Abstract
Objective:
To construct a novel tumor-node-morphology (TNMor) staging system derived from natural language processing (NLP) of pathology reports to predict outcomes of pancreatic ductal adenocarcinoma.
Method:
This retrospective study with 1657 participants was based on a large referral center and The Cancer Genome Atlas Program (TCGA) dataset. In the training cohort, NLP was used to extract and screen prognostic predictors from pathology reports to develop the TNMor system, which was further evaluated with the tumor-node-metastasis (TNM) system in the internal and external validation cohort, respectively. Main outcomes were evaluated by the log-rank test of Kaplan–Meier curves, the concordance index (C-index), and the area under the receiver operating curve (AUC).
Results:
The precision, recall, and F1 scores of the NLP model were 88.83, 89.89, and 89.21%, respectively. In Kaplan–Meier analysis, survival differences between stages in the TNMor system were more significant than that in the TNM system. In addition, our system provided an improved C-index (internal validation, 0.58 vs. 0.54, P<0.001; external validation, 0.64 vs. 0.63, P<0.001), and higher AUCs for 1, 2, and 3-year survival (internal validation: 0.62 vs. 0.54, P<0.001; 0.64 vs. 0.60, P=0.017; 0.69 vs. 0.62, P=0.001; external validation: 0.69 vs. 0.65, P=0.098; 0.68 vs. 0.64, P=0.154; 0.64 vs. 0.55, P=0.032, respectively). Finally, our system was particularly beneficial for precise stratification of patients receiving adjuvant therapy, with an improved C-index (0.61 vs. 0.57, P<0.001), and higher AUCs for 1-year, 2-year, and 3-year survival (0.64 vs. 0.57, P<0.001; 0.64 vs. 0.58, P<0.001; 0.67 vs. 0.61, P<0.001; respectively) compared with the TNM system.
Conclusion:
These findings suggest that the TNMor system performed better than the TNM system in predicting pancreatic ductal adenocarcinoma prognosis. It is a promising system to screen risk-adjusted strategies for precision medicine.
Keywords: natural language processing, pancreatic ductal adenocarcinoma, pathological report, prognosis, stratification
Introduction
Highlights
An innovative Prompt enhanced Generative Machine Reading Comprehension (PGMRC) framework could automatically annotate and extract pathology entities with excellent precision in the training cohort with 1044 participants.
Compared with the tumor-node-metastasis staging system, our new tumor-node-morphology staging system was developed and provided better prognostic prediction accuracy and discriminatory ability for patients with resected pancreatic ductal adenocarcinoma in the training cohort, and the prognostic prediction performance were validated in the internal (448 participants) and external (165 participants) validation cohorts.
Our new tumor-node-morphology staging system might be a potential predictive tool for screening of risk-adjusted treatment strategies in the era of precision medicine.
Pancreatic ductal adenocarcinoma (PDAC) is a highly fatal malignancy1, with a 5-year survival rate of 9–10%2,3. Even after ʻcurativeʼ surgical resection and adjuvant therapy, only ~20% of patients survive 5 years, and less than 4% survive 10 years4–6. Currently, the prognostication and treatment allocation among patients with resected PDAC is based on the 8th edition of the tumor-node-metastasis (TNM) staging system of the American Joint Committee on Cancer (AJCC)7,8. However, this classical prognostic stratification remains to be optimized7,9,10 because of the large prognostic differences among patients with the same TNM stage and receiving the same treatment regimens11. Therefore, the incorporation of novel clinical biomarkers12,13 may be required to improve the existing prognostic system and further guide individualized postoperative management.
Pathology reports have been considered the gold standard for tumor diagnosis and prognosis9. In addition to three key parameters in TNM staging, routine pathology reports contain a wealth of valuable information about the tissue sample or the whole lesion’s location, size, and microscopic morphological features, etc. More importantly, the reports also include the pathologist’s diagnostic impressions or conclusions, which summarize the main findings and attempt to provide answers to clinical queries. Despite their significant clinical implications, current electronic health record (EHR) systems cannot extract and analyze unstructured textual reports due to the time-consuming and labor-intensive manual processing. Natural language processing (NLP) is an important branch of artificial intelligence14–17, which is currently being applied to medical documents18–20, as it could automatically extract and understand unstructured medical textual information21. Previous research mainly focused on general entity extraction in EHRs, such as symptoms and physical signs22. However, the application of NLP for specialized pathology reports is rare due to the limitations of lack of datasets and discontinuities and overlaps in entities23. In terms of improving the performance of named entity recognition (NER), the generative approach is effective in obtaining discontinuous entities but still suffers from exposure bias24. The machine reading comprehension (MRC) approach is helpful to extract overlapping entities25, but its performance is still suboptimal due to its sensitivity to manually constructed query templates26.
Herein, we built a high-quality annotated Pancreatic ductal Adenocarcinoma Named entity recognition (PAN) pathological dataset of resected PDAC. We proposed an innovative Prompt enhanced Generative Machine Reading Comprehension (PGMRC) framework to automatically extract the pathology entities from the PAN dataset. After feature screening, we constructed a morphological classifier (Mor) and then combined it with the classical T and N staging to build a prognosis prediction model, termed the Tumor-Node-Morphology (TNMor) staging system. Subsequently, we evaluated the prognostic prediction accuracy and discriminatory ability of the TNMor system compared with the 8th TNM staging system in both internal and external validation cohorts.
Methods
Study design, participants, and data source
This study was based on a retrospective cohort and an external validation cohort (Table 1). The retrospective cohort was conducted in Shanghai Changhai Hospital, a large referral center. The inclusion criteria of the patients who were admitted into our hospital between 1 January 2012 and 31 December 2018, were as follows: pathologically confirmed PDAC; underwent primary surgical resection. The exclusion criteria of the patients were as follows: intraoperative metastasis (excluding lymph node metastases); macroscopic evidence of margin involvement (R2); received neoadjuvant chemotherapy or radiotherapy; had other malignancies in the past; died within 90 days; and lost to follow-up. The eligible patients were randomly divided into the training and internal validation cohort at a ratio of 7:3, to develop and validate the prognostic model (Supplementary Fig. 1, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Then, the model was independently tested in the external validation cohort, which was derived from the public dataset, pancreatic adenocarcinoma from The Cancer Genome Atlas (TCGA PAAD) cohort, at cBioPortal (https://www.cbioportal.org/), with the same inclusion and exclusion criteria (Supplementary Fig. 1, Supplemental Digital Content 2, http://links.lww.com/JS9/A867).
Table 1.
