Abstract
Olavius crassitunicatus is a small symbiont-bearing worm that occurs at high abundance in oxygen-deficient sediments in the East Pacific Ocean. Using comparative 16S rRNA sequence analysis and fluorescence in situ hybridization, we examined the diversity and phylogeny of bacterial symbionts in two geographically distant O. crassitunicatus populations (separated by 385 km) on the Peru margin (water depth, ∼300 m). Five distinct bacterial phylotypes co-occurred in all specimens from both sites: two members of the γ-Proteobacteria (Gamma 1 and 2 symbionts), two members of the δ-Proteobacteria (Delta 1 and 2 symbionts), and one spirochete. A sixth phylotype belonging to the δ-Proteobacteria (Delta 3 symbiont) was found in only one of the two host populations. Three of the O. crassitunicatus bacterial phylotypes are closely related to symbionts of other gutless oligochaete species; the Gamma 1 phylotype is closely related to sulfide-oxidizing symbionts of Olavius algarvensis, Olavius loisae, and Inanidrilus leukodermatus, the Delta 1 phylotype is closely related to sulfate-reducing symbionts of O. algarvensis, and the spirochete is closely related to spirochetal symbionts of O. loisae. In contrast, the Gamma 2 phylotype and the Delta 2 and 3 phylotypes belong to novel lineages that are not related to other bacterial symbionts. Such a phylogenetically diverse yet highly specific and stable association in which multiple bacterial phylotypes coexist within a single host has not been described previously for marine invertebrates.
Gutless oligochaetes were first discovered 25 years ago in sediments of shallow coral reefs around Bermuda (20). As the name suggests, in these worms the digestive system is completely reduced, and the animals have no gut, anus, or nephridia (excretory system), raising the question of how they gain their nutrition. Shortly after the discovery of these worms, Giere (19) was able to show that they are associated with gram-negative bacteria that are hypothesized to provide a source of nutrition via chemosynthesis (16). To date, 80 gutless oligochaete species have been found in a wide array of habitats throughout the world and have been described. High numbers and great diversity have been found in shallow marine waters of tropical and subtropical coral reefs in the Atlantic, Caribbean, and Pacific oceans (10-13). Recently, gutless oligochaetes were also found in coastal areas of the Mediterranean Sea (21, 22). Only a few species occur in deeper (>100 m), colder waters off the Pacific and Atlantic coasts of North America, off the Pacific coast of South America, and in the eastern part of the Gulf of Mexico (13, 14, 17). Despite their ecological diversity, their worldwide distribution, and the high number of species, gutless oligochaetes are monophyletic (i.e., they descended from a single common ancestor) and belong to only two genera, Inanidrilus and Olavius (15, 36).
The morphologies of the symbiosis are very similar in all gutless oligochaetes examined to date. The bacterial symbionts occur just below the outer cuticle of the worm between extensions of the epidermal cells and are, in most cases, extracellular (19, 24). In all host species examined, two bacterial morphotypes have been described; in some host species, an additional third morphotype can also co-occur. One bacterial morphotype with a diameter of 3 to 5 μm has large intracellular inclusions; a second, smaller morphotype with a diameter of 0.5 to 1 μm has no conspicuous features; and a third morphotype, found in two host species, is very long and thin (length, 10 μm; diameter, 0.3 μm) (23, 24).
Comparative 16S rRNA analyses and fluorescence in situ hybridization (FISH) studies of the larger bacterial morphotype in three gutless oligochaete hosts, Inanidrilus leukodermatus (Bermuda), Olavius algarvensis (Mediterranean Sea), and Olavius loisae (Australia), showed that the symbionts belong to the gamma subclass of the Proteobacteria and are closely related to each other (7-9). The thioautotrophic nature of these symbionts is supported by the presence of sulfur in cytoplasmic globules of the bacteria and by immunocytochemical studies that showed the presence of form I ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), the key CO2-fixing enzyme (28). In contrast to the close evolutionary relationship of the large thiotrophic symbionts, the smaller, rod-shaped bacteria are phylogenetically diverse and can belong to the alpha or delta subclass of the Proteobacteria (7, 9). The phylogeny of the long, thin bacterial morphotype found in some hosts (23, 24) has not been studied yet.
Some of the more unusual habitats of gutless oligochaetes are organic matter-rich sediments in water that is 100 to 400 m deep in upwelling regions off the coast of Peru and Chile. Despite extremely low bottom-water oxygen concentrations (<1 μM), the gutless oligochaete Olavius crassitunicatus FINOGENOVA (17) is the dominant member of the infaunal community, occurring at densities as high as 13,500 individuals m−2 (30, 31). Morphological studies have shown that O. crassitunicatus harbors three structurally distinct types of extracellular bacterial symbionts (23). Large oval bacteria (3.5 by 7.3 μm) with intracellular inclusions fill the major part of the subcuticular space, whereas smaller rod-shaped bacteria (0.7 by 1.9 μm) are found in a peripheral position directly under the cuticle. A third long, thin bacterial morphotype (0.3 to 0.4 by 9.1 μm) occurs in the spaces between the large oval bacteria (23).
In this study, comparative 16S rRNA sequence analysis and FISH were used to examine the diversity and phylogeny of symbionts in two geographically separated (by 385 km) O. crassitunicatus populations from the Peru margin. We discovered a phylogenetically diverse symbiont population consisting of five or six co-occurring bacterial phylotypes that appears to be highly specific and stable in both host populations. The term symbiosis is used here in the nonrestrictive sense as defined by de Bary (5), namely, the “living together of differently named organisms.”
MATERIALS AND METHODS
Specimen collection.
