Abstract
Background
Ebolaviruses Ebola (EBOV), Sudan (SUDV), and Bundibugyo (BDBV) cause severe human disease, which may be accompanied by hemorrhagic syndrome, with high case fatality rates. Monovalent vaccines do not offer cross-protection against these viruses whose endemic areas overlap. Therefore, development of a panebolavirus vaccine is a priority. As a vaccine vector, human parainfluenza virus type 3 (HPIV3) has the advantages of needle-free administration and induction of both systemic and local mucosal antibody responses in the respiratory tract.
Methods
To minimize the antivector immunity, genes encoding the HPIV3 envelope proteins F and HN were removed from the vaccine constructs, resulting in expression of only the ebolavirus envelope protein—glycoprotein. These second-generation vaccine constructs were used to develop a combination vaccine against EBOV, SUDV, and BDBV.
Results
A single intranasal vaccination of guinea pigs or ferrets with the trivalent combination vaccine elicited humoral responses to each of the targeted ebolaviruses, including binding and neutralizing antibodies, as well as Fc-mediated effector functions. This vaccine protected animals from death and disease caused by lethal challenges with EBOV, SUDV, or BDBV.
Conclusions
The combination vaccine elicited protection that was comparable to that induced by the monovalent vaccines, thus demonstrating the value of this combination trivalent vaccine.
Keywords: Ebola virus, intranasal vaccination, vaccine
A single intranasal vaccination of guinea pigs or ferrets with the trivalent combination vaccine based on second-generation human parainfluenza virus type 3 vectors protected animals from death and disease caused by lethal dose of Ebola, Sudan, or Bundibugyo virus.
Viruses Ebola (EBOV), Sudan (SUDV), and Bundibugyo (BDBV), which cause a severe human disease, are members of the genus Ebolavirus of the family of Filoviridae. Because the endemic areas of these viruses in Africa overlap, it would be advantageous to have a vaccine that would protect against all pathogenic ebolaviruses. However, while these 3 viruses are related, monovalent vaccines do not offer effective cross-protection [1]. Because of that, the recent SUDV outbreak in Uganda [2] was not expected to be controlled by any of the 2 approved vaccines against EBOV [3]. Therefore, a panebolavirus vaccine is highly desirable. The most feasible way to develop a panebolavirus vaccine is to use a combination approach. Still, several questions relevant for this approach must be answered. Would combination of 3 components of a polyvalent ebolavirus vaccine result in a sufficient immune response to protect against each of the 3 targeted ebolaviruses? Would this combination result in a skew of the immune response toward one targeted ebolavirus at the expense of the response against another virus? Is respiratory tract delivery feasible for combination ebolavirus vaccines?
Human parainfluenza virus type 3 (HPIV3)-vectored vaccines against ebolaviruses are efficacious in small animal models [4–6] and in nonhuman primates [7–9]. The HPIV3 vaccine platform offers several advantages including the elicitation of mucosal humoral and cellular immunity in the respiratory tract, in addition to the systemic immune response, and a respiratory tract delivery eliminating the need for trained medical personnel. However, preexisting immunity against the vector is a concern because HPIV3 is a common pediatric pathogen that infects the respiratory tract. As HPIV3-specific neutralizing antibodies target the 2 HPIV3 envelope proteins expressed at the viral surface, hemagglutinin-neuraminidase (HN), and the fusion protein (F), a second-generation of ebolavirus HPIV3-based vaccines was designed by substituting the original HPIV3 envelope proteins with the EBOV envelope glycoprotein (GP). This modification resulted in (1) high attenuation of the vaccine construct, (2) resistance of the vaccine particles to neutralizing HPIV3-specific antibodies, (3) elimination of the immune response to HPIV3 HN and F, and consequently (4) enhanced targeting of the added EBOV GP antigen [10]. This second-generation HPIV3-vectored EBOV GP vaccine demonstrated protection against lethal EBOV challenge in guinea pigs [6, 10] and nonhuman primates [9].
Here, we investigated if a combination polyvalent strategy would offer protection against each pathogenic ebolavirus. We generated second-generation HPIV3-vectored monovalent vaccines against SUDV and BDBV and demonstrated their protective efficacy in small animal models. We also used them, along with second-generation HPIV3 EBOV vaccine, as a trivalent vaccine against EBOV, SUDV, and BDBV, and tested the combination vaccine for immunogenicity and efficacy against lethal challenge with each of the 3 viruses in small animal models. Our findings demonstrate that a single immunization with the monovalent vaccines or the trivalent combination elicits homologous binding and neutralizing antibodies as well as Fc-dependent functions, and offers robust protection from death and disease caused by a lethal dose of each of the targeted viruses.
METHODS
Generation of the Vaccine Constructs
The vaccine constructs were based on the following GP sequences: EBOV Mayinga (Genbank NC_002549.1), BDBV Uganda (KU182911.1), and SUDV Gulu (MH121163.1). The construction of a full-length DNA clone for the second-generation EBOV vaccine (HPIV3/ΔF-HN/EboGP) was previously described [10]. To generate the second-generation HPIV3-vectored BDBV and SUDV full-length clones (FLC), EBOV GP open reading frame was replaced with that of SUDV and BDBV extracted from previously described FLCs [11]. The inserts were confirmed by sequence analysis. Viruses were recovered by transfection into BSR-T7 cells with a subsequent 48 hours long incubation at 32°C and 3–5 passages in LLC-MK2 cells (5-day incubation at 32°C). To reach viral stock titers sufficiently high for intranasal immunization of ferrets, viral stocks were concentrated with Centricon Plus 70 devices (EMD Millipore). All vaccine constructs were titrated in LLC-MK2 cells.
