Abstract
Lactobacillus plantarum WCFS1 harbors three plasmids, pWCFS101, pWCFS102, and pWCFS103, with sizes of 1,917, 2,365, and 36,069 bp, respectively. The two smaller plasmids are of unknown function and contain replication genes that are likely to function via the rolling-circle replication mechanism. The host range of the pWCFS101 replicon includes Lactobacillus species and Lactococcus lactis, while that of the pWCFS102 replicon also includes Carnobacterium maltaromaticum and Bacillus subtilis. The larger plasmid is predicted to replicate via the theta-type mechanism. The host range of its replicon seems restricted to L. plantarum. Cloning vectors were constructed based on the replicons of all three plasmids. Plasmid pWCFS103 was demonstrated to be a conjugative plasmid, as it could be transferred to L. plantarum NC8. It confers arsenate and arsenite resistance, which can be used as selective markers.
Lactic acid bacteria are used for the preservation of food and feed raw materials like milk, meat, and vegetables or other plant materials. Certain strains of lactic acid bacteria, in particular, strains from the genus Lactobacillus, have been attributed probiotic activities in humans and animals (30). Several lactic acid bacteria, including lactococci, streptococci, lactobacilli, and pediococci, are known to harbor plasmids. These may encode important traits like resistance to phages or antibiotics, lactose catabolism, and production of proteolytic enzymes or bacteriocins. Lactobacillus plantarum species often harbor several plasmids (49). Several of these have been sequenced (6, 11, 12, 20, 31, 40, 52, 58). Although most of them are of unknown function, one plasmid encoding phage resistance (pMD5057, 10,877 bp) and another plasmid (pLKS, 2,025 bp) probably introduced from another source and coding for tetracycline resistance have been described previously (12, 20). All are smaller than 11 kb and are predicted to replicate via the rolling-circle replication mechanism, except for the largest plasmid, pMD5057, which is predicted to replicate via the theta mechanism (12).
The capacity for conjugal transfer is an important characteristic for plasmids. Self-transmissible conjugative plasmids have the ability to form effective cell-to-cell contact, while mobilizable plasmids are only able to prepare their DNA for transfer (38). Mobilization involves the action of a specific DNA-protein structure called the relaxosome to produce single-stranded cleavage at the nicking site (nic) within the origin of transfer (oriT) of the plasmid (38). To date, there is very little information on conjugation in lactobacilli. Sasaki et al. demonstrated conjugational transfer of the promiscuous theta-replicating plasmid pAMβ1 from Streptococcus faecalis to L. plantarum (51). Ahn et al. described an 8.5-kb chloramphenicol resistance plasmid which had been comobilized with pAMβ1 from L. plantarum to Carnobacterium maltaromaticum (3), previously known as Carnobacterium piscicola (43). To our knowledge, no conjugative L. plantarum plasmids have been reported.
Recently, we determined the complete nucleotide sequence of the genome of L. plantarum WCFS1 (35). The sequences of the three endogenous plasmids of WCFS1 were obtained from this genome sequencing project. Here, we present the nucleotide sequence analyses and characterization of the plasmids from this strain.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli and Bacillus subtilis were grown in Luria-broth-based medium at 37°C (50). L. plantarum and Leuconostoc lactis were grown in MRS broth (Difco Laboratories) at 37 and 30°C, respectively. Lactococcus lactis was grown in M17 broth (Difco Laboratories) supplemented with 0.5% glucose at 30°C. Streptococcus thermophilus was grown in M17 broth supplemented with 0.5% lactose at 42°C. C. maltaromaticum was grown in brain heart infusion medium at 30°C. If appropriate, the media contained chloramphenicol (10 μg/ml), rifampin (50 μg/ml), or erythromycin (5 μg/ml).
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant characteristic(s)a | Reference | ||
|---|---|---|---|---|
| Strains | ||||
| Escherichia coli DH5-α | Plasmid-free strain | 25 | ||
| Lactobacillus plantarum WCFS1 | Single-colony isolate of NCIMB8826, sequenced strain | 35 | ||
| Lactobacillus plantarum WCFS2 | Strain WCFS1 with plasmid pWCFS110 instead of pWCFS103 | This study | ||
| Lactobacillus plantarum WCFS3 | Strain WCFS1 with plasmid pWCFS111 instead of pWCFS103 | This study | ||
| Lactobacillus plantarum WCFS4 | Strain WCFS1 with plasmid pWCFS112 instead of pWCFS103 | This study | ||
| Lactobacillus plantarum NC8 | Plasmid-free strain | 5 | ||
| Lactobacillus plantarum NC8R | Rif derivative of NC8 | This study | ||
| Lactobacillus plantarum NZ7109 | Eryr derivative of WCFS1 | 9 | ||
| Lactobacillus casei LMG6904 | Lactobacillus casei type strain (ATCC 393) | 26 | ||
| Lactobacillus helveticus CNRZ 32 | Plasmid-free strain | 33 | ||
| Lactococcus lactis MG1363 | Plasmid-free strain | 22 | ||
| Leuconostoc lactis NZ6091 | Plasmid-free strain | 13 | ||
| Streptococcus thermophilus ST11 | Plasmid-free strain | 42 | ||
| Carnobacterium maltaromaticum LV17B | Plasmid-free strain | 1 | ||
| Bacillus subtilis 168 | Plasmid-free strain | 53 | ||
| Plasmids | ||||
| pUC18Ery | Ampr Eryr, 3.6-kb pUC18 integration vector carrying the erythromycin resistance gene from pAMβ1 | 57 | ||
| pNZ124 | Cmr, 2.8-kb pSH71 replicon | 46 | ||
| pWCFS101 | 1,917-bp cryptic plasmid | This study | ||
| pWCFS102 | 2,365-bp cryptic plasmid | This study | ||
| pWCFS103 | 36,069-bp conjugative arsenate or arsenite resistance plasmid | This study | ||
| pWCFS104 | Cmr, 3.0-kb, pWCFS101-based cloning vector | This study | ||
| pWCFS105 | Cmr, 3.4-kb, pWCFS102-based cloning vector | This study | ||
| pWCFS106 | Cmr, 4.2-kb, pWCFS103-based cloning vector | This study | ||
| pWCFS107 | Eryr, arsB integration vector | This study | ||
| pWCFS108 | Eryr, cadD integration vector | This study | ||
| pWCFS109 | Eryr, traA integration vector | This study | ||
| pWCFS110 | Eryr, arsB, pWCFS107 integrated into the arsB gene of pWCFS103 | This study | ||
| pWCFS111 | Eryr, cadD, pWCFS108 integrated into the cadD gene of pWCFS103 | This study | ||
| pWCFS112 | Eryr, traA, pWCFS109 integrated into the traA gene of pWCFS103 | This study |
Ampr, ampicillin resistant; Eryr, erythromycin resistant; Tetr, tetracycline resistant, Cmr, chloramphenicol resistant; Rifr, rifampin.
