Abstract
Bacteriophage ΦJL001 infects a novel marine bacterium in the α subclass of the Proteobacteria isolated from the marine sponge Ircinia strobilina. ΦJL001 is a siphovirus and forms turbid plaques on its host. The genome sequence of ΦJL001 was determined in order to better understand the interaction between the marine phage and its sponge-associated host bacterium. The complete genome sequence of ΦJL001 comprised 63,469 bp with an overall G+C content of 62%. The genome has 91 predicted open reading frames (ORFs), and 17 ORFs have been assigned putative functions. ΦJL001 appears to be a temperate phage, and the integrase gene was identified in the genome. DNA hybridization analysis showed that the ΦJL001 genome does not integrate into the host chromosome under the conditions tested. DNA hybridization experiments therefore suggested that ΦJL001 has some pseudolysogenic characteristics. The genome of ΦJL001 contains many putative genes involved in phage DNA replication (e.g., helicase, DNA polymerase, and thymidylate synthase genes) and also contains a putative integrase gene associated with the lysogenic cycle. Phylogeny based on DNA polymerase gene sequences indicates that ΦJL001 is related to a group of siphoviruses that infect mycobacteria. Designation of ΦJL001 as a siphovirus is consistent with the morphology of the phage visualized by transmission electron microscopy. The unique marine phage-host system described here provides a model system for studying the role of phages in sponge microbial communities.
Bacteriophages are important components in many natural microbial ecosystems, and they are known to play an important role in maintaining the composition and structure of microbial communities (13). Viruses are highly abundant in the marine environment, reaching concentrations of 107 to 108 viruses per ml (7, 37). These large numbers of marine viruses can exert significant control in marine bacterial communities with respect to both species composition and genetic transfer (5, 6, 18, 34). To understand the ecological role of viruses in the marine environment, it is necessary to know the infectivity of viruses and the types of interactions that occur between marine viruses and their host bacteria. The potential host bacteria include not only free-living heterotrophic bacteria in the water column but also symbionts of marine invertebrates, including sponges.
Sponges are filter feeders that are capable of filtering thousands of liters of water; for example, a 1-kg sponge can to filter up to 24,000 liters a day (30). The filtering of these enormous amounts of water has the potential for introducing billions of phages into the sponge. Studies of sponge-associated bacterial communities have revealed that several bacterial groups and species are ubiquitous in sponges throughout the world (17). Members of the α subclass of the Proteobacteria are a well-represented group in the complex and the highly diverse sponge microbial communities (17, 19). The host bacterium in the host-phage system described here is a member of the α subclass of the Proteobacteria. Interestingly, representatives of the α subclass have been shown to be dominant members of the culturable bacterial assemblage in some sponges. A member of this subclass dominated the culturable assemblage associated with the sponge Halichondria panicea in the Adriatic Sea, North Sea, and Baltic Sea (2). Analysis of the culturable bacteria present in Rhopaloeides odorabile showed that strain NW001, a member of the α subclass, is the dominant culturable bacterium in this Great Barrier Reef sponge (32). The role of members of the α subclass of the Proteobacteria in sponges remains unknown, although Althoff et al. (2) and Webster and Hill (32) postulated that strains belonging to the α subclass are true sponge symbionts. In two clearly diseased individuals of R. odorabile, strain NW001 could not be isolated, and another member of the α subclass of the Proteobacteria, strain NW4437, dominated the culturable bacterial community (32). Strain NW4437 was shown to be pathogenic for the sponge (33). In the absence of certain strains belonging to the α subclass, the health of sponges may be compromised. In other cases, members of the α subclass of the Proteobacteria appear to be the cause of necrosis (33).
A phage designed to specifically eliminate a particular member of the α subclass of the Proteobacteria could be used as a precise tool for investigating the interaction between these bacteria and sponges. The use of bacteriophages, such as ΦJL001, to specifically target and manipulate microbes in the highly diverse and complex microbial community of sponges should be a invaluable tool for elucidating the roles of sponge symbionts, as well as the roles of phages, in sponge microbial communities. The first description of a complete genome sequence of a marine phage that infects a sponge-associated bacterium is presented here, and the relationship between ΦJL001 and its host, strain JL001 isolated from the sponge Ircinia strobilina, is described below.
MATERIALS AND METHODS
Sample collection and isolation of sponge-associated bacteria.
The sponge I. strobilina was collected at Tennessee Reef just off Key Largo during a research cruise of the Harbor Branch Oceanographic Institution on 24 August 1999. I. strobilina was collected by SCUBA at a depth ca. 10 m, and a 20-liter water sample was taken from the water column immediately surrounding the sponge. All samples were kept at ambient temperature until they were processed (<3 h). The sponge was surface sterilized by washing it briefly with 70% ethanol, followed by rinsing with sterile artificial seawater. By using aseptic techniques, a 1-cm3 section of sponge tissue was excised and homogenized in 10 ml of sterilized seawater with a mortar and pestle. Heterotrophic bacteria were isolated from serial dilutions of processed sponge material spread onto marine agar 2216 plates (Difco, Detroit, Mich.), and cyanobacteria and microalgae were isolated by inoculating dilutions of the sponge tissue into Mn+B12 liquid medium (31). All organisms were grown at 30°C.
Viral concentration.
Prefiltration of the water samples was carried out by two-stage filtration by using no. 3 filters mounted in stainless steel filter holders (Whatman, Clifton, N.J.) and then a 0.2-μm-pore-size polycarbonate filter (Whatman). Viral particles in the water samples were concentrated ca. 200-fold with an S1OY30 Amicon spiral wound cartridge system (Millipore, Bedford, Mass.).
Viral concentrate (1 ml) was added to an algal culture (100 ml) isolated from the I. strobilina sponge.
PFGE and Southern hybridization.
