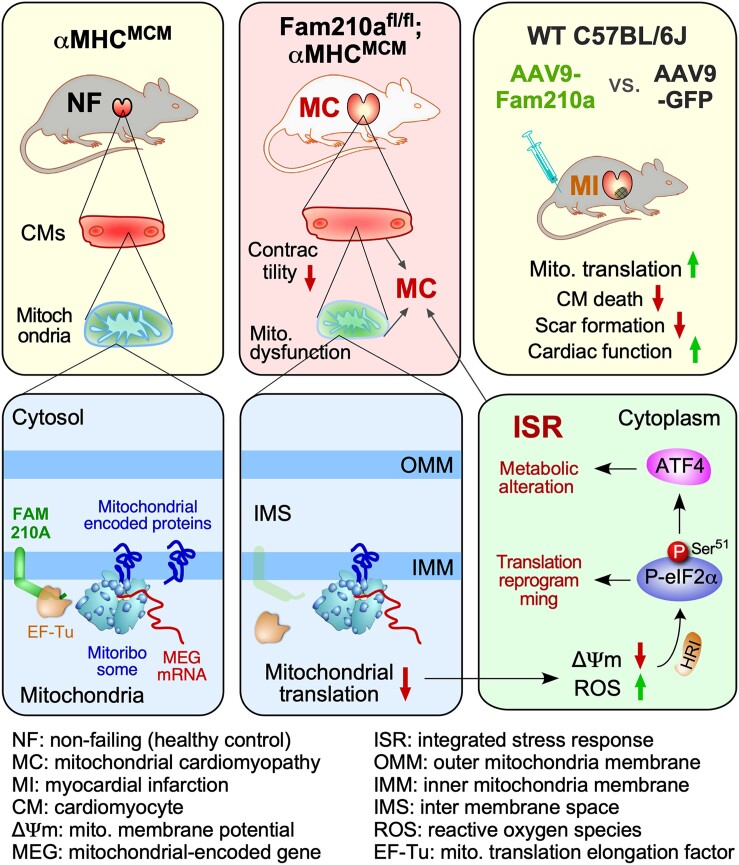
Abstract
Aims
Mitochondria play a vital role in cellular metabolism and energetics and support normal cardiac function. Disrupted mitochondrial function and homeostasis cause a variety of heart diseases. Fam210a (family with sequence similarity 210 member A), a novel mitochondrial gene, is identified as a hub gene in mouse cardiac remodelling by multi-omics studies. Human FAM210A mutations are associated with sarcopenia. However, the physiological role and molecular function of FAM210A remain elusive in the heart. We aim to determine the biological role and molecular mechanism of FAM210A in regulating mitochondrial function and cardiac health in vivo.
Methods and results
Tamoxifen-induced αMHCMCM-driven conditional knockout of Fam210a in the mouse cardiomyocytes induced progressive dilated cardiomyopathy and heart failure, ultimately causing mortality. Fam210a deficient cardiomyocytes exhibit severe mitochondrial morphological disruption and functional decline accompanied by myofilament disarray at the late stage of cardiomyopathy. Furthermore, we observed increased mitochondrial reactive oxygen species production, disturbed mitochondrial membrane potential, and reduced respiratory activity in cardiomyocytes at the early stage before contractile dysfunction and heart failure. Multi-omics analyses indicate that FAM210A deficiency persistently activates integrated stress response, resulting in transcriptomic, translatomic, proteomic, and metabolomic reprogramming, ultimately leading to pathogenic progression of heart failure. Mechanistically, mitochondrial polysome profiling analysis shows that FAM210A loss of function compromises mitochondrial mRNA translation and leads to reduced mitochondrial-encoded proteins, followed by disrupted proteostasis. We observed decreased FAM210A protein expression in human ischaemic heart failure and mouse myocardial infarction tissue samples. To further corroborate FAM210A function in the heart, AAV9-mediated overexpression of FAM210A promotes mitochondrial-encoded protein expression, improves cardiac mitochondrial function, and partially rescues murine hearts from cardiac remodelling and damage in ischaemia-induced heart failure.
Conclusion
These results suggest that FAM210A is a mitochondrial translation regulator to maintain mitochondrial homeostasis and normal cardiomyocyte contractile function. This study also offers a new therapeutic target for treating ischaemic heart disease.
Keywords: Cardiomyopathy, FAM210A, Heart failure, Integrated stress response, Metabolism, Myocardial infarction, Mitochondria, mRNA translation
Graphical Abstract
Graphical Abstract.
Time of primary review: 32 days
1. Introduction
The mitochondrion is an essential cellular organelle to maintain the energy supply for any biochemical reactions in eukaryotic cells.1 The electron transport chain (ETC) complex, located in the mitochondrial inner membrane, generates an electrochemical-proton gradient that drives ATP synthesis. In addition to energetics, mitochondria also play critical roles in calcium signalling, metabolism, and apoptosis. The heart is one of the most mitochondria-rich organs in mammals, as mitochondria account for over 30% of cardiomyocyte (CM) cell mass.2 Mitochondria generate metabolites and ATP via tricarboxylic acid cycle (TCA) and oxidative phosphorylation, fuelling all biological processes, including CM contraction. Loss of function of mitochondrial genes due to genetic mutations can cause mitochondrial cardiomyopathy (MC)3 and other mitochondrial diseases in the skeletal muscle, brain, liver, and kidney, among other organs.4 Moreover, reduced expression of critical mitochondrial proteins and subsequent mitochondrial dysfunction are major drivers of pathogenesis in ischaemic heart disease such as myocardial infarction (MI).5,6 However, the aetiology and effective therapeutic intervention of mitochondria dysfunction related heart disease are still lacking.7,8
Given these crucial roles of mitochondria in CMs, elucidating the molecular functions of critical regulators of mitochondrial homeostasis may provide novel therapeutic targets to combat MC and MI. FAM210A (family with sequence similarity 210 member A) is a mitochondria-localized protein essential for embryonic development and viability.9 Genome-wide association studies have shown multiple FAM210A mutations associated with sarcopenia and osteoporosis.10,11 However, FAM210A is not expressed in the bone but the skeletal muscle and is most highly expressed in the heart.11,12 Skeletal muscle-specific knockout (KO) of Fam210a reduces muscle strength and bone function in vivo,11 and reduced expression of FAM210A impairs myoblast differentiation in vitro.13 One unbiased transcriptome-wide RNA-seq and ribosome profiling analysis in mouse hearts identified FAM210A as a hub gene down-regulated in cardiac pathological hypertrophy and remodelling compared to physiological hypertrophy.14 Recently, our study revealed that the miR-574-FAM210A axis plays a vital role in regulating mitochondrial proteomic homeostasis and cardiac remodelling.12 FAM210A interacts with a critical mitochondrial translation elongation factor EF-Tu in a protein complex involved in regulation of mitochondrial-encoded mRNA translation.12 However, the in vivo physiological function and downstream molecular effector of FAM210A in the heart remain unknown.
Here, we established a novel CM-specific Fam210a conditional KO mouse model to investigate the function of FAM210A in the CM and heart. We observed severe morphological disruption and functional decline in mitochondria, CMs, and the heart at ∼10 weeks post-inducible Fam210a KO. Meanwhile, we only detected reduced mitochondrial membrane potential (Δψm) and increased reactive oxygen species (ROS) accompanied by aberrant mitochondrial function in the absence of any dysfunction in CMs and the heart at the early stage (∼5 weeks post-Fam210a KO). Multi-omics analyses in cKO hearts uncovered chronic activation of the integrated stress response (ISR) as a major component of the gene signature and disease aetiology accompanied by transcriptomic, translatomic, proteomic, and metabolomic reprogramming. Mechanistically, mitochondrial polysome profiling analyses show that FAM210A modulates mitochondrial-encoded mRNA translation and maintains mitochondrial homeostasis. FAM210A deficiency impairs mitochondrial proteomic homeostasis. Overexpression of FAM210A protects the hearts from cardiac injury and pathological remodelling in a mouse MI model. This study demonstrates that FAM210A maintains mitochondrial homeostasis in CMs by regulating the translation of mitochondria-encoded mRNAs and can be targeted for cardiac protection under ischaemic stresses.
2. Methods
A detailed description of materials and methods is available in the supplemental information.
2.1. Mice
All mouse experiments were conducted under protocols approved by the University Committee on Animal Resources (UCAR) of the University of Rochester Medical Center (URMC). All animal procedures conform to the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes or the NIH Guide for the Care and Use of Laboratory Animals. The mice were housed in a 12:12 h light: dark cycle in a temperature-controlled room in the animal housing room of URMC, with free access to water and food. The age and gender were indicated below in each section of experiments using mice. The Fam210a floxed mice (tm1c, Fam210aflox/flox) were generated by the Canadian Mouse Mutant Repository (CMMR, http://www.cmmr.ca/) and purchased through the International Mouse Phenotyping Consortium (https://www.mousephenotype.org/). The tamoxifen-inducible transgenic mouse line αMHCMerCreMer (αMHCMCM/+) was a gift from Dr Eric Small’s lab at URMC. The tamoxifen-induced cardiomyocyte-specific knockout of Fam210a was achieved by crossing Fam210aflox/flox mice with αMHCMCM/+ mice. Age- and gender-matched aMHCMCM/+ mice were used to control Fam210a knockout mice. All the mice are on C57BL/6J background.
Anaesthetic and analgesic agents used in the study are listed below as follows:
| Drug or drug combination | Dose range (mg/kg) and route | Purpose | Frequency | Maximum # doses |
|---|---|---|---|---|
| Buprenorphine SR | 0.5–1 mg/kg, SQ | Relieve pain/stress before MI surgery | 1 time before the operation | 1 |
| Isoflurane | 2%, inhale | Aesthetical treatment | 1 time before the operation | 1 |
| Ketamine/xylazine | 100 mg/kg for ketamine, 10 mg/kg for xylazine, ip | Analgesic treatment | 1 time before harvest | 1 |
After Buprenorphine SR treatment, lab fellows inspected mice once a day for 3 days and registered them in the recording card. Carbon dioxide euthanasia is used for terminating mice. All anaesthetic and analgesic agents used in the study were approved by the UCAR of the URMC.
2.2. Statistical analysis
All quantitative data were presented as mean ± S.e.m. and analysed using Prism 8.3.0 software (GraphPad). Kolmogorov–Smirnov test was used to calculate the normal distribution of the data. For normally distributed data, an unpaired two-tailed Student’s t-test was performed to compare two groups and one-way or two-way ANOVA with Sidak's multiple comparisons test for the comparisons between more than three groups. For data that are not normally distributed, a non-parametric Mann–Whitney test was performed for the comparisons between two groups and a Kruskal–Wallis test with Dunn’s multiple comparisons test for the comparisons between more than three groups. The log-rank test was performed to compare survival rates, and the χ2 test was used to compare cell number counting data. Two-sided P-values of <0.05 were considered to indicate statistical significance. Specific statistical methods were described in the figure legends.