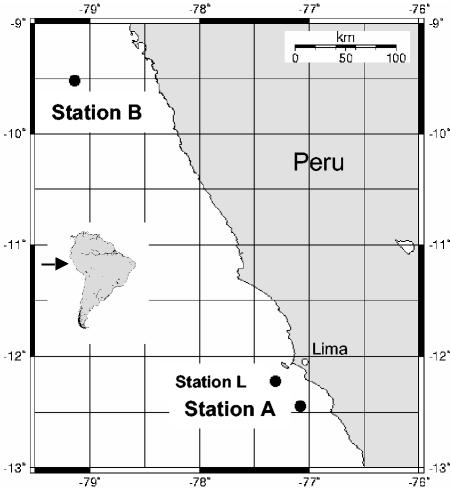
O. crassitunicatus specimens were collected in June 2000 off the Peruvian coast in the Pacific Ocean. Samples were collected from two stations separated by ∼385 km; at station A (12°43.93′S, 77°07.96′W) the water depth was 359 m, and at station B (9°51.52′S, 79° 12.74′W) the water depth was 270 m (Fig. 1). The worms were extracted from the sediment by decantation with seawater and were identified under a microscope. Only active and intact worms were used. Specimens were fixed in 70% ethanol for DNA analyses and for FISH as described previously (8) and were stored at 4°C.
FIG. 1.
Locations of O. crassitunicatus sampling sites on the Peru margin. The worms used in this study were collected at station A at a depth of 359 m and at station B at a depth of 270 m. The two stations are separated from each other by 385 km. Station L is a site sampled by Lisa Levin in 1998 at a depth of 305 m (12°22.7′S, 77°29.1′W); this station was called station A by Levin et al. in reference 30 and station 1 by Levin et al. in reference 31. Specimens from the Levin cruise were used by Giere and Krieger (23) for their ultrastructural studies of O. crassitunicatus.
DNA preparation and PCR amplification.
Three O. crassitunicatus individuals each from stations A and B were prepared individually for PCR. The specimens were rinsed three times in MilliQ water, and DNA was isolated as described by Schizas et al. (40) by a protocol in which proteinase K was used for digestion and the reagent GeneReleaser (BioVentures, Murfreesboro, Tenn.) was used for DNA purification. Amplification was performed with primers specific for the bacterial 16S rRNA gene (primers 8F and 1492R [35]). Template DNA (1 to 2 μl) was added after the PCR mixture (total volume, 100 μl) was heated to 80°C to avoid nonspecific annealing of the primers to nontarget DNA. The following thermocycling conditions were used: one cycle at 80°C for 5 min; 30 cycles at 95°C for 1 min, 40°C for 1 min, and 72°C for 3 min; and one cycle at 72°C for 10 min.
Cloning and sequencing.
PCR products from the six host individuals were cloned separately by using a TA cloning kit (Invitrogen, Breda, The Netherlands) according to the manufacturer's protocol. For screening of 16S rRNA genes, 100 clones per individual were randomly picked and controlled for the correct insert size by PCR with the vector primers M13F and M13R. For template DNA, a small amount of cells from each clone colony was picked with a sterile toothpick and resuspended in 10 μl of sterile water. One to two microliters of this template DNA, after preheating to 95°C for 5 min, was amplified by PCR as described above by using a 30-μl (total volume) mixture. PCR products of the correct size (∼1,500 bp) were screened by partial sequencing of 300 to 500 bp by using the vector primer M13F. Sequencing reactions were performed by using ABI BigDye and an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). Sequences were aligned and compared by using the Bioedit program (www.mbio.ncsu.edu/BioEdit/bioedit.html). Sequences were grouped together in a clone family if they exhibited ≥99% sequence identity (percentage of identical nucleotides). For each host individual, a representative clone from each clone family was prepared by using a QIAprep plasmid kit (QIAGEN, Hilden, Germany) and was nearly fully sequenced in both directions (1,498 bp).
Phylogenetic analysis.
The 16S rRNA symbiont sequences from O. crassituncatus were checked against sequences in the GenBank database by using BLAST (1) for similarity searches. The 16S rRNA sequence data were analyzed by using the ARB software package (www.arb-home.de). Phylogenetic trees were calculated by performing parsimony, distance, and maximum-likelihood analyses with different sets of filters. For tree reconstruction only sequences with more than 1,400 bp were used.
FISH.
Three O. crassitunicatus specimens each from stations A and B were prepared for FISH analysis of bacterial endosymbionts as described previously (8), with the following modifications: in the prehybridization treatments, the tissue sections were incubated for 6 min instead of 10 min in xylene, ethanol, HCl, and proteinase K, and the postfixation step in 4% formaldehyde was omitted. FISH with monolabeled fluorescent oligonucleotide probes was used for the bacterial symbionts belonging to the δ-Proteobacteria by following the general protocol for FISH described by Pernthaler et al. (38). Due to weak signals of the symbionts belonging to the γ-Proteobacteria and Spirochaeta, an enhanced FISH detection method, catalyzed reporter deposition (CARD) FISH with horseradish peroxidase (HRP)-labeled probes and tyramide signal amplification, was used, as described by Schönhuber et al. (41). Briefly, after pretreatment as described above, tissue sections were prehybridized in hybridization buffer (41) for 20 min at 35°C, and this was followed by hybridization with the HRP-labeled probe for 3 h at 35°C. After the sections were washed for 15 min at 35°C in washing buffer (41), they were equilibrated for 15 min at room temperature in 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate; pH 7). The moist tissue sections were incubated with the amplification solution (1× phosphate-buffered saline [pH 7.3], 0.0015% [vol/vol] H2O2, 1% Alexa Fluor 350, 488, or 546 dye [Molecular Probes, Leiden, The Netherlands]) for 20 min at 37°C in the dark and rinsed in 1× SSC for 15 min at room temperature. After air drying, tissue sections were embedded in the mounting fluid Vecta Shield (Vecta Laboratories, Burlingame, Calif.) and stored for microscopic evaluation at −20°C for <1 to 2 days. For dual and triple hybridizations, the CARD FISH protocol was repeated two or three times with the same sections by using different probes and Alexa dyes. To do this, after the last washing step the tissue sections were covered with 0.01 M HCI for 10 min at room temperature to inactivate the HRP. After the tissue sections were washed for 3 min in sterile water, they were hybridized with another probe as described above.