Viral amplification was monitored by pulsed-field gel electrophoresis (PFGE). Supernatants from an algal culture incubated with viral concentrate were prepared for PFGE by using previously described methods (38). PFGE of samples was performed by using a clamped homogeneous electric field system (CHEF DR-III; Bio-Rad, Richmond, Calif.) under the following conditions: 1% (wt/vol) agarose in 1× Tris-borate-EDTA gel buffer (90 mM Tris-borate, 1 mM EDTA; pH 8.0), 0.5× Tris-borate-EDTA tank buffer, 1- to 15-s pulse ramp, 200-V current at a constant temperature of 14°C, and a run time of 22 h.
DNA plugs containing cells of strain JL001 were prepared for PFGE analysis by a previously described procedure, with slight modifications (24). Lysozyme treatment was performed for 4 h at 37°C, and this was followed by 18 h of incubation at 50°C with 1 mg of proteinase K per ml in a solution containing 100 mM EDTA (pH 8.0), 0.2% (wt/vol) sodium deoxycholate, and 1% (wt/vol) sodium lauryl sarcosine. Plugs were rinsed with 20 mM Tris-HCl-50 mM EDTA (pH 8.0) four times for 1 h at room temperature; Phenylmethylsulfonyl fluoride (1 mM) was included in the second rinse solution. PFGE was performed with a CHEF DR-III apparatus under the following conditions: 0.6% (wt/vol) chromosomal-grade agarose in 1× Tris-acetate-EDTA (TAE) gel buffer and 1× TAE tank buffer. Electrophoresis was performed in two blocks. Block 1 was performed by using a switch time of 20 to 24 min 29 s and a current of 200 V with an angle of 106° for 72 h; block 2 was performed by using a switch time of 5.6 s to 2 min 26 s and a current of 200 V with an angle of 120° for 6 h 28 min. Gels were stained with SYBR Green I (PE Applied Biosystems, Foster City, Calif.) used according to the manufacturer's instructions and were visualized with a FluorImager 573 (Molecular Dynamics, Sunnyvale, Calif.).
Viral and bacterial DNA was transferred from pulsed-field gels to H+ Hybond nylon membranes (Amersham Pharmacia Biotech Ltd., Little Chalfont, Bucks, United Kingdom) (39). Membranes were probed with a ΦJL001-specific 32P-radiolabeled probe at a final concentration of ca. 1 × 106 cpm ml−1 (39). The membranes were washed twice for 15 min in 5× SSC-0.5% (wt/vol) sodium dodecyl sulfate (SDS) at 25°C, twice for 15 min in 1× SSC-0.5% (wt/vol) SDS at 37°C, and once for 15 min in 0.1× SSC-1% (wt/vol) SDS) at 37°C (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
Isolation of host bacterium and bacteriophages.
Heterotrophic bacteria present in the microalgal culture of interest were isolated on marine agar 2216 (Difco). Bacterial cultures were then grown in 50 ml of marine broth in 250-ml flasks with shaking at 30°C. Supernatants used to infect the potential hosts were filtered (pore size, 0.2 μm) to ensure that they were bacterium free but could contain any viral particles present in the original nonaxenic culture. The potential hosts (0.1 ml) were incubated with dilutions of the supernatant from the microalgal culture (0.9 ml), and overlays were made with 0.6% marine agar overlaid on marine agar 2216 plates. The phage was isolated and propagated after incubation at 30°C for 3 days.
Identification of bacterial strain JL001.
The DNA of the bacterial isolate was extracted by using a previously described small-scale DNA extraction method (28). PCR amplification of the ca. 1,500-bp 16S rRNA gene fragment from the purified genomic DNA was carried out with primers 8-27F (35) and 1492R (25). Thermal cycling was initiated by denaturation at 94°C for 3 min, and this was followed by denaturation at 92°C for 1.5 min, annealing at 48°C for 1.5 min, and extension at 72°C for 3 min. Thermal cycling was performed in a PTC-200 cycling system (MJ Research, Waltham, Mass.) for 25 cycles of denaturation, followed by a final extension at 72°C for 5 min. The PCR product was subjected to electrophoresis on a 1% agarose gel, stained with ethidium bromide, and visualized by UV excitation. The band of the expected size was excised and purified by using a QIAquick gel extraction kit (QIAGEN, Chatsworth, Calif.). The purified product was sequenced by using an ABI model 373 automated sequencer (Applied Biosystems, Foster City, Calif.). The 16S rRNA gene sequence was aligned manually by using the Phydit software (11), and sequencing data were analyzed by comparison to the sequences of the nearest relatives found by searching the GenBank database with the Basic Local Alignment Search Tool (BLAST) (3). A phylogenetic tree was constructed by using the PHYLIP software and the neighbor-joining method with Jukes-Cantor corrections (27).
Induction of putative lysogens.
Putative lysogens were inoculated into 100 ml of marine broth to obtain an initial optical density at 600 nm of 0.05. Triplicate cultures were incubated with shaking at 30°C. After overnight incubation, the logarithmic-phase cultures were induced with 0.1, 0.25, and 1.0 μg of mitomycin C per ml or by exposure to a wall-mounted germicidal UV light (Phillips Sterilamp G36T6L 39 W) that was 0.75 m above the cultures for 30, 60 or 90 s. Cultures were maintained in the dark during assays to prevent photorepair effects. Phage titers were determined immediately prior to treatment and at 48 and 96 h posttreatment.
Transmission electron microscopy examination of phage morphology.
Lysate was prepared from plaque assays. Plates with confluent lysis were chosen, and phage were collected from the soft agar by shaking the plates gently for 10 min with 10 ml of Mn+B12 liquid medium. The supernatant was centrifuged for 10 min at 5,000 × g to remove agar and filtered through a 0.2-μm-pore-size filter (Millipore) to remove any of the host bacterium present. This lysate was fixed in 2% (wt/vol) glutaraldehyde. The viral particles were harvested directly onto nickel grids by centrifugation (200,000 × g, 30 min) and negatively stained with uranyl acetate (2% [wt/vol] in water).
Phage sequencing.