Baseline characteristics of the patients.
| Characteristic | Total (n=1657) | Training cohort (n=1044) | Internal validation cohort (n=448) | External validation cohort (n=165) |
|---|---|---|---|---|
| Age, median (IQR), years | 65 (58–71) | 65 (58–71) | 66 (59–71) | 64 (56–71) |
| Sex Male (%) | 1017 (61.4) | 628 (60.2) | 289 (64.5) | 100 (60.6) |
| Tumor location (%) | ||||
| head/neck | 1066 (64.3) | 668 (64.0) | 282 (62.9) | 116 (70.3) |
| body/tail | 591 (35.7) | 376 (36.0) | 166 (37.1) | 49 (29.7) |
| Resection type (%) | ||||
| Whipple | 1047 (63.2) | 656 (62.8) | 279 (62.3) | 112 (67.9) |
| Distal pancreatectomy | 567 (34.2) | 360 (34.5) | 160 (35.7) | 47 (28.5) |
| Total pancreatectomy | 43 (2.6) | 28 (2.7) | 9 (2.0) | 6 (3.6) |
| R status (%) | ||||
| R0 | 1303 (78.6) | 827 (79.2) | 361 (80.6) | 115 (69.7) |
| R1 | 354 (21.4) | 217 (20.8) | 87 (19.4) | 50 (30.3) |
| Tumor size, median (IQR), mm | 30 (25–40) | 30 (25–40) | 30 (25–40) | 30 (25–44) |
| T stage-AJCC 8th (%) | ||||
| T1 | 320 (19.3) | 190 (18.2) | 98 (21.9) | 32 (19.4) |
| T2 | 919 (55.5) | 591 (56.6) | 239 (53.3) | 89 (53.9) |
| T3 | 379 (22.9) | 236 (22.6) | 102 (22.8) | 41 (24.8) |
| T4 | 39 (2.4) | 27 (2.6) | 9 (2.0) | 3 (1.8) |
| Harvested lymph nodes, median (IQR) | 18 (13–21) | 18 (13–22) | 18 (12–21) | 18 (12–21) |
| Positive lymph nodes, median (IQR) | 2 (1–4) | 2 (1–4) | 2 (1–4) | 2 (1–4) |
| N stage-AJCC 8th (%) | ||||
| N0 | 350 (21.1) | 220 (21.1) | 89 (19.9) | 41 (24.8) |
| N1 | 772 (46.6) | 467 (44.7) | 233 (52.0) | 72 (43.6) |
| N2 | 535 (32.3) | 357 (34.2) | 126 (28.1) | 52 (31.5) |
| TNM stage-AJCC 8th (%) | ||||
| IA | 87 (5.3) | 50 (4.8) | 25 (5.6) | 12 (7.3) |
| IB | 186 (11.2) | 123 (11.8) | 45 (10.0) | 18 (10.9) |
| IIA | 70 (4.2) | 42 (4.0) | 17 (3.8) | 11 (6.7) |
| IIB | 758 (45.7) | 459 (44.0) | 229 (51.1) | 70 (42.4) |
| III | 556 (33.6) | 370 (35.4) | 132 (29.5) | 54 (32.7) |
| With LVI (%) | 326 (19.7) | 235 (22.5) | 71 (15.8) | 20 (12.1) |
| With PNI (%) | 1341 (80.9) | 871 (83.4) | 361 (80.6) | 109 (66.1) |
| Grade (%) | ||||
| 1 | 25 (1.5) | 15 (1.4) | 6 (1.3) | 4 (2.4) |
| 2 | 1265 (76.3) | 770 (73.8) | 371 (82.8) | 124 (75.2) |
| 3–4 | 367 (22.1) | 259 (24.8) | 71 (15.8) | 37 (22.4) |
| HA (%) | ||||
| I | 118 (7.1) | 68 (6.5) | 40 (8.9) | 10 (6.1) |
| II | 1303 (78.6) | 808 (77.4) | 365 (81.5) | 130 (78.8) |
| III | 236 (14.2) | 168 (16.1) | 43 (9.6) | 25 (15.2) |
| Mor (%) | ||||
| Low | 1075 (64.9) | 632 (60.5) | 333 (74.3) | 110 (66.7) |
| High | 582 (35.1) | 412 (39.5) | 115 (25.7) | 55 (33.3) |
| TNMor stage (%) | ||||
| IA | 229 (13.8) | 140 (13.4) | 65 (14.5) | 24 (14.5) |
| IB | 228 (13.8) | 140 (13.4) | 61 (13.6) | 27 (16.4) |
| II | 509 (30.7) | 304 (29.1) | 162 (36.2) | 43 (26.1) |
| IIIA | 528 (31.9) | 350 (33.5) | 125 (27.9) | 53 (32.1) |
| IIIB | 163 (9.8) | 110 (10.5) | 35 (7.8) | 18 (10.9) |
| Adjuvant therapy | ||||
| Chemotherapy alone | 771 (46.5) | 474 (45.4) | 226 (50.4) | 71 (43.0) |
| Radiotherapy alone | 23 (1.4) | 13 (1.2) | 6 (1.3) | 4 (2.4) |
| Chemoradiation and other | 66 (4.0) | 36 (3.4) | 18 (4.0) | 12 (7.3) |
| None | 733 (44.2) | 475 (45.5) | 184 (41.1) | 74 (44.8) |
| Unknown | 64 (3.9) | 46 (4.4) | 14 (3.1) | 4 (2.4) |
AJCC, the American Joint Committee on Cancer; HA, histological appearances; IQR, interquartile range; LVI, lymphovascular invasion; Mor, morphological classifier; Grade, differentiation grade; PNI, perineural invasion; TNM, tumor-node-metastasis; TNMor, tumor-node-morphology.
The data comprising anonymized structured data (e.g. demographics and diagnosis) and unstructured data (e.g. therapeutic records and pathology reports) were extracted from a clinical data repository and a research data repository27. Follow-up data were collected from the patients’ EHRs or by additional phone interviews. Patients were followed from the date of operation through the last date of oncological surveillance or death, and follow-up was continued until 1 July 2022.
The external validation data were extracted from the public dataset (TCGA PAAD cohort), which included demographics, diagnosis, survival time, structured pathology data (e.g. tumor location, tumor size, tumor grade, lymph node metastasis, distant metastasis, and TNM stage) and pathological images derived from hematoxylin and eosin-stained slides. The pathological images of the external validation were evaluated by three senior pathologists according to the standard process of our center’s pathology reporting. Among them, histological appearance (HA) was classified as type I (tubulopapillary), II (conventional and cribriforming), and III (nests, sheets, cords, and squamous)28, shown in Supplementary Figure 2 (Supplemental Digital Content 2, http://links.lww.com/JS9/A867).
This study was approved by the Institutional Review Board of Shanghai Changhai Hospital. Written informed consent was obtained from all patients before surgery, which contained a statement on the clinical data for scientific research. The study design followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guidelines. This study was registered at the Research Registry (Chinese Clinical Trial) and reported in line with the Strengthening the Reporting of Cohort, Cross-sectional, and Case–control Studies in Surgery (STROCSS) Criteria29 (Supplemental Digital Content 1, http://links.lww.com/JS9/A866).
Processing of pathology reports by NLP
Using the training cohort, we developed an NLP model that could automatically annotate and extract the free text of pathology reports. We first filtered 2831 pathology reports according to our previous report30, the accuracy of the automatically extracted results was not less than 95%. Subsequently, two senior pathologists filtered the uncertain samples, and finally, 1044 high-quality annotated samples were collated and termed the PAN pathological dataset (Fig. 1A).
Figure 1.
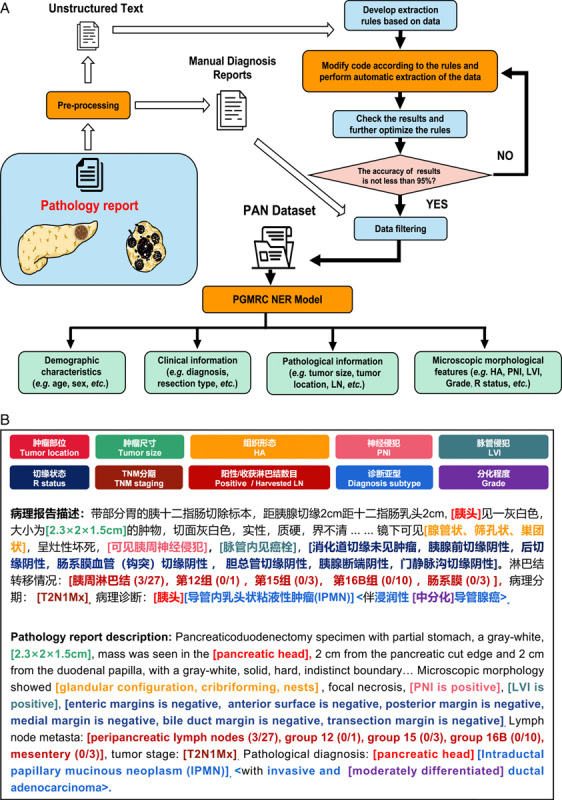
Pipelines of automated NLP technology for processing clinicopathological characteristics of pathology reports. (A) NLP annotation and extraction process of clinicopathological characteristics. (B) Example diagram of an annotated and extracted by PGMRC NER model. NLP, natural language processing; PAN dataset, pancreatic ductal adenocarcinoma named entity recognition dataset; PNI, perineural invasion; LVI, lymphovascular invasion; PGMRC, prompt enhanced generative machine reading comprehension; NER, named entity recognition; TNM, Tumor-node-metastasis; HA, histological appearance.