The oligonucleotide probes designed in this study target 16S rRNA sequences of bacterial symbionts of O. crassitunicatus (Table 1). The probes were checked against sequences in the GenBank database by using BLAST (1) and against small-subunit rRNA sequences in the Ribosomal Database Project by using CHECK-PROBE (37). The specificity of the probes designed for the γ-proteobacterial and δ-proteobacterial symbionts was tested with symbionts from another gutless oligochaete, Olavius ilvae (21), that have one or two mismatches in their 16S rRNA compared to the specific probes for O. crassitunicatus (39). The OcraGAM1, OcraDEL3, and OcraDEL2 probes failed to hybridize to the O. ilvae symbionts even in the absence of formamide, indicating the specificity of these probes at low stringencies. The signals from the OcraDEL1 and OcraGAM2 probes were not visible with 15 and 50% formamide, respectively, while the probe signals were still strong at the same formamide concentrations in hybridizations with O. crassitunicatus symbionts. The specificity of the OcraSPI probe was tested with the reference strain Spirochaeta stenostrepta DSM2028 (one mismatch in the probe target region), and the results showed that there was no signal in the absence of formamide (but there was a strong signal with probe EUB338), indicating the high level of specificity of the OcraSPI probe.
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′-3′) | Positiona | Formamide concn (%, vol/vol)c
|
Reference | |
|---|---|---|---|---|---|---|
| FISH | CARD FISH | |||||
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 338-355 | 35 | 55 | 2 |
| GAM42a | γ-Proteobacteria | GCCTTCCCACATCGTTT | 1027-1043b | 35 | 55 | 33 |
| NON338 | Negative control | ACTCCTACGGGAGGCAGC | 338-355 | 10 | 30 | 43 |
| DSS658 | Desulfosarcina spp., Desulfofaba spp., Desulfococcus spp., Desulfofrigus spp. | TCCACTTCCCTCTCCCAT | 658-685 | 60 | 70 | 34 |
| DSR651 | Desulforhopalus spp. | CCCCCTCCAGTACTCAAG | 651-668 | 35 | 70 | 34 |
| OcraGAM1 | Gamma 1 symbionts | GCATACAGACAGAGGCCC | 200-215 | 40 | This study | |
| OcraGAM2 | Gamma 2 symbionts | CTGCGCTCCCAAAGGCAC | 1024-1041 | 55 | This study | |
| OcraDEL1 | Delta 1 symbionts | CGTCAGCACCTGGTGATA | 467-484 | 20 | This study | |
| OcraDEL2 | Delta 2 symbionts | CATGCAGATTCTTCCCAC | 443-471 | 20 | This study | |
| OcraDEL3 | Delta 3 symbionts | TTTCATAGAGCTTCCCGG | 999-1016 | 20 | This study | |
| OcraSPI | Spirochete symbionts | GCTATCCCCAACCAAAAG | 136-153 | 40 | 39 | |
Position in the 16S rRNA of Escherichia coli unless indicated otherwise.
Position in the 23S rRNA of E. coli.
Formamide concentration in hybridization buffer.
The general Bacteria probe EUB338, the general γ-Proteobacteria probe GAM42a, and the δ-Proteobacteria probes DSS658 and DSR651 were used as positive controls, and the nonsense probe NON338 was used as a negative control. All hybridizations were performed with formamide concentrations that ensured high specificity (Table 1).
Nucleotide sequence accession numbers.
The symbiont 16S rRNA sequences from O. crassitunicatus have been deposited in the GenBank database under accession numbers AJ620507 (Gamma 1 symbiont), AJ620508 (Gamma 2 symbiont), AJ620509 (Delta 1 symbiont), AJ620510 (Delta 2 symbiont), AJ620511 (Delta 3 symbiont), and AJ620512 (spirochete symbiont).
RESULTS
Clone library analysis.
Bacterial 16S rRNA sequences from three O. crassitunicatus specimens each from stations A and B were grouped into six distinct clone families; a total of 573 clones were analyzed (Table 2). Within each clone family, the level of sequence identity was never less than 99.7% (percentage of identical nucleotides). Phylogenetic analyses (see below) revealed that two clone families belong to the γ-Proteobacteria (Gamma 1 and 2), three clone families belong to the δ-Proteobacteria (Delta 1, 2, and 3), and one clone family belongs to the spirochetes.
TABLE 2.
16S rRNA clone libraries from six O. crassitunicatus individuals
| Worm no. | No. of clones (% of total)
|
||||||
|---|---|---|---|---|---|---|---|
| Total | Gamma 1 | Gamma 2 | Delta 1 | Delta 2 | Delta 3 | Spirochaeta | |
| Station A | |||||||
| 1 | 101 | 12 (12) | 25 (25) | 18 (18) | 9 (9) | 33 (33) | 4 (4) |
| 2 | 90 | 18 (20) | 27 (30) | 10 (11) | 14 (16) | 20 (22) | 1 (1) |
| 3 | 90 | 25 (28) | 25 (28) | 19 (21) | 0 (0) | 18 (20) | 3 (3) |
| Station B | |||||||
| 1 | 109 | 3 (3) | 70 (64) | 36 (33) | 0 (0) | 0 (0) | 0 (0) |
| 2 | 77 | 9 (12) | 38 (49) | 21 (27) | 7 (9) | 0 (0) | 2 (3) |
| 3 | 106 | 13 (12) | 20 (19) | 32 (30) | 37 (35) | 0 (0) | 4 (4) |
Sequences belonging to the Gamma 1 and 2 clone families were found in all hosts from stations A and B. For the Delta 1 and 2 clone families, as well as the spirochetes, at least two of three O. crassitunicatus individuals from both stations harbored sequences belonging to these groups. In contrast, sequences belonging to the Delta 3 clone family were found only in individuals from station A.
Phylogenetic analysis.
Parsimony, distance, and maximum-likelihood analyses of the 16S rRNA sequences from the six O. crassitunicatus clone families confirmed that these sequences belonged to two phylogenetically distinct groups of the γ-Proteobacteria, three phylogenetically distinct groups of the δ-Proteobacteria, and one phylogenetically distinct group of the spirochetes. The six O. crassitunicatus sequences are unique to this host and differ from the sequences of symbionts from other host species or free-living bacteria.
