woźniak, D. and Matkowski, A. (2015) Belamcandae chinensis rhizoma–a review of phytochemistry and bioactivity. Fitoterapia, 107, 1–14. [DOI] [PubMed] [Google Scholar]
- Wu, C. , Li, Y. , Chen, Y. , Lao, X. , Sheng, L. , Dai, R. , Meng, W. et al. (2011) Hypoglycemic effect of Belamcanda chinensis leaf extract in normal and STZ‐induced diabetic rats and its potential active faction. Phytomedicine, 18, 292–297. [DOI] [PubMed] [Google Scholar]
- Wu, T. , Zhang, H. , Yuan, B. , Liu, H. , Kong, L. , Chu, Z. and Ding, X. (2022) Tal2b targets and activates the expression of OsF3H03g to hijack OsUGT74H4 and synergistically interfere with rice immunity. New Phytol. 233, 1864–1880. [DOI] [PubMed] [Google Scholar]
- Xie, K. , Chen, R. , Li, J. , Wang, R. , Chen, D. , Dou, X. and Dai, J. (2014) Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius . Org. Lett. 16, 4874–4877. [DOI] [PubMed] [Google Scholar]
- Xie, K. , Chen, R. , Chen, D. , Li, J. , Wang, R. , Yang, L. and Dai, J. (2017) Enzymatic N‐glycosylation of diverse arylamine aglycones by a promiscuous glycosyltransferase from Carthamus tinctorius . Adv. Synth. Catal. 359, 603–608. [Google Scholar]
- Yonekura‐Sakakibara, K. and Hanada, K. (2011) An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 66, 182–193. [DOI] [PubMed] [Google Scholar]
- Zhang, C. , Griffith, B.R. , Fu, Q. , Albermann, C. , Fu, X. , Lee, I.‐K. , Li, L. et al. (2006) Exploiting the reversibility of natural product glycosyltransferase‐catalyzed reactions. Science, 313, 1291–1294. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Zhu, Y. , Ye, J. , Ye, Z. , Zhu, R. , Xie, G. , Zhao, Y. et al. (2021) Iris domestica (iso) flavone 7‐and 3′‐O‐glycosyltransferases can be induced by CuCl2. Front. Plant Sci. 12, 632557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Tectorigenin and its glycosides tectoridin are most abundant in the rhizome of B. chinensis.
Figure S2 The superimposition of Bc7OUGT to UGT74AC2, UGT71G1, UGT85H2 and UGT78G1.
Table S1 Summary of data collection and refinement statistics for Bc7OUGT.
Table S2 Four plant glycosyltransferases with the most similar structure to the Bc7OUGT.
Table S3 Primers used in this work.
Data Availability Statement
Pacbio HiFi Iso‐seq raw reads have been submitted to NCBI as a BioProject under accession PRJNA930589. Sequence information from this article can be found in the GenBank/National Center for Biotechnology Information databases under the following accession numbers: Bch3/12632.p1 (OR260275), Bch3/13400.p1 (OR260277), and Bch3/13211.p1 (OR260279). The coordinates and structure factors for the structures of Bc7OUGT, in complex with UDP‐Glu or with UDP and tectorigenin, have been deposited in the PDB (www.rcsb.org) with PDB ID 8INP and 8ITA.


