Abstract
Background
The filovirus Bundibugyo virus (BDBV) causes severe disease with a mortality rate of approximately 20%–51%. The only licensed filovirus vaccine in the United States, Ervebo, consists of a recombinant vesicular stomatitis virus (rVSV) vector that expresses Ebola virus (EBOV) glycoprotein (GP). Ervebo was shown to rapidly protect against fatal Ebola disease in clinical trials; however, the vaccine is only indicated against EBOV. Recent outbreaks of other filoviruses underscore the need for additional vaccine candidates, particularly for BDBV infections.
Methods
To examine whether the rVSV vaccine candidate rVSVΔG/BDBV-GP could provide therapeutic protection against BDBV, we inoculated seven cynomolgus macaques with 1000 plaque-forming units of BDBV, administering rVSVΔG/BDBV-GP vaccine to 6 of them 20–23 minutes after infection.
Results
Five of the treated animals survived infection (83%) compared to an expected natural survival rate of 21% in this macaque model. All treated animals showed an early circulating immune response, while the untreated animal did not. Surviving animals showed evidence of both GP-specific IgM and IgG production, while animals that succumbed did not produce significant IgG.
Conclusions
This small, proof-of-concept study demonstrated early treatment with rVSVΔG/BDBV-GP provides a survival benefit in this nonhuman primate model of BDBV infection, perhaps through earlier initiation of adaptive immunity.
Keywords: Bundibugyo virus, ebolaviruses, Ervebo, filovirus, postexposure treatment, rVSVΔG/BDBV-GP, recombinant vesicular stomatitis virus, transcriptomics, vaccine
The Ebolavirus genus comprises multiple species including Zaire ebolavirus (representative member Ebola virus [EBOV]), Sudan ebolavirus (representative member Sudan virus [SUDV]), Bundibugyo ebolavirus (representative member Bundibugyo virus [BDBV]), and Taï Forest ebolavirus (representative member Taï Forest virus). Each of these species is associated with a highly fatal disease following infection, referred to as Ebola disease [1] . Members of these species have been responsible for more than half a dozen distinct outbreaks in Central and West Africa over the past decade [2]. The intermittent and unpredictable nature of these outbreaks emphasizes the need for medical interventions that can be effective within a short timeframe. Some success has been achieved for EBOV [1, 3, 4], but additional therapeutic and vaccine options for SUDV and BDBV are needed.
Development of medical countermeasures for filoviruses has relied heavily on animal models of disease, specifically nonhuman primate (NHP) models. Clinical manifestations of ebolavirus-infected humans and NHPs are similar, including high viremia, hypercytokinemia, and consumptive coagulopathy, which may progress to septic shock, multiorgan failure, and death [5]. The virus initially replicates in monocytes and dendritic cells and then spreads to hepatocytes, endothelial cells, and epithelial cells. Therapeutics and vaccines that showed protection when tested in NHP models of Ebola disease have shown efficacy in treatment of human patients.
BDBV is considered less pathogenic than EBOV and SUDV in humans due to the reduced lethality observed during human outbreaks (20%–51% compared to mortality rates of up to 50%–90% seen with SUDV and EBOV, respectively) [6]. BDBV also appears less pathogenic in experimentally infected cynomolgus and rhesus macaques. BDBV infection in these models leads to lethality in approximately 40%–70% of infections [7–9], while EBOV and SUDV infection cause near uniform lethality [5]. The disease course in these NHP models is also slightly extended, which could offer vaccines and therapeutics a wider window to show effectiveness.
Prior studies have shown that the vaccine candidate rVSVΔG/BDBV-GP is highly effective as a preventive vaccine against BDBV when used in a single-injection format [8]. This is similar to other recombinant vesicular stomatitis virus (rVSV)–based vaccines, which have shown protection against EBOV and SUDV infection in NHP models of disease. A distinctive aspect of rVSV-based vaccines against filoviral targets is that in both EBOV and SUDV animal models, early postexposure treatment with these vaccines leads to significant protection. Postexposure vaccination has been used as standard protocol for other fatal viral infections, including rabies virus, and may also have utility following accidental needlesticks or for ring vaccination of contacts of infected individuals during filovirus outbreaks.
To test the hypothesis that rVSVΔG/BDBV-GP can act as a postexposure therapeutic, we undertook a narrowly focused test of this hypothesis. NHPs infected with BDBV were treated with rVSVΔG/BDBV-GP 20–23 minutes after infection and then followed. From animals that survived and those that succumbed to BDBV challenge, we analyzed the clinical pathology, viral loads, humoral response, and circulating transcriptional profiles. rVSVΔG/BDBV-GP postexposure treatment was associated with significant protection from death in this model, with 5 of 6 treated animals surviving. Survivors presented with varying disease severity. Transcriptional analysis revealed that all treated animals had a strong early innate immune response, but in survivors this response appeared to be more robust than in those that succumbed. These results confirm that rVSVΔG/BDBV-GP can offer protection as a postexposure treatment and suggests that innate immune responses might serve as an indication of successful disease mitigation.