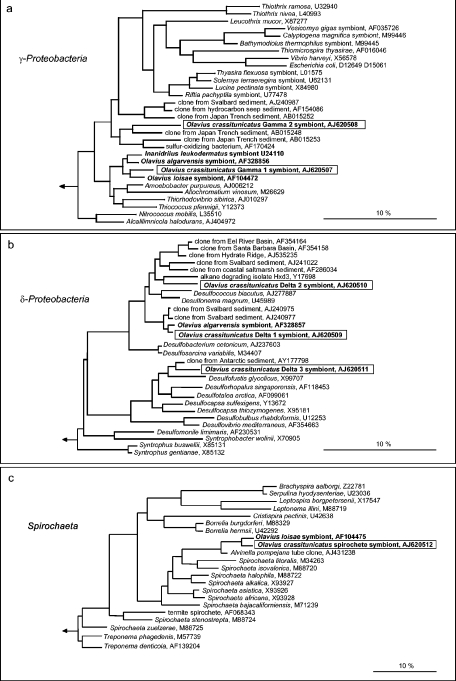
In all phylogenetic analyses, the O. crassitunicatus Gamma 1 sequence consistently fell in a cluster with the sequences of endosymbionts from other gutless oligochaetes, such as Inanidrilus leukodermatus, O. loisae, and O. algarvensis (≥95.6% sequence identity) (Fig. 2a). The closest relatives of this cluster of oligochaete symbionts are a clade of free-living bacteria belonging to the family Chromatiaceae. The second γ-proteobacterial sequence found in O. crassitunicatus, Gamma 2, is not closely related to those of other symbiotic γ-Proteobacteria (Fig. 2a). This sequence was consistently grouped with a clone sequence obtained from deep-sea sediments in the Japan Trench (90.3% sequence identity) by all inference methods. The relationship of this sequence to other 16S rRNA sequences varied depending on the treeing method used. Maximum-likelihood and distance analyses identified a clade containing a sulfur-oxidizing bacterium isolated from a shallow water hydrothermal vent in the Mediterranean Sea (89.8% sequence identity) and a clone sequence from deep-sea sediments in the Japan Trench (89.4% sequence identity) as the closest relatives. In parsimony analyses, the sister group of the Gamma 2 sequence is a cluster of clone sequences isolated from permanently cold sediments off the coast of Svalbard, Japan Trench sediments, and hydrocarbon seep sediments (≥86.2% sequence identity).
FIG.2.
Phylogenetic placement of bacterial symbionts in O. crassitunicatus based on 16S rRNA sequences: maximum-likelihood trees of members of the γ-Proteobacteria (a), δ-Proteobacteria (b), and Spirochaeta (c). Symbionts of gutless oligochaetes are indicated by boldface type, and the O. crassitunicatus symbionts are enclosed in boxes. Bar = 10% estimated sequence divergence.
The three δ-proteobacterial sequences from O. crassitunicatus, Delta 1, 2, and 3, are phylogenetically distinct from each other (Fig. 2b). The Delta 1 sequence is most closely related to the sequence of the sulfate-reducing endosymbiont of the gutless oligochaete O. algarvensis as determined by all treeing methods (98.6% sequence identity). The neighboring clade of these two sequences includes clone sequences isolated from permanently cold sediments off the coast of Svalbard (97.8 to 97.6% sequence identity).
The O. crassitunicatus Delta 2 and 3 sequences are not closely related to other δ-proteobacterial symbiont sequences. The closest relative of the Delta 2 sequence as determined by all inference methods is the sequence of the alkane-degrading bacterial strain Hxd3 isolated from an oil tank (90.9% sequence identity). The neighboring branches of these two sequences were consistently identified as a cluster of clone sequences from sediments obtained above gas hydrates in Eel River, the Santa Barbara Basin, and Hydrate Ridge and a cluster of clone sequences from sediments obtained off the coast of Svalbard and coastal salt marsh sediments (91.2 to 89.9% sequence identity) as determined by all treeing methods. The Delta 3 sequence is most closely related to a clone sequence isolated from Antarctic sediments (94.7% sequence identity) and the sequence of the free-living sulfate-reducing bacterium Desulfofustis glycolicus (92.4% sequence identity).
The O. crassitunicatus spirochete sequence was consistently grouped with the sequence of the spirochete endosymbiont of the gutless oligochaete O. loisae from the Australian Great Barrier Reef in all three phylogenetic analyses (95.4% sequence identity) (Fig. 2c). Peripherally associated with these two sequences is a clone sequence obtained from tubes of the hydrothermal vent polychaete Alvinella pompejana (92.9% sequence identity).
In situ identification.
FISH with oligonucleotide probes confirmed that the six 16S rRNA sequences isolated from O. crassitunicatus originated from bacteria in the symbiont-containing region between the cuticle and the epidermis of the worm (Fig. 3). FISH studies showed that the two γ-proteobacterial symbionts (Gamma 1 and 2), two of the δ-proteobacterial symbionts (Delta 1 and 2), and the spirochete co-occurred in all three specimens from stations A and B. The third δ-proteobacterial symbiont, Delta 3, was found only in the three specimens from station A.
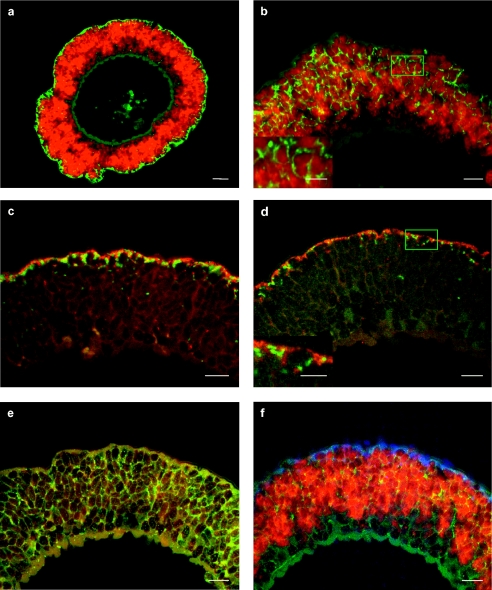
FIG. 3.