Heavy-metal resistance.
The sensitivity of L. plantarum strains toward heavy metals was tested by inoculating MRS broth with a 0- to 50-mg/ml concentration series of arsenite (NaAsO2), arsenate (Na2HAsO4 · 7H2O), cadmium (CdSO4 · 8/3H2O), cobalt (CoCl2), copper (CuCl2 · 2H2O), iron (FeSO4 · 7H2O), lead [Pb(NO3)2], mercury (HgCl2), nickel (NiCl2 · 6H2O), or zinc (ZnCl2) with 2% of an overnight culture and determining the turbidity of the culture at 600 nm after 16 h of incubation.
Conjugation.
Conjugation was performed using filter matings. Cells from overnight cultures of the donor (2 ml) and recipient (200 μl) were mixed, collected on a sterile 0.45-μm-pore-size filter in a plastic filter holder by using a syringe, and washed with 25 ml of sterile H2O. The filters were placed on MRS agar plates and incubated anaerobically at 37°C for 20 h. The cells were resuspended in 5 ml of 0.25× Ringers solution. Dilutions were plated in triplicate on MRS agar plates containing 10 mg of sodium arsenate/ml for determining the donor count and on MRS agar plates containing rifampin and 10 mg of sodium arsenate/ml for determining the transconjugant count. The plates were incubated anaerobically at 37°C for 2 days. Cells from the overnight culture of the donor were treated similarly and plated on MRS agar plates containing rifampin to determine the number of spontaneous rifampin-resistant mutants. The frequency of appearance of spontaneous rifampin-resistant mutants was 20- to 100-fold lower in all conjugation experiments (ranging from 2.0 × 10−8 to 1.8 × 10−9) than the conjugation frequencies. The transconjugant numbers reported here have all been corrected with the spontaneous rifampin-resistant donor numbers.
DNA isolation and manipulation.
Isolation of E. coli plasmid DNA and standard recombinant DNA techniques were performed as described by Sambrook et al. (50). Small-scale isolation of plasmid DNA from the gram-positive bacteria was performed using log-phase cultures as described previously (16). Large-scale plasmid DNA isolation for L. plantarum was performed by the method of Anderson and McKay (4). Electrotransformation was performed using a Gene Pulser apparatus (Bio-Rad). Electrocompetent cells of E. coli (18), L. plantarum (5), Lactococcus lactis (16), Leuconostoc lactis (46), S. thermophilus (42), Lactobacillus helveticus (7), C. maltaromaticum (2), and B. subtilis (36) were prepared as previously described.
Nucleotide sequence analysis.
L. plantarum WCFS1 plasmid sequence data were obtained by a random shotgun approach as was applied for the determination of the chromosomal sequence of this strain (35). Sequence data were assembled and analyzed using the PHRAP assembler (21), and open reading frames were predicted by using Genemark (28) and Glimmer version 2.0 (15) trained on known L. plantarum genes. The SWALL (www.ebi.ac.uk) and EMBL prokaryote libraries were screened for homologies by using the Fasta3 service at the European Bioinformatics Institute website (www.ebi.ac.uk) (45). The multiple-sequence alignment was performed using the ClustalW service at the European Bioinformatics Institute website (55).
Construction of plasmids.
To construct the cloning vectors based on the replicons of the endogenous L. plantarum plasmids, pWCFS101 and pWCFS102 were digested with XbaI and blunted. The replicon from pWCFS103 was obtained by PCR using the high-fidelity Pwo polymerase (Boehringer Mannheim). Cloning sites were introduced in the primers (underlined) with the sequences 5′-GCCGCGGTCGACAAGCCCCTATTCTTCTGTTT-3′ and 5′-GCCGCGCTCGAGTAAGCAAAGCCTGTATGTAA-3′, and the PCR product was digested with XhoI-SalI. A fragment containing the multiple-cloning site and chloramphenicol resistance gene was isolated from pNZ124 as an XhoI-SalI fragment, blunted for pWCFS101 and pWCFS102, and ligated with the three replicons. For each ligation, one orientation of the resulting plasmids was used for further research and designated pWCFS104 (from pWCFS101), pWCFS105 (from pWCFS102), and pWCFS106 (from pWCFS103).
Construction of single-crossover integrants.
To construct single-crossover disruption mutants of arsB, cadD, or traA, internal gene fragments of approximately 0.4 to 0.5 kb were generated by PCR. KpnI and BamHI cloning sites (underlined) were introduced in the primers 5′-CCGCGCGGTACCGTTACTGGGATCGTTTG-3′ and 5′-CCGCGCGGATCCACATTCTTAGGGCATAC-3′ (arsB), 5′-CCGCGCGGTACCATTCTTTATACATCAACTGCAATCGA-3′ and 5′-CCGCGCGGATCCAACAAACAGCGTTACGATTAGCTG-3′ (cadD), and 5′-CCGCGCGGTACCAAAATGGGCGAGTGATCGGGAGA-3′ and 5′-CCGCGCGGATCCCTTGATCGACAAATGATTTTTCGCT-3′ (traA). The PCR fragments were cloned into pUC18Ery (28), and the resulting plasmids were designated pWCFS107 (arsB), pWCFS108 (cadD), and pWCFS109 (traA), respectively. These plasmids were electroporated to L. plantarum WCFS1. Erythromycin-resistant colonies harbored pWCFS103 single-crossover integrant derivatives, the integrity of which was checked by Southern blot hybridization. For each knockout, a single-copy integrant was selected for further research.
