Abstract
The determination of bacterial viability in probiotic products is of economic, technological, and clinical significance. We compared four methods to enumerate three Bifidobacterium strains in fermented oat products during storage. A subpopulation of nonculturable cells retained a functional cell membrane typical of viable cells, indicating that probiotic bacteria become dormant during storage.
Determination of bacterial viability is a complex issue, as illustrated by the numerous scientific papers published on the topic (3, 5, 7). Traditionally, plate counting has been the method of choice for viability assays, but there are obvious disadvantages (2). The use of this method is based on the conceptions that culturability is synonymous with viability and that the replication of a cell on a suitable agar medium is the only direct proof of cell culturability (5). For readily culturable microorganisms, this outlook is justified (1). However, a simple two-value logic system of dividing bacteria into either a viable or nonviable category does not adequately portray the bacterial life cycle. Kell and colleagues (5) have suggested four terms to describe different stages of microorganisms: viable (active and readily culturable), dormant (inactive but ultimately culturable), active but nonculturable, and dead (inactive and nonculturable). Most studies describing dormant bacteria have been conducted using pathogenic microorganisms. The reliable determination of the viability of probiotic bacteria is of technological, clinical, and economic significance. We determined the changes in viability occurring in fermented oat products containing specific Bifidobacterium strains during storage. The four methods used in the experiment were plate counting, fluorescent in situ hybridization (FISH), quantitative real-time PCR, and a commercial LIVE/DEAD BacLight bacterial viability kit (L/D; Molecular Probes). The aim of this study was to define the viabilities of subpopulations of probiotic bacteria in products during storage.
A sterile oat-in-water suspension (“oat milk”) was fermented with Bifidobacterium longum 2C (DSM 14579), B. longum 46 (DSM 14583), or Bifidobacterium lactis Bb-12 (Chr. Hansen, Hørsholm, Denmark). The numbers of bifidobacteria in the fermented products (pHs below 4.5) were monitored during storage at 4°C by using four methods. Plate counts were obtained by plating diluted products on reinforced clostridial medium supplemented with 1.5% agar. FISH analysis was performed by the method described by Langendijk and associates (6). The quantitative real-time PCR methodology described by Gueimonde and colleagues (4) for the quantification of intestinal bifidobacteria was used to analyze Bifidobacterium levels in oat products. The oligonucleotides and PCR conditions previously described (4) were used to quantify B. longum in the products containing this microorganism. To quantify the bacteria in products containing Bb-12, a set of oligonucleotide primers and probes specific for B. animalis and B. lactis was designed (Table 1), and the specificities of the oligonucleotides were tested against an array of different intestinal and food microorganisms (data not shown). To test the cell membrane integrity, a commercial LIVE/DEAD BacLight bacterial viability kit was used according to the manufacturer's instructions. The green fluorescence of the samples was analyzed with a Victor2 multilabel counter (Perkin-Elmer, Turku, Finland) and compared with a previously obtained standard curve (data not shown). Heat-treated and acetone-treated cells were used as controls. A strictly linear relationship between plate counts and L/D counts in a freshly fermented sample was established. After the linearity of the fluorescent response was established and the detection limit was determined, the following equation was formulated to estimate the number of living cells determined by using the L/D assay: cx = [(ax − b)/(a0 − b)] × c0, where cx is the number of living cells (based on L/D staining) in the suspension after x days, ax is the green fluorescence of the suspension after x days, a0 is the green fluorescence of the suspension after 0 days, b is the average background green fluorescence, and c0 is the total number of living cells in fresh suspension, determined by plate counting.
TABLE 1.
Oligonucleotides used for the quantification of B. lactis by real-time PCRa
| Oligonucleotide | Sequence (5′-3′)b |
|---|---|
| Animalis5 | ACCAACCTGCCCTGTGCACCG |
| Animalis3 | CCATCACCCCGCCAACAAGCT |
| Aniprobe | EACCATGCGATGGAGCGGAGCATCCGGTTA |
| Aniquencher | GCTCCATCGCATGGTD |
The annealing temperature of the assay was 67°C.
Bold letters indicate bases that are not complementary to the target. E, 2,2′,2′′,2′′′-{{6,6′-{4′′-[2-(4-isothiocyanatophenyl)ethyl]-1H-pyrazole-1′′,3′′-diyl}bis(pyridine)-2,2′-diyl}bis(methylenenitrilo)}tetrakis(acetato)europium(III). D, dabcyl.