In situ identification of bacterial symbionts in O. crassitunicatus. The epifluorescence images show cross sections through the entire worm (a) (scale bar, 20 μm) and the symbiont-containing region of the worm's body wall (b to f) (scale bars, 10 μm). All worms shown were from station A; the only exception is the worm shown in panel c, which was from station B. (a) Dual hybridization with the GAM42a and DSS658/DSR651 probes, showing γ-proteobacterial symbionts (red) and δ-proteobacterial symbionts (green). (b) Dual hybridization with the OcraGAM1 and OcraGAM2 probes, showing the Gamma 1 symbionts (red) and Gamma 2 symbionts (green). The inset shows an enlargement of the area enclosed by a box (scale bar, 5 μm). (c) Dual hybridization with the OcraDEL1 and OcraDEL2 probes, showing the Delta 1 symbionts (green) and Delta 2 symbionts (red). (d) Dual hybridization with the OcraDEL1/OcraDEL2 and OcraDEL3 probes, showing the Delta 1 and Delta 2 symbionts (red)and the Delta 3 symbionts (green). The inset shows an enlargement of the area enclosed by a box area (scale bar, 5 μm). (e) Monohybridization with the OcraSPI probe, showing spirochete symbionts (green). (f) Triple hybridization with the GAM42a, DSS658/DSR651, and OcraSPI probes, showing the two γ-proteobacterial symbionts (red), the three δ-proteobacterial symbionts (blue), and the spirochete symbionts (yellow). The muscle tissue of the worm at the bottom of the panel appears to be bluish green because of autofluorescence at mixed wavelengths.
The general probe for γ-Proteobacteria, GAM42a, as well as the specific probes OcraGAM1 and OcraGAM2 for the Gamma 1 and 2 symbionts, respectively, hybridized to bacteria throughout the symbiont-containing region (Fig. 3a and b). The hybridization patterns of the two specific probes were distinctly different. The signal from the OcraGAM1 probe was consistent in size, shape, and distribution with the bacteria described as the large morphotype in O. crassitunicatus by Giere and Krieger (23) (Fig. 3b). The OcraGAM2 probe hybridized to much smaller bacteria that were approximately 1 μm long and located in the spaces between the large morphotypes (Fig. 3b).
The hybridization signal from the δ-proteobacterial probes DSS658 and DSR651 was limited to small bacteria in the peripheral area of the symbiont-containing region directly beneath the cuticle (Fig. 3a and f). These general probes are used for identification of δ-Proteobacteria belonging to the genera Desulfosarcina, Desulfofaba, Desulfococcus, Desulfofrigus, and Desulforhopalus and also targeted the δ-proteobacterial sequences Delta 1 to 3 isolated from O. crassitunicatus. A similar hybridization pattern was observed with the specific probes for the three δ-proteobacterial sequences, OcraDEL1, OcraDEL2, and OcraDEL3, confirming that these sequences originated from δ-proteobacterial symbionts beneath the cuticle of the worm. Dual hybridizations with the OcraDEL probes showed that the Delta 1 and 2 symbionts occurred in approximately equal numbers in specimens from both station A and station B (Fig. 3c), while the Delta 3 symbionts were much rarer and were found only in specimens from station A (Fig. 3d).
The specific probe for the spirochete sequence isolated from O. crassitunicatus, OcraSPI, hybridized to long, thin bacteria located between the large Gamma 1 symbionts (Fig. 3e and f). The hybridization signal from this probe was consistent with the shape and distribution pattern of the long, filiform bacterial morphotype described by Giere and Krieger (23) in O. crassitunicatus.
DISCUSSION
This study revealed the coexistence of five to six distinct bacterial phylotypes in the body wall of the gutless marine oligochaete O. crassitunicatus. By using comparative 16S rRNA sequence analysis and fluorescence in situ hybridization, two symbionts belonging to the γ-Proteobacteria, three symbionts belonging to the δ-Proteobacteria, and one spirochete symbiont were identified in this host species. Despite this high diversity, the association between the bacterial endosymbionts and O. crassitunicatus is clearly specific within a given host population, as all individuals from stations A and B harbored the same five or six phylotypes. Furthermore, the association appears to be highly specific and stable in geographically distant host populations, given the co-occurrence of the same five bacterial phylotypes in worms separated from each other by 385 km.
Such great diversity of multiple endosymbiont species has not been observed previously in oligochaete symbioses. In previous studies, either a single phylotype (8) or two or three co-occurring bacterial phylotypes were described for each host species (7, 9). The ability to sequence a much larger number of clones in this study (573 clones versus 60 to 70 clones or direct sequencing in previous studies), in addition to improved FISH techniques, such as CARD FISH, is the most likely explanation for the discovery of this previously unrecognized diversity. Indeed, recent studies have suggested that there is similar diversity in other gutless oligochaete species (Blazejak and Dubilier, unpublished data), including species previously assumed to harbor only one to three bacterial phylotypes (7, 8, 9).
The FISH analyses in this study of the Gamma 1 bacteria in O. crassitunicatus indicate that this symbiont is the same as the large, oval bacterial morphotype found by Giere and Krieger (23) in another O. crassitunicatus population from Peru sediments (station L in Fig. 1). In both the specimens of Giere and Krieger (23) and the worms studied here, these bacteria were unusually large (diameter, 7 to 10 μm), in contrast to other gutless oligochaetes, in which this morphotype is at most only one-half as big (diameter, 3 to 5 μm) (24). The thioautotrophic (i.e., sulfide-oxidizing, CO2-fixing) nature of this symbiont was confirmed by Giere and Krieger (23), who used immunocytochemistry to show the presence of the CO2-fixing enzyme RubisCO and spectroscopy to identify sulfur in intracellular deposits. The close phylogenetic relationship of the O. crassitunicatus Gamma 1 symbiont to chemoautotrophic symbionts of other gutless oligochaetes and the monophyly of this group as determined by all treeing methods are further indications that the O. crassitunicatus Gamma 1 bacteria are autotrophic, sulfide-oxidizing symbionts.