3. Results
3.1. Cardiomyocyte-specific Fam210a knockout causes spontaneous dilated cardiomyopathy
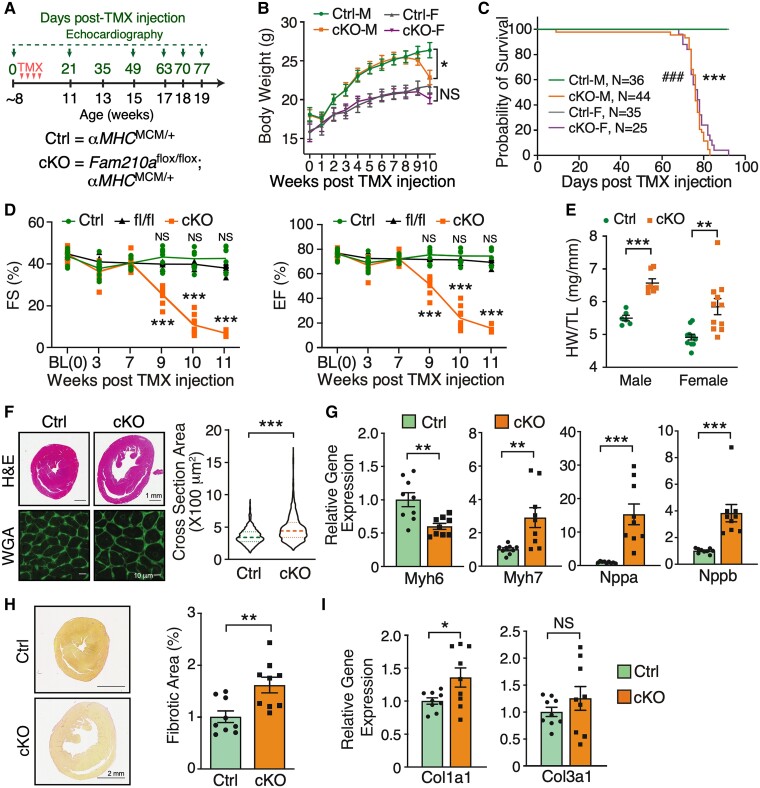
To investigate the role of FAM210A in vivo, we first determined the expression of FAM210A in two major cell types in the heart, CM and cardiac fibroblast (CF). FAM210A is highly expressed in CMs compared with CFs (see Supplementary material online, Figure S1A). Moreover, single-cell RNA-seq data reveal that the highest expression is found in CMs than in any other human cell type (see Supplementary material online, Figure S1B).15 To determine the role of FAM210A in the adult heart, we generated an inducible CM-specific cKO mouse model of Fam210a by cross-breeding Fam210aflox/flox mice with tamoxifen (TMX)-inducible αMHCMCM/+ mice (Figures 1A and S1C and D). After TMX administration, FAM210A mRNA and protein expression was decreased by >90% in the cKO heart (see Supplementary material online, Figure S1E and F). The cKO mice declined in bodyweight 9 weeks post-TMX-induced KO (Figure 1B). Both male and female cKO mice showed complete penetrance of lethality after TMX injection for ∼70–80 days (Figure 1C), with a life span of 77.20 ± 1.07 days in females and 74.89 ± 1.58 days in males. Echocardiographic examinations suggest that Fam210a cKO mice experienced progressive dilated cardiomyopathy with decreased fractional shortening, ejection fraction (Figure 1D), and induced chamber dilation (see Supplementary material online, Figure S1G–J and Supplementary material online, Table S1) compared to αMHCMCM/+ (Ctrl) and Fam210a fl/fl mice. As we did not observe any significant difference in cardiac functions between αMHCMCM/+ and Fam210a fl/fl mice in the echocardiographic outcome within 11 weeks post-TMX injections (see Supplementary material online, Table S1), we used αMHCMCM/+ mice as the control group in the following phenotyping and multi-omics studies. The cardiac function remained normal until 7 weeks post-KO and started to drop severely after ∼9 weeks. Therefore, we defined the early (1–7 weeks) and late stages (8–11 weeks) of this cardiomyopathy disease model using 7 weeks post-KO as the cut-off time point of transition from normal to abnormal cardiac function. We first characterized the disease phenotypes at the late stage at ∼10 weeks post-KO. The increased ratio of heart weight to tibia length (by 19.56% in males, by 18.93% in females) and myocyte cross-section area by wheat germ agglutinin (WGA) staining confirmed spontaneous dilated cardiomyopathy with hypertrophied CMs in cKO mice (Figure 1E and F). Consistent with the marked CM hypertrophy, hypertrophy marker gene expression (Myh7, Nppa, and Nppb) was drastically induced, while Myh6 expression was reduced in cKO hearts compared with control hearts (Figure 1G). Furthermore, a slight increase in cardiac fibrosis was indicated by picrosirius red staining and confirmed by a modest rise in fibrosis marker gene expression (Col1a1 and Col3a1) (Figure 1H and I). Altogether, these results suggest that CM-specific Fam210a KO in adult mice leads to progressive HF with enlarged CMs and dilated left ventricle chamber, and ultimately causes mortality at ∼75 days after Fam210a KO.
Figure 1.
FAM210A deficiency in cardiomyocytes leads to dilated cardiomyopathy and heart failure. (A) Experimental timeline for phenotypic examination of CM-specific Fam210a cKO mice. (B) The body weight was measured weekly post-TMX-induced Fam210a KO in male and female mice. n = 11 for Ctrl and cKO male (M). n = 7 for Ctrl female (F) and n = 10 for cKO female. (C) The survival rate in male and female mice post-KO. ###P < 0.001 for M and ***P < 0.001 for F by log-rank test. (D) Fractional shortening (FS) and ejection fraction (EF) were measured by echocardiography in cKO and control mice. n = 5M + 4F for Ctrl and cKO. n = 5M + 2F for Fam210a fl/fl control mice. NS, not significant for fl/fl vs. Ctrl by two-way ANOVA with Sidak's multiple comparisons. (E) Heart weight/tibia length (HW/TL) ratio in male and female cKO mice. n = 6 and 7 for male Ctrl and cKO. n = 11 for female Ctrl and cKO. (F and H) Wheat germ agglutinin (WGA) staining for the cross-sectional area of CMs (F; n = 3 hearts with >1000 CMs quantified per heart) and picrosirius red staining of collagen deposition (H; n = 5M + 4F for Ctrl and cKO) in the hearts of control and cKO mice at ∼65 days post-KO. (G and I) Cardiac hypertrophy (G) and fibrosis (I) marker gene expression at ∼65 days post-KO (n = 5M + 4F). *P < 0.05; **P < 0.01; ***P < 0.001 by two-way ANOVA with Sidak's multiple comparisons test (B and D), Student’s t-test (E and G–I), and Mann–Whitney test (F and Nppb in G).
3.2. FAM210A deletion leads to myofilament disarray and contractile dysfunction of cardiomyocytes
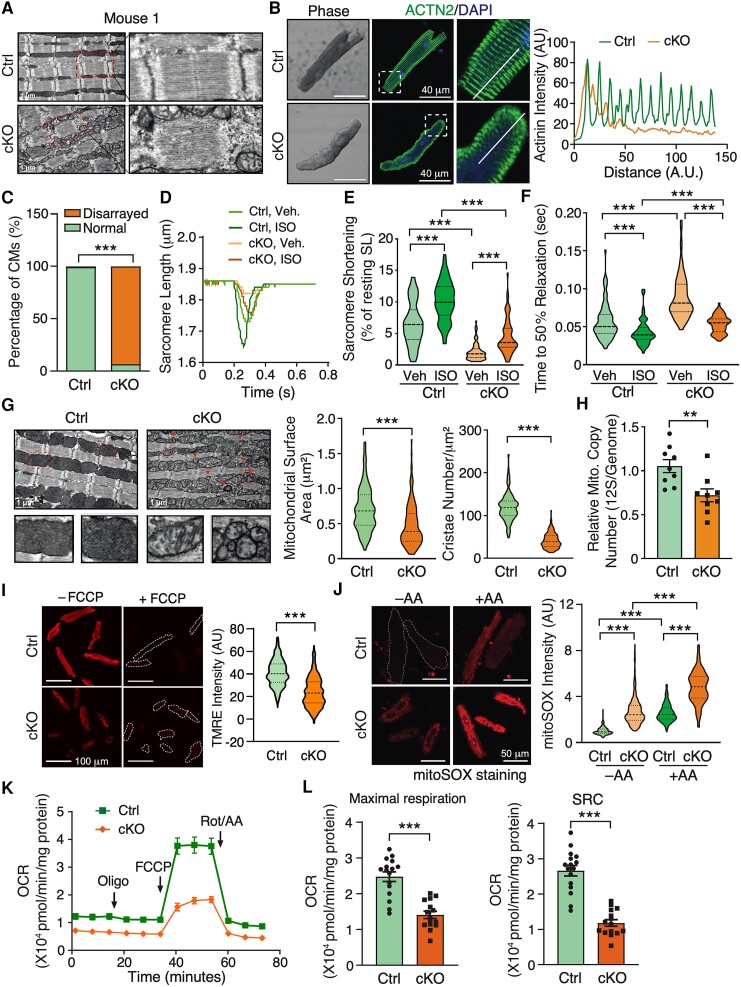
To dissect the pathological effects of FAM210A deletion on CM cells, we first performed transmission electron microscopy to visualize the morphological changes in diseased CMs in the whole heart tissue sections of Fam210a cKO mice at the late stage of cardiomyopathy (∼10 weeks post-KO). The sarcomeric architecture was remarkably disrupted, as indicated by the destruction of the organized Z disk, I band, M line (Figures 2A and S2A), and the loss of intercalated disc integrity between CMs in the cKO hearts (see Supplementary material online, Figure S2B). The observation of myofilament disarray was confirmed in isolated primary CMs from cKO and control mice by immunostaining of Z disk and thin filament marker protein α-actinin 2 and cardiac troponin T, respectively (Figures 2B and C and S2C). In addition, quantitative mass spectrometry analysis was performed to evaluate the proteomic changes in the whole heart tissues of Fam210a cKO and control mice. The sarcomeric protein levels of Troponin C1, Troponin T2, Troponin I3, Myosin heavy chain 6 and 3, Myosin light chain 2, and Tropomyosin 1 (log2FC = −0.66) were decreased while Myosin heavy chain 7, Myosin light chain 4 and 7, α-actinin 1 and 4, and Tropomyosin 2/3/4 (log2FC = 0.95, 0.88, and 0.53) were increased (see Supplementary material online, Figure S2D and Supplementary material online, Table S3), suggesting a proteomic homeostatic imbalance of myofilament proteins in cKO hearts. Immunoblot experiments verified the same trend of changes of α-actinin (increased by 2.66-folds) and cardiac Troponin T (decreased by 40.9%) in the whole heart lysate of cKO hearts (see Supplementary material online, Figure S2E). To examine whether dysregulation of sarcomeric protein expression affects CM contractility, we measured the dynamic change in sarcomere length during the contraction cycle of CMs isolated from control and cKO mice at the late stage using the myocyte contractility recording system. In agreement with dysregulated sarcomeric structure proteins, Fam210a KO CMs exhibited significantly compromised contractile function compared to control CMs in the presence or absence of isoproterenol (ISO) stimulation (Figures 2D–F and S2F–H). The sarcomere shortening (Figure 2D and E) and maximal shortening velocity (see Supplementary material online, Figure S2F) were reduced in KO CMs. As a result, time to 50% sarcomere shortening was increased in KO CMs (see Supplementary material online, Figure S2G). Meanwhile, we noticed that maximal relengthening velocity was reduced (see Supplementary material online, Figure S2H), and time to 50% relaxation was increased accordingly (Figure 2F). Taken together, these results suggest that genetic deletion of FAM210A in CMs leads to dysregulated expression of myofilament proteins and pronounced disruption of the sarcomeric structure, thereby driving contractile dysfunction in CMs.
Figure 2.