METHODS
Ethics Statement
Animal studies were conducted in compliance with the Animal Welfare Act and other federal statutes and regulations relating to animals and experiments involving animals. All experiments adhered to principles stated in the eighth edition of the “Guide for the Care and Use of Laboratory Animals” (National Research Council, 2011). The Galveston National Laboratory (GNL) where this research was conducted at the University of Texas Medical Branch (UTMB) is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International and has an approved Office of Laboratory Animal Welfare (OLAW) Assurance (No. A3314-01). Animal studies were performed in biosafety level 4 (BSL-4) biocontainment at the UTMB and the protocol was approved by the UTMB Institutional Biosafety Committee.
rVSV Vaccine Vector and Challenge Virus
The replicating rVSV expressing the BDBV glycoprotein (GP; rVSVΔG/BDBV-GP) was recovered from cDNA as previously described [10]. The BDBV used in this study, strain 200706291 (GenBank accession No. MK028856.1), was isolated from a fatal human case in western Uganda during the outbreak in 2007 [8]. The virus was kindly provided by Dr Thomas G. Ksiazek. The challenge stock of BDBV was propagated on Vero E6 cells twice. The FASTA consensus sequence and variant frequencies are provided in the Supplementary Material. Notably, the consensus sequence of our virus seed stock is identical to the parental sequence (GenBank MK028856.1). Maximum variances of our stock included indel mutations found at positions 11952 and 11027, but these mutations accounted for less than 18% and 7% of the virus population, respectively. All other low frequency (LoFreq) variants constituted less than 6% of the virus population. Both vaccine candidate and BDBV stocks tested negative for endotoxin.
LoFreq Variant Identification
The library for sequencing the BDBV challenge seed was prepared with an NEBNext Ultra II RNA Prep Kit (New England BioLabs, Inc) following the manufacturer's protocol. Briefly, approximately 100 ng of RNA was fragmented for 15 minutes, followed by cDNA synthesis, end repair, and adapter ligation. After 5 rounds of polymerase chain reaction (PCR), the resulting library was analyzed on an Agilent Bioanalyzer and quantified by qPCR. Samples were pooled and sequenced with a paired-end 75 base protocol on an Illumina NextSeq 550 using the High-Output kit.
Reads were processed with Trimmomatic version 0.36 [11] to remove low-quality base calls and any adapter sequences. The de novo assembly program ABySS [12] was used to assemble the reads into contigs, using several different sets of reads, and kmer values from 20 to 40. Contigs greater than 400 bases long were compared against the National Center for Biotechnology Information (NCBI) nucleotide collection using BLAST. A nearly full-length BDBV-Uganda viral contig was obtained and all the remaining contigs mapped to either host cell ribosomal RNA or mitochondria. The trimmed reads from each sample were mapped to the sample consensus sequence BWA version 0.7.17 [13] and visualized with the Integrated Genomics Viewer [14] to confirm a correct assembly.
For single-nucleotide variant and insertion/deletion calling the trimmed reads from each sample were mapped to the reference sequence with BWA. The LoFreq version 2.1.3.1 [15] call and call-indels commands were used for variant calling, after the mapped reads were preprocessed with the LoFreq viterbi and indelqual commands to fix alignments at the read ends and insert indel quality scores, respectively (Supplementary Material). Variant calls were filtered at a level of 0.05%.
Immunization and Treatment
Seven, healthy, adult filovirus-naive cynomolgus macaques (Macaca fascicularis) of Chinese origin weighing 3.56–6.70 kg and ranging from 4 to 7 years of age were used in this study (4 females and 3 males) (Worldwide Primates). All 7 animals were challenged by intramuscular (IM) injection in the left quadricep (0.5 mL) with a 1000 plaque-forming unit (PFU) target dose of BDBV challenge stock (the actual dose was 913 PFU). Approximately 20–23 minutes postchallenge, 6 animals were treated with approximately 2 × 107 PFU of rVSVΔG/BDBV-GP via IM injection. The 1-mL inoculation was equally distributed between the left and right quadriceps. Animals were monitored for viremia and clinical signs of illness (temperature, weight loss, changes in blood count, and blood chemistries) during the treatment and BDBV challenge portions of the study. Blood was collected on days 0, 1, 5, 7, 9, 12, 15, 21, and 28 postchallenge (Figure 1). Samples were also collected on the day of euthanasia. An internal scoring protocol was implemented to track disease progression in challenged animals. Animals were checked at least twice daily after challenge for scoring criteria such as behavior and posture/activity level (score of 0–9), appetite (score of 0–2), respiration (score of 0–9), and the presence of hemorrhagic manifestations (score of 0–9). Subjects that reached a clinical score ≥ 9 were promptly euthanized with a pentobarbital solution.
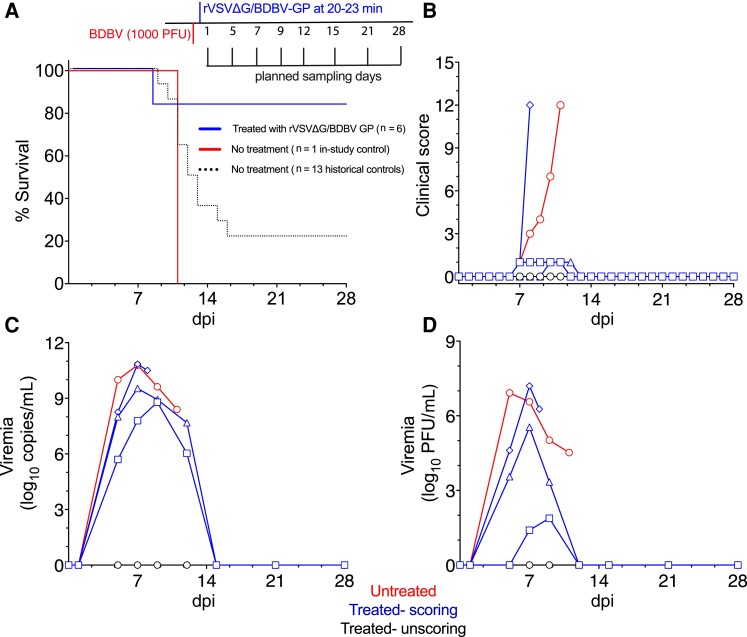
Figure 1.