Plasmid stability tests.
Plasmid stability of pWCFS104, pWCFS105, and pWCFS106 was tested in the erythromycin-resistant WCFS1 derivative NZ7109. Strains were cultured by serial transfer (1/1,000) in MRS broth supplemented with erythromycin. After approximately 110 generations (11 transfers), dilution series were plated on MRS plates supplemented with erythromycin. From each strain, 100 colonies were transferred to MRS plates supplemented with erythromycin and erythromycin plus chloramphenicol.
Relative-copy-number determination.
Plasmid copy number was assessed using real-time PCR detection. For that purpose, plasmids pWCFS104, pWCFS105, and pWCFS106 were transformed to L. plantarum strain NZ7109, which harbors a single copy of a chromosomally inserted erythromycin resistance gene and was obtained via a standard two-step homologous recombination strategy (9). This chromosomal marker was used as a chromosomal copy number reference to which all plasmid copy numbers were compared. Real-time PCR amplification was performed using exponentially grown L. plantarum containing pWCFS104, pWCFS105, or pWCFS106. Cells from 1 ml of culture were harvested by centrifugation, washed with water, and pelleted by centrifugation. Cells were lysed by microwave treatment (3 min, 800 W) and resuspended in 400 μl of water. Appropriate solutions of these suspensions were used as templates for PCRs. These reactions contained the primer pairs designed on the chromosomal marker ery (TM-ery-F99, 5′-TTCACCGAACACTAGGGTTGC-3′; TM-ery-R100, 5′-ATTCCGCTGGCAGCTTAAG-3′) combined with the FAM (6-carboyfluorescein) reporter and TAMRA (6-carboxytetramethylrhodamine) quencher dye containing and ery probe (TM-ery-FAM, 5′-FAM-TGCACACTCAAGTCTCGATTCAGCA-TAMRA-3′) and the primers designed for detection of the plasmid-derived chloramphenicol resistance gene cat (TM-cat-F96, 5′-TCAAATACAGCTTTTAGAACTGG-3′; TM-cat-R97, 5′-ACCATCAAAAATTGTATAAAGTGGC-3′) combined with the VIC reporter and TAMRA quencher dye containing cat probe (TM-cat-VIC98, 5′-VIC-GCGACGGAGAGTTAGGTTATTGGG-TAMRA-3′). Reactions were performed with the TaqMan core reagent kit (Applied Biosystems, Nieuwekerk a/d IJsel, The Netherlands). The threshold cycle number (Ct) was determined (27) using ABI Prism 7700 sequence detection system software and was used to calculate the relative gene copy number (Nrelative) for each plasmid in relation to the chromosomal copy number with the formula Nrelative = 2(Ct,cat − Ct,ery), where Ct,cat is the Ct for the reaction with cat and Ct,ery is the Ct for the reaction with ery. These experiments were performed in triplicate.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of plasmids pWCFS101, pWCFS102, and pWCFS103 have been submitted to the EMBL database and are available under accession numbers CR377164, CR377165, and CR377166, respectively.
RESULTS AND DISCUSSION
Sequence analysis of pWCFS101, pWCFS102, and pWCFS103.
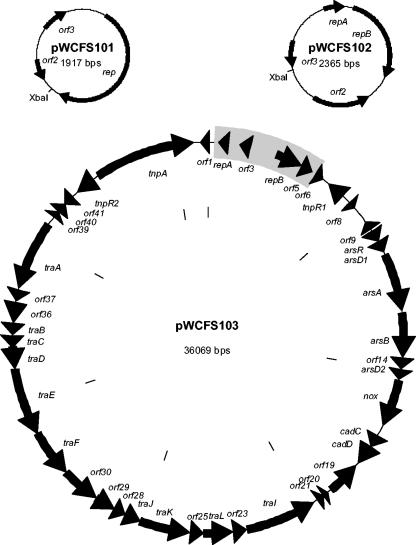
The organization of the three plasmids found in L. plantarum WCFS1 is shown Fig. 1. The average G+C contents were 39.5, 34.3, and 40.8% for pWCFS101, pWCFS102, and pWCFS103, respectively, compared to 44.5% for the chromosome of strain WCFS1 (35). All open reading frames larger than 40 amino acids were compared to those in the SWALL database (www.ebi.ac.uk). The results are depicted in Table 2. Plasmids pWCFS101 and pWCFS102 are cryptic, while pWCFS103 contains several genes for heavy-metal resistance, NADH-oxidase activity, and conjugation and a complete transposon.
FIG. 1.
Physical and genetic maps of plasmids pWCFS101, pWCFS102, and pWCFS103. The unique XbaI sites used to construct pWCFS104 and pWCFS105 are indicated. The replicon region of pWCFS103 used to construct pWCFS106 is boxed grey.
TABLE 2.