Establishment of an NLP extraction model
In the PAN dataset, some entities consist of sequences of discrete words or overlap with others (Fig. 1B). To identify these overlapped and discontinuous entities31, we trained a PGMRC NER model to extract as much detail as possible from the PAN dataset (Fig. 1A). Specifically, we formulated NER as an MRC task and employed a pretrained encoder–decoder module and a bidirectional and auto-regressive transformer to generate entity span sequences according to the query. In this way, we employed queries to guide the model to focus on the answer entities of the context. Then, we introduced a continuous prompt sequence of key-value pairs to the attention mechanism in the transformer to enhance the attention layer. Finally, we used the pointer mechanism to generate the entity and tag index sequence. After training, the PGMRC NER model could automatically annotate and extract a large number of entity types30.
Screening of prognostic predictors for morphological features
With the morphological features annotated and extracted by the PGMRC model, all the morphological features in the training cohort were incorporated into univariate Cox regression analyses for overall survival (OS). Variables with P<0.05 were selected for the multivariate Cox regression analyses. Forward stepwise regression was used to screen the independent predictors. Finally, by combining the professional suggestions of three senior oncologists, four predictors (LVI, PNI, HA, and tumor grade) were selected to construct a morphological classification algorithm (Supplementary Table 1, Supplemental Digital Content 2, http://links.lww.com/JS9/A867, Supplementary Fig. 3, Supplemental Digital Content 2, http://links.lww.com/JS9/A867).
Construction of the morphological classifier
Using these four predictors, we constructed a logistic regression (LR)-based morphological classifier (Mor) to predict the OS in the training cohort. To increase the interpretability of the classifier, we proposed a feature crossing method, which comprises a feature importance module and a weighted summation module. As shown in Figure 2, the feature importance module trains an LR model based on the morphological data and the user’s survival label to obtain the weight coefficient of the corresponding feature in the model as the feature importance . The weighted summation module first converts the feature importance into the corresponding feature weight distribution by softmax:
Figure 2.
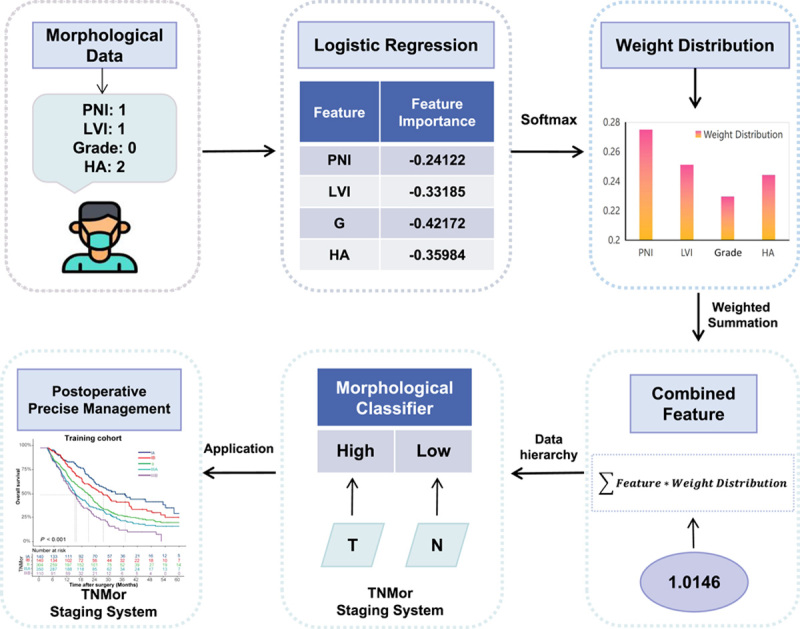
Overview of the construction of the Morphological Classifier. Four predictors were chosen to construct the morphological classifier by the logistic regression method in the training cohort. To increase the interpretability of the classifier, a feature crossing method which concludes a feature importance module and a weighted summation module was used. Then, the median value of combined feature was used as the cut-off value of the classifier, and divide the patients into high-Mor and low-Mor groups. Finally, the morphological classifier was incorporated into T and N staging to contruct the TNMor staging system. PNI, perineural invasion; LVI, lymphovascular invasion; HA, histological appearance; TNMor, tumor-node-morphology.
where is the exponential value of the feature importance corresponding to each type of feature. Then, based on the distribution classification of expert knowledge, the weighted summation is performed between the distribution and the corresponding feature values to obtain the new combined feature values :
where is the calculated value corresponding to the different classes of the features in the data. According to the above process, the different eigenvalues of each sample in the data can be finally combined into a new score (termed the Mor value) for the survival risk prediction task.
Then, we calculated the cut-off value of Mor by the minimum P-value approach and divided the patients into high-Mor and low-Mor groups (Fig. 2).
Construction of the TNMor staging system
We then incorporated the established Mor, other pathological information, and demographic characteristics into the univariate Cox regression analyses of the OS in the training cohort. Variables with P<0.05 were selected for the multivariate Cox regression analyses. Finally, the Mor and the classical T and N staging were chosen to build a novel TNMor staging system (Supplementary Table 2, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). A total of 24 survival prediction classifications were generated based on four categories of T, three categories of N32, and two categories of Mor (Supplementary Fig. 4A, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Using Kaplan–Meier analysis, the median OS for each classification was calculated and ranked. Referring to the five classifications of TNM staging, we reduced the 24 classifications of new system into five classifications based on the ranking results of the median OS and expert opinions (Supplementary Fig. 4, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Moreover, to achieve a more even sample distribution across the stages, we divided the TNMor system into stages I (A, B), II, and III (A, B), corresponding to 280 (140, 140), 304, and 460 (350, 110) patients in respective stages (Supplementary Fig. 4B, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). The staging criteria following the TNMor and TNM staging system are presented in Table 2.
Table 2.
Prognostic groups of the TNM and TNMor staging system.
| TNM staging system | TNMor staging system | ||||||
|---|---|---|---|---|---|---|---|
| T | N | M | T | N | Mor | ||
| Stage IA | T1 | N0 | M0 | Stage IA | T1 | N0 | any Mor |
| T2 | N0 | Mor1 | |||||
| Stage IB | T2 | N0 | M0 | Stage IB | T2 | N0 | Mor2 |
| T3 | N0 | any Mor | |||||
| T1 | N1 | Mor1 | |||||
| Stage IIA | T3 | N0 | M0 | Stage II | T1 | N1 | Mor2 |
| T1 | N2 | Mor1 | |||||
| T2, T3 | N1 | Mor1 | |||||
| T4 | N0 | Mor1 | |||||
| Stage IIB | T1, T2, T3 | N1 | M0 | Stage IIIA | T1 | N2 | Mor2 |
| T2, T3 | N1 | Mor2 | |||||
| T2 | N2 | any Mor | |||||
| T4 | N0 | Mor2 | |||||
| Stage III | T1, T2, T3 | N2 | M0 | Stage IIIB | T3 | N2 | any Mor |
| T4 | any N | T4 | N1, N2 | any Mor | |||
Mor1, low-Mor; Mor2, high-Mor; TNM, tumor-node-metastasis; TNMor, tumor-node-morphology.