Cardiomyocyte-specific deletion of Fam210a causes myofilament disarray and mitochondrial dysfunction in cardiomyocytes. (A) A representative electron microscopy image shows a disturbed sarcomeric structure in CMs from whole heart tissue sections of a late-stage Fam210a cKO heart (∼65 days post-KO). (B) Immunofluorescence staining of ACTN2 indicates myofilament disarray in isolated CMs from cKO hearts at ∼65 days post-KO. Right panels: quantification of fluorescence intensity distribution (AU, arbitrary unit) along the white line in the IF image. (C) Quantification of disarrayed CM percentile from IF staining in isolated CMs from hearts of control or cKO mice. n > 2500 CMs were quantified from four hearts. (D–F) FAM210A deficiency reduces sarcomere length and attenuates the contractility of isolated CMs from cKO hearts at ∼65 days post-KO with or without ISO stimulation. n = 82/79/77/68 CMs were quantified for Ctrl-Veh, Ctrl-ISO, cKO-Veh, and cKO-ISO from four hearts. (G) Electron microscopy shows decreased cristae and mitochondrial size in Fam210a cKO CMs from whole heart tissue sections. Triangles indicate fragmented mitochondria. The mitochondrial surface area and cristae number are quantified in >200 mitochondria from n = 3 hearts (cKO vs. Ctrl). (H) The mtDNA copy number in control and cKO hearts was measured by qPCR using primers targeting the mitochondrial 12S rDNA locus (n = 5M + 4F). qPCR of the nuclear genomic DNA was used as a normalizer. (I) Mitochondrial membrane potential (Δψm) was determined by TMRE staining in isolated CMs from late-stage Fam210a cKO hearts. For the FCCP treatment, 10 μM FCCP was added into the medium for 5 min before imaging. Greater than 600 CMs from four hearts were quantified. The TMRE intensity was normalized by the intensity after FCCP treatment. (J) The ROS level in isolated CMs from late-stage cKO hearts. Greater than 200 CMs from three hearts were quantified. AA, antimycin A (2 mM for 30 min). (K and L) The mitochondrial respiratory activity in isolated CMs from control and cKO hearts at ∼65 days post-KO was measured by the Seahorse assay. n = 16 biological replicates for Ctrl and n = 15 for cKO from CM isolations in three hearts. SRC, spare respiratory capacity (OCRMax − OCRBas). *P < 0.05, **P < 0.01, ***P < 0.001 by χ2 test (C), Kruskal–Wallis test with Dunn’s multiple comparisons test (E, F, and J), Student’s t-test (H and L), and Mann–Whitney test (G and I).
3.3. FAM210A maintains cardiac mitochondrial function in cardiomyocytes
Next, we ask the question of the effect of Fam210a KO on cardiac mitochondrial function. Electron microscopy imaging showed that mitochondrial morphology was dramatically altered in cKO hearts at the late stage of HF. Cristae were greatly reduced and sparsely present in the mitochondria of KO CMs compared to control CMs (Figure 2G, left panel). Also, an increased number of small-sized mitochondria were observed in KO CMs but not in control CMs. Quantification results from electron microscopy images indicated that mitochondrial surface area and cristae number were reduced by 32.6% and 65.6%, respectively (Figure 2G, middle and right panels). In addition, the mitochondrial DNA (mtDNA) copy number was decreased by 31.60% and 36.67% as measured by PCR using specific primers targeting 12S rDNA or Cox1 locus in the mitochondrial genome, respectively, in cKO hearts compared to control hearts at the late stage (Figures 2H and S3A). To determine the cause of reduced mtDNA copy number, we measured the expression of critical genes involved in mitochondrial biogenesis and mitophagy in the cKO hearts. Although Pgc1α and Pgc1β mRNA was decreased by 36.1% and 44.7%, respectively (see Supplementary material online, Figure S3B), PGC1α protein expression was slightly increased, indicating that mitochondrial biogenesis may stay unchanged or modestly enhanced (see Supplementary material online, Figure S3C). As a consequence of decreased mtDNA copy number in KO CMs, the protein expression of the mitochondrial localized transcription factor TFAM was reduced by half (see Supplementary material online, Figure S3C). In other aspects, the expression of autophagy marker protein LC3B II was increased by ∼3–4-folds (see Supplementary material online, Figure S3C) and the autophagic flux was enhanced in the presence of autophasome and lysosome fusion inhibitor, bafilomycin A1 (Baf A1), at the late stage (see Supplementary material online, Figure S3D), suggesting that enhanced autophagic activity may contribute partially to the reduced mtDNA copy number at the late stage through mitochondria degradation. Furthermore, blue native gel assays showed that ETC assembly was compromised to some extent in Fam210a cKO hearts compared to control hearts at the late stage (see Supplementary material online, Figure S3E). ETC assembly defect or dysfunction is often associated with compromised Δψm and increased mitochondrial ROS production.16 In isolated KO CMs, the Δψm was reduced by 37.3% (Figure 2I). ROS production was increased by 1.87 and 2.72-folds in the absence or presence of the Complex III inhibitor Antimycin A, respectively (Figure 2J). More importantly, seahorse measurement showed that the mitochondrial respiratory activity was significantly compromised in FAM210A deficient CMs (Figure 2K), as indicated by a ∼50% decrease in the maximal oxygen consumption rate and spare respiratory capacity (Figure 2L). Consistent with aberrant mitochondrial function, we observed a significantly increased (∼2.5%) CM cell death at the late stage of Fam210a cKO hearts (see Supplementary material online, Figure S3F). These results suggest that FAM210A is required to maintain cardiac mitochondrial homeostasis, and its deficiency causes severe mitochondrial dysfunction and cardiomyopathy.
3.4. Disrupted mitochondrial membrane potential and enhanced ROS production are early events in Fam210a cKO hearts
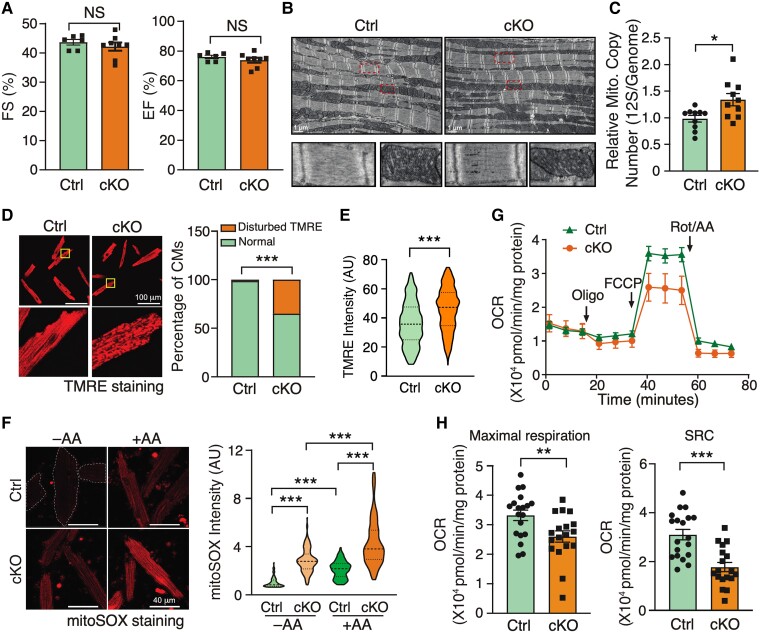
To tease out the disease-causing events from the consequent secondary effects of HF, we examined the functional changes in early-stage heart tissues of Fam210a cKO mice. We did not observe any functional abnormality of hearts and morphological change of CMs with a minimal expressional change of CM hypertrophy marker genes at the early stage (∼5 weeks post-KO) (Figures 3A and S4A–C). Additionally, cardiac fibrosis was absent, and fibrosis marker gene expression was unaltered in cKO hearts (see Supplementary material online, Figure S4D and E). Electron microscopy analysis indicated no significant disruption in the overall morphology of the CM sarcomeric network or mitochondrial structure (Figure 3B). Consistently, we did not identify any remarkable difference in contractile functions (see Supplementary material online, Figure S4F–L) between KO and control CMs in the presence or absence of ISO stimulation. These results reveal no significant cardiac dysfunction in Fam210a cKO hearts at the early stage.
Figure 3.
Disturbed mitochondrial membrane potential and increased ROS production are early events in FAM210A deficient CMs. (A–C) Cardiac function (A, n = 6 for Ctrl and n = 8 for cKO) and mitochondrial morphology (B, n = 3) stay unchanged while mtDNA copy number is slightly increased as quantified by PCR using primers targeting 12S rDNA (C, n = 10 for Ctrl and cKO) at 5 weeks post-KO. (D and E) Mitochondrial membrane potential (Δψm) was determined by TMRE staining in isolated CMs from early-stage Fam210a cKO hearts (D). Right panel: >1500 CMs were quantified from four hearts. For Δψm measurement with the FCCP treatment in (E), 10 μM FCCP was added into the medium for 5 min before imaging. Greater than 280 CMs were quantified from four hearts. The TMRE intensity was normalized by the intensity after FCCP treatment. Representative images are shown in Figure S5H. (F) Increased mitochondrial ROS (>80 CMs were quantified from three hearts) was observed in isolated CMs from early-stage cKO hearts. (G and H) Respiratory activity is attenuated in isolated CMs from early-stage KO hearts. n = 19 biological replicates for Ctrl and n = 18 for cKO in (G). Oligo, oligomycin; FCCP, carbonyl cyanide-p-trifluoromethoxyphenyl-hydrazone; Rot/AA, rotenone/antimycin A. *P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t-test (A, C, and H), χ2 test (D), Mann–Whitney test (E), and Kruskal–Wallis test with Dunn’s multiple comparisons test (F).
Given the localization of FAM210A in mitochondria, we assume that the primary disease-causing events are associated with mitochondria at the molecular level before any noticeable phenotypic changes occur in overall mitochondrial morphology, CMs, and the heart. To test this hypothesis, we first measured the mitochondrial surface area and cristae number and width, and noticed a slight increase in the cristae number and a modest decrease in the cristae width (see Supplementary material online, Figure S5A). This altered cristae structure driven by loss of FAM210A can be partially explained by the previously reported interaction of FAM210A (C18ORF19) with ATAD3A17 that controls cristae structure.18 However, we did not observe a significant change in ATAD3A protein expression in Fam210a cKO hearts at the early stage (see Supplementary material online, Figure S5B). Additionally, given that mtDNA copy number was slightly increased at the early stage in cKO hearts compared to control hearts (Figures 3C and S5C), we measured the expression of mitochondria biogenesis factor genes and found that the expression of Pgc1α and Pgc1β mRNA was indeed reduced by 36.1% and 27.9%, respectively (see Supplementary material online, Figure S5D). However, the protein expression of neither the nuclear nor mitochondrial localized mitochondria biogenesis factor, PGC1α and TFAM, was changed (see Supplementary material online, Figure S5E). In contrast, autophagic activity was unchanged or slightly decreased as indicated by LC3B II protein expression with or without of Baf A1 treatment (see Supplementary material online, Figure S5F), implying that autophagy-mediated mitochondria degradation pathway may have a minimal effect on the slightly increased mtDNA copy number at the early stage in Fam210a cKO hearts. Moreover, blue native gel assays showed that ETC assembly was modestly affected in Fam210a cKO hearts compared to control hearts at the early stage (see Supplementary material online, Figure S5G). Then, we examined Δψm and ROS production in mitochondria of isolated CMs. TMRE staining showed disturbed Δψm in part of the mitochondria in ∼40% of KO CMs at the early stage (Figure 3D) and an overall increase of Δψm in Fam210a KO CMs (Figures 3E and S5H). This heterogeneity in defects of Δψm was not observed in control CMs. Moreover, ROS production was significantly increased at the early stage in KO CMs (Figure 3F).
Finally, although we observed a modest overall increase in the intensity of TMRE staining in FAM210A deficient CMs (Figure 3E), seahorse measurement revealed that the mitochondrial respiratory activity was moderately compromised due to disruption of Δψm in part of the mitochondria (Figure 3G), with a 21.7% decrease of the maximal oxygen consumption rate and a 42.9% decrease of spare respiratory capacity (Figure 3H) in KO CMs compared to control cells at the early stage. Collectively, these results suggest that reduced Δψm and respiratory activity accompanied by increased ROS production are primary disease-driving events, which contribute to the progression of dilated cardiomyopathy in Fam210a cKO mice.