Putative genes and their products, deduced from the plasmid nucleotide sequences
| Plasmid and gene | Codon
|
No. of amino acids | Best homolog (organism, GenBank accession no.) | % Identity (no. of amino acids overlapping) | Proposed function of gene product | |
|---|---|---|---|---|---|---|
| Start | Stopa | |||||
| pWCFS101 | ||||||
| rep | 163 | 1122 | 319 | Rep protein pLAB1000 (Lactobacillus hilgardii, E973099) | 71 (314) | Replication protein |
| orf2 | 1406 | 1260 C | 48 | No hits | Hypothetical protein | |
| orf3 | 1627 | 1809 | 60 | No hits | Hypothetical protein | |
| pWCFS102 | ||||||
| repB | 102 | 761 | 219 | RepB-like protein pLH2 (Lactobacillus helveticus, Q48562) | 88 (219) | Replication protein |
| orf2 | 1393 | 953 C | 146 | No hits | Hypothetical protein | |
| orf3 | 1889 | 1686 C | 67 | No hits | Hypothetical protein | |
| repA | 2242 | 35 | 52 | CopA pPSC22 (Lactobacillus plantarum, Q48820) | 60 (50) | Copy number control protein |
| pWCFS103 | ||||||
| orf1 | 51 | 35839 C | 93 | DinJ (Escherichia coli, Q47150) | 30 (80) | DNA-damage-inducible protein |
| repA | 610 | 302 C | 102 | RepA-like protein pSAK1 (Lactobacillus sakei, Q48860) | 53 (76)b | Replication protein |
| orf3 | 925 | 1257 C | 110 | No hits | Hypothetical protein | |
| repB | 2061 | 2861 | 266 | RepB pAD1 (Enterococcus faecalis, Q52229) | 39 (276) | Copy number control protein |
| orf5 | 2863 | 3213 | 116 | Hypothetical protein (Lactobacillus sakei, Q9AED4) | 33 (113) | Hypothetical protein |
| orf6 | 3344 | 3586 | 80 | No hits | Hypothetical protein | |
| tnpR1 | 4370 | 3816 C | 184 | Resolvase-like protein pMD136 (Pediococcus pentosus, Q9WW47) | 83 (184) | Putative resolvase |
| orf8 | 4649 | 4855 | 68 | No hits | Hypothetical protein | |
| orf9 | 5641 | 5378 C | 87 | No hits | Hypothetical protein | |
| arsR | 5713 | 6072 | 119 | ArsR-like protein pL1100 (Listeria innocua, Q926M3) | 48 (119) | Regulator of arsenical resistance operon |
| arsD1 | 6059 | 6421 | 120 | ArsD-like protein pL1100 (Listeria innocua, Q926M4) | 42 (123) | Repressor of arsenical resistance operon |
| arsA | 6505 | 8235 | 576 | ArsA-like protein pL1100 (Listeria innocua, Q926M2) | 65 (568) | Arsenical pump-driving ATPase |
| arsB | 8294 | 9589 | 431 | ArsB-like protein (Listeria monocytogenes, P96678) | 73 (427) | Arsenical pump membrane protein |
| orf14 | 9606 | 9935 | 109 | No hits | Hypothetical protein | |
| arsD2 | 9958 | 10257 | 99 | ArsD-like protein pL1100 (Listeria innocua, Q926M4) | 43 (95) | Repressor of arsenical resistance operon |
| nox | 10281 | 11681 | 466 | Hypothetical protein pL1100 (Listeria innocua, Q926L9) | 36 (450) | Putative NADH oxidase |
| cadC | 12031 | 12399 | 122 | Transcriptional regulator ArsR family (Chlorobium tepidum, Q8KE78) | 37 (97) | Putative positive regulator of cadmium resistance |
| cadD | 12401 | 13015 | 204 | CadD pUB101 (Staphylococcus aureus, Q8GNY9) | 50 (205) | Putative cadmium resistance protein |
| orf19 | 14131 | 13100 C | 344 | Transposase (Lactobacillus delbrueckii, Q93L10) | 41 (338) | Putative transposase |
| orf20 | 14404 | 14240 C | 54 | LtrC pMRC01 (Lactococcus lactis, O87224) | 61 (49)c | Hypothetical protein |
| orf21 | 14623 | 14408 C | 71 | Hypothetical 8.0-kDa protein pMRC01 (Lactococcus lactis, O87223) | 57 (70) | Hypothetical protein |
| tra1 | 16881 | 14746 C | 711 | Trs1 pMRC01 (Lactococcus lactis, O87222) | 60 (722) | DNA topoisomerase |
| orf23 | 17298 | 16888 C | 136 | No hits | Hypothetical protein | |
| traL | 18170 | 17313 C | 285 | TrsL pMRC01 (Lactococcus lactisO87221) | 44 (277) | Conjugation protein |
| orf25 | 18565 | 18176 C | 129 | Hypothetical 14.7-kDa protein pMRC01 (Lactococcus lactis, O87220) | 32 (126) | Hypothetical protein |
| traK | 20076 | 18565 C | 503 | TraK pMRC01 (Lactococcus lactis, O87219) | 76 (503) | Conjugation protein |
| traJ | 20548 | 20078 C | 156 | TrsJ pMRC01 (Lactococcus lactis, O87218) | 48 (150) | Conjugation protein |
| orf28 | 20917 | 20549 C | 122 | Hypothetical protein pMRC01 (Lactococcus lactis, O87217) | 42 (118) | Hypothetical protein |
| orf29 | 21521 | 20904 C | 205 | Hypothetical 23.3-kDa protein pMRC01 (Lactococcus lactis, O87216) | 48 (204) | Hypothetical protein |
| orf30 | 22690 | 21536 C | 384 | Hypothetical protein pMRC01 (Lactococcus lactis, O87215) | 54 (395) | Hypothetical protein |
| traF | 24109 | 22691 C | 472 | TrsF pMRC01 (Lactococcus lactis, O87214) | 50 (471) | Conjugation protein |
| traE | 26120 | 24102 C | 672 | TrsE pMRC01 (Lactococcus lactis, O87213) | 82 (671) | Conjugation protein |
| traE | 26791 | 26132 C | 219 | TrsD pMRC01 (Lactococcus lactis, O87212) | 80 (219) | Conjugation protein |
| traC | 27122 | 27760 C | 120 | TrsC pMRC01 (Lactococcus lactis, O87211) | 73 (115) | Conjugation protein |
| traB | 27478 | 27143 C | 111 | TrsB pMRC01 (Lactococcus lactis, O87210) | 72 (107) | Conjugation protein |
| orf36 | 28094 | 27480 C | 204 | Hypothetical 23.2-kDa protein pMRC01 (Lactococcus lactis, O97209) | 51 (201) | Hypothetical protein |
| orf37 | 28471 | 28133 C | 112 | Hypothetical 13.1-kDa protein pMRC01 (Lactococcus lactis, O87208) | 46 (90) | Hypothetical protein |
| traA | 30584 | 28524 C | 686 | TraA pMRC01 (Lactococcus lactis, O87207) | 49 (677) | Nickase |
| orf39 | 30856 | 31065 | 69 | Hypothetical 10.9-kDa protein pMRC01 (Lactococcus lactis, O87206) | 56 (66) | Hypothetical protein |
| orf40 | 31088 | 31366 | 92 | Hypothetical 10.6-kDa protein pMRC01 (Lactococcus lactis, O87204) | 50 (88) | Hypothetical protein |
| orf41 | 31356 | 31670 C | 104 | No hits | Hypothetical protein | |
| tnpR2 | 32466 | 31880 C | 195 | TnpR transposon Tn551 (Staphylococcus aureus, TNR7_ENTFA) | 40 (181) | Putative resolvase |
| tnpA | 32645 | 35638 | 997 | Transposase pAM373 (Enterococcus faecalis, Q9F117) | 64 (987) | Putative transposase |
C, complementary sequence.