Statistical analysis
The precision, recall, and F1 scores were used to evaluate the performance of the NLP model. A multivariate Cox proportional hazards model with forward stepwise regression was used to select the variables. The criterion of P<0.05 was used for entry and P>0.10 for removal of variables, and the hazard ratio with a 95% CI was calculated.
OS was calculated from the date of surgery to the last date of surveillance or death. Patients who did not experience the main endpoint were censored at the last available follow-up. Survival analysis was conducted by Kaplan–Meier curves. The discriminatory ability of the prognostic models was evaluated through pairwise comparison between stages analyzed by the log-rank test and median OS. Prognostic accuracy was assessed using the concordance index (C-index) and the time-dependent area under the receiver operating characteristic (ROC) curve (AUC). For a given survival threshold, ROC was plotted as sensitivity versus 1-specificity for defining the binary all-risk score cut-off. Performance metrics included the plotted ROC, the associated AUC, and randomized P-values to test for significance. All tests were two-tailed and P<0.05 was considered significant. Statistical analysis was performed using SAS version 9.4 (SAS Institute Inc.) and R version 4.0.4 (R Foundation for Statistical Computing).
Results
Characteristics of the participants
A total of 1657 participants were included to train and validate the prognostic model; they were divided into the training (1044), internal validation (448), and external validation (165) cohorts (Supplementary Fig. 1, Table 1, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). The median (IQR) follow-up duration in the training cohort was 36.6 (34.1–39.0) months, with 5-year survival rates being 19.7% (Supplementary Fig. 5A, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). In internal and external validation cohorts, the median (IQR) follow-up duration was 41.2 (35.3–47.2) months and 32.5 (31.0–33.9) months, respectively. The 5-year survival rates were 13.2 and 22.8%, respectively (Supplementary Fig. 5B, C, Supplemental Digital Content 2, http://links.lww.com/JS9/A867).
Association of the Mor with prognosis
The pathology entities were annotated and extracted by the PGMRC model, with a precision of 88.83%, a recall of 89.89%, and an F1 score of 89.21%. Based on the Cox regression analysis (Supplementary Table 1, Supplemental Digital Content 2, http://links.lww.com/JS9/A867), we incorporated HA, PNI, LVI, and tumor grade to construct the Mor in the training cohort33. As shown in Supplementary Fig. 3 (Supplemental Digital Content 2, http://links.lww.com/JS9/A867), significant differences in the estimated median OS were observed between groups Grade 1/2 and Grade 3/4 (22.8 months vs. 13.5 months, P<0.001), between groups with and without PNI (19.1 months vs. 26.9 months, P<0.001), between groups with and without LVI (21.9 months vs. 15.7 months, P=0.008), and among groups HA I, II, and III (30.8 months vs. 21.9 months vs. 13.0 months, P<0.001).
The Mor formula was presented in the Methods section, 0.7 was calculated as the cut-off value for the Mor classification by the minimum P-value approach. Kaplan–Meier curves showed (Supplementary Fig. 6, Supplemental Digital Content 2, http://links.lww.com/JS9/A867) the 5-year survival rate in the high-Mor group were significantly lower than that in the low-Mor group (training cohort, 22.0 vs. 15.8%, P<0.001; internal validation cohort, 14.1 vs. 9.9%, P=0.007; external validation cohort, 19.4 vs. 24.2%, P=0.19). Then, prognostic predictions of the Mor were further evaluated after stratification in the total cohort (Supplementary Fig. 7, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Our results showed the Mor remained a significant prognostic indicator after stratification by various clinicopathological variables (e.g. age, sex, tumor location, and R status), indicating an independent association between the Mor and prognosis.
Establishment of the TNMor staging system
Five stages following the TNMor system were developed by incorporating the classical T and N staging and Mor (Supplementary Fig. 4B, Supplemental Digital Content 2, http://links.lww.com/JS9/A867, Table 2). As shown in Table 3, using the TNM system, stage IA was found in 50 patients (4.8%), stage IB in 123 patients (11.8%), stage IIA in 42 patients (4.0%), stage IIB in 459 patients (44.0%), and stage III in 370 patients (35.4%) in the training cohort. In comparison, using the TNMor system, stage IA was found in 140 patients (13.4%), stage IB in 140 patients (13.4%), stage II in 304 patients (29.1%), stage IIIA in 350 patients (33.5%), and stage IIIB in 110 patients (10.5%). Using the TNMor system, 349 patients (33.4%) migrated to different stages, of whom 121 (11.6%) were assigned to lower stages and 228 (21.8%) to higher stages.
Table 3.
Cross-tabulation of the TNMor and eighth edition of the TNM staging system in the training cohort.
| TNM Staging, AJCC 8th , No. (%) | ||||||
|---|---|---|---|---|---|---|
| TNMor Staging | IA | IB | IIA | IIB | III | Total |
| IA | 50 (4.8) | 90 (8.6) | 0 | 0 | 0 | 140 (13.4) |
| IB | 0 | 33 (3.2) | 42 (4.0) | 65 (6.2) | 0 | 140 (13.4) |
| II | 0 | 0 | 0 | 273 (26.1) | 31 (3.0) | 304 (29.1) |
| IIIA | 0 | 0 | 0 | 121 (11.6) | 229 (21.9) | 350 (33.5) |
| IIIB | 0 | 0 | 0 | 0 | 110 (10.5) | 110 (10.5) |
| Total | 50 (4.8) | 123 (11.8) | 42 (4.0) | 459 (44.0) | 370 (35.4) | 1044 (100) |
AJCC, the American Joint Committee on Cancer; TNM, tumor-node-metastasis; TNMor, tumor-node-morphology.
Comparison with the TNM staging system
First, we compared the discriminatory ability of the TNMor system with the TNM system for OS by comparison of P-value of the log-rank test of various stages. The results showed that there were significant discriminatory differences between stages of the two systems, and the discriminatory ability of the TNMor system was more significant than that of the TNM system (Supplementary Table 3, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Meanwhile, survival analysis demonstrated that the TNMor system had better discriminatory power for prognostic prediction, with the median OS of 36.7, 27.3, 20.9, 15.3, and 15.0 months for stages IA, IB, II, IIIA, and IIIB, respectively. In comparison, the median OS of the TNM system for stages IA, IB, IIA, IIB, and III were 36.7, 32.5, 29.0, 20.2, and 16.6 months, respectively (Fig. 3A, D). These results were validated in both internal (Fig. 3B, E) and external validation cohorts (Fig. 3C, F). Taken together, our system provided better discriminatory ability for the prognosis prediction of resected PDAC.
Figure 3.