Survival of macaques and comparison of viral loads after BDBV challenge and postexposure treatment with rVSVΔG/BDBV-GP. A, Study design and survival curve of BDBV-exposed cynomolgus macaques left untreated (n = 1) or treated with rVSVΔG/BDBV-GP (n = 6). Historical untreated controls are also depicted (n = 13). All animals were infected intramuscularly with a target dose of 1000 PFU of BDBV-Uganda and then 6 animals were treated with rVSVΔG/BDBV-GP 20-23 minutes later. B, Clinical scores of BDBV-infected macaques; criteria include behavior, posture and activity level, appetite, respiration, and the presence of hemorrhagic manifestations. C, Circulating viral RNA. Measured by RT-qPCR in whole blood and reported as log10 copies/mL. The limit of detection for this assay was 1000 copies/mL. D, Viral loads were measured in plasma samples by standard plaque assay and reported as log10 PFU/mL. The limit of detection for this assay was 25 PFU/mL. Individual treated subjects are denoted by the following symbols: fatal treated, diamond; survivor 3, triangle; survivor 4, square. Abbreviations: BDBV, Bundibugyo virus; dpi, days postinfection; GP, glycoprotein; PFU, plaque-forming unit; RT-qPCR, quantitative real-time polymerase chain reaction; rVSV, recombinant vesicular stomatitis virus.
Blood Processing and Peripheral Blood Mononuclear Cell Isolation
Blood was collected by femoral venipuncture into EDTA, heparin, and clot activating vacutainer tubes (BD Biosciences). For isolation of peripheral blood mononuclear cells (PBMCs), heparin-treated blood and the spun EDTA pellet were diluted with phosphate-buffered saline (PBS) and carefully layered onto a Histopaque cushion within Accuspin tubes (Sigma). The tubes were centrifuged at approximately 800g at room temperature for 15 minutes and the resulting buffy coat was collected. Cells were washed once in R10 (RPMI media [Gibco] supplemented with 10% fetal bovine serum [FBS], 100 U/mL penicillin, 100 g/mL streptomycin solution, and 1% L-glutamine) and treated briefly with ammonium-chloride-potassium (ACK) lysing buffer (Gibco) to remove any contaminating erythrocytes. PBMCs were then centrifuged at approximately 250g for 10 minutes to eliminate residual platelets, washed twice with R10 media, and enumerated with a TC20 Automated Cell Counter (Bio-Rad). Cells were cryopreserved in 10% dimethyl sulfoxide (DMSO) in FBS. At least 3 million PBMC were inactivated with Trizol LS buffer (Thermo Fisher Scientific) for isolation of RNA and subsequent RNAseq analyses.
Detection of Viremia
RNA was isolated from whole blood utilizing the Viral RNA mini-kit (Qiagen) using 100 µL of blood into 600 µL of viral lysis buffer AVL (Qiagen). Primers/probe targeting the GP gene of BDBV were used for quantitative real-time PCR (qRT-PCR) with the probe used here being 6-carboxyfluorescein (6FAM)-59 AGGCTTCCCTCGCTGCCGTTATG 39-6 carboxytetramethylrhodamine (TAMRA) (Life Technologies). Assays were run using the CFX96 detection system (BioRad Laboratories) in One-Step probe qRT-PCR kits (Qiagen). The number of viral copies in a sample was calculated using a genome equivalent (GEq) standard. To create the GEq standard, RNA from our BDBV viral stock was extracted, and the number of strain-specific genomes was calculated using Avogadro's number and the molecular weight of the viral genome.
Determination of infectious virus titers was performed by plaque assay with Vero E6 cells from all serum samples. Briefly, increasing 10-fold dilutions of the samples were adsorbed to Vero E6 monolayers in duplicate wells (200 mL); the limit of detection for this assay is 25 PFU/mL.
Hematology and Serum Biochemistry Analysis
EDTA-treated blood was analyzed using a VetScan HM5 hematology analyzer (Abaxis) to determine total white blood cell counts, white blood cell differentials, red blood cell counts, platelet counts, hematocrit values, mean cell volumes, total hemoglobin concentrations, mean corpuscular volumes, and mean corpuscular hemoglobin concentrations. A Piccolo point-of-care analyzer and biochemistry panel plus analyzer discs (Abaxis) were used to test for serum concentrations of albumin, amylase, alanine aminotransferase, alkaline phosphatase, γ-glutamyltransferase, aspartate aminotransferase, glucose, cholesterol, total protein, blood urea nitrogen, creatinine, uric acid, and C-reactive protein.