Lysate was harvested and phage DNA was extracted by using methods described previously (28). The viral DNA was randomly sheared by passage through a 26-gauge needle. Passages through the needle were experimentally determined to give good yields of sheared DNA in the 1- to 5-kb size range (results not shown). Sheared DNA was blunt ended by using a DNA terminator end repair kit (Lucigen, Middleton, Wis.). Blunt-ended fragments were gel purified on a 1× TAE-1% (wt/vol) agarose gel. Fragments of the desired size (1 to 5 kb) were excised and extracted with a QIAquick gel extraction kit (QIAGEN). The resulting fragments were ligated into the pSMART vector and electroporated into Ecloni 10G electrocompetent cells (Lucigen). Transformed cells were spread onto Terrific broth plates (Lucigen) and incubated overnight at 37°C. Colonies were subcultured the following day.
Sequencing was performed by Agencourt (Beverly, Mass.) and with an model ABI 373 automated sequencer (Applied Biosystems, Perkin-Elmer) at the BioAnalytical Services Laboratory of the Center of Marine Biotechnology. Each clone was sequenced with plasmid forward and reverse primers SL1 (5′ CAGTCCAGTTACGCTGGAGTC 3′) and SR1 (5′ CTTTCTGCTGGAGGGGTCAGGTATG 3′). Consensus sequences were assembled by using the AssemblyLIGN (Accelrys, San Diego, Calif.) and PhredPhrap software. For the larger cloned fragments, internal primers were designed from the original sequence by using MacVector (Accelrys) and were synthesized by Sigma-Genosys (The Woodlands, Tex.). The BLAST programs Blastn, Blastx, Blastp, and PSI-BLAST were used to compare continuous sequences (contigs) to nucleotide and amino acid databases (3). Searches for open reading frames (ORFs), were performed online with the WebGeneMark.htm software (4). The predicted genes were compared with sequences in the GenBank database. A hit was considered significant if it had an E value of <0.001.
Phylogenetic analysis of a functional gene.
The translated amino acid sequence for the putative genes encoding DNA polymerase and integrase was used to construct phylogenetic trees. The amino acid sequence was aligned with the sequences of viruses found in the GenBank database with the program CLUSTAL W by using MacVector 7.1. A Blosum 30 matrix was calculated with a gap penalty of 10.0. Evolutionary distances were calculated by the Jukes-Cantor method, and a distance tree was constructed with the neighbor-joining algorithm. The bootstrap method was employed with 1,000 replicates to estimate the robustness of the tree topologies.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequence of strain JL001 has been deposited in the GenBank database under accession no. AY584527, and the complete genome sequence of ΦJL001 has been deposited in the GenBank database under accession no. AY576273.
RESULTS
Isolation of host bacterium.
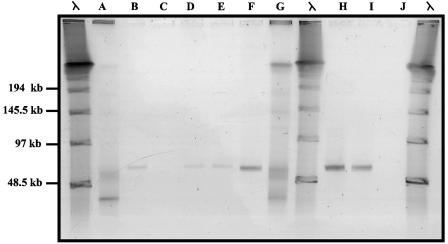
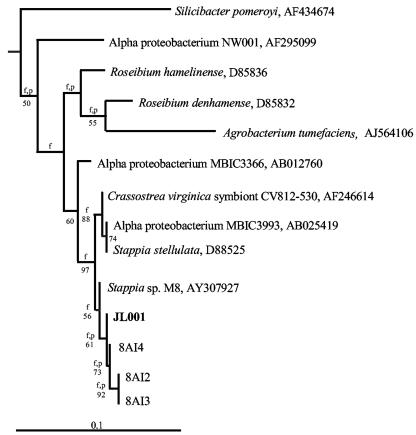
The microalgal culture isolated from the sponge I. strobilina infected with the viral concentrate was monitored biweekly by using PFGE to detect viral amplification. A band indicating a possible increase in the level of a specific virus was detected after 8 weeks (Fig. 1). There were no visible signs of lysis of the microalgal culture. Several transfers of the algal culture were made, and a band suggesting viral amplification was consistently observed by PFGE after 8 weeks. Since no lysis of the microalgal culture was observed, we considered the possibility that this band indicated the presence of a bacteriophage infecting a heterotrophic bacterium from the original sample of I. strobilina that was still present in the algal culture. The algal culture was plated onto marine agar 2216, and four morphotypes of heterotrophic bacteria were isolated. These bacteria were all closely related α-Proteobacteria, and their phylogenic relationship is shown in Fig. 2. Marine agar overlays inoculated with putative phage-containing supernatant from the algal culture were prepared with each of these bacteria. Plaques were observed on the strain designated JL001, indicating that this strain was the host of a phage present in the algal culture. The phage, designated ΦJL001, formed turbid plaques after 3 days of incubation. Strain JL001 had a distinctive light brown colony morphology after 3 days of growth on marine agar 2216 and was estimated to have comprised ca. 8% of the total culturable bacteria originally isolated from the sponge, based on counts of this colony morphology on the initial isolation plates.
FIG. 1.
PFGE analysis of microalgal cultures, showing the appearance of an approximately 60-kb band 8 weeks after addition of viral concentrates. Lanes A and G, viral concentrates; lanes B, D, E, and F, cultures to which concentrate from lane A was added; lanes H and I, cultures to which concentrate from lane G was added; lanes C and J, control cultures to which no viral concentrate was added. Molecular size markers are λ concatemer ladders.
FIG. 2.
Neighbor-joining tree based on the 16S rRNA gene sequence, showing the phylogenetic relationship between strain JL001 and its close relatives. Levels of bootstrap support greater than 50% based on a neighbor-joining analysis of 1,000 resampled data sets are indicated at the nodes. Branches that were also found when the Fitch-Margoliash and maximum-parsimony methods were used are indicated by f and p, respectively. Scale bar = 0.1 substitution per nucleotide position.
Phage biology and lysogenic characteristics.