3.5. Transcriptomic and proteomic reprogramming occurs at early and late stages of Fam210a cKO hearts
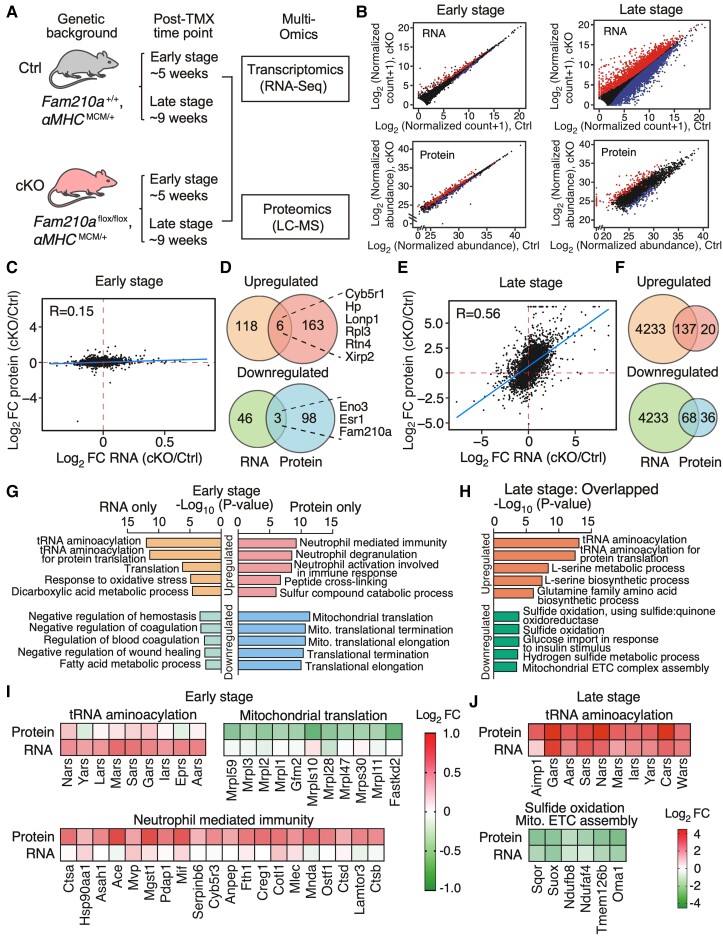
We noticed no obvious pathological phenotypes in CMs and the heart of Fam210a cKO mice at the early stage (Figures 3A and S4). The cardiac dilation and failure occur progressively during the late stage (Figure 1). To dissect global gene expression changes during HF development and understand the aetiology at the molecular level, bulk RNA-seq and quantitative protein mass spectrometry were performed for whole heart tissues harvested from cKO and control mice at ∼5 weeks (early-stage) and ∼9 weeks (late stage) post-KO (Figure 4A and B and Supplementary material online, Tables S2 and S3). At the early stage, 124 mRNAs were significantly up-regulated, while 49 mRNAs were significantly down-regulated in the cKO hearts (Figure 4B and Supplementary material online, Table S2). In parallel, 169 proteins were increased while 101 proteins were decreased in the early-stage cKO hearts (Figure 4B; Supplementary material online, Table S3). Moreover, no strong correlation was observed between mRNA and protein dysregulated expression at the early stage (Figure 4C). Consistently, only a few genes were overlapped between differentially expressed mRNAs and proteins at the early stage (six up-regulated and three down-regulated genes) (Figure 4D and Supplementary material online, Table S4). Fam210a is present as one of the three down-regulated genes as a positive control. In contrast, a large number of mRNAs were differentially regulated at the RNA level, with 4370 significantly up-regulated genes and 4301 significantly down-regulated genes in the late-stage cKO hearts (Figure 4B; Supplementary material online, Table S2), probably driven by the combined effect of FAM210A deficiency and massive secondary effects of HF at the late stage. In parallel, within detectable proteins by mass spectrometry, 157 proteins were up-regulated while 104 proteins were down-regulated (Figure 4B; Supplementary material online, Table S3). Intriguingly, a strong positive correlation (R = 0.56) between the expression of mRNAs and proteins was observed at the late stage (Figure 4E), indicating a co-ordinated regulation of the steady-state mRNA and protein expression at this stage. In agreement with this observation, most of the significantly dysregulated proteins in the proteomic analysis were also significantly dysregulated at the mRNA level. Within overlapped dysregulated genes, 137 genes were up-regulated, and 68 genes were down-regulated at the late stage (Figure 4F). Notably, the most prominent enriched gene pathways uncovered by gene ontology (GO) analysis in up-regulated mRNAs at the early stage include tRNA aminoacylation and translation (Figure 4G and I and Supplementary material online, Table S4), while down-regulated mRNAs were enriched in coagulation and metabolic pathways at the early stage (Figure 4G and Supplementary material online, Table S4). At the protein level, the up-regulated genes were enriched in neutrophil-mediated immunity, while down-regulated genes were enriched in mitochondrial translation machinery at the early stage (Figure 4G and I and Supplementary material online, Table S4). In the overlapped genes at the late stage, tRNA aminoacylation and amino acid metabolism were the most highly enriched pathways in up-regulated genes, while sulfide oxidation and mitochondrial ETC assembly were enriched in down-regulated genes at the late stage (Figure 4H and J and Supplementary material online, Table S4).
Figure 4.
Transcriptomic and proteomic profiles in FAM210A deficient hearts. (A) Experimental timeline of sample collection for transcriptomic and proteomic analyses in Fam210a cKO hearts. (B) Differentially expressed genes at RNA and protein levels in cKO hearts at the early and late stages. Red: significantly increased genes; blue: significantly increased genes. (C and E) The Pearson correlation between changes of RNA and protein in cKO hearts at the early (C) and late stages (E). (D and F) Venn diagrams illustrate the overlap of dysregulated genes at RNA and protein levels in cKO hearts at the early (D) and late stages (F). (G) Gene ontology (GO) analyses of the biological process for dysregulated genes from (D) at the RNA or protein level in cKO hearts at the early stage. Enrichr web tool was used to conduct the GO analysis. (H) GO analyses of biological process for overlapped genes from (F) at the RNA level in cKO hearts at the late stage. Only the top five GO terms were shown in (G) and (I). (I) Expression heat map of a selected group of RNA-only up-regulated genes, protein-only up-regulated genes, and protein-only down-regulated genes at the early stage from top GO terms of (G). (J) Expression heat map of select co-regulated genes at the RNA and protein levels at the late stage from top GO terms in (H).
To explore the global translation profile upon Fam210a cKO in vivo, we performed ribosome profiling (Ribo-seq) using sucrose cushion (1 M sucrose, 34.2% w/v)19,20 with whole heart tissue lysates of control and Fam210a cKO hearts at the early stage (see Supplementary material online, Figure S5I and J and Supplementary material online, Table S5). This method can capture cytosolic monosomes and high molecular weight mitochondrial monosomes complexed with their ribosome-protected footprints (RPF) upon RNase I digestion. We discovered that 5 mRNAs (P adj < 0.05) and 130 mRNAs (P < 0.05) had increased RPF density as calculated by Ribo-seq normalized reads divided by RNA-seq normalized reads using the RiboDiff web tool.21 GO results also highlighted the respiratory electron transport chain-related processes, mainly the mitochondrial-encoded genes (MEGs), as a major pathway affected among the top RPF density-up-regulated mRNAs (see Supplementary material online, Figure S5K, left). Among these RPF density-increased mRNAs, we noticed multiple MEGs, including mt-Nd6 (Complex I gene; P = 1.17 × 10−8; log2FC = 1.41), mt-Cytb (Complex III gene; P = 1.39 × 10−3; log2FC = 0.99), mt-Co1 (or Cox1; Complex IV gene; P = 9.30 × 10−6; log2FC = 1.04), and mt-Co3 (or Cox3; Complex IV gene; P = 0.012; log2FC = 1.51). In addition, we observed that mt-Nd5 (Complex I gene; P = 0.040; log2FC = 0.50) and mt-Nd2 (Complex I gene; P = 0.057; log2FC = 1.29) show increased RPF density with borderline statistical significance. In contrast, the mRNA expression of these MEGs remained unchanged in the corresponding RNA-seq dataset (see Supplementary material online, Table S5). It is widely accepted that RPF density is inversely proportional to translation efficiency (TE) if translation is regulated at the elongation step.22 Thus, FAM210A loss of function may lead to reduced mitochondrial translation elongation and accumulated ribosomes on the MEG mRNAs due to possible ribosome pausing or stalling (see Supplementary material online, Figure S5L). Therefore, these findings support our idea of FAM210A-mediated regulation of translation elongation of MEG mRNAs12 (see Supplementary material online, Figure S5L). On the other hand, we found that 7 mRNAs (P adj < 0.05) and 110 mRNAs (P < 0.05) showed decreased RPF density in our Ribo-seq data (see Supplementary material online, Figure S5J and Supplementary material online, Table S5). All of these mRNAs, enriched in multiple gene ontology terms related to regulation of JNK cascade, elastic fibre assembly, protein ubiquitination, and TORC1 signalling, are encoded by the nuclear genome and translated inside the cytoplasm by cytosolic ribosomes (see Supplementary material online, Figure S5K, right). It is well known that RPF density is proportional to TE if translation is regulated at the initiation step22 (see Supplementary material online, Figure S5L). These results suggest a limited number of mRNAs are down-regulated at the translation initiation step as a secondary effect following Fam210a cKO at the early stage. This secondary effect is likely resulting from ISR, a common signature of mitochondrial cardiomyopathy, driven by phosphorylation and inactivation of eukaryotic translation initiation factor 2α (eIF2α).23
3.6. ISR is persistently activated in Fam210a deficient hearts
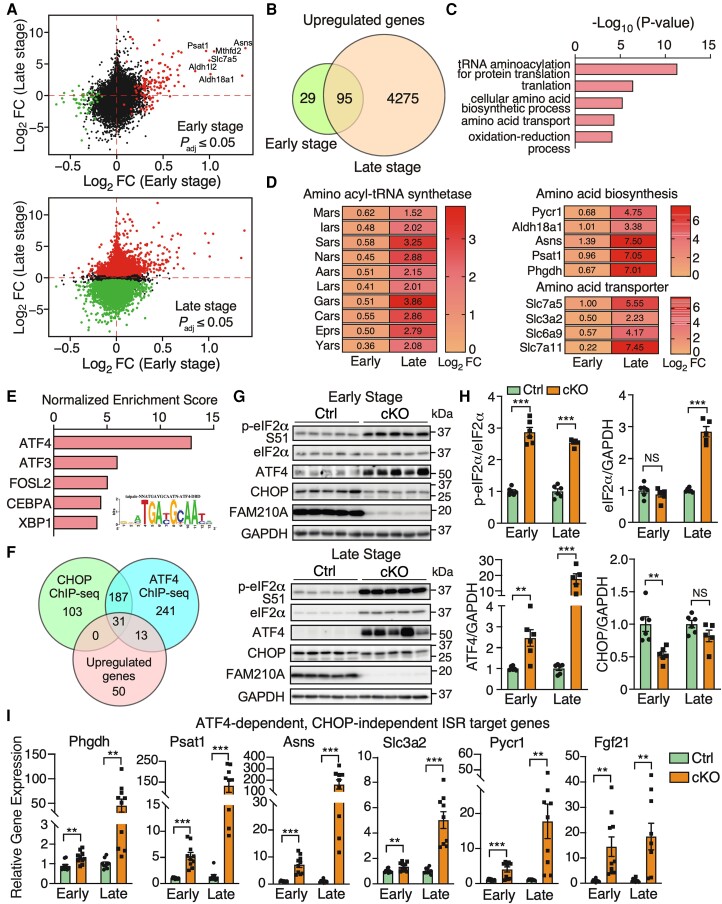
Since RNA-seq analysis provides much deeper genome-wide information about adaptive response and gene regulation than proteomics, we sought to determine the dysregulated gene signature from early and late stages (Figure 5A and Supplementary material online, Table S2). Overlapping significantly dysregulated genes from RNA-seq datasets revealed 95 up-regulated genes and 29 down-regulated genes at both stages (Figures 5B and S6A and Supplementary material online, Table S6). In comparison, a large number of unique dysregulated genes (4275 up-regulated and 4272 down-regulated genes) were uncovered at the late stage compared to a unique cohort of 29 up-regulated and 21 down-regulated genes at the early stage (Figures 5B and S6A and Supplementary material online, Table S6). GO analysis of overlapped up-regulated genes identified that aminoacyl-tRNA synthetases, amino acid synthases, and amino acid transporters were enriched, indicating a persistent activation of the ISR in Fam210a cKO hearts at both stages (Figure 5C and D and Supplementary material online, Table S6). Meanwhile, GO analysis of unique dysregulated mRNAs at the late stage showed that proteasome and ribosome pathways were up-regulated (see Supplementary material online, Figure S6B and C and Supplementary material online, Table S6), suggesting a futile cycle of increased protein degradation and synthesis machinery, which may maintain the proteostasis in KO CMs as a compensatory response. On the other hand, the ETC pathway was down-regulated (see Supplementary material online, Figure S6B and D). We also observed decreased expression of most fatty acid β-oxidation related metabolic enzymes and increased expression of more than half the glycolysis related metabolic enzymes (see Supplementary material online, Figure S6E), which is consistent with the fatty acid-to-glycolysis metabolic switch during HF.