Histopathology and Immunohistochemistry
Necropsy was performed on all subjects in the BSL-4 facility. Tissue samples for histopathologic and immunohistochemical (IHC) examination were immersed in 10% neutral buffered formalin for at least 21 days, followed by a change of formalin, before removal from the BSL-4 laboratory. Inactivated tissue samples were processed in a BSL-1 laboratory. Tissue sections were deparaffinized and rehydrated through xylene and graded ethanols. Slides went through heat antigen retrieval in a steamer at 95°C for 20 minutes in Sigma citrate buffer, pH 6.0, 10 × (Sigma Aldrich). The tissue sections were processed for IHC using the Thermo Autostainer 360 (ThermoFisher). Specific anti-BDBV immunoreactivity was detected using an anti-BDBV GP primary antibody at a 1:2000 dilution for 60 minutes (IBT BioServices). Secondary antibody used was biotinylated goat anti-rabbit IgG (No. BA-1000; Vector Laboratories) at 1:200 for 30 minutes followed by Vector Streptavidin Alkaline Phosphatase at a dilution of 1:200 for 20 minutes (No. SA-5100; Vector Laboratories). Slides were developed with Bio-Red (No. BP-100-FR; Biopath Laboratories) for 7 minutes and counterstained with hematoxylin for 45 seconds.
Humoral Immune Response
Sera collected at the indicated time points were tested for BDBV GP-specific immunoglobulin M (IgM) and IgG antibodies by enzyme-linked immunosorbent assay (ELISA). MaxiSorp 96-well plates (catalog No. 44–204; Thermo Fisher) were coated overnight with 15 ng/well (0.15 mL) of recombinant BDBV GP lacking the transmembrane region (GPΔTM; Integrated Biotherapeutics) in a sodium carbonate/bicarbonate solution (pH 9.6). Antigen-adsorbed wells were subsequently blocked with 2% bovine serum antigen (BSA) in 1× PBS for at least 2 hours. Irradiated sera samples were initially diluted 1:100 and then 2-fold through 1:6400 in ELISA diluent (2% BSA in 1× PBS and 0.2% Tween 20). After a 1-hour incubation, cells were washed 6 times with wash buffer (1× PBS with 0.2% Tween 20) and incubated for an hour with a 1:5000 dilution of horseradish peroxidase-conjugated anti-monkey IgM or IgG (Fitzgerald Industries International). SigmaFast O-phenylenediamine (OPD) substrate (product No. P9187; Sigma) was added to the wells after 6 additional washes to develop the colorimetric reaction. The reaction was stopped with 3 M sulfuric acid approximately 5 minutes after the addition of OPD, and absorbance values were measured at a wavelength of 492 nm on a spectrophotometer (Cytation 5, BioTek). Absorbance values were normalized by subtracting the values for uncoated wells from the values for antigen-coated wells at the corresponding serum dilution. Average end-point titers were defined as the reciprocal of the last adjusted serum dilution with a value of ≥ 0.36 for IgM and ≥ 0.20 IgG, respectively.
RNA-Sequencing Library Preparation
PBMC RNA was extracted using a Direct-zol RNA Miniprep kit (Zymo Research) according to the vendor’s instructions. An Illumina Ribo-Zero Stranded kit was used to deplete ribosomal RNA (rRNA) and construct cDNA libraries according to the protocol provided by the manufacturer. RNA was fragmented, converted to double-stranded cDNA, and adapters ligated to each strand. The resulting base-pair cDNA fragments were then amplified by PCR. Each library was prepared with a unique indexed adapter for multiplexing. Multiplexed libraries were subjected to paired-end 75 base pair sequencing using the Illumina NextSeq550 PE platform.
Bioinformatic Analyses
The RNA sequencing (RNA-seq) workflow module of Bioconductor's systemPipeR open-source software was used to perform the bioinformatic analysis. Sequencing quality of each sequenced sample was analyzed with fastQC (version 0.11.7). Raw fastq files were trimmed with trimmomatic (version 0.36) with “LEADING” and “TRAILING” set to cut sequences below quality Phred score of 3 from the beginning or end of the read, respectively. Sequences below 15 bp were automatically rejected. The STAR RNA-seq aligner [16] version 2.5.3a was then used to create an index containing the cynomolgus macaque (version 5.0), BDBV, and VSV genomes. Trimmed reads were aligned to these genomes using STAR. The number of reads associated with each transcript was then counted with featureCounts (subread package version 1.6.2). These counts and metadata were then imported into R (version 4.1.2). Genes with multiple possible transcripts were combined to get the number of reads per gene using DESeq2 (version 1.32.0). DESeq2 was used to normalize library sizes and measure differential expression between preinfection and postinfection samples. Volcano plots were produced with ggplot2 (version 3.4.1).
Statistical Analysis
Statistical analysis was carried out in R (version 4.1.2) or GraphPad Prism version 9.3.1. Survival analysis was calculated with a Fisher exact test (2-tailed test). DESeq2 was used to measure differential expression between preinfection and postinfection samples. Adjusted P values (−log10) are reported for transcription data.
RESULTS
Seven cynomolgus macaques were inoculated IM with a 1000 PFU target dose of BDBV. Prior iterations of this model of BDBV in cynomolgus macaques have shown a survival rate of approximately 23% [8, 17]. In this iteration, the single sham-treated animal succumbed to BDBV infection on 11 days postinfection (dpi). Five of the 6 animals that were treated with rVSVΔG/BDBV-GP 20–23 minutes after challenge survived (83%, Figure 1A and Supplementary Table 1); 1 animal succumbed at 8 dpi. This demonstrated that postexposure treatment with the rVSV-based vaccine provided significant (log-rank [Mantel-Cox] test, 2-tailed P value .0272) but not complete protection.