The occurrence of a second γ-proteobacterial symbiont, like Gamma 2 in O. crassitunicatus, has not been described previously for other oligochaete species, which so far have been found to harbor only a single Gamma 1-like symbiont (7-9). Endosymbiotic associations with more than one γ-proteobacterial phylotype have been observed in the wood-boring mussel Lyrodus pedicellatus (6) and in the cold-seep thyasirid clam Maorithyas hadalis (18). The function of these multiple symbionts in these bivalves is unclear, just as the metabolism of the novel O. crassitunicatus Gamma 2 phylotype remains to be determined. The Gamma 2 symbiont belongs to a phylogenetic group that includes clone sequences from cold-seep communities in the Japan Trench (32), as well as a chemoautotrophic, sulfur-oxidizing isolate obtained from a shallow hydrothermal vent in the Mediterranean Sea (42), suggesting that the Gamma 2 symbiont might also participate in chemosynthetic pathways. Studies of functional genes involved in this symbiosis, such as genes coding for RubisCO forms I and II and enzymes involved in sulfur metabolism, such as adenosine-5′-phosphosulfate reductase (aprA), as well as in dissimilatory nitrate reduction (nitrite reductase; nirK), are in progress.
Three phylogenetically distinct δ-proteobacterial symbionts were found in O. crassitunicatus. The close evolutionary relationship of these symbionts to free-living and symbiotic sulfate-reducing bacteria and the predominance of this type of metabolism within the δ-Proteobacteria suggest that these symbionts also use sulfate as an electron acceptor. The first chemoautotrophic host known to harbor a δ-proteobacterial symbiont was the gutless oligochaete O. algarvensis obtained from sediments in the Mediterranean Sea. In this species, only a single δ-proteobacterial symbiont was found, and it was identified as a sulfate reducer based on molecular and physiological data (9). In O. crassitunicatus, the 16S rRNA sequence of the Delta 1 symbiont is very closely related to that of the sulfate-reducing symbiont of O. algarvensis (98.6% sequence identity) (9). In contrast, the Delta 2 and 3 symbionts belong to novel lineages not previously known to occur in symbiotic associations.
In the Mediterranean Sea sediments in which O. algarvensis occurs, sulfide is not detectable by smell, and measurements have shown that the sulfide concentrations are very low (<1 μM) (9). This led to the suggestion that the sulfate-reducing symbiont of O. algarvensis could provide the thioautotrophic symbiont of this host with an internal source of sulfide (9). In this study, sulfide concentrations were not measured at the two sampling sites where O. crassitunicatus was obtained, but at station A the sediments clearly smelled of sulfide. At another site off the coast of Peru where O. crassitunicatus was reported to occur in high numbers together with the free-living, sulfide-oxidizing bacteria Thioploca spp. (station L in Fig. 1), sulfide was also detected by smell (30). The presence of sulfide in the habitat of O. crassitunicatus suggests that the role of the sulfate-reducing symbionts in these worms is not restricted to supplying sulfide for the thioautotrophic symbionts. One distinct difference between the O. algarvensis symbiosis and the O. crassitunicatus symbiosis is the distribution of the sulfate reducers. In O. algarvensis, the sulfate-reducing bacteria occur throughout the entire symbiont-containing region and are in close contact with the thioautotrophic symbionts. In contrast, in O. crassitunicatus the sulfate reducers occur almost exclusively in the outer part of the symbiont-containing region, just below the cuticle of the worm, and have little contact with the thioautotrophic symbionts. This suggests that the uptake of substrates such as organic carbon, or hydrogen if the sulfate reducers are autotrophic, from the environment may play an important role in the O. crassitunicatus symbiosis. This could also explain the presence of two or three δ-proteobacterial phylotypes in these hosts. Sulfate-reducing bacteria are known to metabolize a wide variety of electron donors and carbon sources, including low-molecular-weight compounds from the fermentative breakdown of biomolecules, various aromatic compounds (44), and even hydrocarbons (26). Multiple sulfate-reducing symbionts with different preferences for carbon compounds and electron donors could provide nutritional versatility that broadens the spectrum of nutritional sources available to the O. crassitunicatus association.
A symbiotic association between a spirochete and a gutless oligochaete was first suggested to occur in O. loisae from the Australian Great Barrier Reef (7). However, the authors were not able to unambiguously confirm that the spirochete 16S rRNA sequence isolated from O. loisae originated from symbiotic bacteria because the specific probes developed for the spirochete sequence did not hybridize to bacteria in the body wall of O. loisae. In this study a spirochete was successfully identified in the symbiont-containing region of a gutless oligochaete by using CARD FISH and the spirochete-specific probe OcraSPI. The hybridization signal from the spirochete probe was consistent with the shape and distribution pattern of the long, thin bacterial morphotype described in the ultrastructural study of O. crassitunicatus specimens from station L (Fig. 1) (23), indicating that spirochetes consistently occur as symbionts in these hosts.
The spirochete symbionts of O. crassitunicatus and O. loisae are closely related to each other (95.4% sequence identity) and form a monophyletic group as determined by all three treeing methods used. The close relationship of these two symbionts despite the great geographic distance between the two hosts and the differences in their habitats (deep-water slope sediments versus shallow coral reef sediments) indicates that the spirochete symbiosis is integral to the oligochaete hosts and independent of geographic or environmental factors. Indeed, studies of other gutless oligochaetes have shown that spirochetes regularly occur as symbionts in these hosts (39).