Characterization of GP Expression by Western Blotting
Monolayers of LLC-MK2 cells in 6-well plates were infected with either wild-type HPIV3 (control), or each of the monovalent vaccines, or HPIV3/ΔF-HN/Trivalent containing all 3 monovalent vaccine constructs at a multiplicity of infection (MOI) of 2 plaque-forming unit (PFU)/cell for 24 hours at 32°C. Then, cells were lysed in RIPA buffer (ThermoFisher Scientific) and run on a 4%–12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in the presence of purified GP proteins as controls (EBOV GP, IBT Bioservices, No. 0501-015; BDBV GP, IBT Bioservices, No. 0505-015; SUDV GP, IBT Bioservices, No. 0502-015). After transfer to nitrocellulose, the western blots were stained with the same antibody combinations as previously described [11]. Western blots were visualized with the LI-COR Odyssey Fc imaging system.
Characterization of GP Expression by Flow Cytometry
Monolayers of LLC-MK2 cells in 6-well plates were infected with HPIV3 (control) or each monovalent vaccine construct at a MOI of 1.0 PFU/cell, or a mixture of the 3 monovalent vaccines each at a MOI of 1.0 PFU/cell, for 48 hours at 32°C. The cells were washed twice with phosphate-buffered saline (PBS) and treated with a Cellstripper solution (Corning) for 15 minutes at 37°C. Suspensions of cells were transferred to fluorescence-activated cell sorting (FACS) tubes, pelleted, and washed with PBS twice. Cells were stained with Live/Dead Fixable Aqua (ThermoFisher Scientific) for 20 minutes at 4°C and washed 2 more times with PBS and with PBS supplemented with 2% fetal bovine serum (FBS). Cells were stained for 30 minutes using 1 of the following antibodies: human mAb EBOV90 specific for EBOV GP, human mAb BDBV52 specific for BDBV GP (both provided by Dr James Crowe, Vanderbilt Medical Center), or mouse mAb specific for SUDV GP (IBT Bioservices, No. 0202-029) at a dilution of 1:200. After staining, the cells were washed with PBS/2% FBS twice, and 1 of the following secondary antibodies was added at 1:200 dilution: goat anti-human immunoglobulin G (IgG) fluorescein isothiocyanate (FITC; Invitrogen, No. A18830) or and goat anti-mouse phycoerythrin (Invitrogen, No. 31861). Cells were incubated for 30 minutes at 4°C, washed twice with PBS/2% FBS and fixed with BD CytoFix solution (BD Biosciences) for 30 minutes at 4°C. Cells were washed with PBS/2% FBS and PBS and analyzed by flow cytometry using a BD FACSymphony (BD Biosciences); 3 × 104 events were collected.
Analysis of Replication Kinetics of the Vaccine Constructs
Monolayers of LLC-MK2 cells in 24-well plates were inoculated with HPIV3, each individual vaccine construct, or the trivalent mixture comprising of equal dose of the individual vaccine constructs at 1.0 or 0.05 PFU/cell. Following absorption for 1 hour at 32°C, monolayers were washed twice with PBS, fresh Opti-MEM medium (ThermoFisher Scientific) with 2% FBS was added, and cells were incubated at 32°C. Combined supernatants and cell samples were harvested at 24, 48, 72, or 96 hours postinoculation and stored at −80°C. Samples were titrated in duplicates, transferred to LLC-MK2 cell monolayers in 48-well plates, and overlayed with MEM/0.5% methylcellulose/2% FBS. The plates were incubated at 32°C for 5 days, and the monolayers were fixed with 80% acetone. Plates were immunostained with primary antibodies for 1 hour at 37°C. The following primary antibodies were used: polyclonal rabbit antibodies specific for EBOV GP (IBT Bioservices, No, 0301-015), human mAb BDBV52 (provided by Dr James Crowe), or mouse mAb specific for SUDV GP (IBT Bioservices, No. 0202-029) at a 1:2000 dilution in PBS with 5% milk for 1 hour at 37°C. Plates were washed 3 times with PBS and incubated with secondary antibodies specific for human, rabbit, or mouse IgG, respectively, conjugated with horseradish peroxidase (KPL) diluted 1:2000 in PBS with 5% milk for 1 hour at 37°C. The viral plaques were visualized with ImmPACT AEC substrate (Vector Laboratories) for 20–30 minutes. Plaques were counted using a microscope.
Vaccination and Ebolavirus Challenge
Eight-week-old HPIV3-naive Dunkin-Hartley female guinea pigs were acquired from Charles Rivers Laboratories (Kingston, NY). For blood collections and vaccine inoculations, animals were anesthetized with 5% isoflurane. On day 0, guinea pigs were inoculated with 4 × 105 PFU of each of the monovalent vaccine constructs or a mixture of the 3 constructs at 4 × 105 PFU/construct resulting in the total dose of 1.2 × 106 PFU in 200 µL PBS intranasally (100 µL per nostril). On days −1 (1 day prior the first vaccine inoculation) and 28, retro-orbital blood collections were performed. On day 33, vaccinated and control animals were exposed to the targeted dose of 103 PFU of guinea pig-adapted EBOV Mayinga or SUDV Boneface delivered by intraperitoneal injection. Animals were monitored at least 1 time per day for weight loss and signs of disease using the following parameters: score of 1 (healthy), score of 2 (ruffled fur, weight loss >10%, hunched), score of 3 (score of 2 plus 1 additional sign: lethargy, orbital tightening, and/or weight loss >15%), and score of 4 (score of 3 plus 1 additional sign: neurologic sign, refusal to move upon stimulation, or weight loss >20%). Guinea pigs with a score of 4 were moribund and were euthanized as per the Institutional Animal Care and Use Committee (IACUC)-approved protocol. Retro-orbital blood collections were performed from surviving animals at days 3, 6, 9, 13, and 28 postchallenge. All remaining guinea pigs were euthanized at 28 days postchallenge.