The control animal developed clinical signs typical of BDBV infection. Clinical signs of fever, anorexia, a macular rash, and depression drove the increased clinical scoring criteria (Figure 1B and Supplementary Table 1). These indicators of illness were also seen in the single NHP that received postexposure treatment but succumbed. Minor signs that led to clinical scoring over multiple days were observed in 2 of the 5 NHPs that ultimately survived. These signs resolved by 12 dpi in both survivors. Three animals showed no evidence of unusual distress or classifiable disease.
As expected, the 2 animals that succumbed following BDBV challenge showed levels of detectable viremia, with each showing peak whole blood viral RNA values above 10 log10 copies/mL (Figure 1C). Surprisingly, 2 of the 5 NHPs that received treatment and survived challenge also showed very significant levels of circulating viral RNA, with peaks of 8 and 9 log10 copies/mL. Each of these animals had shown low-grade signs of disease in clinical scoring. Three of the 4 animals that showed viral genome accumulation also showed levels of replication-competent virus in the plasma at titers above 4 log10 PFU/mL (Figure 1D). One animal showed very low levels of replicating virus in serum (< 2 log10 PFU/mL) that appeared to be a marked contrast from the observable 8 log10 copies/mL of viral RNA.
Gross Pathology Findings
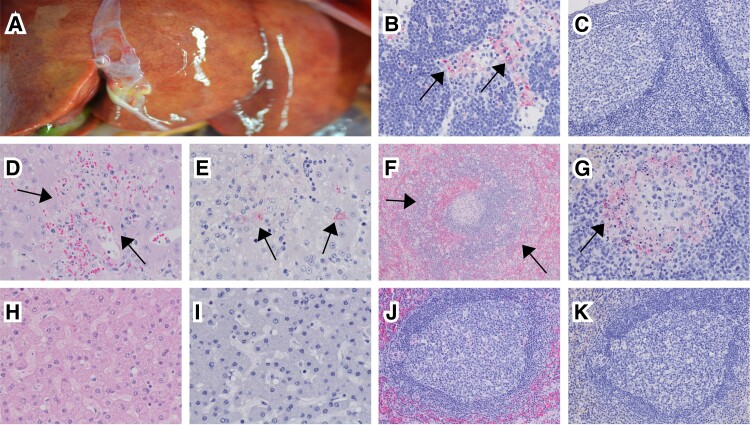
Both animals that succumbed to disease were examined at euthanasia to determine whether they displayed typical signs of BDBV pathogenesis. One or more of the following gross lesions were noted: petechial rash, necrotizing hepatitis (Figure 2A), splenomegaly, lymphadenomegaly, hemorrhagic adrenalitis, ascites, cystitis, and hemorrhagic orchitis. Surviving animals did not show notable pathology except for 1 animal that had minimal unilateral clouding of the eye (oculus sinister) and necrotizing dermatitis of the tip of the tail. This NHP had displayed both strong accumulation of viral RNA in the serum and levels of infectious virus in excess of 5 log10 PFU/mL at 7 dpi.
Figure 2.
Gross pathology and histopathology following BDBV challenge. A, Necrotizing hepatitis in 1 NHP that succumbed. B, BDBV GP-positive mononuclear cells (arrows) within the subcapsular sinus of the lymph node. C, Lack of significant BDBV immunolabeling in the lymph node of a surviving animal. D, Necrotizing hepatitis (arrows) seen in an NHP that succumbed. E, BDBV GP-positive hepatocytes (arrows) and mononuclear cells. F, Splenitis with lymphocytolysis (arrows). G, BDBV GP-positive mononuclear cells within germinal centers (arrow). H–K, Examination of a VSV-BDBV–treated survivor: (H) liver, no appreciable lesions; (I) liver, no appreciable immunolabeling; (J) spleen, no appreciable lesions; (K) spleen, no appreciable immunolabeling. B, D, E, G, H, and I, 40 × magnification. C, J, and K, 20 × magnification. F, 10 × magnification. Abbreviations: BDBV, Bundibugyo virus; GP, glycoprotein; NHP, nonhuman primate.
Multiple tissues representing the major target organs of ebolaviruses (lymphoid tissues, liver, adrenal gland, lung, and pancreas) and immune-privileged sites (brain, urogenital tract, and eyes) were examined histologically. Both macaques that succumbed to disease had similar histologic lesions. In these animals there was an influx of histiocytes and associated necrosis noted in organs that were IHC positive for anti-BDBV GP. This included lymphoid histiocytosis with lymphocytolysis of axillary and inguinal lymph nodes (Figure 2B and 2C), necrotizing hepatitis (Figure 2D and 2E), necrotizing splenitis with lymphocytosis (Figure 2F and 2G), interstitial pneumonia, nephritis, adrenalitis, pancreatitis, cystitis, orchitis, prostatitis, conjunctivitis, and uveitis.
Occasionally, IHC-positive endothelium and/or epithelial cells, such as hepatocytes, renal tubular epithelium, adrenal cortical cells, pancreatic beta cells, and transitional epithelium were noted. NHPs that survived BDBV challenge had mild lymphocytic infiltrates in the kidney and lung. No other histologic lesions were noted and no IHC labeling was appreciated in NHPs that survived (eg, Figure 2H–2K).