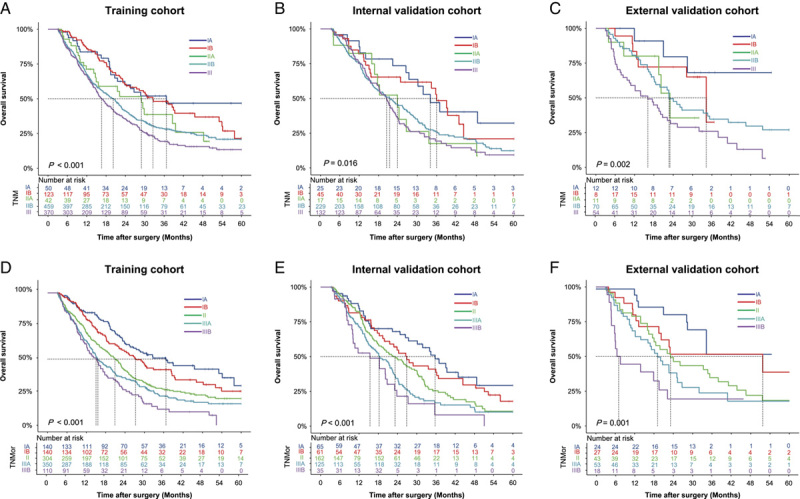
Kaplan–Meier survival curves according to the TNM and TNMor staging system. The OS rates of the TNM staging system in the training cohort (A), the internal validation cohort (B) and the external validation cohort (C), respectively. The OS rates of the TNMor staging system in the training cohort (D), the internal validation cohort (E) and the external validation cohort (F), respectively. OS, overall survival; TNM, tumor-node-metastasis; TNMor, tumor-node-morphology.
We further evaluated the OS predictive accuracy of the new system and found it also displayed a significantly improved C-index compared with the TNM system (Supplementary Table 4, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Furthermore, we compared the predictive accuracy of the systems in terms of 1-year, 2-year, and 3-year survival, and found the TNMor system exhibited a moderately higher AUC than the TNM system (Fig. 4, Supplementary Table 5, Supplemental Digital Content 2, http://links.lww.com/JS9/A867) across the cohorts. All these results indicated TNMor system could provide additional prognostic value for PDAC compared with the TNM system.
Figure 4.
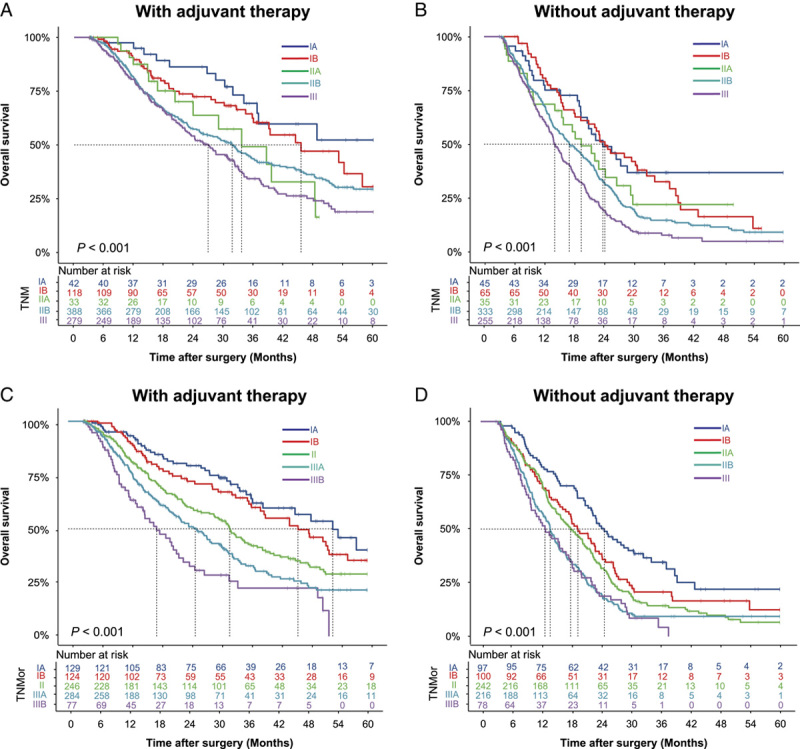
Kaplan–Meier survival curves according to the TNM and TNMor staging systems for patients with or without adjuvant therapy in the total cohort. The OS rates of the TNM staging system in patients receiving (A) or not receiving (B) adjuvant therapy. The OS rates of the TNMor staging system in patients receiving (C) or not receiving (D) adjuvant therapy. OS, overall survival; TNM, tumor-node-metastasis; TNMor, tumor-node-morphology.
Prognostic predictive value of the TNMor staging system for patients with adjuvant therapy
To assess whether the predictive value of the TNMor system was affected by adjuvant therapy, we compared the predictive ability of the system for prognosis in PDAC patients who did or not receive adjuvant therapy (Table 1). The clinicopathological characteristics of patients after stratification according to adjuvant therapy status are presented in Supplementary Table 6, Supplemental Digital Content 2, http://links.lww.com/JS9/A867. The discriminatory of two systems was also evaluated in patients with or without adjuvant therapy by comparison of the P-value of the log-rank test of all stages (Supplementary Table 7, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). For patients with adjuvant therapy, there were nine pairs of stages with a significant difference (P<0.05) on survival predicted by TNMor system, compared with six pairs by the TNM system. For patients without adjuvant therapy, eight pairs versus six pairs of stages with significant difference (P<0.05) for TNMor system and the TNM system, respectively. These results showed the TNMor had a higher discriminatory ability, especially for patients with adjuvant therapy. Similarly, survival curves also supported that the TNMor staging system had a better discriminatory power compared with the TNM system for patients receiving adjuvant therapy (Fig. 5A, C). However, for patients without adjuvant therapy, the TNMor system failed to provide additional discriminatory advantages compared with the TNM system (Fig. 5B, D).
Figure 5.
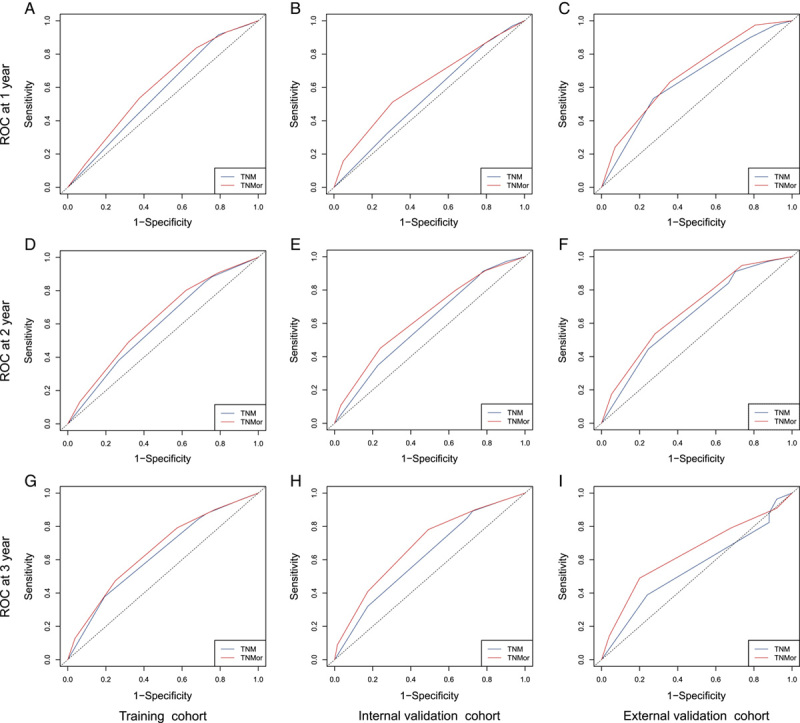
Comparison of ROCs of the TNM and TNMor staging systems for postoperative survival prediction over time. Comparison of 1-year, 2-year, and 3-year ROCs for the TNM and TNMor staging systems in the training cohort (A, D, G), the internal validation cohort (B, E, H), and the external validation cohort (C, F, I), respectively. ROC, receiver operating curve; TNMor, tumor-node-morphology; TNM, tumor-node-metastasis; TNMor, tumor-node-morphology.