The oligochaete spirochetes fall on a neighboring branch with a clone sequence (accession no. AJ431238) isolated from the tubes of the hydrothermal vent polychaete Alvinella pompejana (M. A. Cambon-Bonavita, unpublished data). (Another spirochete sequence that also originated from A. pompejana tubes [accession no. AF180309] and was previously suggested to be most closely related to the O. loisae spirochete [4] does not fall within the oligochaete clade but rather is related to Spirochaeta alkalica based on BLAST and treeing analyses; this sequence was not included in the trees shown here due to its short length, 474 bp.) The free-living marine spirochetes Spirochaeta isovalerica and Spirochaeta litoralis consistently form a neighboring clade of the oligochaete spirochetes. These bacteria were isolated from sulfidic muddy sediments and are obligate anaerobes that ferment carbohydrates mainly to acetate, ethanol, CO2, and H2 (25, 27). While fermentation is one possible metabolic pathway of the oligochaete spirochetes, they could also have a completely different metabolism, just as the spirochete symbionts in termites do not have properties of their closest free-living relatives in the genus Treponema (3). Instead, termite spirochetes were recently found to be chemoautotrophic and to use H2 and CO2 to produce acetate (29). This type of metabolism should clearly be beneficial to the oligochaete hosts, providing them with an additional chemoautotrophic symbiont as a source of carbon and energy.
Of the six different bacterial phylotypes that coexist in O. crassitunicatus, three belong to clades in which only symbionts occur: (i) the Gamma 1 symbionts, (ii) the Delta 1 symbionts, and (iii) the spirochetes. Within these three clades, the host species are separated by large geographic distances (Bermuda, Mediterranean Sea, Australia, and Peru) and come from very different habitats (coral reef sediments, coarse-grained coastal sands, and deep-water silty muds). The symbionts in these three clades, however, are closely related to each other (>95 to 98% sequence identity), indicating that within each clade the symbionts descended from a common ancestor. Three bacterial phylotypes found in O. crassitunicatus belong to novel lineages that have not been found previously in symbiotic associations (Gamma 2, Delta 2, and Delta 3).
All symbiont phylotypes except Delta 3 appear to be highly specific and stable both within a given population and between host populations. However, variation can clearly occur between populations, as seen in the Delta 3 phylotype, which occurred only in hosts from station A. There are two explanations for the intraspecific variation of the Delta 3 symbiont: (i) this symbiont originally occurred in both populations and was lost by hosts at station B, or (ii) this symbiont never occurred in the population at station B, so that the association was established independently by the hosts at station A. Future studies of other O. crassitunicatus populations from the Peru and Chile margin, as well as a better understanding of the acquisition of symbionts, should help answer these and other questions concerning the establishment and evolution of symbioses in oligochaete hosts.
Acknowledgments
We are grateful to Daniela Riechmann for collecting the worms and to Julia Herglotz for technical assistance.
This work was supported by the Max Planck Society, Munich, Germany.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breznak, J. A. 2002. Phylogenetic diversity and physiology of termite gut spirochetes. Intergr. Comp. Biol. 42:313-318. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, B. J., and S. C. Cary. 2001. Characterization of a novel spirochete associated with the hydrothermal vent polychaete annelid, Alvinella pompejana. Appl. Environ. Microbiol. 67:110-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Bary, A. 1879. Die Erscheinung der Symbiose. Verlag von Karl J. Trubner, Strassburg, Germany.
- 6.Distel, D. L., D. J. Beaudoin, and W. Morrill. 2002. Coexistence of multiple proteobacterial endosymbionts in the gills of the wood-boring bivalve Lyrodus pedicellatus (Bivalvia: Teredinidae). Appl. Environ. Microbiol. 68:6292-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubilier, N., R. Amann, C. Erséus, G. Muyzer, S. Y. Park, O. Giere, and C. M. Cavanaugh. 1999. Phylogenetic diversity of bacterial endosymbionts in the gutless marine oligochaete Olavius loisae (Annelida). Mar. Ecol. Prog. Ser. 178:271-280. [Google Scholar]
- 8.Dubilier, N., O. Giere, D. L. Distel, and C. M. Cavanaugh. 1995. Characterization of chemoautotrophic bacterial symbionts in a gutless marine worm (Oligochaeta, Annelida) by phylogenetic 16S rRNA sequence analysis and in situ hybridization. Appl. Environ. Microbiol. 61:2346-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubilier, N., C. Mülders, T. Ferdelman, D. de Beer, A. Pernthaler, M. Klein, M. Wagner, C. Erséus, F. Thiermann, J. Krieger, O. Giere, and R. Amann. 2001. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411:298-302. [DOI] [PubMed] [Google Scholar]
- 10.Erséus, C. 1992. A generic revision of the Phallodrilinae (Oligochaeta, Tubificidae). Zool. Scr. 21:5-48. [Google Scholar]
- 11.Erséus, C. 2003. The gutless Tubificidae (Annelida, Oligochaeta) of the Bahamas. Meiofauna Mar. 12:59-84. [Google Scholar]
- 12.Erséus, C. 1990. The marine Tubificidae (Oliogchaeta) of the barrier reef ecosystems at Carrie Bow Cay, Belize, and other parts of the Caribbean Sea, with descriptions of twenty-seven new species and revision of Heterodrilus, Thalassodrilides and Smithsonidrilus. Zool. Scr. 19:243-303. [Google Scholar]
- 13.Erséus, C. 1984. Taxonomy and phylogeny of the gutless Phallodrilinae (Oligochaeta, Tubificidae), with descriptions of one new genus and twenty-two new species. Zool. Scr. 13:239-272. [Google Scholar]