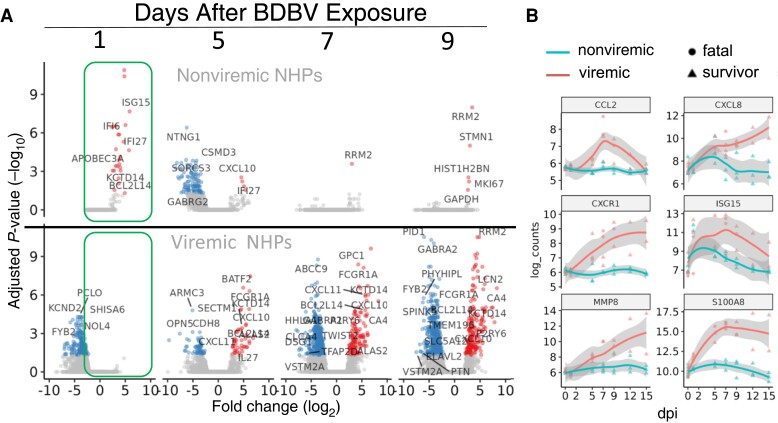
Transcriptomic Signature of Positive Response
Analysis of the circulating immune response showed that there was a clear circulating signal of protection from signs of disease. When compared to their preinfection transcriptional baseline in a volcano plot (Figure 3A, top), animals that showed no clinical signs of infection showed strong and statistically significant induction of interferon-stimulated genes (ISG) at 1 dpi, that subsided by 7 dpi with no other major signs of illness. In contrast, animals that had evidence of BDBV infection at any severity had a broader variation in the observed early induction of interferon-like immune response that was followed by major transcriptional responses consistent with filovirus infection (Figure 3A, bottom). When the ISG response was examined further, it was apparent that at 1 dpi all NHP that had been challenged with rVSVΔG/BDBV-GP had a robust ISG response, while the BDBV-only challenged NHP did not show an ISG response (Supplementary Figure 1).
Figure 3.
Transcriptomic analysis of viremic and nonviremic animals following Bundibugyo virus (BDBV) challenge and postexposure treatment. A, Volcano plot showing mRNA accumulation changes for animals that did not show signs of disease (top) and for those that had viremia (bottom). Boxed section shows responses at 1 dpi. B, Time course plots of selected mRNAs showing differences in expression over the first 15 days of the experimental plan.
The transcriptional differences between the nonviremic and viremic animals were also clear when specific mRNA levels in the blood were tracked over the acute phase of illness. We selected 6 mRNAs that have been identified as being strongly upregulated in response to filovirus infection in earlier publications. These included mRNAs encoding the early responding interferon (IFN)-responsive mRNA ISG15 [18]; the chemokine, CXCR1, and cytokines CCL2 (MCP-1) and CXCL8 (strongly upregulated in subjects with clinical signs); S100A8, (selectively upregulated in BDBV-infected NHPs that succumbed to infection in other studies [7]); and MMP8 (a marker of neutrophils, which has been seen as a late-stage transcriptomic biomarker of severe disease [19]).
Individuals with clinical signs showed increasing expression of S100A8, MMP8, CCL2, and CXCR1 over time, while nonviremic animals did not (Figure 3B). There was significant variation in the timing of the initial appearance of these markers of severe infection (e.g., MMP8 lagged compared to S100A8) as well as expression changes at later times of infection, that correlated with disease severity. Both nonviremic and viremic animals showed ISG15 and CXCL10 responses during infection, with the nonviremic responses more highly upregulated at 1 dpi and only transiently upregulated compared to viremic animals.
Development of Humoral Response
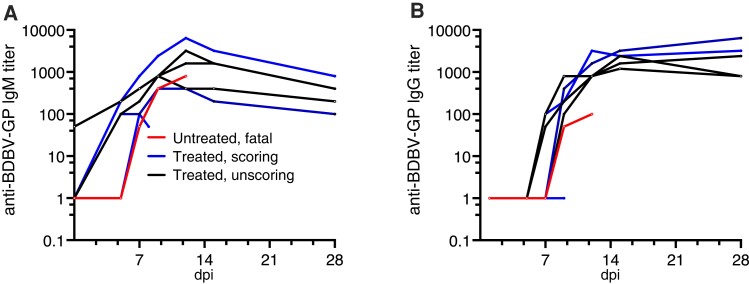
To understand how humoral responses developed in these animals, we performed anti-BDBV GP IgM (Figure 4A) and IgG (Figure 4B) ELISAs on serum samples collected over the course of the experiment. Both animals that succumbed developed low IgM titers only after 5 dpi. Of these 2 animals, only the untreated animal that succumbed showed IgG titers at 9 and 12 dpi. Survivors formed BDBV GP-specific IgM and IgG responses, with IgM appearing as early as day 5. All survivors developed IgG titers against BDBV GP between 5 and 7 dpi. There was no clear difference in either IgM or IgG titers when comparing animals that developed clinical signs and survived versus animals that did not develop clinical signs. The single animal that was treated and succumbed differed from those that survived in not showing substantial production of IgG antibodies.
Figure 4.