In terms of the OS prediction, the TNMor system also displayed a significantly improved C-index (0.61 vs. 0.57, P<0.001, Supplementary Table 8, Supplemental Digital Content 2, http://links.lww.com/JS9/A867) for PDAC patients with adjuvant therapy, whereas the patients without adjuvant therapy had a moderately improved C-index (0.59 vs. 0.57, P <0.001, Supplementary Table 9, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Similarly, in terms of 1-year, 2-year, or 3-year survival prediction, the TNMor system exhibited a significantly higher AUC than the TNM system for patients with adjuvant therapy (Fig. 6A, C, E; Supplementary Table 9, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). However, for patients without adjuvant therapy, the TNMor staging system failed to provide an improved AUC in predicting 2-year or 3-year survival (Fig. 6D, F; Supplementary Table 9, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). All these results indicated TNMor system was particularly beneficial for the precise management of patients receiving adjuvant therapy.
Figure 6.
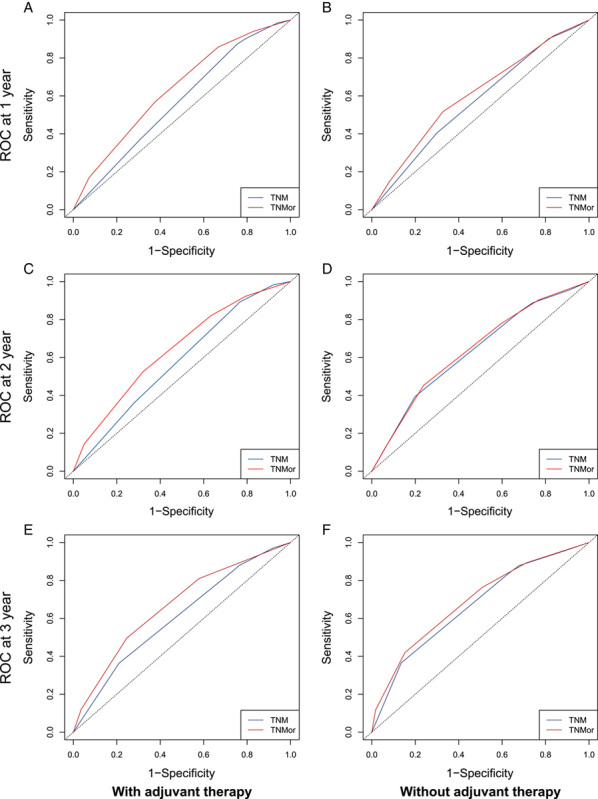
Comparison of ROCs of the TNM and TNMor staging systems for postoperative survival prediction over time in the total cohort. Comparison of 1-year, 2-year, and 3-year ROCs for the TNM and TNMor staging systems for patients with (A, C, E) or without (B, D, F) adjuvant therapy. ROC, receiver operating curve; TNM, tumor-node-metastasis; TNMor, tumor-node-morphology.
An example of applying the TNMor staging system
To facilitate the generalized application of this system, we proposed a structured pathology report template (Supplementary Table 10, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Then, we designed an online version of the application software (http://101.35.227.214/), which could directly output the Mor, TNMor stage, and other prediction results using the required pathological features. An example of how to use the TNMor system was shown in Fig. 7. A 68-year-old male who underwent PDAC resection, with T3, N2, LVI positive, PNI positive, squamous morphological feature, poor differentiation in the pathological report. The system identified the patient as T3N2Mor2, Stage IIIB. The estimated median survival time for this stage was 15 months, and the 1-year, 2-year, and 3-year survival probability were 61.0, 26.8, and 12.0%, respectively.
Figure 7.
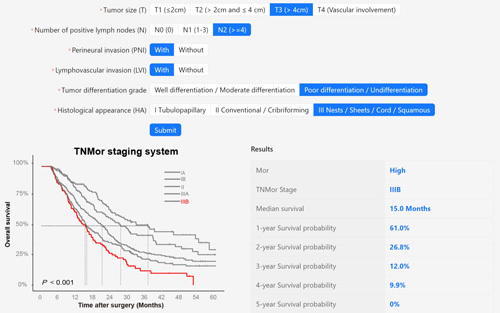
Layout of an online version of the developed TNMor staging system. TNMor, tumor-node-morphology; PNI, perineural invasion; LVI, lymphovascular invasion; HA, histological appearance; Mor, morphological classifier.
Discussion
Herein, we present an NLP model that could automatically annotate and extract the details of pathology reports. Due to the lack of readily available pathology datasets, we first built a high-quality PAN dataset consisting of 1044 annotated samples. To the best of our knowledge, this is one of the large-scale PDAC pathology datasets up to date34. In addition, the quality of the database is very high because the annotation was based on manual supervision35. After that, we trained the PGMRC NER model to extract as much detail as possible from the PAN dataset (Fig. 1), with the precision of 88.83%, recall of 89.89% and F1 score of 89.21%. Compared with traditional NLP models24–26,36, our state-of-the-art model overcomes the limitations of entity discontinuity and overlap, thus leading to higher entity recognition accuracy. Then, we employed Cox regression analyses to screen morphological predictors. As shown in Figure 2, PNI contributed the most to prognosis prediction, followed by LVI, and then HA. Consistent with many previous studies33,37–39, our study also supported that PNI, LVI and poor differentiation grade represented common hallmarks of PDAC and correlated with poor prognosis. More interestingly, our study found HA40 to be a very important prognostic factor, with type III having the worst prognosis (Supplementary Fig. 3D, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). These morphological features suggest the complex biological alterations of tumors, and therefore may be alternative prognostic factors in the era of precision medicine. Considering the convenience of clinical application, we incorporated these four morphological features into a morphological classifier (Mor). Furthermore, we demonstrated that Mor is an independent prognostic factor (Supplementary Fig. 6, Supplemental Digital Content 2, http://links.lww.com/JS9/A867), even across subgroups of clinicopathological variables (Supplementary Fig. 7, Supplemental Digital Content 2, http://links.lww.com/JS9/A867).
Despite its limited performance for OS prediction, the TNM system remains a cornerstone for predicting the prognosis of PDAC. Thus, we combined T, N, and Mor to construct the TNMor staging system. Using the novel system, 33.4% of patients migrated to different stages of the TNM staging (Table 3). Also, we found that the re-staged TNMor system could provide a more reasonable risk stratification (Fig. 3, Supplementary Table 3, Supplemental Digital Content 2, http://links.lww.com/JS9/A867) and significant improvements in the C-index and AUC in terms of survival prediction (Supplementary Tables 4, 5, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). More importantly, we further demonstrated that the TNMor system was particularly beneficial for the precise stratification of PDAC patients receiving adjuvant therapy (Fig. 4, Supplementary Table 7, Supplemental Digital Content 2, http://links.lww.com/JS9/A867). Currently, NCCN guidelines for PDAC recommended that all postoperative patients without neoadjuvant therapy should undergo adjuvant therapy, but the selection priority of therapeutic regimens is still controversial41,42. Our refined staging system may allow for screening of risk-adjusted treatment strategies in the era of precision medicine43,44. To make it easier understood, we designed an online version of the application software of TNMor staging system and provide an application example (Fig. 7). Through this example, we can clearly see that the TNMor staging system could provide personalized prognostic prediction (e.g. median OS, the 1-year, 2-year, and 3-year survival probability, etc.) for postoperative PDAC patients, which may help doctors screen and optimize postoperative therapeutic regimens, and ultimately improve therapeutic responses.