After infection of turbid cultures of the host, strain JL001, with high titers (ca. 109 phage particles per ml of culture), lysis of the liquid culture was not observed. It was not possible to make high-titer preparations of ΦJL001 from liquid cultures of the host in the logarithmic growth phase. Instead, high-titer preparations of ΦJL001 were harvested from lawns of confluent lysis in sloppy overlays of strain JL001. These characteristics suggested that ΦJL001 might be a temperate phage rather than a lytic phage. In order to determine if the phage entered into a lysogenic state with its host, strain JL001 was examined for prophage induction by using mitomycin C and UV radiation treatments. The greatest induction occurred 96 h after treatment with mitomycin C at a concentration of 0.1 μg/ml (2.2 × 1014 ± 0.4 × 1014 PFU/ml, compared with the control containing 3.8 × 1013 ± 1 × 1013 PFU/ml) and UV treatment for 90 s (5.5 × 1012 ± 4 × 1012 PFU/ml, compared with the control containing 1.0 × 1012 ± 0.4 × 1012 PFU/ml). Induction resulted in increases in phage counts of ca. 0.5 to 1 order of magnitude compared with uninduced controls.
The phage produced several plaque morphologies on its host, including turbid plaques and plaques with haloes, from which three putative lysogens were isolated, which were designated strains JL002, JL003, and JL004. The possibility of a mixed lysate was discounted since phage preparations were prepared from single plaques and each plaque morphology on replating once again gave several different plaque morphologies. 16S rRNA gene sequencing confirmed that these isolates had that same 16S rRNA sequence as the original host bacterium, strain JL001. The colony morphology of the three putative lysogens differed from the colony morphology of strain JL001, and the colonies were more raised colonies and were white rather than light brown. The three putative lysogens had a colony morphology that differed from the colony morphology of the original host strain, which formed smooth and rounded colonies. The putative lysogens formed colonies that were slightly smaller with a rough, crinkled texture and irregular edges. Small white colonies formed on agar overlays but were not isolated.
Homoimmunity of lysogens.
When phage ΦJL001 was tested with strains JL002, JL003, and JL004, no plaques or clearing was observed on the bacterial lawns, suggesting homoimmunity of these lysogens to phage ΦJL001, although the possibility that the phenotype was due to cells that were resistant to infection but were not true lysogens cannot be ruled out. Control overlays with strain JL001 showed a high level of plaque formation when they were challenged with ΦJL001.
Transmission electron microscopy examination of phage morphology.
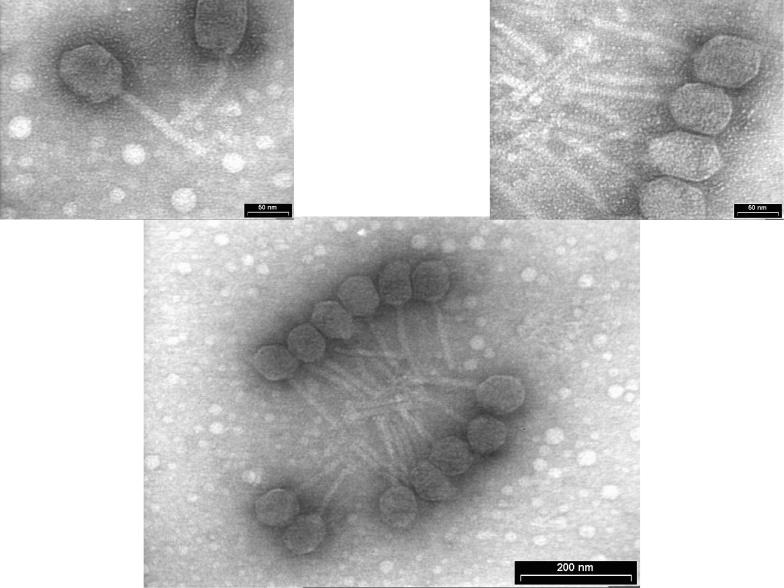
Morphological characteristics of ΦJL001 were compiled from multiple micrographs of phage particles in order to minimize size or shape anomalies (Fig. 3). The phage head diameter was ca. 75 nm, while the tail length was ca. 125 nm. The tail morphology is typical of phages in the family Siphoviridae.
FIG. 3.
Morphological characteristics of ΦJL001. The phage head diameter is ca. 75 nm, and the tail length is ca. 125 nm. The top left image shows a phage with a slightly bent tail, indicating that the tail is flexible, which is characteristic of phage in the family Siphoviridae.
Integration of ΦJL001 into the genome of strain JL001.
The results of Southern hybridization studies of total DNA preparations of strain JL001 and the putative lysogen strains JL002, JL003, and JL004 probed with a radiolabeled fragment of ΦJL001 are shown in Fig. 4. PFGE of preparations of the putative lysogens revealed bands at sizes that corresponded approximately (but not exactly) to the size observed for ΦJL001. The size differences for the band at ca. 60 kb for the putative lysogens and the original phage lysate may have been due to overloaded wells, binding of protein present in the chromosomal preparations, or rearrangements of the phage genome. The ca. 60-kb bands in preparations from the putative lysogens hybridized strongly with the 1,500-bp radiolabeled probe from ΦJL001, while the original host strain did not contain a corresponding band (Fig. 4). No hybridization signal was detected at the position corresponding to the chromosomal DNA bands, indicating that phage ΦJL001 was not integrated into the chromosomes of the putative lysogens.
FIG. 4.
PFGE (left gel) and Southern hybridization (right gel). Lane 1, ΦJL001; lanes 2, 3, 4, putative lysogen strains JL002, JL003, and JL004, respectively; lane 5, strain JL001.
Phage sequence.