Figure 5.
Cardiomyocyte-specific FAM210A deficiency leads to persistent activation of ATF4-dependent CHOP-independent integrated stress response. (A) Differentially expressed genes at the transcriptomic level at the early (top) and late stages (bottom). (B) The Venn diagram illustrates the overlap of up-regulated genes at the RNA level in Fam210a cKO hearts at the early and late stages. (C) GO analysis using the Enrichr web tool suggests enhanced amino acid metabolism-related gene expression in cKO hearts. (D) Expression heat map of the differentially expressed genes from top three GO terms in (C). (E) iRegulon analyses identified ATF4 as the top transcription factor with the most enriched regulatory motif in all the 95 genes [from (B)] in the inset. (F) Overlapping up-regulated genes with CHOP and ATF4 ChIP-Seq databases reveals ATF4-regulated target genes in CMs. (G) Western blot detection of phospho-eIF2α, ATF4, and CHOP at the early and late stages of cKO hearts. (H) Quantification of protein expression as shown in (G). n = 6 for Ctrl and cKO at the early stage. n = 6 for Ctrl and n = 5 for cKO at the late stage. (I) Relative RNA expression of ATF4-dependent, CHOP-independent ISR downstream targets in cKO hearts at the early and late stages. β-Actin was used as a normalizer. n = 5M + 5F for Ctrl and cKO at the early stage. n = 5M + 4F for Ctrl and cKO at the late stage. NS, not significant; **P < 0.01; ***P < 0.001 by Student’s t-test (H and I).
Consistent with the activation of ISR, transcription factor analysis of overlapped up-regulated genes using the iRegulon tool24 revealed that activating transcription factor 4 (ATF4) was the dominant transcription factor potentially involved in the transcriptional activation of the ISR target genes (Figure 5E). Activation of ISR often induces the expression of ATF4 and CCAAT-enhancer-binding protein homologous protein (CHOP), which is responsible for initiating cell apoptosis.25 To distinguish the transcriptional effects from ATF4 or CHOP, we overlapped persistently up-regulated genes with existing CHOP and ATF4 ChIP-Seq data.26 We found that some ISR targets were unique for ATF4, while no CHOP-specific targets were found (Figure 5F and Supplementary material online, Table S7). Western blot analysis confirmed activation of ISR as indicated by phosphorylation of translation initiation factor eIF2α and increased protein and mRNA expression of ATF4 (Figures 5G and H and S7A). Intriguingly, the CHOP protein level was reduced at the early stage (Figure 5G and H), suggesting a CM-specific ATF4-driven, CHOP-independent ISR program in the cKO hearts. We confirmed that the CHOP-independent, ATF4 target genes were significantly increased in KO CMs at both stages, including phosphoglycerate dehydrogenase (Phgdh), phosphoserine aminotransferase 1 (Psat1), asparagine synthetase (Asns), solute carrier family 3 member 2 (Slc3a2), pyrroline-5-carboxylate reductase 1 (Pycr1), and fibroblast growth factor 21 (Fgf21) (Figure 5I). In addition, the shared target genes of ATF4 and CHOP were also remarkably increased at both stages, including activating transcription factor 5 (Atf5), solute carrier family 7 member 5 (Slc7a5), phosphoserine phosphatase (Psph), methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2/methenyltetrahydrofolate cyclohydrolase (Mthfd2), aldehyde dehydrogenase 1 family member L2 (Aldh1l2), aldehyde dehydrogenase 18 family member A1 (Aldh18a1), serine hydroxymethyltransferase 2 (Shmt2), and growth differentiation factor 15 (Gdf15) (see Supplementary material online, Figure S7A). In contrast, we only observed an increase of mRNA of three mitochondrial unfolded protein response (UPRmt) marker genes at the late stage, including heat shock protein family D member 1 (Hsp60), heat shock protein family A member 4 (Hsp70), and mitochondrial lon peptidase 1 (LonP1) (see Supplementary material online, Figure S7B). Only LONP1 protein expression was increased at the late stage (see Supplementary material online, Figure S7C and D), suggesting that UPRmt is not a major downstream effector pathway caused by FAM210A deficiency at the early stage in the heart. Four kinases are known to phosphorylate eIF2α and activate the ISR upon various stresses.27 In FAM210A deficient hearts, we only observed a band upshift of Heme-Regulated Inhibitor (HRI) kinase while the other three kinases (GCN2, PEKR, and PKR) were unchanged in their phosphorylation at the early stage (see Supplementary material online, Figure S7E and F), suggesting that HRI may be the kinase activating the ISR caused by Fam210a deficiency and mitochondrial translation dysregulation. This observation supports the idea that HRI kinase senses mitochondrial damage28 and activates the ISR upon mitochondrial stresses in the heart. However, all four kinases were activated at the late stage, probably due to various stresses in HF. These results suggest that chronic ISR is activated in Fam210a KO CMs, which may contribute to the aetiology of HF.
3.7. Early-stage metabolomic alterations are present before cardiac dysfunction in Fam210a cKO hearts
Our transcriptomic and proteomic analyses demonstrate that a persistent ISR is activated as indicated by the up-regulation of multiple ISR downstream target genes involved in amino acid biosynthesis (Asns, Aldh18a1, Pycr1, Phgdh, and Psat1), nucleotide synthesis (Ctps and Pck1), and amino acid transport (Slc38a1, Slc3a2, Slc6a9, Slc7a11, and Slc7a5) in the Fam210a cKO hearts (Figure 5 and Supplementary material online, Table S2). Therefore, we ask whether transcriptomic and proteomic reprogramming of these metabolism-related pathways in Fam210a cKO hearts may remodel the overall metabolomic profile. To address this question, we used liquid chromatography-tandem mass spectrometry (LC-MS/MS) to evaluate the metabolomic profile of the hearts of cKO and control mice. Principal component analysis suggests that the overall metabolic profile in cKO hearts (red) is different from that in controls (green) at the early stage of MC (30-day post-KO) (see Supplementary material online, Figure S8A and Supplementary material online, Table S8) as well as at the late stage (60-day post-KO) (see Supplementary material online, Figure S8B and Supplementary material online, Table S8). Consistent with the alteration in ISR downstream metabolic enzymes at the early stage (Figure 5A and D), Metabolite Set Enrichment Analysis (see Supplementary material online, Figure S8C) and top dysregulated metabolites in heat maps revealed that amino acid and nucleotide biosynthetic pathways are among the top changed metabolic pathways, including multiple dysregulated amino acids and nucleotide precursors (see Supplementary material online, Figure S8D). These metabolic pathways stayed dysregulated throughout the late stage of cKO hearts (see Supplementary material online, Figure S8E and F), such as reduced histidine and glutamine, increased phenylalanine and ornithine, and increased nucleotide synthesis (see Supplementary material online, Figure S8G). Additionally, several other metabolic pathways were also remarkably changed at the late stage (see Supplementary material online, Figure S8E and F), probably due to massive gene dysregulation at the mRNA and protein levels caused by HF (Figure 5A), such as the glycolysis pathway (see Supplementary material online, Figure S6E), arachidonic acid metabolism, and oestrogen metabolism. Interestingly, we observed a persistent increase of glutathione and glutathione disulfate at early and late stages (see Supplementary material online, Figure S8G), indicating a compensatory response to maintain the cellular redox balance in response to enhanced mitochondrial ROS production in cKO hearts. Collectively, these metabolomic analyses indicate that FAM210A deficiency in CMs remodels glycolysis- and citric acid cycle-centred amino acids and nucleotide metabolism in the heart, which contributes to the pathogenesis underlying cardiomyopathy and HF.
3.8. FAM210A depletion reduces mitochondrial-encoded mRNA translation in murine hearts
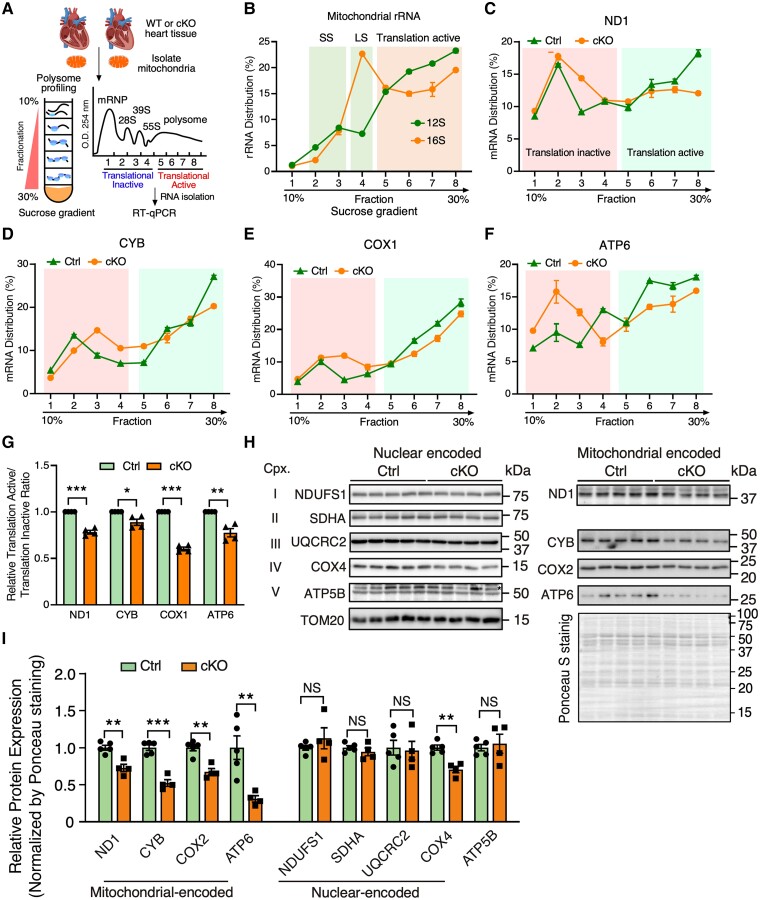
Our prior studies indicate that FAM210A interacts with mitochondrial translation elongation factor EF-Tu and positively regulates mitochondrial-encoded protein expression.12 However, whether FAM210A regulates mitochondrial-encoded proteins at the translational level in the heart in vivo is unclear. Considering mitochondrial translational defects as a common trigger of ISR,29–31 we sought to examine any potential mitochondrial translation defects in the FAM210A deficient hearts. To prove that loss of FAM210A influences mitochondrial translation, we performed mitochondrial polysome profiling using 10–30% sucrose gradient centrifugation coupled with RT–qPCR in isolated mitochondria from the early-stage Fam210a cKO and control hearts (Figure 6A and B). TE of mitochondrial-encoded gene mRNAs was significantly reduced in cKO hearts compared to controls, as indicated by decreased mRNA association with translationally active fractions of MEGs (Figure 6C–G). Consistently, we observed moderate but significant reductions at the steady-state levels in multiple mitochondrial-encoded ETC proteins in isolated mitochondria from Fam210a cKO hearts compared to control hearts at the early stage, including ND1, CYB, COX2, and ATP6 using global protein loading as a normalizer (Figure 6H and I). In contrast, we did not observe significant changes in the nuclear-encoded mitochondrial proteins (NDUFS1, SDHA, UQCRC2, and ATP5B) except COX4. Considering the role of regulating mitochondrial protein expression by FAM210A,12 we reason that a slightly increased mitochondrial DNA copy number (Figure 3C) is unable to compensate for any reduced TE of mitochondrial-encoded proteins caused by loss of FAM210A. Consistent with this idea, when mitochondrial DNA copy number was reduced at the late stage (Figure 2H), we observed that the steady-state level of mitochondrial-encoded proteins and nuclear-encoded proteins was significantly reduced in Fam210a cKO hearts, as shown by mass spectrometry (see Supplementary material online, Figure S9A and Supplementary material online, Table S3) and confirmed by western blot analyses (see Supplementary material online, Figure S9B and C). Taken together, these results suggest that loss of FAM210A compromises the translation of multiple mitochondrial-encoded ETC component genes in vivo as the trigger for initiating mitochondrial dysfunction.