Notably, because the TNMor system is derived from pathology reports, the system can be easily applied to clinical practice without additional financial burden. Therefore, we hope that our system could become one of the routine clinical decision support tools45. Firstly, for pathologists, NLP applications could be used to assist in generating accurate pathology reports, which is a tedious and time-consuming. Our system; however, could quickly process pathology data of PDAC, automatically extract important features, and analyze the data in a visual way, ultimately providing a comprehensive and easy-to-use interpretation of pathology reports. Secondly, for oncologists, our prognostic tool could provide valuable information for individualized postoperative management46. Although adjuvant therapy was recommended to apply to patients who underwent primary surgery in the updated NCCN guideline for PDAC, the best regimens have not been clearly defined, and comparisons between different treatment strategies are limited by the heterogeneity of study populations42,47,48. Therefore, there is an urgent need for tools that predict postoperative survival to help guide postresection management. Hence, we hope that our TNMor staging system may facilitate oncologists to develop tailored therapies, and improve the adjuvant therapy responses49,50. Thirdly, for PDAC patients, our staging system can help patients better understand their disease risk stratification, and improve their compliance of therapy51. Pathology reports are highly specialized and difficult to comprehend for patients. Our system additionally provides risk stratification information, which helps patients make an accurate assessment of their disease and cooperate more actively with their doctors.
Despite these encouraging results, the present research still has some limitations. First, due to the retrospective experimental design, our research has some inherent bias. Second, our external verification data were relatively small, and data distribution of several characteristics was not exactly equal between the training and external validation cohort. Therefore, it is necessary to verify the clinical effectiveness of the system in prospective randomized trials. Finally, although the Mor was found to be a potential prognostic predictor for patients with resected PDAC, it is still limited by the sampling problem and tumor heterogeneity. In the future, these morphological features could potentially be evaluated in conjunction with laboratory indices or imaging parameters to provide more objective, multidimensional, and function-related diagnostic and therapeutic outputs.
Conclusion
In this study, we constructed a morphological classifier using NLP-based pathology reports and found a significant association between Mor and the prognosis of resected PDAC. Then, we developed and validated the TNMor system and demonstrated its additional prognostic value compared with the TNM system. Finally, we proved that the novel system was particularly beneficial for the precise stratification of PDAC patients receiving adjuvant therapy, which may allow for screening of risk-adjusted treatment strategies in the era of precision medicine.
Ethical approval
The study was approved by the Institutional Review Board of Shanghai Changhai Hospital Ethics Committee.
Consent
Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request.
Sources of funding
This work was funded by Shanghai Science and Technology Committee Program (grant number 20511101200, 20511101205) and Clinical Research Plan of Shanghai Hospital Development Center (grant number SHDC2020CR2001A).
Author contribution
B.L., B.W., P.Z., S.W., Y.Y., and G.J.: were involved in the study design, data collection, and analysis; B.L., B.W., P.Z., H.C.,S.W., Z.T., S.Ga., P.L., W.J., Z.S., L.W., B.G., Z.K., Y.W., H.J., and S.Gu.: collected and processed data; B.L. and B.W.: drafted the paper; G.J., Y.Y. and L.H.: designed and supervised the study. All authors approved the paper.
Conflicts of interest disclosure
The authors declare that they have no conflicts of interest.
Research registration unique identifying number (UIN)
Research on the construction of multidisciplinary precision and standardized diagnosis and treatment system for pancreatic cancer in Chinese Clinical Trial (ChiCTR2000037163).
Guarantor
Gang Jin.
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Provenance and peer review
Our paper was not invited.
Supplementary Material
Footnotes
B.L., B.W., and P.Z. contributed equally to the work.
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.lww.com/international-journal-of-surgery.
Published online 11 August 2023
Contributor Information
Bo Li, Email: percybob@sina.com.
Beilei Wang, Email: beileiwang@smmu.edu.cn.
Pengjie Zhuang, Email: 51215901094@stu.ecnu.edu.cn.
Hongwei Cao, Email: chw777@163.com.
Shengyong Wu, Email: wu_sheng_yong@163.com.
Zhendong Tan, Email: 51205901060@stu.ecnu.edu.cn.
Suizhi Gao, Email: gaosuizhi@163.com.
Penghao Li, Email: 839066946@qq.com.
Wei Jing, Email: jingwei7777@163.com.
Zhuo Shao, Email: szlaugh@163.com.
Kailian Zheng, Email: zhengkl828@126.com.
Lele Wu, Email: lele_wu2021@126.com.
Bai Gao, Email: gaobaish@126.com.
Yang Wang, Email: docwy@sina.com.
Hui Jiang, Email: jianghui5131@163.com.
Shiwei Guo, Email: gestwa@163.com.
Liang He, Email: lhe@cs.ecnu.edu.cn.
Yan Yang, Email: yanyang@cs.ecnu.edu.cn.
Gang Jin, Email: jingang@smmu.edu.cn.
References
- 1.Park W, Chawla A, O’Reilly EM. Pancreatic cancer: a review. JAMA 2021;326:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Wagle NS, et al. Cancer statistics, 2023. CA Cancer J Clin 2023;73:17–48. [DOI] [PubMed] [Google Scholar]
- 3.Mizrahi JD, Surana R, Valle JW, et al. Pancreatic cancer. Lancet 2020;395:2008–2020. [DOI] [PubMed] [Google Scholar]
- 4.Hank T, Hinz U, Reiner T, et al. A Pretreatment prognostic score to stratify survival in pancreatic cancer. Ann Surg 2022;276:e914–e922. [DOI] [PubMed] [Google Scholar]
- 5.Fong ZV, Chang DC, Hur C, et al. Variation in long-term oncologic outcomes by type of cancer center accreditation: an analysis of a SEER-medicare population with pancreatic cancer. Am J Surg 2020;220:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kindler HL. A glimmer of hope for pancreatic cancer. N Engl J Med 2018;379:2463–2464. [DOI] [PubMed] [Google Scholar]
- 7.Schouten TJ, Daamen LA, Dorland G, et al. Nationwide validation of the 8th American Joint Committee on Cancer TNM staging system and five proposed modifications for resected pancreatic cancer. Ann Surg Oncol 2022;29:5988–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang H, Kim SS, Sung MJ, et al. Evaluation of the 8th edition AJCC staging system for the clinical staging of pancreatic cancer. Cancers (Basel) 2022;14:4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Roessel S, Kasumova GG, Verheij J, et al. International validation of the eighth edition of the American Joint Committee on Cancer (AJCC) TNM staging system in patients with resected pancreatic cancer. JAMA Surg 2018;153:e183617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malleo G, Maggino L, Qadan M, et al. Reassessment of the optimal number of examined lymph nodes in pancreatoduodenectomy for pancreatic ductal adenocarcinoma. Ann Surg 2022;276:e518–e526. [DOI] [PubMed] [Google Scholar]
- 11.Hammad AY, Hodges JC, AlMasri S, et al. Evaluation of adjuvant chemotherapy survival outcomes among patients with surgically resected pancreatic carcinoma with node-negative disease after neoadjuvant therapy. JAMA Surg 2023;158:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshioka Y, Shimomura M, Saito K, et al. Circulating cancer-associated extracellular vesicles as early detection and recurrence biomarkers for pancreatic cancer. Cancer Sci 2022;113:3498–3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Wang Y, Wang J, et al. Negative p53 expression confers worse prognosis in patients with resected pancreatic ductal adenocarcinoma: research focused on reinterpretation of immunohistochemical staining. Pancreas 2022;51:1217–1224. [DOI] [PubMed] [Google Scholar]