Antibody titers in BDBV-infected subjects. A, BDBV glycoprotein-specific IgM in serum samples of all animals. B, BDBV glycoprotein-specific IgG in serum samples of all animals. Abbreviations: BDBV, Bundibugyo virus; dpi, days postinfection; GP, glycoprotein; Ig, immunoglobulin.
DISCUSSION
Here we demonstrate rapid postexposure administration of a rVSV-vectored BDBV vaccine candidate elicited protection against death following BDBV challenge. The exhibition of a protective effect bears similarity to prior examinations of the postexposure efficacy of other rVSV-based filovirus vaccines [20–23]. Administration of rVSVΔG/EBOV-GP at 20–23 minutes postchallenge with EBOV also showed the 3 different responses (no effect, no disease, and intermediate and resolving disease) seen here [23]. A similar approach with Marburg virus (MARV) challenge and rVSVΔG/MARV-GP administration also showed varying penetrance of protection [20]. This is an accordant observation across multiple similar but distinct models that suggests that rVSV-vectored vaccines consistently act to protect against death but do not completely eliminate disease in a postexposure context.
One hypothesis that we formed prior to the experiment was that the longer disease course of BDBV in cynomolgus macaques compared to EBOV, SUDV, and MARV would lead to increased postexposure efficacy because the innate and adaptive immune responses following treatment with rVSVΔG/BDBV-GP would have a longer time to develop. That this was not the case, and the observation of different types of protection afforded by rVSV-vectored vaccines (from significant clinical scoring and viremia to no clinical scoring and the absence of detectable viremia), may indicate that the protection evoked is determined by the strength of response to this postexposure prophylaxis or to the effective induction of downstream immune maturation events more than the timing of its initiation.
An obvious host response to rVSVΔG/BDBV-GP was early and strong induction of ISG. This interferon response was observable 1 dpi faster than is normally observed following filovirus challenge [18, 24]. The response is consistent with a similar ISG response seen following rVSVΔG/EBOV-GP treatment in humans and is likely a response to vaccine vector replication [25]. This response has previously been associated with protection from infection [26], but ISG expression was not a consistent indicator of protection in this model. Failure to develop additional transcriptional signals of infection, including cytokine mRNA expression (Figure 3B), did effectively indicate a lack of detectable infection in this model.
In contrast to the lack of clear transcriptional indicators of outcome in this investigation, the effective development and class-switching of anti-BDBV GP-specific IgG antibodies indeed correlates with outcome, with the 1 treated animal that succumbed to infection never forming substantial BDBV GP-specific IgM titers. This result, although in need of additional support from further testing, is consistent with the idea that the generation of antibody responses is a key determinant of outcome in BDBV infection. This is similar to prior work showing that the strength of antibody development in BDBV-infected rhesus macaques also correlates with outcome [7]. Future work should examine antibody dynamics to VSV antigens (eg, matrix protein) or other BDBV immunogens besides BDBV GP (eg, VP40, nucleoprotein) to discern the importance of specific versus nonspecific immune activation in mediating rVSV postexposure protection.
This study further expands the evidence that rVSV-based vectors can effectively serve as a postexposure prophylaxis agent for many filoviral diseases. Postexposure prophylaxis using rVSV-based vectors provides protection in SUDV, EBOV, and MARV models of disease [23, 27, 28]. This study showed that this can be extended to protection against BDBV exposure if postexposure treatment is rapid.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Courtney Woolsey, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Jamie Strampe, National Emerging Infectious Diseases Laboratories, Boston University, Boston, Massachusetts, USA; Department of Microbiology, Boston University School of Medicine, Boston, Massachusetts, USA.
Karla A Fenton, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Krystle N Agans, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Jasmine Martinez, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Viktoriya Borisevich, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Natalie S Dobias, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Daniel J Deer, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Joan B Geisbert, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Robert W Cross, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
John H Connor, National Emerging Infectious Diseases Laboratories, Boston University, Boston, Massachusetts, USA; Department of Microbiology, Boston University School of Medicine, Boston, Massachusetts, USA.
Thomas W Geisbert, Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, University of Texas Medical Branch, Galveston, Texas, USA.
Notes
Author contributions. C. W., J. H. C., and T. W. G. conceived and designed the study. D. J. D., J. B. G., and T. W. G. performed the challenge experiment. D. J. D., J. B. G., R. W. C., and T. W. G. performed the animal procedures and clinical observations. K. N. A. and V. B. performed the clinical pathology assays. V. B. performed the plaque assays. K. N. A. performed the PCR assays. J. M. performed the ELISAs. K. A. F. performed the necropsies and gross pathology analysis and interpreted the gross and histological data. N. S. D. performed the IHC assays. J. S. performed the bioinformatic analyses. J. H. C. wrote the article with additions from C. W. and K. A. F; and C. W., J. S., K. A. F., R. W. C., J. H. C., and T. W. G. edited the article. All authors had access to the data and approved the final version of the manuscript.
Acknowledgments. We thank the University of Texas Medical Branch Animal Resource Center staff for husbandry support of laboratory animals; Kevin Melody, Chad Mire, and Matt Hyde for assistance with animal observations and scoring; Corri Levine for assistance with blood measurements; Tera Sorvillo for assistance with PBMC separations; Steven Widen for performing the LoFreq analysis and sequencing of cDNA libraries; and Jill Thompson for preparing the cDNA library prep.