Twelve-week-old female ferrets were acquired from Marshall BioResources (North Rose, NY). For blood collections and vaccine inoculations, animals were anesthetized with 5% isoflurane. On day 0, ferrets were inoculated with 1 × 107 PFU of each of the monovalent vaccine constructs or a mixture of the 3 constructs at 1 × 107 per construct resulting in the total dose of 3 × 107 PFU in 1.0 mL PBS intranasally (500 µL per nostril). On day −1 (1 day prior to the vaccine inoculation) and on day 28, blood collections were performed. On day 33, vaccinated and control animals were exposed to the targeted dose of 103 PFU of BDBV Uganda by intramuscular injection. Animals were monitored up to 3 times daily for weight loss and signs of disease: score of 1 (healthy), score of 2 (ruffled fur, lethargic, hunched), score of 3 (score of 2 plus 1 additional sign: increased respiratory rate >50 breaths per minute [BPM], petechiae, dark feces or diarrhea, vomiting, dehydration), and score of 4 (score of 3 plus 1 additional sign: reluctance to ambulate, visible ecchymosis, severe dehydration, respiratory rate >80 BPM). Animals with a score of 4 were moribund and were euthanized as per the IACUC-approved protocol. Blood collections were performed from surviving animals on days 3, 6, 9, 13, and 28 postchallenge. All remaining ferrets were euthanized 28 days after BDBV challenge. The control group had 2 ferrets, and 3 control ferrets from a previous study [11] were added as historic controls to reach n = 5. For these historic controls, IgG enzyme-linked immunosorbent assay (ELISA) binding data, virus neutralization data, and challenge data were taken from the original study, which was performed in an identical manner to this study, while the Fc effector assays in the current study were run with sera from the 3 historic control serum samples analyzed in parallel with the current samples.
Enzyme Linked Immunosorbent Assays
Sera collected from animals were tested for their ability to bind GP of the 3 ebolaviruses by ELISA. Detection of GP-specific IgG was performed as previously described [11]. GP-specific IgA response was measured in the same manner except that the following secondary antibodies were used, horseradish peroxidase-conjugated sheep anti-guinea pig IgA (ICL, No. SA-60P-Z) and alkaline phosphatase goat anti-ferret IgA (Sigma, No. SAB3700788-1MG) at dilution 1:500.
Plaque Reduction Assay
Sera collected from animals were tested for virus neutralizing capabilities against EBOV Mayinga, BDBV Uganda, and SUDV Gulu as previously described [11].
Antibody-Mediated Neutrophil Phagocytosis
Recombinant GP proteins were biotinylated and coupled to 1 µm FITC+ NeutrAvidin beads (Life Technologies) as previously described [12].
Antibody-Dependent Cellular Phagocytosis by Human Monocytes
Recombinant GPs were biotinylated and coupled to 1 µm FITC+ NeutrAvidin beads. Serum samples from vaccinated animals were diluted 1:500 in culture medium and incubated with GP-coated beads for 2 hours at 37°C followed by addition of THP-1 human monocytic cells for 18 hours. Cells were fixed with 4% paraformaldehyde and analyzed on a Sartorius iQue flow cytometer, and a minimum of 10 000 events were recorded and analyzed. The phagocytic score was determined as previously [12].
Antibody-Mediated Complement Deposition
Recombinant GPs were biotinylated and coupled to 1 µm red fluorescent NeutrAvidin beads and antibody-mediated complement deposition (ADCD) was measured as previously described [12].
Viremia
Viremia was determined by titrating serum samples obtained after ebolavirus challenge by plaque assay on Vero E6 cells as previously described [11].
Statistics
All statistical calculations were performed using GraphPad Prism version 9.5.1. A P value of <.05 was considered significant.
Biosafety
All work with ebolaviruses was performed in the Galveston National Laboratory biosafety level 4 laboratories. All staff had the appropriate training and US Government permissions and registrations for work with ebolaviruses.
Ethics Statement
This study was carried out in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal work was approved by the University of Texas Medical Branch Institutional Animal Care and Use Committee. All efforts were made to minimize animal suffering and all procedures involving potential pain were performed with the appropriate anesthetic or analgesic. The number of animals used in this study was scientifically justified based on statistical analyses of virological and immunological outcomes.
RESULTS
Development of Second-Generation HPIV3-Vectored Ebolavirus Vaccine Constructs
The second-generation HPIV3-vectored EBOV vaccine was developed previously as follows. The F and HN genes were removed from the HPIV3 FLC. The transcriptional cassette for expression of EBOV GP was inserted in the FLC between the P and M genes resulting in the subsequent recovery of the replication-competent yet highly attenuated vaccine construct HPIV3/ΔF-HN/EboGP [10]. Here, we replaced the open reading frame of EBOV GP with that of SUDV or BDBV in the FLC to generate HPIV3/ΔF-HN/SUDV-GP and HPIV3/ΔF-HN/BDBV-GP vectored vaccine constructs, respectively (Figure 1A). The resulting replication-competent chimeric viruses were recovered as previously described [4]. As expected, the constructs were deficient in F and HN, so that the only glycoprotein expressed on the viral surface was the SUDV or BDBV GP (Figure 1B). Expression of the GP antigens by the individual vaccine constructs and the trivalent combination (HPIV3/ΔF-HN/Trivalent) was assessed in LLC-MK2 cells by western blot (Figure 1C) and flow cytometry (Supplementary Figure 1). Expression of GP of each of the viruses was greater in cells infected with each individual construct than of the same GP the mixture. Next, growth kinetics of the vaccine constructs in LLC-MK2 cells was compared (Supplementary Figure 2). These data demonstrated (1) attenuation of each of the vaccine constructs as compared to the HPIV3 vector; and (2) reduced growth of the EBOV and BDBV vaccine constructs when added alone as compared to the mixture, with the effect detectable at the high MOI only (1 PFU/cell) at a late time (72 hours postinfection). The different behavior of the SUDV vaccine construct is likely related to some unique features of the SUDV GP sequence.