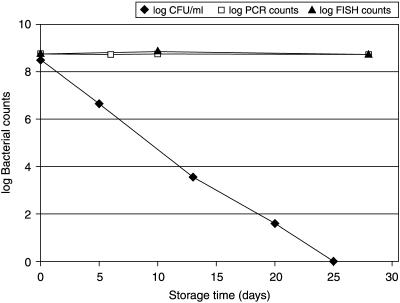
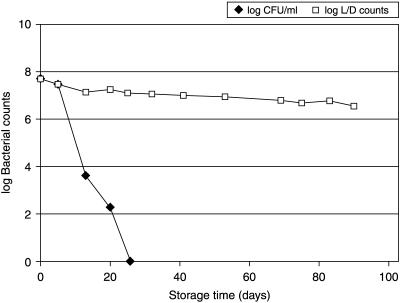
Plate counts decreased approximately 2 log units per week for B. longum (Fig. 1) but remained stable for B. lactis. FISH and real-time PCR counts remained unchanged for all bacteria, suggesting that bacterial DNA was intact and no cell lysis had occurred (Fig. 1). However, viability counts of B. longum determined by the LIVE/DEAD kit remained relatively stable, decreasing 1.2 log units in 3 months (Fig. 2). The striking difference between plate count results and L/D count results indicates that a subpopulation of the B. longum strains may have entered a dormant stage or possibly an active but nonculturable stage. Such a conception is further supported by the fact that the L/D counts continued to change for more than 2 months after cells had lost their culturability. If the two methods measured the same subpopulation of cells, one would expect the L/D counts to have remained more or less unchanged after plate counts had reached the detection limit. However, L/D count results and plate count results appeared to be independent of each other. We suggest that this is because the two methods measure different subpopulations of bacteria. The plate count method counts cells which are viable and culturable on nutrient agar, whereas the L/D assay counts the readily culturable cells and also cells which have an intact and functional cell membrane typical of viable cells but do not form colonies on conventional growth media. The data obtained here do not allow us to determine whether cells are ultimately culturable. However, the results indicate that although the bacteria are not readily culturable, they are not necessarily dead as defined by Kell and colleagues (5).
FIG. 1.
Plate count, real-time PCR, and FISH results of the products containing B. longum.
FIG. 2.
Plate count and L/D assay results of the products containing B. longum.
These findings highlight the need for further assessment of the methods applied to determine the viabilities of probiotic products. The evaluation of cell viabilities in stored probiotic foods is of vital importance economically and technologically, and it is also important for efficacy. In addition, the reliable determination of viability is a prerequisite to the much-needed regulation of and legislation on the quality of probiotic products. To our knowledge, this study is the first to report a subpopulation of potentially probiotic cells entering a state of reduced culturability while maintaining a functional cell membrane during prolonged storage. Further work will be required to determine whether these bacteria are truly viable and ultimately culturable or the observed dormant-like state of the bacteria is merely the gateway to imminent cell death.
Acknowledgments
We thank Riikka Parhiala for her skillful assistance.
S.J.L. was supported by the Applied Biosciences Graduate School.
REFERENCES
- 1.Barer, M. R., R. J. Smith, R. P. Cooney, and P. T. Kimmitt. 2000. Relationships between culturability, activity and virulence in pathogenic bacteria. J. Infect. Chemother. 6:108-111. [DOI] [PubMed] [Google Scholar]
- 2.Breeuwer, P., and T. Abee. 2000. Assessment of viability of microorganisms employing fluorescence techniques. Int. J. Food Microbiol. 55:193-200. [DOI] [PubMed] [Google Scholar]
- 3.Colwell, R. R. 2000. Viable but nonculturable bacteria: a survival strategy. J. Infect. Chemother. 6:121-125. [DOI] [PubMed] [Google Scholar]
- 4.Gueimonde, M., S. Tölkkö, T. Korpimäki, and S. Salminen. 2004. New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples. Appl. Environ. Microbiol. 70:4165-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kell, D. B., A. S. Kaprelyants, D. H. Weichart, C. R. Harwood, and M. R. Barer. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Leeuwenhoek 73:169-187. [DOI] [PubMed] [Google Scholar]
- 6.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nystrom, T. 2001. Not quite dead enough: on bacterial life, culturability, senescence, and death. Arch. Microbiol. 176:159-164. [DOI] [PubMed] [Google Scholar]