Disclaimer. The opinions expressed by the authors contributing to this article do not necessarily reflect the opinions of the University of Texas Medical Branch or the institutions with which the authors are affiliated.
Financial support. This work was supported in part by the Department of Health and Human Services, National Institutes of Health (grant number U19AI142785 to T. W. G.); and in part by the University of Texas Medical Branch (UTMB), Department of Microbiology and Immunology funds to T. W. G. Support for biosafety level 4 operations at the Galveston National Laboratory was provided by National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant number UC7AI094660 to UTMB).
Supplement sponsorship . This article appears as part of the supplement “10th International Symposium on Filoviruses.”
Availability of data and materials. The RNA-seq datasets generated from this study are available in the NCBI Gene Expression Omnibus (GEO) repository [accession number GSE237547]. All other study data are included in the article.
References
- 1. Feldmann H, Sprecher A, Geisbert TW. Ebola. N Engl J Med 2020; 382:1832–42. [DOI] [PubMed] [Google Scholar]
- 2. Jacob ST, Crozier I, Fischer WA II, et al. Ebola virus disease. Nat Rev Dis Primers 2020; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wolf J, Jannat R, Dubey S, et al. Development of pandemic vaccines: eRVEBO case study. Vaccines (Basel) 2021; 9:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Woolsey C, Geisbert TW. Current state of Ebola virus vaccines: a snapshot. PLoS Pathog 2021; 17:e1010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Geisbert TW, Strong JE, Feldmann H. Considerations in the use of nonhuman primate models of Ebola virus and Marburg virus infection. J Infect Dis 2015; 212(Suppl 2):S91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roddy P, Howard N, Van Kerkhove MD, et al. Clinical manifestations and case management of Ebola haemorrhagic fever caused by a newly identified virus strain, Bundibugyo, Uganda, 2007–2008. PLoS One 2012; 7:e52986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woolsey C, Borisevich V, Agans KN, Fenton KA, Cross RW, Geisbert TW. Bundibugyo ebolavirus survival is associated with early activation of adaptive immunity and reduced myeloid-derived suppressor cell signaling. mBio 2021; 12:e0151721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mire CE, Geisbert JB, Marzi A, Agans KN, Feldmann H, Geisbert TW. Vesicular stomatitis virus–based vaccines protect nonhuman primates against Bundibugyo ebolavirus. PLoS Negl Trop Dis 2013; 7:e2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falzarano D, Feldmann F, Grolla A, et al. Single immunization with a monovalent vesicular stomatitis virus–based vaccine protects nonhuman primates against heterologous challenge with Bundibugyo ebolavirus. J Infect Dis 2011; 204(Suppl 3):S1082–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Garbutt M, Liebscher R, Wahl-Jensen V, et al. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol 2004; 78:5458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res 2009; 19:1117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011; 29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilm A, Aw PP, Bertrand D, et al. Lofreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 2012; 40:11189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gilchuk P, Mire CE, Geisbert JB, et al. Efficacy of human monoclonal antibody monotherapy against Bundibugyo virus infection in nonhuman primates. J Infect Dis 2018; 218:S565–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Speranza E, Bixler SL, Altamura LA, et al. A conserved transcriptional response to intranasal Ebola virus exposure in nonhuman primates prior to onset of fever. Sci Transl Med 2018; 10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cilloniz C, Ebihara H, Ni C, et al. Functional genomics reveals the induction of inflammatory response and metalloproteinase gene expression during lethal Ebola virus infection. J Virol 2011; 85:9060–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Woolsey C, Geisbert JB, Matassov D, et al. Postexposure efficacy of recombinant vesicular stomatitis virus vectors against high and low doses of Marburg virus variant Angola in nonhuman primates. J Infect Dis 2018; 218:S582–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cross RW, Mire CE, Feldmann H, Geisbert TW. Post-exposure treatments for Ebola and Marburg virus infections. Nat Rev Drug Discov 2018; 17:413–34. [DOI] [PubMed] [Google Scholar]
- 22. Geisbert TW, Hensley LE, Geisbert JB, et al. Postexposure treatment of Marburg virus infection. Emerg Infect Dis 2010; 16:1119–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feldmann H, Jones SM, Daddario-DiCaprio KM, et al. Effective post-exposure treatment of Ebola infection. PLoS Pathog 2007; 3:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woolsey C, Jankeel A, Matassov D, et al. Immune correlates of postexposure vaccine protection against Marburg virus. Sci Rep 2020; 10:3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santoro F, Donato A, Lucchesi S, et al. Human transcriptomic response to the VSV-vectored Ebola vaccine. Vaccines (Basel) 2021; 9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Menicucci AR, Jankeel A, Feldmann H, Marzi A, Messaoudi I. Antiviral innate responses induced by VSV-EBOV vaccination contribute to rapid protection. mBio 2019; 10. doi: 10.1128/mbio.00597-00519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geisbert TW, Daddario-DiCaprio KM, Williams KJ, et al. Recombinant vesicular stomatitis virus vector mediates postexposure protection against Sudan Ebola hemorrhagic fever in nonhuman primates. J Virol 2008; 82:5664–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daddario-DiCaprio KM, Geisbert TW, Ströher U, et al. Postexposure protection against Marburg haemorrhagic fever with recombinant vesicular stomatitis virus vectors in nonhuman primates: an efficacy assessment. Lancet 2006; 367:1399–404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.