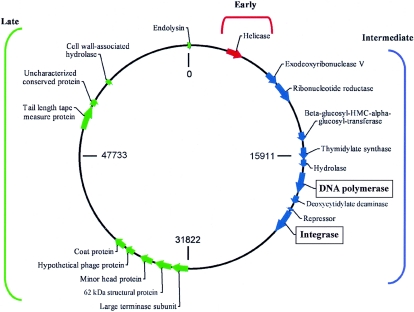
The genome of ΦJL001 is comprised of 63,469 bp and is circularly permuted. ΦJL001 has a G+C content of 62%. The overall coverage was 6× to 10×, and a few areas with lower coverage were checked by sequencing of PCR products covering these areas. GeneMark predicted 91 putative ORFs. Putative functions were assigned to 17 of the 91 ORFs, which are indicated in the circular map shown in Fig. 5. On this map, putative genes are transcribed in a clockwise direction, and they appear to be clustered in three regions: early genes, presumably for establishing virus infection (0 to 7,500 bp); intermediate genes, involved in DNA synthesis, modification, and replication (7,500 to 32,000 bp); and late genes, for the assembly and release of new virus particles (32,000 to 63,000 bp). A putative origin of replication was predicted in the early region by detection of a region with a low G+C content. The designations of the ORFs are shown in Table 1.
FIG. 5.
Circular genomic map of ΦJL001. The predicted genes are clustered into three regions; early genes are indicated by red, intermediate genes are indicated by blue, and late genes are indicated by green.
TABLE 1.
Bacteriophage ΦJL001 ORFs and putative genes
| ORF | Positions (bp) | Protein size (amino acids) | Accession no. | E value | Significant match | Conserved domain | Function |
|---|---|---|---|---|---|---|---|
| 1 | 1-375 | 125 | NSa | ||||
| 2 | 372-854 | 161 | NS | ||||
| 3 | 904-1020 | 39 | NS | ||||
| 4 | 1017-1277 | 87 | NP_772739 | 2.00E-06 | Bradyrhizobium japonicum genome | ||
| 5 | 1294-2931 | 546 | NS | ||||
| 6 | 3070-3249 | 60 | NS | ||||
| 7 | 32534773 | 507 | ZP_00060797 | 8.00E-14 | Hypothetical protein (Clostridium thermocellum) | COG0553.1 | HepA, SNF2 family of DNA/RNA helicases |
| 8 | 4770-5126 | 119 | NS | ||||
| 9 | 5243-5479 | 79 | NS | ||||
| 10 | 5472-6032 | 187 | NS | ||||
| 11 | 6078-6878 | 267 | NS | ||||
| 12 | 6974-7471 | 166 | NS | ||||
| 13 | 7468-8673 | 402 | NP_539536 | 3.00E-94 | Exodeoxyribonuclease V alpha chain (Brucella melitensis) | COG0507.1 | RecD, exonuclease V |
| 14 | 8670-10478 | 603 | ZP_00005311 | 1.00E-135 | Hypothetical protein (Rhodobacter sphaeroides) | COG0209.1 | NrdA, ribonucleotide reductase |
| 15 | 10494-10916 | 141 | NS | ||||
| 16 | 10929-11507 | 193 | NP_817561 | 7.00E-08 | gp114 (mycobacteriophage CJW1) | ||
| 17 | 11578-11778 | 67 | NS | ||||
| 18 | 11832-12287 | 152 | NS | ||||
| 19 | 12271-13137 | 289 | NP_821395 | 6.00E-10 | Hypothetical protein (Streptomyces avermitilis) | ||
| 20 | 13134-13490 | 119 | NS | ||||
| 21 | 13497-13622 | 42 | NS | ||||
| 22 | 13637-14455 | 273 | Q06718 | 1.00E-04 | Beta-glucosyl-hydroxymethyl cytosine-alpha-glucosyl-transferase (phage T6) | ||
| 23 | 14472-15071 | 200 | NS | ||||
| 24 | 15068-16120 | 351 | P31654 | 6.00E-32 | Deoxyuridylate hydroxymethyltransferase (phage SPO1) | COG0207.1 | ThyA, thymidylate synthase |
| 25 | 16095-16688 | 198 | NS | COG1896.1 | Predicted hydrolases of HD superfamily | ||
| 26 | 16685-17113 | 143 | NS | ||||
| 27 | 17110-17361 | 84 | NS | ||||
| 28 | 17361-19256 | 632 | NP_229419 | 6.00E-33 | DNA-directed DNA polymerase 1 (Thermotoga maritima) | COG0749.1 | PolA, DNA polymerase 1 |
| 29 | 19253-19492 | 80 | NS | ||||
| 30 | 19509-20243 | 245 | NP_635521 | 4.00E-26 | Deoxycytidylate deaminase (Xanthomonas campestris) | COG2131.1 | ComEB, deoxycytidylate deaminase |
| 31 | 20240-20683 | 148 | NS | ||||
| 32 | 20742-20972 | 77 | NS | cd00093.2 | Helix-turn-helix XRE family-like proteins | ||
| 33 | 21078-23321 | 748 | ZP_00048756 | 5.00E-42 | Hypothetical protein (Magnetospirillum magnetotacticum) | pfam05272.1 | VirE, virulence-associated protein E |
| 34 | 23407-23775 | 123 | NS | ||||
| 35 | 23861-24127 | 89 | NS | ||||
| 36 | 24124-24435 | 104 | NS | ||||
| 37 | 24432-24911 | 160 | NS | ||||
| 38 | 24908-24997 | 30 | NS | ||||
| 39 | 25072-25272 | 67 | NS | ||||
| 40 | 25269-25643 | 125 | NS | ||||
| 41 | 25654-26037 | 128 | NS | ||||
| 42 | 26037-26282 | 82 | NS | ||||
| 43 | 26285-26467 | 61 | NS | ||||
| 44 | 26552-26797 | 82 | NS | ||||
| 45 | 26809-27453 | 215 | NS | ||||
| 46 | 27658-27921 | 88 | NS | ||||
| 47 | 27965-28141 | 59 | NS | ||||
| 48 | 28155-28322 | 56 | NS | ||||
| 49 | 28331-28582 | 84 | NS | ||||
| 50 | 28579-28941 | 121 | NS | ||||