Homologous to C-terminal part of protein.
Homologous to N-terminal part of protein.
Replicons.
Plasmids pWCFS101 and pWCFS102 are homologous to rolling-circle replicating plasmids. pWCFS101 has a pC194-type replicon (34) with a single replication gene, repA, encoding a 37-kDa protein that was very similar to the Rep proteins from Lactobacillus hilgardii plasmid pLAB1000 (29) and L. plantarum plasmid pLP1 (8). Downstream of the repA gene at position 1798, there began a sequence (5′-TTCTTATCTTGATA-3′) which was identical to the double-stranded origin of pC194 (24). Plasmid R1162 carries a set of three and a half 20-bp repeats that are essential for the expression of incompatibility and copy number control (41). Likewise, plasmid pLP1 contains a region with 13 contiguous direct repeats of 17 bp that are suggested to have a similar function (8). pWCFS101 has a region with five and a half direct repeats of 17 bp (5′-AGTGCGCATTATCATGT-3′) that are identical to those of pLP1 and may serve the same function.
pWCFS102 has a pMV158-type replicon (34) and carries two replication genes. The repA gene encodes a 6-kDa protein that is homologous to copy control proteins like RepC of pWV01 (39). The repB gene encodes a 25-kDa protein that is highly homologous to the Rep proteins from L. helveticus plasmid pLH2 and Lactobacillus curvatus plasmid pLC2 (37, 47). A putative double-stranded origin is formed by a sequence starting at position 2003, with high similarity to the double-stranded origins of pWV01, pFX2, and pLC2 (Fig. 2) (23, 37). These sequences are able to form a stem-loop structure and contain the nick site at which the Rep protein cleaves the DNA. The Rep binding region of pMV158-type replicons is composed of a set of two or three iterons which are separated from the nick site by an intervening region of 13 to 91 bp (34). Unlike the nick regions, the Rep binding regions are not conserved in their nucleotide sequence (34). We could not detect any clear direct repeats in the region close to the putative nick site nor elsewhere on pWCFS102.
FIG. 2.
Alignment of the double-stranded origins of plasmids pWV01/pFX2 (23) and pLC2 (37) and the putative double-stranded origin of pWCFS102. Dashed arrows indicate the inverted repeats. The arrowheads indicate the nick sites in pFX2 (23). Alignment mismatches are boxed in grey.
Plasmid pWCFS103 has a replicon that is homologous to theta replicons. The repA gene encodes a 12-kDa protein that shares homology with the C-terminal part of the replication proteins from the Lactobacillus sakei plasmid pSAK1, the Lactococcus lactis plasmid pCI2000, and the L. helveticus plasmids pLJ1 and pLH1 (32, 54, 56). Compared to those replication proteins, RepA lacks approximately 260 amino acids at its N terminus, the region that has strongest conservation and includes the helix-turn-helix motif that is considered to be involved in DNA binding (56). The repB gene encodes a 30-kDa protein and is homologous to proposed plasmid copy control proteins from the Enterococcus faecalis plasmids pAD1 and pAM373 and the Bacillus thuringiensis plasmid pAW63 (14, 60, 61). Downstream of repB starts orf4, of which the start codon overlaps with the repB stop codon, suggesting transcriptional linkage of orf4 to repB and a role for orf4 in replication. For pAD1 and pAW63, a similar small gene that partly overlaps repB was found (60, 61). In between the repA and repB genes, an iteron region with 11 and 10 copies of imperfect repeats of the sequence 5′-TGTATCCT-3′ spaced by 37 nucleotides was found (Fig. 3). Plasmid pAD1 also has a series of 25 8-bp direct repeats located in between repA and repB, which is predicted to function as the origin of replication (60).
FIG. 3.
Iteron region of pWCFS103. Dashed arrows indicate the direct and inverted repeats. Mismatches from the consensus sequence are boxed grey. The ribosome binding site (boldface) and start codon (underlined) of repB are indicated.
Host range.
Based on the three replicons, three cloning vectors that contain the chloramphenicol resistance gene and multiple-cloning site from pNZ124 were constructed (see Materials and Methods). For pWCFS106, the 3.2-kb fragment with repA, repB, and orf5 contained the complete replicon and could be stably maintained in L. plantarum NC8. To gain insight into the host range of the three cloning vectors, plasmid DNA isolated from L. plantarum NC8 was transformed to various gram-positive organisms and E. coli (Table 3). The theta-type pWCFS106 demonstrated a narrow host range and seemed restricted to L. plantarum. Rolling-circle-type pWCFS104 and pWCFS105 had a broader but limited host range, as both were able to replicate in L. helveticus, Lactobacillus casei, and Leuconostoc lactis and pWCFS105 was able to replicate also in C. maltaromaticum and B. subtilis.
TABLE 3.
Host range of L. plantarum WCFS1 cloning vectorsa
| Host | pWCFS104 | pWCFS105 | pWCFS106 |
|---|---|---|---|
| Lactobacillus plantarum NC8 | + | + | + |
| Lactobacillus helveticus CNRZ 32 | + | + | − |
| Lactobacillus casei LMG6904 | + | + | − |
| Leuconostoc lactis NZ6091 | + | + | − |
| Lactococcus lactis MG1363 | − | − | − |
| Streptococcus thermophilus ST11 | − | − | − |
| Carnobacterium maltaromaticum LV17B | − | + | − |
| Bacillus subtilis 168 | − | + | − |
| Escherichia coli DH5-α | − | − | − |
Transformants observed (+) or not observed (−).