Figure 6.
FAM210A regulates mitochondrial translation in the heart. (A) Schematic of the workflow of polysome profiling analysis of cardiac mitochondrial lysates from WT and cKO mouse hearts. (B) RT–qPCR detection of mitochondrial 12S and 16S rRNA. SS, small mitoribosome subunit; LS, large mitoribosome subunit (C–G) RT–qPCR analysis of mitochondrial-encoded mRNA distributions of Nd1, Cyb, Cox1, and Atp6 across different mitochondrial polysome profiling fractions (C–F). The quantification of the mRNA distribution ratio between translation active and inactive fractions was shown in (G) (n = 4 biological replicates). (H and I) Western blot measurement of nuclear- and mitochondrial-encoded ETC protein levels using isolated mitochondria from early-stage Ctrl or Fam210a cKO hearts at 5-week post-gene knockout (H). The quantification results are shown in (I). n = 5 for Ctrl and n = 4 for cKO. *P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t-test (G and I).
3.9. FAM210A overexpression alleviates cardiac pathological remodelling in myocardial infarction
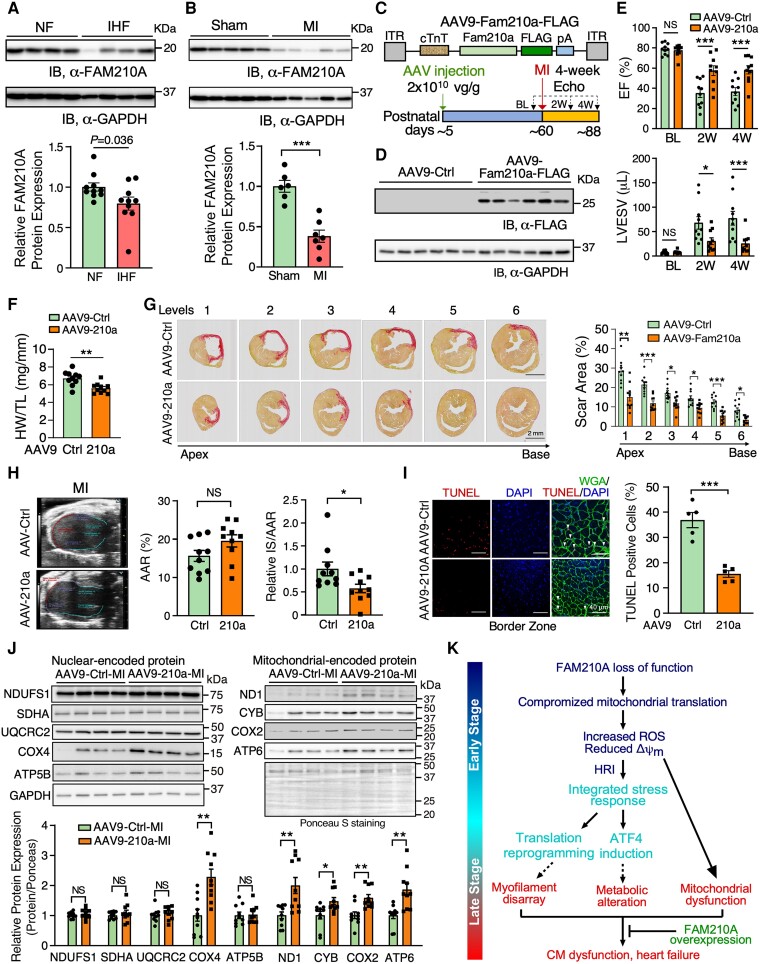
Compromised mitochondrial activity is a common feature in hearts under ischaemic stresses, and an increase in mitochondrial biogenesis is considered a promising therapeutic strategy to treat ischaemic heart disease.7 FAM210A was previously reported to be reduced by 34% in murine hearts at the mRNA level under MI vs. sham surgical conditions by a microarray-based transcriptome screening.32 After examining the expression of FAM210A in human HF samples, we found that FAM210A protein expression was significantly decreased in the hearts of ischaemic HF (IHF) patients compared to non-failing human hearts (Figure 7A). However, FAM210A mRNA showed a reduction trend without statistical significance (see Supplementary material online, Figure S9D). Moreover, the FAM210A protein level was also reduced in the mouse MI hearts compared to sham-controlled ones (Figure 7B). The mRNA level was significantly reduced in the MI compared to sham hearts (see Supplementary material online, Figure S9E). In addition, we observed reduced expression of mitochondrial-encoded proteins in human IHF and mouse MI heart tissue samples compared to their corresponding non-failing controls (see Supplementary material online, Figure S9F and G). As we uncovered the essential role of FAM210A in maintaining mitochondrial mRNA translation (Figures 6 and S9) and observed reduced FAM210A protein expression in human and mouse IHF samples, we sought to determine whether overexpression of FAM210A would enhance the mitochondrial protein expression and reduce cardiac damage using an MI mouse model (Figure 7C). To test this hypothesis, we subcutaneously injected control GFP and FAM210A-overexpressing adeno-associated viruses (AAV9) in WT mice at around post-natal day P5. MI surgery was performed in Fam210a AAV9 overexpression (OE) and control mice at ∼8 weeks followed by left anterior descending artery (LAD) ligation for 4 weeks, and the cardiac function was monitored biweekly by echocardiography. At the endpoint, FAM210A OE was confirmed by immunoblot in the hearts at 4 weeks post-LAD ligation (Figure 7D), and the cardiac function was significantly improved in MI mice with FAM210A OE compared to the control AAV treatment (Figure 7E and Supplementary material online, Table S9). The HW/TL ratio of MI hearts with FAM210A OE was decreased by 16.6% (Figure 7F). Moreover, the scar area was reduced by FAM210A OE, as indicated by picrosirius red staining (Figure 7G). We quantified infarct size normalized to area at risk (AAR) using an echocardiographic analysis. We did not notice any difference in AAR while the infarct size was significantly decreased after normalization by AAR (Figure 7H). Consistently, CM apoptosis was also significantly reduced by 57.9% at the border zone, as indicated by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining (Figure 7I). Western blot analysis showed that mitochondrial-encoded protein expression was enhanced by FAM210A overexpression, including ND1, CYB, COX2, and ATP6 (Figure 7J). In contrast, nuclear-encoded mitochondrial protein expression was not affected except COX4, indicating a translation regulatory function of FAM210A in promoting mitochondrial-encoded protein expression in vivo. Moreover, ATP production and mitochondrial respiratory activity were increased in FAM210A OE hearts (see Supplementary material online, Figure S10A and B), suggesting an improved bioenergetics and mitochondrial function in MI hearts. As a result, ISR activation was compromised after FAM210A OE in MI mice, as indicated by reduced p-eIF2α/eIF2α ratio at the protein level (see Supplementary material online, Figure S10C). Taken together, these data suggest that FAM210A overexpression protects hearts from cardiac damage and functional decline under ischaemic stress.
Figure 7.
FAM210A overexpression improved cardiac function after myocardial infarction. (A) FAM210A protein expression in human ischaemic heart failure (IHF) vs. non-failing (NF) hearts. n = 10 for NF; n = 10 for IHF. (B) FAM210A protein expression in murine hearts from MI (2 weeks) vs. sham-treated mice. n = 6 for sham; n = 7 for MI. (C) Workflow of the AAV9-FAM210A-FLAG overexpression in an MI model. AAV9-GFP was used as a negative control treatment. (D) Western blot detection of FAM210A protein at 4-week post-MI. (E) Left ventricular function was measured by echocardiography in Fam210a overexpression and control mice with MI surgery. EF, ejection fraction; LVESV, left ventricle end-systolic volume; LVEDV, left ventricle end-diastolic volume. n = 10 for Ctrl (6M + 4F) and overexpression (3M + 7F). (F) HW/TL ratio in Fam210a overexpression mice. n = 10 for Ctrl (6M + 4F) and overexpression (3M + 7F). (G) Picrosirius red staining of the scar area in the hearts of Ctrl and Fam210a overexpression mice at 4 weeks post-MI. Right panel: quantification of the scar area at each level. (H) Measurement of AAR and infarct areas using echocardiographic analysis. n = 10 for AAV-Ctrl and AAV-210a. NS, not significant; AAR, area at risk; IS, infarct size. (I) Representative images of CM apoptosis in hearts of mice subjected to MI operation. Red colour indicates TUNEL-positive (apoptotic) nuclei, green indicates CM staining by WGA, and blue (DAPI) represents nuclei. The percentile of TUNEL-positive nuclei was quantified in the right panel. (J) Western blot analysis of nuclear-encoded mitochondrial ETC proteins and mitochondrial-encoded ETC proteins. (K) Schematic working model of the impact and consequence of FAM210A loss-of-function (cKO) and gain-of-function (AAV9 OE) in the heart. *P < 0.05; **P < 0.01; ***P < 0.001 by two-way ANOVA with Sidak's multiple comparisons test (E and G), Student’s t-test (B, F, and H–J), and Mann–Whitney test (A).
4. Discussion
This study demonstrates a novel molecular function of FAM210A in mitochondria, CMs, and the heart using genetic knockout mouse models. FAM210A regulates mitochondrial mRNA translation, and loss of FAM210A causes reduced mitochondrial protein synthesis. The mitochondrial translational defect causes disrupted Δψm and increased ROS that activates ISR and subsequent remodelling of the cellular transcriptome, translatome, proteome, and metabolism in CMs, leading to dysfunction of CM contraction, eventually causing HF and organismal mortality. We also observed reduced FAM210A protein expression in IHF patients’ whole hearts and MI mouse hearts. The overexpression of FAM210A in mouse CMs enhances mitochondrial protein expression and protects the heart from MI-induced cardiac damage and dysfunction. These results suggest that FAM210A is a novel mitochondrial translation regulator to maintain mitochondrial homeostasis in cardiomyocytes and offers a new therapeutic target for cardiac disease (Figure 7K).