| 51 | 29059-29277 | 73 | NS | ||||
| 52 | 29274-29558 | 95 | NS | ||||
| 53 | 29555-29785 | 77 | NS | ||||
| 54 | 29808-29990 | 61 | NS | ||||
| 55 | 30058-30264 | 69 | NS | ||||
| 56 | 30261-30464 | 68 | NS | ||||
| 57 | 30461-31132 | 224 | NS | ||||
| 58 | 31302-31382 | 27 | NS | ||||
| 59 | 31498-32127 | 210 | NS | ||||
| 60 | 32124-33653 | 510 | ZP_00045650 | 6.00E-27 | Bacteriophage terminase large subunit (Mesorhizobium loti) | ||
| 61 | 33653-35212 | 520 | CAB53859 | 1.00E-15 | 62-kDa structural protein (bacteriophage MB78) | ||
| 62 | 35202-35438 | 79 | NS | ||||
| 63 | 35428-36639 | 404 | ZP_00127008 | 7.00E-35 | Hypothetical protein (Pseudomonas syringae) | pfam04233.3 | Phage Mu F-like protein, minor head protein |
| 64 | 36636-36863 | 76 | NS | ||||
| 65 | 36860-36976 | 39 | NS | ||||
| 66 | 37041-37214 | 58 | NS | ||||
| 67 | 37281-38150 | 290 | NP_405657 | 4.00E-06 | Hypothetical phage protein (Yersinia pestis) | ||
| 68 | 38296-39420 | 375 | NP_690674 | 2.00E-41 | Coat protein (bacteriophage SPP1) | ||
| 69 | 39514-40329 | 272 | NS | ||||
| 70 | 40346-41026 | 227 | NS | ||||
| 71 | 41030-42331 | 434 | NS | ||||
| 72 | 42328-42693 | 122 | NS | ||||
| 73 | 42696-43718 | 341 | NS | ||||
| 74 | 43718-45445 | 576 | NS | ||||
| 75 | 45445-45633 | 63 | NS | ||||
| 76 | 45617-46516 | 300 | NS | ||||
| 77 | 46513-46905 | 131 | NS | ||||
| 78 | 46902-47312 | 137 | NS | ||||
| 79 | 47309-48967 | 553 | NS | ||||
| 80 | 48977-49108 | 44 | NS | ||||
| 81 | 49171-49584 | 138 | NS | ||||
| 82 | 49909-50349 | 147 | NP_791903 | 7.00E-04 | Hypothetical protein (Pseudomonas syringae) | ||
| 83 | 50351-52609 | 753 | NP_536371 | 6.00E-23 | Putative tail length tape measure protein | COG5281.1 | Mu-like prophage protein |
| 84 | 52609-53229 | 207 | ZP_00004532 | 2.00E-31 | Hypothetical protein (Rhodobacter sphaeroides) | COG5448.1 | Uncharacterized conserved protein |
| 85 | 53231-55000 | 590 | ZP_00004533 | 2.00E-31 | Hypothetical protein (Rhodobacter sphaeroides) | COG5449.1 | Uncharacterized conserved protein |
| 86 | 55006-55434 | 143 | NP_421571 | 9.00E-06 | Hypothetical protein (Caulobacter crescentus CB15) | COG0791.1 | Cell wall-associated hydrolases |
| 87 | 55435-58893 | 1153 | ZP_00011489 | 2.00E-35 | Hypothetical protein (Rhodopseudomonas palustris) | ||
| 88 | 58897-60360 | 488 | NS | ||||
| 89 | 60365-62989 | 875 | ZP_00055704 | 6.00E-05 | Hypothetical protein (Magnetospirillum magnetotacticum) | ||
| 90 | 63040-63306 | 89 | NS | ||||
| 91 | 63312-63632 | 107 | NP_543082 | 7.00E-26 | Putative endolysin (bacteriophage P27) |
NS, not significant.
Phylogenetic analysis of a functional gene.
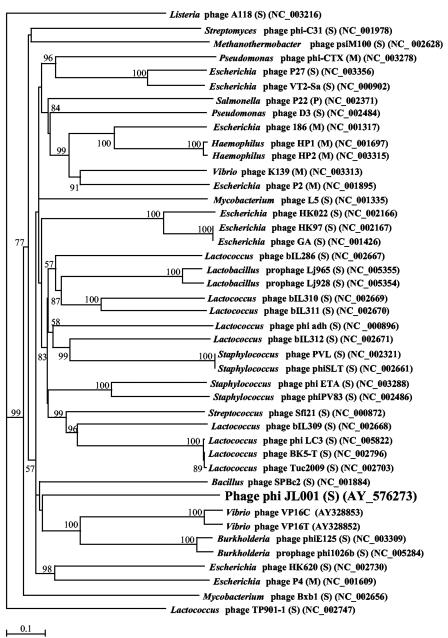
Comparison of the putative genes encoding DNA polymerase and integrase found in the genomic sequence of ΦJL001 to other phage DNA polymerase and integrase gene sequences deposited in the GenBank database suggested that ΦJL001 is affiliated with the family Siphoviridae. Phylogenetic trees based on the DNA polymerase and integrase genes are shown in Fig. 6 and 7, respectively. Both trees show that ΦJL001 is deeply rooted, suggesting that it is quite different from previously described phages. The polymerase gene of ΦJL001 appears to share an origin with a group of siphoviruses that infect mycobacteria.
FIG. 6.
Unrooted neighbor-joining tree based on the aligned amino acid sequences encoded by the DNA polymerase gene from ΦJL001 and 28 other phages. M, Myoviridae; S, Siphoviridae; P, Podoviridae. A total of 659 residues of the aligned region, including gaps, were used for phylogenetic reconstruction. A Blosum 30 matrix was calculated with a gap penalty of 10.0. The numbers at the nodes are bootstrap values based on 1,000 resamplings. Bootstrap values less than 50 are not shown. Scale bar = 0.2 amino acid substitution per residue.
FIG. 7.
Unrooted neighbor-joining tree based on the aligned amino acid sequences encoded by the integrase gene from ΦJL001 and 42 other phages. M, Myoviridae; S, Siphoviridae; P, Podoviridae. A total of 405 aligned residues, including gaps, were used for phylogenetic reconstruction. A Blosum 30 matrix was calculated with a gap penalty of 10.0. The numbers at the nodes are bootstrap values based on 1,000 resamplings. Bootstrap values less than 50 are not shown. Scale bar = 0.1 amino acid substitution per residue.