Plasmid stability.
The plasmid stability of the three cloning vectors pWCFS104, pWCFS105, and pWCFS106 in rifampin-resistant L. plantarum NZ7109 was determined after approximately 110 generations of growth without selection pressure. One hundred colonies were replica plated to plates supplemented with erythromycin and with erythromycin and chloramphenicol. All grew on the erythromycin plates, and 97 (pWCFS104), 100 (pWCFS105), and 98 (pWCFS106) colonies grew on the chloramphenicol-containing plates, illustrating that all three replicons are stably maintained in L. plantarum.
Plasmid copy numbers.
Relative plasmid copy numbers were determined by real-time PCR analysis as described in the Materials and Methods section. These experiments revealed that the relative copy numbers of pWCFS104, pWCFS105, and pWCFS106 were 11.9 ± 1.2, 2.8 ± 0.3, and 4.7 ± 0.4, respectively. Therefore, these plasmids all generate a different gene dosage of genetic elements cloned on these vectors.
Mobilization.
Plasmid pWCFS103 carries a large gene cluster that has high homology with the mobilization gene cluster of the Lactococcus lactis plasmid pMRC01 (17). The Lactococcus lactis Tra region comprises an origin of transfer (oriT) sequence flanked by a gene cluster of 18 genes (traA, orf6, orf7, traB, traC, traD, traE, traF, orf13, orf14, orf15, traJ, traK, orf18, traL, traI, orf21, and ltrC) and a gene cluster of two genes in the opposite orientation (orf4 and orf3) (17). The pWCFS103 Tra region runs from orf40 (orf3 in pMRC01) to orf20 (ltrC in pMRC01) and is flanked by two transposase genes. Compared to pMRC01, it has an additional open reading frame, orf23, of unknown function inserted between traL and traI. Although there is no clear homology in the oriT region, the CGAAG sequence, conserved among several conjugative plasmids (17, 59) and preceded by a TTAAG sequence, is found upstream of traA and may function as the oriT.
To study the conjugal transfer of pWCFS103, L. plantarum WCFS1 and NC8R, a rifampin-resistant derivative of NC8, were used in filter matings. Conjugants were selected on plates containing rifampin to select for the recipient and containing sodium arsenate to select for plasmid pWCFS103 (see below). Typical frequencies of conjugation were between 4.4 × 10−6 and 3.1 × 10−7 per donor. Conjugation of the erythromycin-resistant derivatives of pWCFS103, pWCFS110, pWCFS111, and pWCFS112 resulted in similar frequencies of transfer. For pWCFS112, this was unexpected, as this derivative contains a single-crossover integration in traA, thought to be involved in conjugation by encoding the nicking enzyme. As TraA is a large protein, the possibility that an additional start site still gives rise to a functional nicking enzyme cannot be ruled out.
Heavy-metal resistance.
Plasmid pWCFS103 contains two gene clusters that are predicted to be involved in arsenate and/or arsenite resistance and cadmium resistance (Table 2). Bacterial arsenite transporters provide resistance to salts of the metalloids arsenic and antimonite (44). Arsenic mainly occurs as As(V) in arsenate and As(III) in arsenite. Arsenate is structurally related to phosphate and interferes with phosphate metabolism. It will be reduced to arsenite and exported (44). There are two types of arsenic resistance (ars) operons described to date. One consists of three genes, arsR, arsB, and arsC, coding for a regulator ArsR, the arsenate reductase ArsC, and a secondary transporter, ArsB, that couples extrusion of anions to the transmembrane electrochemical potential (19, 48). The other ars operon contains two additional genes, arsD and arsA, inserted between arsR and arsB (19, 48). ArsD is a second transcriptional regulator, and ArsA is an ATPase that associates with ArsB and converts it into a primary ATP-coupled arsenite transporter with improved resistance (19). The pWCFS103 ars operon is such a high-resistance gene cluster but lacks the arsenate reductase gene arsC and contains two copies of the arsD regulator gene. The arsC gene appeared to be present on the L. plantarum WCFS1 chromosome (35), thereby completing the set of ars genes in this strain. Moreover, the mobilization data suggest that the chromosomally encoded arsC function found in L. plantarum WCFS1 is conserved in other strains of this species. This makes the organization of the L. plantarum ars gene cluster unique compared to those of other bacteria where arsC is always present.
The pWCFS103 cadD gene is homologous to the cadD gene from Staphylococcus aureus encoded on plasmid pRW001 (10). S. aureus cadD confers low-level resistance to cadmium (below 10 μg per ml), which is increased approximately 10-fold when provided with a functional cadC-like gene, cadX, encoding a transcription regulator of the cadmium operon (10). The pWCFS103 cadC gene is homologous to transcription regulators, including cadC of S. aureus.
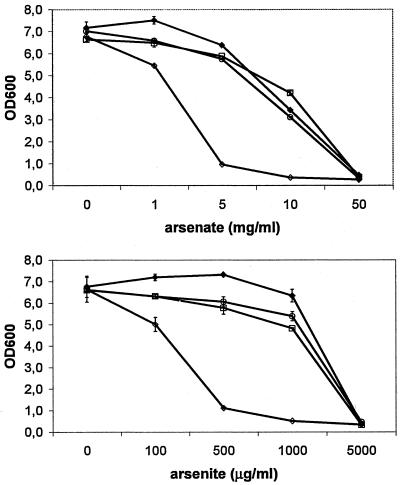
To test whether pWCFS103 confers heavy-metal resistance and if the ars and cad genes play a role in this resistance, L. plantarum WCFS1, L. plantarum WCFS2 (arsB), L. plantarum WCFS3 (cadD), and L. plantarum WCFS4 (traA) were incubated in media containing various concentrations of arsenite, arsenate, cadmium, cobalt, copper, iron, mercury, or nickel salts. Strain WCFS2 (with arsB disrupted) displayed retarded growth at lower arsenate and arsenite concentrations relative to the growth of strains WCFS1, WCFS3, and WCFS4 (Fig. 4). At 500 μg of arsenite per ml, WCFS2 barely showed growth after 16 h, while for the other strains, this result occurred at 5 mg of arsenite per ml. For the other heavy metals tested, no differences in final turbidity were observed for the four strains. However, a very limited effect on resistance toward iron and mercury was observed during growth. At close-to-lethal concentrations (10 mg per ml and 25 μg per ml for iron and mercury, respectively), the growth of WCFS4 was slightly better than that of WCFS3 (cadD disrupted) (data not shown). No effect of pWCFS103 on cobalt, copper, iron, or nickel resistance was observed (data not shown). From these experiments, we conclude that arsB is involved in arsenate or arsenite resistance. We have no evidence that the cadD gene is involved in cadmium resistance, but this may be obscured by the presence of another, more efficient cadmium resistance system encoded on the chromosome of WCFS1. In agreement with this suggestion is the prediction that several chromosomally encoded cation transporters have a predicted substrate range that includes cadmium (lp_1919, lp_3327, and lp_3435 [35]).