Figure 1.
Vaccine constructs and study design. A, Vaccine candidates were designed by inserting the GP gene of EBOV, SUDV, or BDBV between the P and M genes of HPIV3. B, The F and HN HPIV3 genes were removed so that the filovirus glycoprotein of interest is expressed as the sole transmembrane envelope protein in the vaccine constructs. C, Expression of filovirus GP proteins by cells infected with the monovalent or trivalent vaccine constructs evaluated by western blotting. Purified GP proteins were used as positive controls, and actin or GAPDH were used as loading controls. The experiment was performed independently twice with essentially similar results. D, Guinea pigs and ferrets were vaccinated intranasally on day 0 with the monovalent vaccines or with the trivalent combination, and the control group received the empty HPIV3 vector. The filovirus challenge occurred on day 33, via the intraperitoneal route for the guinea pig model and via the intramuscular route for the ferret model. Abbreviations: BDBV, Bundibugyo virus; EBOV, Ebola virus; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GP, glycoprotein; HPIV3, human parainfluenza virus type 3; SUDV, Sudan virus; WT, wild type.
Individual Vaccines and Their Combination Induce Robust Ebolavirus Binding and Neutralizing Antibody Responses
The vaccines were evaluated in the guinea pig model against EBOV and SUDV and in the ferret model against BDBV (Figure 1D). On day 0, Dunkin-Hartley guinea pigs were vaccinated by a single intranasal dose of the individual EBOV or SUDV monovalent vaccines (4 × 105 PFU/animal), or HPIV3/ΔF-HN/Trivalent (4 × 105 PFU/construct, total vaccine dose of 1.2 × 106 PFU/animal). The control group received 4 × 105 PFU of wild-type HPIV3. On day 33, all guinea pigs were challenged intraperitoneally with lethal doses of 103 PFU of guinea pig-adapted EBOV [13] or SUDV [14]. The 2 monovalent vaccines and HPIV3/ΔF-HN/Trivalent elicited IgG binding to the homologous EBOV GP or SUDV GP (Figure 2A). HPIV3/ΔF-HN/Trivalent elicited EBOV and SUDV GP-binding antibodies at lower levels compared to the respective monovalent vaccines, but the differences were not statistically significant. IgA-specific homologous antibody titers resulted in similar profiles (Figure 2B). Serum samples from the EBOV and SUDV monovalent vaccine-immunized guinea pigs effectively neutralized EBOV Mayinga [15] and SUDV Gulu [16], respectively (Figure 2C). The immune sera from HPIV3/ΔF-HN/Trivalent vaccinated animals also elicited antibodies neutralizing SUDV, but not EBOV or BDBV Uganda [17] (Figure 2C). This distinct neutralization profile may be due to the different antigen expression level in HPIV3/ΔF-HN/Trivalent compared to the monovalent vaccines (Figure 1C).
Figure 2.
Antibody responses to HPIV3/ΔF-HN-EboGP, HPIV3/ΔF-HN/SUDV-GP, and HPIV3/ΔF-HN/Trivalent in guinea pigs. A, Binding IgG evaluated by ELISA. B, Binding IgA evaluated by ELISA. C, Virus-neutralizing antibody titers determined by plaque reduction assay. The GP antigens for ELISA or viruses for neutralization are indicated at the top of the figure, and the vaccines or vector control are indicated at the bottom. The dotted line indicates the level of nonspecific binding in pooled samples collected before vaccination. n = 5 except the HPIV3 group in EBOV and BDBV studies where n = 4. P values were determined by Kruskal-Wallis test followed by Dunn postcomparisons test. Abbreviations: BDBV, Bundibugyo virus; EBOV, Ebola virus; ELISA, enzyme-linked immunosorbent assay; GP, glycoprotein; HPIV3, human parainfluenza virus type 3; Ig, immunoglobulin; SUDV, Sudan virus.
Ferrets were vaccinated on day 0 by a single intranasal dose of the BDBV monovalent vaccine (1 × 107 PFU/animal) or with the combination of EBOV, SUDV, and BDBV individual vaccines in the HPIV3/ΔF-HN/Trivalent (1 × 107 PFU/construct, total vaccine dose of 3 × 107 PFU/ferret) (Figure 1D). The control group received 1 × 107 PFU of wild-type HPIV3. On day 33, ferrets were challenged intramuscularly with a lethal dose of 103 PFU BDBV Uganda. The HPIV3/ΔF-HN/Trivalent elicited IgG binding antibodies to EBOV, SUDV, and BDBV GPs while the monovalent BDBV vaccine induced high IgG binding titers to BDBV GP and low IgG binding antibody titers to EBOV and SUDV GPs (Figure 3A). The trivalent and the BDBV monovalent vaccines also elicited IgA-binding antibodies to SUDV and BDBV GPs but not EBOV GP (Figure 3B). HPIV3/ΔF-HN/Trivalent and the BDBV monovalent vaccines elicited antibodies neutralizing BDBV (Figure 3C). HPIV3/ΔF-HN/Trivalent elicited neutralizing antibodies against EBOV and against SUDV in 3 out of 5 ferrets (Figure 3C). In addition, the BDBV monovalent vaccine elicited EBOV neutralizing antibodies in all vaccinated ferrets (Figure 3C).
Figure 3.