DISCUSSION
Phage ΦJL001 is the first marine phage that infects a sponge-associated bacterium that has been sequenced. Eleven marine phages have been sequenced previously. These phages are the roseophage SI01 (26), cyanophage P60 (10), the lipid-containing Pseudoalteromonas espejiana phage PM2 (20), Vibrio harveyi phage VHML (22), three Vibrio parahaemolyticus phages (VpV262, VP16T [14], and VP16C [29]), three Prochlorococcus phages (P-SSP7, P-SSM2, and P-SSM4 [18a]), and the broad-host-range vibriophage KVP40 (21). Only one of these phages, VHML, exhibits temperate characteristics (22), and none has been described as pseudolysogenic. Genomic analysis of uncultured marine viral communities indicated that the diversity of these communities is extremely high, with the number of viral types ranging from several hundred to several thousand in the two communities that were studied (8). Since it is clear that virioplankton are an active and important component of marine microbial communities (12, 37), it is certainly important to sequence genomes of representative marine viruses. Phage ΦJL001 is of interest in this regard since it shows some pseudolysogenic characteristics; pseudolysogenic phage-host interactions may be shared by many marine bacteria (36).
Morphological characteristics, sequence homology to previously described phages, and phylogenic analysis based on the putative DNA polymerase and integrase genes supported affiliation of ΦJL001 with the family Siphoviridae. Comparison of the 91 predicted ORFs with currently available sequences revealed the following relationships. First, there is a high proportion of unique genes (>50%) in the genome of ΦJL001 that are unrelated to genes of other bacteriophages or any other previously sequenced organism. Second, predicted genes with significant hits to genes encoding previously described proteins showed sequence homology to genes of several types of viruses, several groups of bacteria, and even higher eukaryotic organisms. Studies of phage evolution show that double-stranded DNA phages and prophages are mosaics that arose by horizontal transfer of genetic material from a global phage pool. The mosaic nature of ΦJL001 is consistent with findings for previously sequenced phages that indicate that phages are highly mosaic (9, 15, 16, 23).
Basic life histories of marine phages can be elucidated by comparison of their complete genomes to the genomes of other extensively studied phages (26). The genomic sequence of ΦJL001 is consistent with some known aspects of its biology. Many siphoviruses form lysogenic relationships with their hosts (29). Interestingly, a gene that encodes a putative integrase is found in most lysogenic siphovirus and myovirus genomes, suggesting that the phages are able to integrate their genomes into the host genome and become lysogenized (10). We detected a gene with homology to known integrases in the genome of ΦJL001. Lytic phages typically contain a DNA polymerase gene and some other genes (e.g., primase and helicase genes) associated with DNA replication. Most of the lysogenic phage genomes of members of the Siphoviridae and Myoviridae do not contain these DNA replication genes. The presence of both a putative DNA polymerase gene and an integrase gene in ΦJL001 is noteworthy considering that ΦJL001 displayed some pseudolysogenic characteristics. Including ΦJL001, only 7 of 84 known siphophage genomes contain both the DNA polymerase and integrase genes.
Lysogeny is characterized by homoimmunity to superinfection, physical and chemical induction, and integration of the phage genome into the host genome (1). Induction of ΦJL001 and the homoimmunity characteristics of ΦJL001 resemble true lysogeny. However, unlike what is observed in true lysogeny, the phage ΦJL001 genome does not integrate into host cellular replicons. This lack of chromosomal integration is consistent with pseudolysogeny (36).
Homoimmunity in phage is often the result of excess repressor molecules that render the lysogenic bacterium immune to superinfection. The continual synthesis of repressor molecules maintains the phage genome as a prophage and prevents the transcription of phage genes, leading to initiation of the lysogenic cycle (1). This interaction may be explained by two factors. First, the lysogen may exhibit certain characteristics of pseudolysogeny due to weak or poor repression of phage DNA transcription. A partially defective repressor protein may cause this effect. Pseudolysogenic characteristics include high host cell abundance along with a high rate of phage production. An unstable repressor would allow a high rate of spontaneous induction along with high host cell abundance. Alternatively, this relationship between phages and bacteria in the marine environment may be the result of a mixture of sensitive and resistant host cells and/or a mixture of virulent and temperate phages.
Phage ΦJL001 is the first phage isolated that infects a sponge-associated host bacterium. The characteristics of ΦJL001 suggest that this virus may have the potential to exert significant control on the population of its host within the sponge microbial community. Further studies of this host-phage system should allow us to better understand the relationship between the host and the phage and, at another level, the role of phages within the very complex sponge microbial community. The use of phages may allow workers to manipulate populations of key microbes in the community of a sponge and could lead to a clearer picture of factors that keep the diversity of microbial communities in balance. This unique host-phage system may provide a model for elucidating interactions that occur within sponge microbial communities.
Acknowledgments
Funding for this study was provided by the VIRTUE Program (Wallenberg Foundation) and by the Microbial Observatories Program, National Science Foundation (grant MCB-0238515). This support is gratefully acknowledged.
Shirley Pomponi and Harbor Branch Oceanographic Institution are thanked for enabling participation of R.T.H. in a research cruise. We thank Steven Miller and Otto Rutten for providing sampling opportunities at Key Largo through the UNC-Wilmington National Undersea Research Center. We are grateful to Gunnar Bratbak and Mikal Heldal for electron microscopy. Frank Robb and Olivier Peraud are thanked for helpful discussions.
Footnotes
Contribution no. 04-611 from the Center of Marine Biotechnology.
REFERENCES
- 1.Ackermann, H. W., and M. S. DuBow. 1987. General properties of bacteriophages, p. 49-85. In H. W. Ackermann and M. S. DuBow (ed.), Viruses of prokaryotes. CRC Press, Boca Raton, Fla.