FIG. 4.
Arsenate and arsenite resistance in L. plantarum. Filled diamonds, WCFS1; open diamonds, WCFS2 (arsB); open circles, WCFS3 (cadD); open squares, WCFS4 (traA). Each data point is the mean of duplicate cultures. Error bars indicate the deviation from the mean for single cultures. OD600, optical density at 600 nm.
Acknowledgments
We are grateful to Bernadette Renckens for assistance with annotation of gene functions.
REFERENCES
- 1.Ahn, C., and M. E. Stiles. 1990. Plasmid-associated bacteriocin production by a strain of Carnobacterium piscicola from meat. Appl. Environ. Microbiol. 56:2503-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, C., and M. E. Stiles. 1992. Mobilization and expression of bacteriocin plasmids from Carnobacterium piscicola isolated from meat. J. Appl. Bacteriol. 73:217-228. [Google Scholar]
- 3.Ahn, C., D. Collins-Thompson, C. Duncan, and M. E. Stiles. 1992. Mobilization and location of the genetic determinant of chloramphenicol resistance from Lactobacillus plantarum caTC2R. Plasmid 27:169-176. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aukrust, T., and H. Blom. 1992. Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int. 25:253-261. [Google Scholar]
- 6.Bates, E. E. M., and H. J. Gilbert. 1989. Characterization of a cryptic plasmid from Lactobacillus plantarum. Gene 85:253-258. [DOI] [PubMed] [Google Scholar]
- 7.Bhowmik, T., and J. L. Steele. 1993. Development of an electroporation procedure for gene disruption in Lactobacillus helveticus CNRZ 32. J. Gen. Microbiol. 139:1433-1439. [Google Scholar]
- 8.Bouia, A., F. Bringel, L. Frey, B. Kammerer, A. Belarbi, A. Guyonvarch, and J. C. Hubert. 1989. Structural organization of pLP1, a cryptic plasmid from Lactobacillus plantarum CCM 1904. Plasmid 22:185-192. [DOI] [PubMed] [Google Scholar]
- 9.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crupper, S. S., V. Worrell, G. C. Stewart, and J. J. Iandolo. 1999. Cloning and expression of cadD, a new cadmium resistance gene of Staphylococcus aureus. J. Bacteriol. 181:4071-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daming, R., W. Yinyu, W. Zilai, C. Jun, L. Hekui, and Z. Jingye. 2003. Complete DNA sequence and analysis of two cryptic plasmids isolated from Lactobacillus plantarum. Plasmid 50:70-73. [DOI] [PubMed] [Google Scholar]
- 12.Danielsen, M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed] [Google Scholar]
- 13.David, S., G. Simons, and W. M. De Vos. 1989. Plasmid transformation by electroporation of Leuconostoc paramesenteroides and its use in molecular cloning. Appl. Environ. Microbiol. 55:1483-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Boever, E. H., D. B. Clewell, and C. M. Fraser. 2000. Enterococcus faecalis conjugative plasmid pAM373: complete nucleotide sequence and genetic analyses of sex pheromone response. Mol. Microbiol. 37:1327-1341. [DOI] [PubMed] [Google Scholar]
- 15.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Vos, W. M., P. Vos, H. de Haard, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis subsp. cremoris SK11 gene encoding an extracellular serine proteinase. Gene 85:169-176. [DOI] [PubMed] [Google Scholar]
- 17.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29:1029-1038. [DOI] [PubMed] [Google Scholar]
- 18.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Driessen, A. J. M., B. P. Rosen, and W. N. Konings. 2000. Diversity of transport mechanisms: common structural principles. Trends Biochem. Sci. 25:397-401. [DOI] [PubMed] [Google Scholar]
- 20.Eguchi, T., K. Doi, K. Nishiyama, S. Ohmomo, and S. Ogata. 2000. Characterization of a phage resistance plasmid, pLKS, of silage-making Lactobacillus plantarum NGRI0101. Biosci. Biotechnol. Biochem. 64:751-756. [DOI] [PubMed] [Google Scholar]
- 21.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 22.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grohmann, E., M. Moscoso, E. L. Zechner, G. del Solar, and M. Espinosa. 1998. In vivo definition of the functional origin of leading strand replication on the lactococcal plasmid pFX2. Mol. Gen. Genet. 260:38-47. [DOI] [PubMed] [Google Scholar]
- 24.Gros, M. F., H. te Riele, and S. D. Ehrlich. 1987. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 6:3863-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, P. A., and E. F. Lessel. 1971. Lactobacillus casei (Orla-Jensen) comb. nov. Int. J. Syst. Bacteriol. 21:69-71. [Google Scholar]
- 27.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 28.Isono, K., J. D. McIninch, and M. Borodovsky. 1994. Characteristic features of the nucleotide sequences of yeast mitochondrial ribosomal protein genes as analyzed by computer program GeneMark. DNA Res. 1:263-269. [DOI] [PubMed] [Google Scholar]
- 29.Josson, K., P. Soetaert, F. Michiels, H. Joos, and J. Mahillon. 1990. Lactobacillus hilgardii plasmid pLAB1000 consists of two functional cassettes commonly found in other gram-positive organisms. J. Bacteriol. 172:3089-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalliomaki, M., S. Salminen, H. Arvilommi, P. Kero, P. Koskinen, and E. Isolauri. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076-1079. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko, Y., H. Kobayashi, P. Kiatpapan, T. Nishimoto, R. Napitupulu, H. Ono, and Y. Murooka. 2000. Development of a host-vector system for Lactobacillus plantarum L137 isolated from a traditional fermented food produced in the Philippines J. Biosci. Bioeng. 89:62-67. [DOI] [PubMed] [Google Scholar]