Antibody responses to HPIV3/ΔF-HN/BDBV-GP and HPIV3/ΔF-HN/Trivalent in ferrets. A, Binding IgG evaluated by ELISA. B, Binding IgA evaluated by ELISA. C, Virus-neutralizing antibody titers determined by plaque reduction assay. The GP antigens for ELISA or viruses for neutralization are indicated at the top of the figure, and the vaccines or vector control are indicated at the bottom. Pooled serum samples collected prior to vaccination demonstrated no detectable binding. n = 5 animals per group except for IgA assays where n = 2 for HPIV3. P values were determined by Kruskal-Wallis test followed by Dunn postcomparisons test. Abbreviations: BDBV, Bundibugyo virus; EBOV, Ebola virus; ELISA, enzyme-linked immunosorbent assay; GP, glycoprotein; HPIV3, human parainfluenza virus type 3; Ig, immunoglobulin; SUDV, Sudan virus.
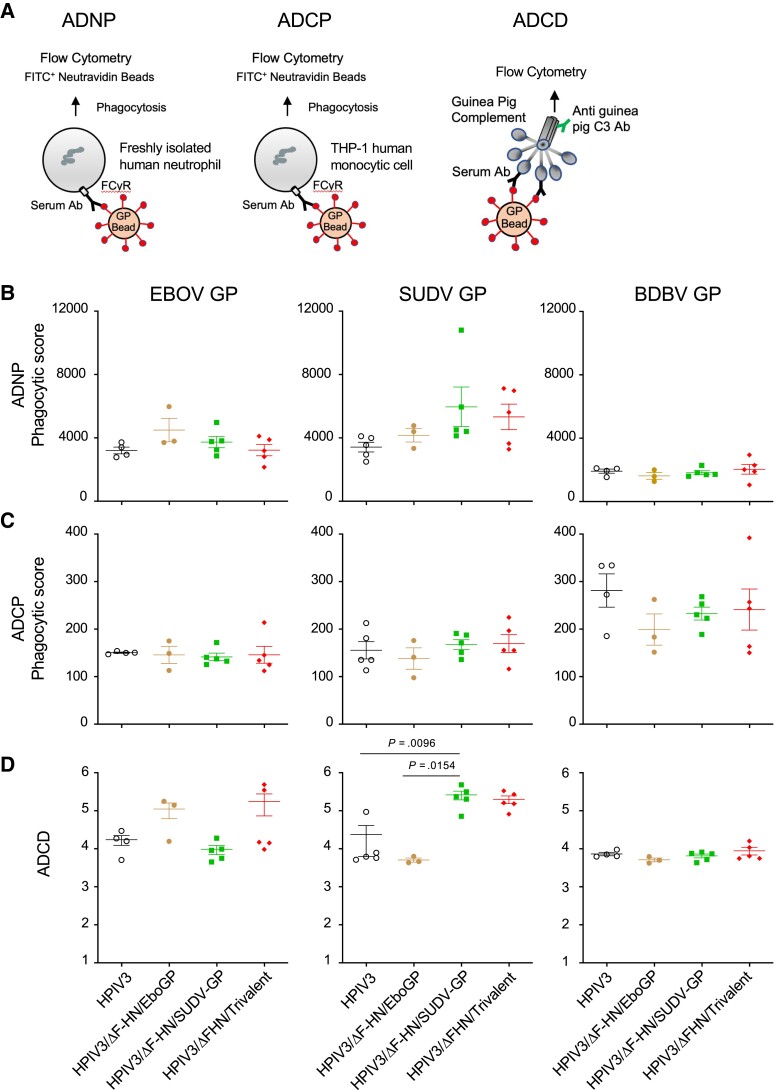
Vaccines Elicit Antibody-Dependent Complement Deposition
Antibody Fc-mediated protective effects can play an essential role in vaccine-induced protection against ebolaviruses [9]. We investigated whether the monovalent and trivalent vaccines induced antibody-dependent neutrophil phagocytosis (ADNP) (Figure 4A and 4B, Figure 5A, and Supplementary Figure 3) and antibody-dependent cellular phagocytosis (ADCP) (Figure 4A and 4C, Figure 5B, and Supplementary Figure 3) in guinea pigs and ferrets. In addition, we examined if the vaccines activated the complement system by assessing C3 deposition (ADCD) in vaccinated animals (Figure 4A and 4D, Figure 5C, and Supplementary Figure 3). For guinea pigs, it has been shown that human antibodies can bind guinea pig Fc-receptors, suggesting structural and functional similarities [18]; however, guinea pig IgG likely only interacts with a subset of human Fc receptors [19]. While ferret IgG subclasses and Fc receptors are not well studied, the IgG structure is conserved between the species with 1 identified IgG subclass in ferrets [20]. Moreover, ferret and human antibodies show similar N-glycan structures, indicating that their Fc-receptor activation can be achieved in a similar fashion [21]. The assays we used have been shown to successfully detect ferret antibody responses against influenza, suggesting cross-species detection [22, 23]. Therefore, measuring the ability of ferret and guinea pig antibodies to elicit Fc-effector functions on human cells can be used as an approximation of activity.
Figure 4.
Fc-mediated responses to HPIV3/ΔF-HN-EboGP, HPIV3/ΔF-HN/SUDV-GP, and HPIV3/ΔF-HN/Trivalent in guinea pigs. A, Layout of the ADNP, ADCP, and ADCD assays. B, ADNP data. C, ADCP data. D, ADCD data. The GP proteins used in the assays are indicated at the top of panel B, and the vaccines or vector control are indicated at the bottom of the figure. n = 5 except for the EBOV challenge study where n = 4 for the HPIV3 group and n = 3 for HPIV3/ΔF-HN-EboGP group. P values were determined by Kruskal-Wallis test followed by Dunn postcomparisons test. The figures show representative data from at least 2 independent donors. The values indicate the mean from 2 technical replicates. Error bars are standard error of the mean (SEM). Abbreviations: Ab, antibody; BDBV, Bundibugyo virus; EBOV, Ebola virus; GP, glycoprotein; HPIV3, human parainfluenza virus type 3; SUDV, Sudan virus.
Figure 5.