- 2.Althoff, K., C. Schütt, R. Steffen, R. Batel, and W. E. G. Müller. 1998. Evidence for a symbiosis between bacteria of the genus Rhodobacter and the marine sponge Halichondria panicea: harbor also for putatively toxic bacteria? Mar. Biol. 130:529-536. [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bratbak, G., M. Heldal, F. Thingstad, B. Riemann, and O. H. Haslund. 1992. Incorporation of viruses into the budget of microbial C-transfer. A first approach. Mar. Ecol. Prog. Ser. 83:273-280. [Google Scholar]
- 6.Bratbak, G., M. Heldal, F. Thingstad, and P. Tuomi. 1996. Dynamics of virus abundance in coastal seawater. FEMS Microbiol. Ecol. 19:263-269. [Google Scholar]
- 7.Bratbak, G., F. Thingstad, and M. Heldal. 1994. Viruses and the microbial loop. Microb. Ecol. 28:209-221. [DOI] [PubMed] [Google Scholar]
- 8.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 99:14250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brussow, H., and F. Desiere. 2001. Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39:213-222. [DOI] [PubMed] [Google Scholar]
- 10.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun, J. 1995. Computer-assisted classification and identification of actinomycetes. Ph.D. thesis. University of Newcastle upon Tyne, Newcastle upon Tyne, United Kingdom.
- 12.Fuhrman, J. A. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541-548. [DOI] [PubMed] [Google Scholar]
- 13.Fuhrman, J. A., and C. A. Suttle. 1993. Viruses in marine planktonic systems. Oceanography 6:51-63. [Google Scholar]
- 14.Hardies, S. C., A. M. Comeau, P. Serwer, and C. A. Suttle. 2003. The complete sequence of marine bacteriophage VpV262 infecting Vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology 310:359-371. [DOI] [PubMed] [Google Scholar]
- 15.Hendrix, R. W. 2003. Bacteriophage genomics. Curr. Opin. Microbiol. 6:506-511. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix, R. W., M. C. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hentschel, U., J. Hopke, M. Horn, A. B. Friedrich, M. Wagner, J. Hacker, and B. S. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, S. C., and J. H. Paul. 1994. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine environment. Mar. Ecol. Prog. Ser. 104:163-172. [Google Scholar]
- 18a.Lindell, D., M. B. Sullivan, Z. I. Johnson, A. C. Tolonen, F. Rohwer, and S. W. Chisholm. 2004. Transfer of photosynthetic genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA 101:11013-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez, J. V., P. J. McCarthy, K. E. Janda, R. Willoughby, and S. A. Pomponi. 1999. Molecular techniques reveal wide phyletic diversity of heterotrophic microbes associated with Discodermia spp. (Porifera: Demospongia). Mem. Queensl. Mus. 44:329-341. [Google Scholar]
- 20.Mannisto, R. H., H. M. Kivela, L. Paulin, D. H. Bamford, and J. K. Bamford. 1999. The complete genome sequence of PM2, the first lipid-containing bacterial virus to be isolated. Virology 262:355-363. [DOI] [PubMed] [Google Scholar]
- 21.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oakey, H. J., B. R. Cullen, and L. Owens. 2002. The complete nucleotide sequence of the Vibrio harveyi bacteriophage VHML. J. Appl. Microbiol. 93:1089-1098. [DOI] [PubMed] [Google Scholar]
- 23.Pedulla, M. L., M. E. Ford, J. M. Houtz, T. Karthikeyan, C. Wadsworth, J. A. Lewis, D. Jacobs-Sera, J. Falbo, J. Gross, N. R. Pannunzio, W. Brucker, V. Kumar, J. Kandasamy, L. Keenan, S. Bardarov, J. Kriakov, J. G. Lawrence, W. R. Jacobs, Jr., R. W. Hendrix, and G. F. Hatfull. 2003. Origins of highly mosaic mycobacteriophage genomes. Cell 113:171-182. [DOI] [PubMed] [Google Scholar]
- 24.Ravel, J., H. Schrempf, and R. T. Hill. 1998. Mercury resistance is encoded by transferable giant linear plasmids in two Chesapeake Bay Streptomyces strains. Appl. Environ. Microbiol. 64:3383-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reysenbach, A.-L., L. J. Giver, G. S. Wickham, and N. R. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohwer, F., A. Segall, G. Steward, V. Seguritan, M. Breitbart, F. Wolven, and F. Azam. 2000. The complete genomic sequence of the marine phage Roseophage SI01 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408-418. [Google Scholar]
- 27.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Seguritan, V., I. W. Feng, F. Rohwer, M. Swift, and A. M. Segall. 2003. Genome sequences of two closely related Vibrio parahaemolyticus phages, VP16T and VP16C. J. Bacteriol. 185:6434-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogel, S. 1977. Current-induced flow through living sponges in nature. Proc. Natl. Acad. Sci. USA 74:2069-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waterbury, J. B., and R. Y. Stanier. 1978. Patterns of growth and development in pleurocapsalean cyanobacteria. Microbiol. Rev. 42:2-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webster, N. S., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-proteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 33.Webster, N. S., A. P. Negri, R. I. Webb, and R. T. Hill. 2002. A spongin-boring α-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 232:305-309. [Google Scholar]
- 34.Weinbauer, M. G., and C. A. Suttle. 1996. Potential significance of lysogeny to bacteriophage production and bacterial mortality in coastal waters of the Gulf of Mexico. Appl. Environ. Microbiol. 62:4374-4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williamson, S. J., M. R. McLaughlin, and J. H. Paul. 2001. Interaction of the PhiHSIC virus with its host: lysogeny or pseudolysogeny? Appl. Environ. Microbiol. 67:1682-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 64:69-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wommack, K. E., J. Ravel, R. T. Hill, J. Chun, and R. R. Colwell. 1999. Population dynamics of Chesapeake Bay virioplankton: total-community analysis by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 65:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wommack, K. E., J. Ravel, R. T. Hill, and R. R. Colwell. 1999. Hybridization analysis of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 65:241-250. [DOI] [PMC free article] [PubMed] [Google Scholar]