4.1. Translation regulatory function of FAM210A in cardiac mitochondria in vivo
Our prior in vitro cell culture studies show that FAM210A regulates mitochondrial-encoded protein expression, possibly through interacting with EF-Tu and mitoribosome, anchoring the mitochondrial translation machinery in the inner mitochondrial membrane.12 To our knowledge, the current study provides the first evidence to support the idea that FAM210A influences mitochondrial translation in vivo using the CM-specific cKO mouse model and AAV9-mediated CM-specific overexpression in the MI mouse model. αMHCMCM and Fam210a fl/fl mice were used as controls to rigorously support our phenotypic characterizations (Figures 1D and S1G–J). Our Ribo-seq and polysome profiling data show that mitochondrial translation was decreased in FAM210A deficient CMs at the early stage before HF (see Supplementary material online, Figures S5I–L and 6A–G). Consistently, the steady-state level of mitochondrial-encoded proteins was moderately and significantly reduced at this early stage without apparent changes in nuclear-encoded mitochondrial proteins (Figure 6H and I). However, this proteomic imbalance status was worsened with time, as indicated by the significant decrease of mitochondrial-encoded proteins and even nuclear-encoded mitochondrial proteins at the late stage as cardiac dysfunction occurs at ∼9 weeks post-TMX-induced Fam210a KO (see Supplementary material online, Figure S9A–C). Thus, the primary events in mitochondria before cardiac dysfunction are likely the disease-driving factors, including disrupted Δψm, increased ROS production, and decreased respiratory rate (Figure 3). Before these mitochondrial defects, the initial trigger of such early events is the loss of FAM210A, which leads to an aberrant mitochondrial translation (Figure 7K). These findings imply that reduced FAM210A in IHF patients’ hearts or MI mouse hearts may contribute to compromised mitochondrial translation, disrupted proteostasis, and mitochondrial dysfunction. Given the fact that FAM210A and mitoribosome proteins were the top binding partners of mitochondrial translation-related protein ATAD3A17 and EF-Tu,12 we speculate that FAM210A may form a complex with ATAD3A, which acts as a mitochondrial inner membrane-associated scaffold or adaptor to increase the local concentration of EF-Tu as well as mitoribosomes to enhance TE at the elongation step.12 Fam210a cKO in CMs may reduce EF-Tu docked on the mitochondrial inner membrane, leading to impaired channelling of aminoacyl-tRNA to the mitoribosome decoding centre. This slows down the translocation and movement of mitoribosome (reduced elongation rate) and accumulates more mitoribosomes on mitochondrial-encoded mRNAs, thereby increasing RPF density (more ribosome footprints) (see Supplementary material online, Figure S5I–L). Consequently, Fam210a cKO activates the ISR that inhibits cytosolic translation initiation due to p-eIF2α, leading to reduced loading of cytoribosome on cytoplasmic mRNAs, thereby decreasing RPF density (fewer ribosome footprints) (see Supplementary material online, Figure S5I–L). Interestingly, we also observed dysregulated cristae structure in Fam210a cKO hearts at the early stage, which further supported its functional association with ATAD3A.17,18 Loss of FAM210A did not alter the expression of ATAD3A at the early stage (see Supplementary material online, Figure S5B), indicating that the FAM210A-ATAD3A complex may exert dual functions in mitochondria, including recruiting EF-Tu and mitoribosome for mitochondrial translation, and regulating the mitochondrial cristae structure. Further experiments are required to elucidate the detailed molecular and functional connections between FAM210A with ATAD3A. The FAM210A gene is conserved in more than 213 organisms, including humans, mice, zebrafish, and Caenorhabditis elegans.33 Fam210a mRNA is expressed in all organs and is enriched in the heart, brain, skeletal muscle, and testis of humans.34 Therefore, the biological function and molecular mechanism we discover here may be generalizable, and further studies are required to prove FAM210A function across multiple cell types and other species.
4.2. ISR contributes to disease progression of mitochondrial cardiomyopathy
The mammalian target of rapamycin (mTOR) pathway is often associated with cardiac pathological hypertrophy.35 We measured the phosphorylation of mTOR kinase and its two substrate proteins, P70S6K and 4EBP1, and found Fam210a KO activated neither (data not shown). Moreover, we did not observe a strong UPRmt activation. At the same time, ISR was markedly activated in Fam210a cKO hearts at the early stage (Figures 5G and H and S7E and F), suggesting that ISR is a central pathway activated by FAM210A deficiency. ISR is a major translational control pathway to promote the survival of cells under stress conditions in the acute phase and can lead to cell apoptosis after chronic activation.27 Our multi-omics results reveal that ISR is persistently activated at early and late stages in Fam210a cKO hearts (Figure 7K). Intriguingly, we observed limited but significantly increased (∼2.5%) CM cell death at the late stage of Fam210a cKO hearts (see Supplementary material online, Figure S3F), as indicated by TUNEL staining. This may cause chamber dilation over time and explains the slightly increased cardiac fibrosis at the late stage. However, as downstream effectors of ISR, only ATF4 but not CHOP was up-regulated at the protein level (Figure 5G and H). ATF4 activates pro-survival gene expression without triggering a pro-apoptosis gene program mainly activated by CHOP.27 Therefore, this ISR seems to be anti-apoptotic for a prolonged time to minimize CM death, as indicated by increased expression of an anti-apoptotic marker gene Mcl1 and no drastic change in pro-apoptotic genes such as Bcl2l11 and Apaf1 (see Supplementary material online, Figure S10D). This ‘double-edged sword’ effect of ISR activated in Fam210a cKO hearts can partially explain the delayed onset of severe cardiac pathological remodelling and heart failure (∼7 weeks post-TMX treatment) in Fam210a cKO hearts in addition to the relatively long half-life of FAM210A protein.36 However, the ISR-mediated adaptive translational response may protect against cell death at the expense of disrupted proteomic homeostasis, leading to aberrant sarcomeric protein expression and contractile dysfunction in CMs (Figures 2 and 4). We observed that robust activation of two effectors reprograms the cellular transcriptome, translatome, proteome, and metabolism, in response to stresses in Fam210a cKO hearts, including activated phosphorylation of eIF2α and increased protein expression of ATF4. Phosphorylated eIF2α inhibits cap-dependent mRNA translation.37 This translational defect inhibits the synthesis of myofilament proteins required for contractile function and nuclear-encoded mitochondrial proteins (see Supplementary material online, Figures S2D and S9A), which causes CM contractile dysfunction. On the other hand, increased ATF4 activates the transcription of multiple metabolic enzymes (Figure 5). As a consequence, amino acid and nucleotide metabolism is altered, and this change may contribute to the progression of HF. Thus, it is worthwhile to test if targeting the ISR pathway provides any beneficial effect in treating MC and MI based on the fact that genetic deletion of ISR kinase PKR protects hearts from pressure overload-induced HF38 and that chemical inhibition of ISR by ISRIB prevents atrial fibrillation post-MI.39 Our integrated Ribo-seq and RNA-seq analysis in both cKO and control αMHCMCM hearts at the early stage supports the role of FAM210A in regulating mitochondrial mRNA translation elongation in vivo (see Supplementary material online, Figure S5I–L). It is well known that compromised cytosolic translation elongation triggers ISR activation.40,41 Here, we provide evidence to support the notion that impaired mitochondrial translation elongation (see Supplementary material online, Figure S5I–L) can also activate the ISR pathway possibly via HRI kinase (see Supplementary material online, Figure S7E and F). Heme or iron deficiency enhances phosphorylation of eIF2α through HRI and activates ISR, thereby affecting cytosolic translation.42 The ISR activation affects the synthesis of mitochondrial ribosomal proteins, which are translated in cytosol, thereby regulating the protein synthesis inside mitochondria during erythropoiesis.42 In our study, HRI is activated by mitochondrial translation deficiency at the early stage in Fam210a cKO hearts. Similar to the effect in erythroblasts, HRI may indirectly influence mitochondrial translation via a secondary effect to some extent in Fam210a deficient CMs, as indicated by enriched mitochondrial translation elongation and termination factors in down-regulated proteins in our early-stage mass spectrometry data (Figure 4G).
4.3. FAM210A overexpression protects hearts during myocardial infarction
In contrast to reduced mitochondrial-encoded mRNA translation by genetic knockout of Fam210a in murine hearts, we show that overexpression of FAM210A positively regulates mitochondrial translation and protects hearts from pathological remodelling triggered by ischaemic stress during MI (Figure 7). We assume that reduced FAM210A expression in human IHF and mouse MI hearts may contribute to a compromised compensatory boost in mitochondrial mRNA translation and protein synthesis in the mitochondrial matrix,43 disrupt mitochondrial homeostasis, and exaggerate downstream mitochondrial and CM dysfunction. Therefore, overexpression of FAM210A can enhance mitochondrial translation in CMs, improve mitochondrial respiratory activity and energetics (ATP production), and protect hearts under ischaemic stresses by promoting mitochondrial activities (see Supplementary material online, Figures S10A–C and 7K). This phenotypic rescue by overexpression of FAM210A is consistent with the proposed mechanism and benefits of maintaining a balance of cytosolic and mitochondrial translatomes and translation programs in yeast and human cells.44,45 A prior multi-omics study in animal models shows an increased FAM210A expression in swim-activated physiological hypertrophy compared to pressure overload-induced cardiac pathological hypertrophy,14 supporting the potential cardiac protective function of FAM210A overexpression. Further studies are required to examine whether FAM210A overexpression shows any cardiac protection in non-ischaemic HF models since we observed exaggerated cardiac pathological hypertrophy in a pressure overload HF mouse model with induced FAM210A expression upon genetic ablation of miR-574.12
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contributions
P.Y. obtained the funding and launched the study. P.Y. and J.W. conceived the ideas, designed the experiments, analysed the data, and wrote the manuscript. J.W. and K.C.V.S. carried out the experimental work. O.H. and S.C. provided technical assistance and performed specific experiments. J.M., W.H.W.T., and C.Y. provided conceptual feedback, technical support, and research platforms for metabolomic analysis, human sample examination, and cardiomyocyte functional characterization, respectively. All the authors discussed the results and had the opportunity to comment on the manuscript.
Supplementary Material
Acknowledgements
We are grateful to Jared Hollinger, Eng-Soon Khor (Aab CVRI), and Bin Liu (Mississippi State University) for their critical reading of the manuscript and Qiuqing Wang (Cleveland Clinic) for biostatistical consulting. We appreciate the technical assistance from Erika Flores Medina, Xenia Schafer, Mohan Amy, and Deanne Mickelsen (Aab CVRI) in histology, mass spectrometry-based metabolomics, surgical, and echocardiography operations (AAR image analysis), respectively. RNA-seq and primary data analysis were performed by Jason R. Myers from the Genomics Research Center at the University of Rochester. Genomics Research Center at the University of Rochester and Novogene company performed Ribo-seq (RNA-seq) and data analysis. We appreciate the bioinformatic data analysis for Ribo-seq and RNA-seq performed by Dalia Ghoneim. Kevin Welle performed protein mass spectrometry analysis from the Mass Spectrometry Resource Lab at the University of Rochester. Chad Galloway and Karen de Mesy Bentley conducted the electron microscopic imaging of heart sections in the Electron Microscopy Core at the University of Rochester. Eric Small provided us with the tamoxifen-inducible transgenic mouse line αMHCMerCreMer (αMHCMCM/+). Hani Awad and Emma Gira contributed to testing the potential changes in bone morphology and function in Fam210a cKO mice. None of the authors have any financial conflict of interest related to the research described in this manuscript.
Contributor Information
Jiangbin Wu, Aab Cardiovascular Research Institute, Department of Medicine, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA.
Kadiam C Venkata Subbaiah, Aab Cardiovascular Research Institute, Department of Medicine, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA.