Fc-mediated responses to HPIV3/ΔF-HN/BDBV-GP and HPIV3/ΔF-HN/Trivalent in ferrets. A, ADNP data. B, ADCP data. C, ADCD data. The GP proteins used in the assays are indicated at the top of the figure, and the vaccines or vector control are indicated at the bottom. n = 5 per group. P values were determined by Kruskal-Wallis test followed by Dunn postcomparisons test. The figures show representative data from at least 2 independent donors. The values indicate the mean from 2 technical replicates. Error bars are standard error of the mean (SEM). Abbreviations: BDBV, Bundibugyo virus; EBOV, Ebola virus; GP, glycoprotein; HPIV3, human parainfluenza virus type 3; SUDV, Sudan virus.
The monovalent EBOV vaccine and the HPIV3/ΔF-HN/Trivalent did not elicit EBOV GP-specific ADNP, ADCP, or ADCD responses in immunized guinea pigs (Figure 4) or ferrets (HPIV3/ΔF-HN/Trivalent only; Figure 5). While monovalent vaccine-induced ADCP responses were not significant in guinea pigs (Figure 4C), monovalent BDBV vaccine elicited significantly higher ADCP than HPIV3/ΔF-HN/Trivalent in vaccinated ferrets (Figure 5B). Overall, the strongest vaccine-induced Fc-dependent responses were ADCD in both guinea pigs and ferrets (Figure 4D and Figure 5C), as observed with significantly higher homologous GP-specific responses by the monovalent SUDV vaccine in guinea pigs (Figure 4C), as well as the monovalent EBOV vaccine (Figure 4D) and the HPIV3/ΔF-HN/Trivalent vaccine in ferrets (SUDV GP response; Figure 5C). Taken together, these results show that 1 immunization with the trivalent or monovalent vaccines induced detectable levels of ADCD, which was more pronounced in animals vaccinated with the trivalent vaccine, while ADNP and ADCP were not detected in most of the animals.
A Single Dose of the Monovalent and Trivalent Vaccines Protects Animals From Disease and Death Caused by Ebolaviruses
Four weeks after vaccination, guinea pigs were challenged with guinea pig-adapted EBOV Mayinga [13] or SUDV Boneface [14], while ferrets were challenged with wild-type BDBV Uganda [24] (Figure 6). All guinea pigs vaccinated with the EBOV monovalent vaccine and 80% of animals vaccinated with HPIV3/ΔF-HN/Trivalent survived (Figure 6A) while all control animals developed the disease and viremia and 80% of them succumbed. Only 2 animals in the combination group showed signs of disease (Figure 6B) and displayed loss of weight (Figure 6C) but no vaccinated animals had viremia during the entire observation period (Figure 6D). In the SUDV challenge study, all guinea pigs vaccinated with the combination vaccine survived, and 1 animal vaccinated with the SUDV monovalent vaccine did not (Figure 6A), while all control animals developed the disease and viremia and 80% of them succumbed. One animal in each of the monovalent and the combination groups showed signs of disease (Figure 6B) and displayed temporary loss of weight (Figure 6C) but none of the vaccinated animals had viremia (Figure 6D). All ferrets vaccinated with the monovalent BDBV vaccine or the combination survived (Figure 6A). All control animals had viremia and displayed signs of disease (Figure 6B and 6D); they all reach moribund status and were euthanized (Figure 6A). In contrast, none of the BDBV-vaccinated ferrets showed any loss of weight or other signs of the disease during the observation period and none had any viremia (Figure 6B–D). Taken together, our data show that a single dose of the monovalent vaccines and HPIV3/ΔF-HN/Trivalent offer robust protection from death and disease caused by EBOV, SUDV, and BDBV.
Figure 6.
Protection against lethal EBOV, SUDV, or BDBV challenge induced by the homologous vaccines or HPIV3/ΔF-HN/Trivalent in guinea pigs or ferrets. A, Survival. B, Disease scores. C, Changes in body weight. D, Viremia; horizontal line indicates limit of detection. n = 5 animals per group except n = 4 guinea pigs for HPIV3 group in EBOV challenge study. For survival, P values are indicated for comparisons against the respective vector-control group based on log-rank Mantel-Cox statistical analyses. Abbreviations: BDBV, Bundibugyo virus; EBOV, Ebola virus; HPIV3, human parainfluenza virus type 3; PFU, plaque-forming unit; SUDV, Sudan virus.
DISCUSSION
Combination polyvalent vaccine formulation is an effective approach to provide broad protection against ebolaviruses, as we demonstrated with the first-generation HPIV3-vectored EBOV vaccine [11]. However, the first-generation HPIV3-vectored vaccines are partially sensitive to HPIV3-neutralzing antibodies [4, 5], which are present in a significant part of the adult human population [25]. In contrast, the second-generation vaccine vector, which lacks the HPIV3 envelope proteins HN and F, is resistant to HPIV3-specific neutralizing antibodies [10]. We developed second-generation vaccines against SUDV and BDBV and tested them, along with the similar vaccine against EBOV, for protection against EBOV and SUDV in guinea pigs and against BDBV in ferrets. Our data show that a single immunization with the monovalent vaccines or HPIV3/ΔF-HN/Trivalent elicited homologous binding and neutralizing antibodies as well as modest levels of Fc-dependent responses, mostly ADCD. Importantly, monovalent and combination vaccines offered a robust protection from death and disease caused by each of the 3 lethal ebolavirus challenges.
EBOV infection routes include transmission through fomites, biological fluid droplets, and contact with mucosal surface of the respiratory tract [26]. Therefore, a strong local immune response in the respiratory tract would be beneficial. IgA plays an important immunological role in the mucosa, where it can interact with pathogens before they establish a systemic infection. While we did not test IgA responses in the respiratory tract, serum IgA responses to acute respiratory tract infection can be an indirect measure of a mucosal immune response [27, 28]. We previously demonstrated that the first-generation HPIV3-vestored vaccine against EBOV does induce mucosal EBOV-specific IgA response in the respiratory tract of vaccinated nonhuman primates [8]. We also demonstrated that systemic EBOV-specific IgA induced by this vaccine in nonhuman primates is one of the most important correlates of protection [9]. Therefore, the detected IgA antibody response is likely to provide an additional layer of protection against ebolavirus exposure through the respiratory tract.