- 14.Erséus, C. 1991. Two new deep-water species of the gutless genus Olavius (Oligochaeta: Tubificidae) from both sides of North America. Proc. Biol. Soc. Wash. 104:627-630. [Google Scholar]
- 15.Erséus, C., M. Källersjö, M. Ekman, and R. Hovmöller. 2002. 18S rDNA phylogeny of Tubificidae and its constituent taxa (Clitellata): dismissal of the Naididae. Mol. Phylogenet. Evol. 22:414-422. [DOI] [PubMed] [Google Scholar]
- 16.Felbeck, H., G. Liebezeit, R. Dawson, and O. Giere. 1983. CO2 fixation in tissues of marine oligochaetes (Phallodrilus leukodermatus and P. planus) containing symbiotic, chemoautotrophic bacteria. Mar. Biol. 75:187-191. [Google Scholar]
- 17.Finogenova, N. 1986. Six new species of marine Tubificidae (Oligochaeta) from the continental shelf off Peru. Zool. Scr. 15:45-51. [Google Scholar]
- 18.Fujiwara, Y., C. Kato, N. Masui, K. Fujikura, and S. Kojima. 2001. Dual symbiosis in the cold-seep thyasirid clam Maorithyas hadalis from the hadal zone in the Japan Trench, western Pacific. Mar. Ecol. Prog. Ser. 214:151-159. [Google Scholar]
- 19.Giere, O. 1981. The gutless marine oligochaete Phallodrilus leukodermatus. Structural studies on an aberrant tubificid associated with bacteria. Mar. Ecol. Prog. Ser. 5:353-357. [Google Scholar]
- 20.Giere, O. 1979. Studies on marine Oligochaeta from Bermuda, with emphasis on new Phallodrilus species (Tubificidae). Cah. Biol. Mar. 20:301-314. [Google Scholar]
- 21.Giere, O., and C. Erséus. 2002. Taxonomy and new bacterial symbioses of gutless marine Tubificidae (Annelida, Oligochaeta) from the Island of Elba (Italy). Org. Divers. Evol. 2:289-297. [Google Scholar]
- 22.Giere, O., C. Erséus, and F. Stuhlmacher. 1998. A new species of Olavius (Tubificidae, Phallodrilinae) from the Algarve Coast in Portugal, the first East Atlantic gutless oligochaete with symbiotic bacteria. Zool. Anz. 237:209-214. [Google Scholar]
- 23.Giere, O., and J. Krieger. 2001. A triple bacterial endosymbiosis in a gutless oligochaete (Annelida): ultrastructural and immunocytochemical evidence. Invertebr. Biol. 120:41-49. [Google Scholar]
- 24.Giere, O., C. Nieser, R. Windoffer, and C. Erséus. 1995. A comparative structural study on bacterial symbioses of Caribbean gutless Tubificidae (Annelida, Oligochaeta). Acta Zool. 76:281-290. [Google Scholar]
- 25.Harwood, C. S., and E. Canale-Parola. 1983. Spirochaeta isovalerica sp. nov., a marine anaerobe that forms branched-chain fatty acids as fermentation products. Int. J. Syst. Bacteriol. 33:573-579. [Google Scholar]
- 26.Heider, H., A. M. Spormann, H. R. Beller, and F. Widdel. 1999. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev. 22:459-473. [Google Scholar]
- 27.Hespell, R. B., and E. Canale-Parola. 1973. Glucose and pyruvate metabolism of Spirochaeta litoralis, an anaerobic marine spirochete. J. Bacteriol. 116:931-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krieger, J., O. Giere, and N. Dubilier. 2000. Localization of RubisCO and sulfur in endosymbiotic bacteria of the gutless marine oligochaete Inanidrilus leukodermatus (Annelida). Mar. Biol. 137:239-244. [Google Scholar]
- 29.Leadbetter, J. R., T. M. Schmidt, J. R. Graber, and J. A. Breznak. 1999. Acetogenesis from H2 plus CO2 by spirochetes from termite guts. Science 283:686-689. [DOI] [PubMed] [Google Scholar]
- 30.Levin, L., D. Gutiérrez, A. Rathburn, C. Neira, J. Sellanes, P. Muñoz, V. Gallardo, and M. Salamanca. 2002. Benthic processes on the Peru margin: a transect across the oxygen minimum zone during the 1997-98 El Niño. Prog. Oceanogr. 53:1-27. [Google Scholar]
- 31.Levin, L. A., A. E. Rathburn, D. Gutiérrez, P. Muñoz, and A. Shankle. 2003. Bioturbation by symbiont-bearing annelids in near-anoxic sediments: implications for biofacies models and paleo-oxygen assessments. Palaeogeogr. Palaeoclimatol. Palaeoecol. 199:129-140. [Google Scholar]
- 32.Li, L., C. Kato, and K. Horikoshi. 1999. Microbial diversity in sediments collected from the deepest cold-seep area, the Japan trench. Mar. Biotechnol. 1:391-400. [DOI] [PubMed] [Google Scholar]
- 33.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 34.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. 1998. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25:43-61. [Google Scholar]
- 35.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 36.Nylander, J. A. A., C. Erséus, and M. Källersjö. 1999. A test of monophyly of the gutless Phallodrilinae (Oligochaeta, Tubificidae) and the use of a 573 bp region of the mitochondrial cytochrome oxidase I gene in analysis of annelid phylogeny. Zoo. Scr. 28:305-313. [Google Scholar]
- 37.Olsen, G. J., R. Overbeck, N. Larson, T. L. Marsh, M. J. McCaughey, M. A. Maciukenas, W. M. Kuan, T. J. Macke, Y. Xing, and C. R. Woese. 1992. Ribosomal Database Project. Nucleic Acids Res. 20:2199-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pernthaler, J., F. O. Glöckner, W. Schönhuber, and R. Amann. 2001. Fluorescence in situ hybridization with rRNA-targeted oligonucleotides probes. Methods Microbiol. 30:207-226. [Google Scholar]
- 39.Rühland, C., A. Blazejak, A. Loy, M. Wagner, C. Erséus, R. Amann, and N. Dubilier. Unpublished data.
- 40.Schizas, N. V., G. T. Street, B. C. Coull, G. T. Chandler, and J. M. Quattro. 1997. An efficient DNA extraction method for small metazoans. Mol. Mar. Biol. Biotechnol. 6:381-383. [PubMed] [Google Scholar]
- 41.Schönhuber, W., B. Zarda, S. Eix, R. Rippka, M. Herdman, W. Ludwig, and R. Amann. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sievert, S. M., G. Muyzer, and J. Küver. 1999. Novel sulfur-oxidizing bacteria from a shallow submarine hydrothermal vent, most closely related to obligate symbionts of invertebrates. Ph.D. dissertation. University of Bremen, Bremen, Germany.
- 43.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ-hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 44.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulfate-reducing bacteria, p. 3352-3389. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, vol. IV. Springer-Verlag New York, N.Y.