Omar Hedaya, Aab Cardiovascular Research Institute, Department of Medicine, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA; Department of Biochemistry & Biophysics, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA.
Si Chen, Aab Cardiovascular Research Institute, Department of Medicine, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA.
Joshua Munger, Department of Biochemistry & Biophysics, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA.
Wai Hong Wilson Tang, Department of Cardiovascular Medicine, Cleveland Clinic, 9500 Euclid Avenue, Cleveland, OH 44195, USA.
Chen Yan, Aab Cardiovascular Research Institute, Department of Medicine, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA.
Peng Yao, Aab Cardiovascular Research Institute, Department of Medicine, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA; Department of Biochemistry & Biophysics, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA; The Center for RNA Biology, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA; The Center for Biomedical Informatics, University of Rochester School of Medicine & Dentistry, 601 Elmwood Avenue, Rochester, NY 14642, USA.
Funding
This work was supported in part by National Institutes of Health grants R01 HL132899, R01 HL147954, R01 HL164584 and R01 HL169432 (to P.Y.), start-up funds from Aab Cardiovascular Research Institute of the University of Rochester Medical Center (to P.Y.), American Heart Association Postdoctoral Fellowship 19POST34400013 and Career Development Award 848985 (to J.W.), NIH T32 Fellowship (T32 GM068411 to O.H.), and National Institutes of Health grant R01 HL154318 (to C.Y.).
Data availability
RNA-seq data produced and used in this study were deposited on Gene Expression Omnibus (GEO) database under accession number GSE195957.
Translational Perspective.
Mitochondrial homeostasis is critical for maintaining healthy cardiac function. Disruption of mitochondrial function causes severe cardiomyopathy and heart failure. In the present study, we show that FAM210A is a mitochondrial translation regulator required for maintaining cardiac mitochondrial homeostasis in vivo. Cardiomyocyte-specific FAM210A deficiency leads to mitochondrial dysfunction and spontaneous cardiomyopathy. Moreover, our results indicate that FAM210A is down-regulated in human and mouse ischaemic heart failure samples and overexpression of FAM210A protects hearts from myocardial infarction-induced heart failure, suggesting that FAM210A-mediated mitochondrial translation regulatory pathway can be a potential therapeutic target for ischaemic heart disease.
References
- 1. Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature 2012;491:374–383. [DOI] [PubMed] [Google Scholar]
- 2. Sun N, Finkel T. Cardiac mitochondria: a surprise about size. J Mol Cell Cardiol 2015;82:213–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. El-Hattab AW, Scaglia F. Mitochondrial cardiomyopathies. Front Cardiovasc Med 2016;3:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Suomalainen A, Battersby BJ. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cell Biol 2018;19:77–92. [DOI] [PubMed] [Google Scholar]
- 5. Bauer TM, Murphy E. Role of mitochondrial calcium and the permeability transition pore in regulating cell death. Circ Res 2020;126:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramachandra CJA, Hernandez-Resendiz S, Crespo-Avilan GE, Lin YH, Hausenloy DJ. Mitochondria in acute myocardial infarction and cardioprotection. EBioMedicine 2020;57:102884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown DA, Perry JB, Allen ME, Sabbah HN, Stauffer BL, Shaikh SR, Cleland JG, Colucci WS, Butler J, Voors AA, Anker SD, Pitt B, Pieske B, Filippatos G, Greene SJ, Gheorghiade M. Expert consensus document: mitochondrial function as a therapeutic target in heart failure. Nat Rev Cardiol 2017;14:238–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou B, Tian R. Mitochondrial dysfunction in pathophysiology of heart failure. J Clin Invest 2018;128:3716–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LM, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A; International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center; MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon AM, Henkelman RM, Brown SD, Adams DJ, Lloyd KC, McKerlie C, Beaudet AL, Bućan M, Murray SA. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, Gonzalez-Macias J, Kahonen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren O, Lorenc RS, Marc J, Mellstrom D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NL, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gomez C, Palsson ST, Reppe S, Rotter JI, Sigurdsson G, van Meurs JB, Verlaan D, Williams FM, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw KT, Lehtimaki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HA, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AW, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JP, Kiel DP, Rivadeneira F. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet 2012;44:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka KI, Xue Y, Nguyen-Yamamoto L, Morris JA, Kanazawa I, Sugimoto T, Wing SS, Richards JB, Goltzman D. FAM210A is a novel determinant of bone and muscle structure and strength. Proc Natl Acad Sci U S A 2018;115:E3759–E3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu J, Venkata Subbaiah KC, Jiang F, Hedaya O, Mohan A, Yang T, Welle K, Ghaemmaghami S, Tang WHW, Small E, Yan C, Yao P. MicroRNA-574 regulates FAM210A expression and influences pathological cardiac remodeling. EMBO Mol Med 2020;13:e12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanaka KI, Kanazawa I, Richards JB, Goltzman D, Sugimoto T. Modulators of Fam210a and roles of Fam210a in the function of myoblasts. Calcif Tissue Int 2020;106:533–540. [DOI] [PubMed] [Google Scholar]
- 14. Boileau E, Doroudgar S, Riechert E, Jurgensen L, Ho TC, Katus HA, Volkers M, Dieterich C. A multi-network comparative analysis of transcriptome and translatome identifies novel hub genes in cardiac remodeling. Front Genet 2020;11:583124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karlsson M, Zhang C, Mear L, Zhong W, Digre A, Katona B, Sjostedt E, Butler L, Odeberg J, Dusart P, Edfors F, Oksvold P, von Feilitzen K, Zwahlen M, Arif M, Altay O, Li X, Ozcan M, Mardinoglu A, Fagerberg L, Mulder J, Luo Y, Ponten F, Uhlen M, Lindskog C. A single-cell type transcriptomics map of human tissues. Sci Adv 2021;7:eabh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao RZ, Jiang S, Zhang L, Yu ZB. Mitochondrial electron transport chain, ROS generation and uncoupling (review). Int J Mol Med 2019;44:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He J, Cooper HM, Reyes A, Di Re M, Sembongi H, Litwin TR, Gao J, Neuman KC, Fearnley IM, Spinazzola A, Walker JE, Holt IJ. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res 2012;40:6109–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peralta S, Goffart S, Williams SL, Diaz F, Garcia S, Nissanka N, Area-Gomez E, Pohjoismaki J, Moraes CT. ATAD3 controls mitochondrial cristae structure in mouse muscle, influencing mtDNA replication and cholesterol levels. J Cell Sci 2018;131:jcs217075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McGlincy NJ, Ingolia NT. Transcriptome-wide measurement of translation by ribosome profiling. Methods 2017;126:112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 2014;8:1365–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhong Y, Karaletsos T, Drewe P, Sreedharan VT, Kuo D, Singh K, Wendel HG, Ratsch G. Ribodiff: detecting changes of mRNA translation efficiency from ribosome footprints. Bioinformatics 2017;33:139–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lian X, Guo J, Gu W, Cui Y, Zhong J, Jin J, He QY, Wang T, Zhang G. Genome-wide and experimental resolution of relative translation elongation speed at individual gene level in human cells. PLoS Genet 2016;12:e1005901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuhl I, Miranda M, Atanassov I, Kuznetsova I, Hinze Y, Mourier A, Filipovska A, Larsson NG. Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. Elife 2017;6:e30952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V, Hulselmans G, Herten K, Naval Sanchez M, Potier D, Svetlichnyy D, Kalender Atak Z, Fiers M, Marine JC, Aerts S. Iregulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 2014;10:e1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The role of the PERK/eIF2alpha/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr Mol Med 2016;16:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, Kilberg MS, Sartor MA, Kaufman RJ. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 2013;15:481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep 2016;17:1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fessler E, Eckl EM, Schmitt S, Mancilla IA, Meyer-Bender MF, Hanf M, Philippou-Massier J, Krebs S, Zischka H, Jae LT. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 2020;579:433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dogan SA, Pujol C, Maiti P, Kukat A, Wang S, Hermans S, Senft K, Wibom R, Rugarli EI, Trifunovic A. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab 2014;19:458–469. [DOI] [PubMed] [Google Scholar]
- 30. Agnew T, Goldsworthy M, Aguilar C, Morgan A, Simon M, Hilton H, Esapa C, Wu Y, Cater H, Bentley L, Scudamore C, Poulton J, Morten KJ, Thompson K, He L, Brown SDM, Taylor RW, Bowl MR, Cox RD. A Wars2 mutant mouse model displays OXPHOS deficiencies and activation of tissue-specific stress response pathways. Cell Rep 2018;25:3315–3328.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Quiros PM, Prado MA, Zamboni N, D'Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J Cell Biol 2017;216:2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nishihama N, Abe Y, Kubota K, Nagayama T, Hirai T, Ueda K, Nakamaru K, Hirata T, Fukuda K, Koishi R, Makino S. Identification of fat storage-inducing transmembrane proteins 1 and 2 as putative therapeutic targets for heart failure by integrated analysis of proteome and transcriptome. J Proteomics Bioinform 2018;11:173–182. [Google Scholar]
- 33. Kang J, Zhou H, Sun F, Chen Y, Zhao J, Yang WJ, Xu S, Chen C. Caenorhabditis elegans homologue of Fam210 is required for oogenesis and reproduction. J Genet Genomics 2020;47:694–704. [DOI] [PubMed] [Google Scholar]
- 34. GTEx Consortium . The genotype-tissue expression (GTEx) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sciarretta S, Forte M, Frati G, Sadoshima J. New insights into the role of mTOR signaling in the cardiovascular system. Circ Res 2018;122:489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim TY, Wang D, Kim AK, Lau E, Lin AJ, Liem DA, Zhang J, Zong NC, Lam MP, Ping P. Metabolic labeling reveals proteome dynamics of mouse mitochondria. Mol Cell Proteomics 2012;11:1586–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Costa-Mattioli M, Walter P. The integrated stress response: from mechanism to disease. Science 2020;368:eaat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang H, Xu X, Fassett J, Kwak D, Liu X, Hu X, Falls TJ, Bell JC, Li H, Bitterman P, Bache RJ, Chen Y. Double-stranded RNA-dependent protein kinase deficiency protects the heart from systolic overload-induced congestive heart failure. Circulation 2014;129:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang T, Wu Y, Hu Z, Xing W, Kun LV, Wang D, Hu N. Small-molecule integrated stress response inhibitor reduces susceptibility to postinfarct atrial fibrillation in rats via the inhibition of integrated stress responses. J Pharmacol Exp Ther 2021;378:197–206. [DOI] [PubMed] [Google Scholar]
- 40. Ishimura R, Nagy G, Dotu I, Chuang JH, Ackerman SL. Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. Elife 2016;5:e14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vind AC, Genzor AV, Bekker-Jensen S. Ribosomal stress-surveillance: three pathways is a magic number. Nucleic Acids Res 2020;48:10648–10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S, Macias-Garcia A, Ulirsch JC, Velazquez J, Butty VL, Levine SS, Sankaran VG, Chen JJ. HRI coordinates translation necessary for protein homeostasis and mitochondrial function in erythropoiesis. Elife 2019;8:e46976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gudbjarnason S, Deschryver C, Chiba C, Yamanaka J, Bing RJ. Protein and nucleic acid synthesis during the reparative processes following myocardial infarction. Circ Res 1964;15:320–326. [DOI] [PubMed] [Google Scholar]
- 44. Couvillion MT, Soto IC, Shipkovenska G, Churchman LS. Synchronized mitochondrial and cytosolic translation programs. Nature 2016;533:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soto I, Couvillion M, Hansen KG, McShane E, Moran JC, Barrientos A, Churchman LS. Balanced mitochondrial and cytosolic translatomes underlie the biogenesis of human respiratory complexes. Genome Biol 2022;23:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data produced and used in this study were deposited on Gene Expression Omnibus (GEO) database under accession number GSE195957.