Immunization with a combination of antigenically related vaccines can skew immune responses and change the epitope hierarchy, as shown with multivalent vaccines against polio or dengue viruses, resulting in an imbalanced response to individual vaccine components [29–32]. Multivalent approaches have been tested with Vesicular Stomatitis Virus (VSV)– or HPIV3-based ebolavirus vaccines and have also shown antigenic bias. A trivalent VSV-vectored vaccine with EBOV, SUDV, and Marburg virus antigens protected macaques from death caused by each of the 3 filoviruses, but animals exposed to SUDV showed signs of disease before recovering [33], likely due to a more dominant response to the EBOV and Marburg virus vaccine components. In our study, mammalian cells infected with the monovalent vaccines expressed higher levels of GP antigens compared to the combination vaccine (Figure 1C and Supplementary Figure 1), similar to what we observed with the first-generation HPIV3 vector expressing EBOV GP [11]. This discrepancy in expression is most likely due to the difference in replication efficiency (observed at high MOI; Supplementary Figure 2), which caused the lower EBOV- and BDBV-specific binding and neutralizing antibodies induced in animals immunized with the vaccine combination compared to their monovalent counterparts. A potential solution to this antigenic imbalance would be to fine-tune the doses of individual components in the combination vaccine. Notably, despite these expression level differences, the vaccine protection efficacy between the monovalent and the combination vaccine was comparable, thus suggesting that the protection was likely mediated both by virus neutralization and ADCD. This is consistent with our previous study with several EBOV vaccines, which demonstrated correlation of survival with ADCD, in addition to some other parameters [9]. In addition, while ADCD can work through lysis via formation of the membrane attack complex, it can also increase phagocytosis via complement receptor CR3 on phagocytes [34]. Furthermore, complement-mediated neutralization of EBOV and SUDV was recently identified as potential mechanism of protection [35]. However, we cannot rule out the role of T-cell immunity in the protection, which was not evaluated in the present study due to the limited availability of reagents specific for immune cells of guinea pigs and ferrets. Despite this limitation, our data show that 1 dose of the combination vaccine offers robust protection against lethal challenge with EBOV, SUDV, and BDBV, an important feature for vaccines to be deployed in emergency situations to fight ebolavirus outbreaks.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Delphine C Malherbe, Department of Pathology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, Galveston, Texas, USA.
J Brian Kimble, Department of Pathology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, Galveston, Texas, USA.
Caroline Atyeo, Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard, Cambridge, Massachusetts, USA.
Stephanie Fischinger, Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard, Cambridge, Massachusetts, USA.
Michelle Meyer, Department of Pathology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, Galveston, Texas, USA.
S Gabrielle Cody, Department of Pathology, University of Texas Medical Branch, Galveston, Texas, USA.
Matthew Hyde, Galveston National Laboratory, Galveston, Texas, USA.
Galit Alter, Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard, Cambridge, Massachusetts, USA.
Alexander Bukreyev, Department of Pathology, University of Texas Medical Branch, Galveston, Texas, USA; Galveston National Laboratory, Galveston, Texas, USA; Department of Microbiology and Immunology, University of Texas Medical Branch, Galveston, Texas, USA.
Notes
Acknowledgments. We are grateful to Dr Peter L. Collins (NIAID, NIH) for providing the HPIV3 reverse genetics system and Ms Keziah Hernandez for work on the figures. We thank the Animal Resources Center of the Galveston National Laboratory for assisting with all animal work; Dr Khaled S. Mohammed for technical assistance with the ELISA assays; Dr Thomas G. Ksiazek (University of Texas Medical Branch) for his gift of polyclonal goat anti-EBOV serum; Dr James E. Crowe (Vanderbilt University) for his gift of monoclonal antibodies BDBV43, BDBV52, and EBOV90; Dr Ryan McNamara for providing the Supplementary Figure 3; and Ms Dora Salinas for editing the manuscript.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant number 1R01AI102887-01A1).
Supplement sponsorship . This article appears as part of the supplement “10th International Symposium on Filoviruses.”
References
- 1. Jones SM, Feldmann H, Ströher U, et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat Med 2005; 11:786–90. [DOI] [PubMed] [Google Scholar]
- 2. Centers for Disease Control and Prevention . Uganda Ebola Outbreak, September 2022.https://www.cdc.gov/vhf/ebola/outbreaks/uganda/2022-sep.html. Accessed 18 October 2022.