- 14.Yim WW, Yetisgen M, Harris WP, et al. Natural language processing in oncology: a review. JAMA Oncol 2016;2:797–804. [DOI] [PubMed] [Google Scholar]
- 15.López-Úbeda P, Martín-Noguerol T, Aneiros-Fernández J, et al. Natural language processing in pathology: current trends and future insights. Am J Pathol 2022;S0002-9440:00244–00249. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen Wenker T, Natarajan Y, Caskey K, et al. Using natural language processing to automatically identify dysplasia in pathology reports for patients with barrett’s esophagus. Clin Gastroenterol Hepatol 2023;21:1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song G, Chung SJ, Seo JY, et al. Natural language processing for information extraction of gastric diseases and its application in large-scale clinical research. J Clin Med 2022;11:2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019;20:e253–e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Fu S, Wen A, et al. Assessment of electronic health record for cancer research and patient care through a scoping review of cancer natural language processing. JCO Clin Cancer Inform 2022;6:e2200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee I, Bozkurt S, Caswell-Jin JL, et al. Natural language processing approaches to detect the timeline of metastatic recurrence of breast cancer. JCO Clin Cancer Inform 2019;3:1–12. [DOI] [PubMed] [Google Scholar]
- 21.Yao K, Singh A, Sridhar K, et al. Artificial intelligence in pathology: a simple and practical guide. Adv Anat Pathol 2020;27:385–393. [DOI] [PubMed] [Google Scholar]
- 22.Datta S, Bernstam EV, Roberts K. A frame semantic overview of NLP-based information extraction for cancer-related EHR notes. J Biomed Inform 2019;100:103301. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Lian JW, Chin YH, et al. Assessing the prognostic significance of tumor-infiltrating lymphocytes in patients with melanoma using pathologic features identified by natural language processing. JAMA Netw Open 2021;4:e2126337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Feng Y, Meng F, et al. Bridging the gap between training and inference for neural machine translation. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics (ACL) 2019:4334–4343. [Google Scholar]
- 25.Li X, Feng J, Meng Y, et al. A unified MRC framework for named entity recognition. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics (ACL) 2020:5849–5859. [Google Scholar]
- 26.Shin T, Razeghi Y, IV, Wallace RLL, et al. Eliciting knowledge from language models with automatically generated prompts. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP) 2020:4222–35. [Google Scholar]
- 27.Wang B, Wei W, Shao Z, et al. Establishment of a machine learning model for early and differential diagnosis of pancreatic ductal adenocarcinoma using laboratory routine data. Adv Intell Syst 2021;3:2100033. [Google Scholar]
- 28.Kalimuthu NS, Wilson GW, Grant RC, et al. Morphological classification of pancreatic ductal adenocarcinoma that predicts molecular subtypes and correlates with clinical outcome. Gut 2020;69:317–328. [DOI] [PubMed] [Google Scholar]
- 29.Mathew G, Agha R, Albrecht J, et al. STROCSS 2021: strengthening the reporting of cohort, cross-sectional and case-control studies in surgery. Int J Surg 2021;96:106165. [DOI] [PubMed] [Google Scholar]
- 30.Tan Z, Yang Y, Li B, et al. Prompt enhanced generative mrc framework for pancreatic cancer NER. Paper presented at: 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) IEEE 2022:817–20. [Google Scholar]
- 31.Yan H, Gui T, Dai J, et al. A unified generative framework for various ner subtasks Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (ACL-IJCNLP). 2021:5808–22. [Google Scholar]
- 32.Nagtegaal ID, Odze RD, Klimstra D, et al. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020;76:182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epstein JD, Kozak G, Fong ZV, et al. Microscopic lymphovascular invasion is an independent predictor of survival in resected pancreatic ductal adenocarcinoma. J Surg Oncol 2017;116:658–664. [DOI] [PubMed] [Google Scholar]
- 34.Homeyer A, Geißler C, Schwen LO, et al. Recommendations on compiling test datasets for evaluating artificial intelligence solutions in pathology. Mod Pathol 2022;35:1759–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobritz M, Patel V, Rindskopf D, et al. Practice-based learning and improvement: improving morbidity and mortality review using natural language processing. J Surg Res 2023;283:351–356. [DOI] [PubMed] [Google Scholar]
- 36.Van Vleck TT, Farrell D, Chan L. Natural language processing in nephrology. Adv Chronic Kidney Dis 2022;29:465–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selvaggi F, Melchiorre E, Casari I, et al. Perineural invasion in pancreatic ductal adenocarcinoma: from molecules towards drugs of clinical relevance. Cancers (Basel) 2022;14:5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang G, Li B, Yin X, et al. Systemic therapy and perioperative management improve the prognosis of pancreatic ductal adenocarcinoma: a retrospective cohort study of 2000 consecutive cases. Int J Surg 2022;104:106786. [DOI] [PubMed] [Google Scholar]
- 39.Liffers ST, Godfrey L, Frohn L, et al. Molecular heterogeneity and commonalities in pancreatic cancer precursors with gastric and intestinal phenotype. Gut 2023;72:522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rasmussen LG, Verbeke CS, Sørensen MD, et al. Gene expression profiling of morphologic subtypes of pancreatic ductal adenocarcinoma using surgical and EUS-FNB specimens. Pancreatology 2021;21:530–543. [DOI] [PubMed] [Google Scholar]
- 41.Chase M, Friedman HS, Joo S, et al. Adjuvant and neoadjuvant treatment patterns among resectable pancreatic cancer patients in the USA. Future Oncol 2022;18:3929–3939. [DOI] [PubMed] [Google Scholar]
- 42.Conroy T, Castan F, Lopez A, et al. Five-year outcomes of FOLFIRINOX vs Gemcitabine as adjuvant therapy for pancreatic cancer: a randomized clinical trial. JAMA Oncol 2022;8:1571–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X, Zhang G, Liang T. Subtyping for pancreatic cancer precision therapy. Trends Pharmacol Sci 2022;43:482–494. [DOI] [PubMed] [Google Scholar]
- 44.O’Kane GM, Lowery MA. Moving the needle on precision medicine in pancreatic cancer. J Clin Oncol 2022;40:2693–2705. [DOI] [PubMed] [Google Scholar]
- 45.Junet V, Matos-Filipe P, García-Illarramendi JM, et al. A decision support system based on artificial intelligence and systems biology for the simulation of pancreatic cancer patient status. CPT Pharmacometrics Syst Pharmacol 2023;12:916–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Savani M, Shroff RT. Decision-making regarding perioperative therapy in individuals with localized pancreatic adenocarcinoma. Hematol Oncol Clin North Am 2022;36:961–978. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Huang J, Jiang C, et al. Comparison the efficacy and safety of different neoadjuvant regimens for resectable and borderline resectable pancreatic cancer: a systematic review and network meta-analysis. Eur J Clin Pharmacol 2023;79:323–340. [DOI] [PubMed] [Google Scholar]
- 48.Akdeniz N, Kaplan MA, İnanç M, et al. The efficacy and safety of treatment regimens used in the first-line setting in metastatic pancreatic cancer patients: a multicenter real-life study. Pancreas 2022;51:1153–1159. [DOI] [PubMed] [Google Scholar]
- 49.Seo YD, Katz MHG. Preoperative therapy for pancreatic adenocarcinoma-precision beyond anatomy. Cancer 2022;128:3041–56. [DOI] [PubMed] [Google Scholar]
- 50.De Jong EJM, Janssen QP, Simons TFA, et al. Real-world evidence of adjuvant gemcitabine plus capecitabine vs gemcitabine monotherapy for pancreatic ductal adenocarcinoma. Int J Cancer 2022;150:1654–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reif de Paula T, Gorroochurn P, Haas EM, et al. A national evaluation of the predictors of compliance and survival from adjuvant chemotherapy in high-risk stage II colon cancer: a national cancer database (NCDB) analysis. Surgery 2022;172:859–868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