- 32.Kearney, K., G. F. Fitzgerald, and J. F. Seegers. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khalid, M. N., and E. H. Marth. 1990. Purification and partial characterization of a prolyl-dipeptidyl aminopeptidase from Lactobacillus helveticus CNRZ 32. Appl. Environ. Microbiol. 56:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan, S. A. 1997. Rolling-circle replication of bacterial plasmids. Microbiol. Mol. Biol. Rev. 61:442-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Klein Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Nierop Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleerebezem, M., R. Bongers, G. Rutten, W. M. de Vos, and O. P. Kuipers. 2004. Autoregulation of subtilin biosynthesis in Bacillus subtilis: the role of the spa-box in subtilin-responsive promoters. Peptides 25:1415-1424. [DOI] [PubMed] [Google Scholar]
- 37.Klein, J. R., C. Ulrich, and R. Plapp. 1993. Characterization and sequence analysis of a small cryptic plasmid from Lactobacillus curvatus LTH683 and its use for construction of new Lactobacillus cloning vectors. Plasmid 30:14-29. [DOI] [PubMed] [Google Scholar]
- 38.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 39.Leenhouts, K. J., B. Tolner, S. Bron, J. Kok, G. Venema, and J. F. Seegers. 1991. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 26:55-66. [DOI] [PubMed] [Google Scholar]
- 40.Leer, R. J., N. van Luijk, M. Posno, and P. H. Pouwels. 1992. Structural and functional analysis of two cryptic plasmids from Lactobacillus pentosus MD353 and Lactobacillus plantarum ATCC 8014. Mol. Gen. Genet. 234:265-274. [DOI] [PubMed] [Google Scholar]
- 41.Lin, L. S., Y. J. Kim, and R. J. Meyer. 1987. The 20 bp, directly repeated DNA sequence of broad host range plasmid R1162 exerts incompatibility in vivo and inhibits R1162 DNA replication in vitro. Mol. Gen. Genet. 208:390-397. [DOI] [PubMed] [Google Scholar]
- 42.Mollet, B., J. Knol, B. Poolman, O. Marciset, and M. Delley. 1993. Directed genomic integration, gene replacement, and integrative gene expression in Streptococcus thermophilus. J. Bacteriol. 175:4315-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mora, D., M. Scarpellini, L. Franzetti, S. Colombo, and A. Galli. 2003. Reclassification of Lactobacillus maltaromicus (Miller et al. 1974) DSM 20342(T) and DSM 20344 and Carnobacterium piscicola (Collins et al. 1987) DSM 20730(T) and DSM 20722 as Carnobacterium maltaromaticum comb. nov. Int. J. Syst. Evol. Microbiol. 53:675-678. [DOI] [PubMed] [Google Scholar]
- 44.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 45.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 46.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pridmore, D., T. Stefanova, and B. Mollet. 1994. Cryptic plasmids from Lactobacillus helveticus and their evolutionary relationship. FEMS Microbiol. Lett. 124:301-305. [DOI] [PubMed] [Google Scholar]
- 48.Rosen, B. P. 1999. Families of arsenic transporters. Trends Microbiol. 7:207-212. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz-Barba, J. L., J. C. Piard, and R. Jiménez-Díaz. 1991. Plasmid profiles and curing of plasmids in Lactobacillus plantarum strains isolated from green olive fermentations. J. Appl. Bacteriol. 71:417-421. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 51.Sasaki, Y., N. Taketomo, and T. Sasaki. 1988. Factors affecting transfer frequency of pAMβ1 from Streptococcus faecalis to Lactobacillus plantarum. J. Bacteriol. 170:5939-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skaugen, M. 1989. The complete nucleotide sequence of a small cryptic plasmid from Lactobacillus plantarum. Plasmid 22:175-179. [DOI] [PubMed] [Google Scholar]
- 53.Spizizen, J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. USA 44:1072-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takiguchi, R., H. Hashiba, K. Aoyama, and S. Ishii. 1989. Complete nucleotide sequence and characterization of a cryptic plasmid from Lactobacillus helveticus subsp. jugurti. Appl. Environ. Microbiol. 55:1653-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson, J. K., S. Foley, K. J. McConville, C. Nicholson, M. A. Collins, and R. D. Pridmore. 1999. Complete sequence of plasmid pLH1 from Lactobacillus helveticus ATCC15009: analysis reveals the presence of regions homologous to other native plasmids from the host strain. Plasmid 42:221-235. [DOI] [PubMed] [Google Scholar]
- 57.van Kranenburg, R., J. D. Marugg, I. I. van Swam, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 58.Vujcic, M., and L. Topisirovic. 1993. Molecular analysis of the rolling-circle replicating plasmid pA1 of Lactobacillus plantarum A112. Appl. Environ. Microbiol. 59:274-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, A., and F. L. Macrina. 1995. Characterization of six linked open reading frames necessary for pIP501-mediated conjugation. Plasmid 34:206-210. [DOI] [PubMed] [Google Scholar]
- 60.Weaver, K. E., D. B. Clewell, and F. An. 1993. Identification, characterization, and nucleotide sequence of a region of Enterococcus faecalis pheromone-responsive plasmid pAD1 capable of autonomous replication. J. Bacteriol. 175:1900-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilcks, A., L. Smidt, O. A. Økstad, A.-B. Kolstø, J. Mahillon, and L. Andrup. 1999. Replication mechanism and sequence analysis of the replicon of pAW63, a conjugative plasmid from Bacillus thuringiensis. J. Bacteriol. 181:3193-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]