- 3. World Health Organization . Safety of two Ebola virus vaccines,2020.https://www.who.int/groups/global-advisory-committee-on-vaccine-safety/topics/ebola-virus-vaccines. Accessed 18 October 2022. [Google Scholar]
- 4. Bukreyev A, Yang L, Zaki SR, et al. A single intranasal inoculation with a paramyxovirus-vectored vaccine protects guinea pigs against a lethal-dose Ebola virus challenge. J Virol 2006; 80:2267–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yang L, Sanchez A, Ward JM, Murphy BR, Collins PL, Bukreyev A. A paramyxovirus-vectored intranasal vaccine against Ebola virus is immunogenic in vector-immune animals. Virology 2008; 377:255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meyer M, Yoshida A, Ramanathan P, et al. Antibody repertoires to the same Ebola vaccine antigen are differentially affected by vaccine vectors. Cell Rep 2018; 24:1816–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bukreyev A, Rollin PE, Tate MK, et al. Successful topical respiratory tract immunization of primates against Ebola virus. J Virol 2007; 81:6379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meyer M, Garron T, Lubaki NM, et al. Aerosolized Ebola vaccine protects primates and elicits lung-resident T cell responses. J Clin Invest 2015; 125:3241–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meyer M, Gunn BM, Malherbe DC, et al. Ebola vaccine-induced protection in nonhuman primates correlates with antibody specificity and Fc-mediated effects. Sci Transl Med 2021; 13:eabg6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bukreyev A, Marzi A, Feldmann F, et al. Chimeric human parainfluenza virus bearing the Ebola virus glycoprotein as the sole surface protein is immunogenic and highly protective against Ebola virus challenge. Virology 2009; 383:348–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kimble JB, Malherbe DC, Meyer M, et al. Antibody-mediated protective mechanisms induced by a trivalent parainfluenza virus-vectored ebolavirus vaccine. J Virol 2019; 93:e01845-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malherbe DC, Domi A, Hauser MJ, et al. A single immunization with a modified vaccinia Ankara vectored vaccine producing Sudan virus-like particles protects from lethal infection. NPJ Vaccines 2022; 7:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Connolly BM, Steele KE, Davis KJ, et al. Pathogenesis of experimental Ebola virus infection in guinea pigs. J Infect Dis 1999; 179:S203–17. [DOI] [PubMed] [Google Scholar]
- 14. Wong G, He S, Wei H, et al. Development and characterization of a guinea pig-adapted Sudan virus. J Virol 2015; 90:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Towner JS, Paragas J, Dover JE, et al. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology 2005; 332:20–7. [DOI] [PubMed] [Google Scholar]
- 16. Sanchez A, Rollin PE. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res 2005; 113:16–25. [DOI] [PubMed] [Google Scholar]
- 17. Towner JS, Sealy TK, Khristova ML, et al. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog 2008; 4:e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mao C, Near R, Gao W. Identification of a guinea pig Fcgamma receptor that exhibits enhanced binding to afucosylated human and mouse IgG. J Infect Dis Med 2017; 1:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gormus BJ, Vessella RL, Martin LN, Kaplan ME. Heterogeneity of human lymphocyte Fc receptors: studies using heat-aggregated and antigen-complexed IgG from human, rabbit, guinea pig, horse and goat. Comp Immunol Microbiol Infect Dis 1982; 5:483–99. [DOI] [PubMed] [Google Scholar]
- 20. Wong J, Layton D, Wheatley AK, Kent SJ. Improving immunological insights into the ferret model of human viral infectious disease. Influenza Other Respir Viruses 2019; 13:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zou G, Kosikova M, Kim SR, et al. Comprehensive analysis of N-glycans in IgG purified from ferrets with or without influenza A virus infection. J Biol Chem 2018; 293:19277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chromikova V, Tan J, Aslam S, et al. Activity of human serum antibodies in an influenza virus hemagglutinin stalk-based ADCC reporter assay correlates with activity in a CD107a degranulation assay. Vaccine 2020; 38:1953–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nachbagauer R, Miller MS, Hai R, et al. Hemagglutinin stalk immunity reduces influenza virus replication and transmission in ferrets. J Virol 2015; 90:3268–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cross RW, Mire CE, Borisevich V, Geisbert JB, Fenton KA, Geisbert TW. The domestic ferret (Mustela putorius furo) as a lethal infection model for 3 species of ebolavirus. J Infect Dis 2016; 214:565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karron RA, Collins PL. In: Knipe DM, Howley PM, eds. Fields virology, Vol. 1. Alphen aan den Rijn (Netherlands): Wolters Kluwer, Lippincott Williams & Wilkins, 2013:996–1023. [Google Scholar]
- 26. Osterholm MT, Moore KA, Kelley NS, et al. Transmission of Ebola viruses: what we know and what we do not know. mBio 2015; 6:e00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown TA, Murphy BR, Radl J, Haaijman JJ, Mestecky J. Subclass distribution and molecular form of immunoglobulin A hemagglutinin antibodies in sera and nasal secretions after experimental secondary infection with influenza A virus in humans. J Clin Microbiol 1985; 22:259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Burlington DB, Clements ML, Meiklejohn G, Phelan M, Murphy BR. Hemagglutinin-specific antibody responses in immunoglobulin G, A, and M isotypes as measured by enzyme-linked immunosorbent assay after primary or secondary infection of humans with influenza A virus. Infect Immun 1983; 41:540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Plotkin SA, Koprowski H, Stokes J Jr. Clinical trials in infants of orally administered attenuated poliomyelitis viruses. Pediatrics 1959; 23:1041–62. [PubMed] [Google Scholar]
- 30. Perkins FT, Yetts R, Gaisford W. Response of 3-months-old infants to 3 doses of trivalent oral poliomyelitis vaccine. Br Med J 1963; 1:1573–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guy B, Barban V, Mantel N, et al. Evaluation of interferences between dengue vaccine serotypes in a monkey model. Am J Trop Med Hyg 2009; 80:302–11. [PubMed] [Google Scholar]
- 32. Anderson KB, Gibbons RV, Edelman R, et al. Interference and facilitation between dengue serotypes in a tetravalent live dengue virus vaccine candidate. J Infect Dis 2011; 204:442–50. [DOI] [PubMed] [Google Scholar]
- 33. Geisbert TW, Geisbert JB, Leung A, et al. Single-injection vaccine protects nonhuman primates against infection with Marburg virus and three species of Ebola virus. J Virol 2009; 83:7296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang ZY, Hunter S, Chien P, et al. Interaction of two phagocytic host defense systems: Fcgamma receptors and complement receptor 3. J Biol Chem 2011; 286:160–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mellors J, Tipton T, Fehling SK, et al. Complement-mediated neutralisation identified in Ebola virus disease survivor plasma: implications for protection and pathogenesis. Front Immunol 2022; 13:857481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.