Abstract
The transcription factor AflR is required for up-regulation of specific pathway genes involved in aflatoxin biosynthesis in the filamentous fungus Aspergillus. nor-1 encodes an early aflatoxin pathway enzyme; its promoter contains a consensus AflR binding site (AflR1). Proteins in Aspergillus parasiticus cell extracts and AflR expressed in Escherichia coli do not bind to A. parasiticus AflR1 in vitro, so it was not clear if this site was required for nor-1 expression or if other transcription factors contributed to gene regulation. In this study we defined the role of AflR1 in nor-1 expression in A. parasiticus and identified additional cis-acting sites required for maximum nor-1 transcriptional activation. Deletion and substitution of AflR1 in the nor-1 promoter in A. parasiticus nor-1::GUS reporter strains showed that this site is required for nor-1 transcriptional activation in vivo. Substitution of a putative TATA box in the nor-1 promoter resulted in nondetectable β-glucuronidase (GUS) activity, demonstrating that this TATA box is functional in vivo. We also identified a novel cis-acting site, designated NorL, between residues −210 and −238 that was required for maximum nor-1 transcriptional activation in A. parasiticus grown in liquid medium and on solid medium. Using an electrophoretic mobility shift assay, we identified a specific NorL-dependent DNA-protein complex that relies on a functional AflR, either directly or indirectly, for maximum binding capacity. Because the NorL site appears only once in the aflatoxin gene cluster, its association with the nor-1 promoter may have important implications for the overall regulatory scheme for the aflatoxin pathway.
Aflatoxins, which are mycotoxins that are produced predominantly by Aspergillus parasiticus and Aspergillus flavus, frequently contaminate economically important crops, such as corn, cotton, peanuts, and tree nuts (31). Aflatoxin B1, the most abundant of the aflatoxins, is also the most toxic and carcinogenic (21). Animal studies have demonstrated that aflatoxin is a potent hepatocarcinogen, and human epidemiological data have linked aflatoxin exposure with liver cancer (reviewed in references 11 and 17). As a result, susceptible crops are monitored for aflatoxin contamination in the United States and throughout the world, resulting in a large economic cost to growers and marketers of commodities. Our long-term goal is to reduce or eliminate aflatoxin from the food chain.
By elucidating the molecular mechanisms that regulate aflatoxin biosynthesis, we hope to generate novel approaches and targets for inhibition of aflatoxin gene expression. Aflatoxin biosynthesis is a complex process that requires at least 18 different enzymatic activities (3). The genes involved in aflatoxin biosynthesis reside in a 70-kb cluster and appear to be coregulated (30). Analysis of the regulatory mutant A. flavus strain 650 (2) first identified AflR as a pathway regulator (23). Subsequently, aflR homologs were identified in A. parasiticus (6) and Aspergillus nidulans (33).
Several independent lines of evidence confirmed that AflR has a key regulatory role in aflatoxin biosynthesis (6, 12, 13, 15, 16, 23, 32, 33). AflR cis-acting sites are required for transcriptional activation of three aflatoxin biosynthetic genes in vivo, including stcU (15), avnA (4), and pksA (14). Although AflR is a key regulator of aflatoxin synthesis, it is not clear that AflR is the only transcription factor required for transcriptional activation of all aflatoxin genes or if all of the consensus AflR cis-acting sites in the aflatoxin cluster are functionally significant.
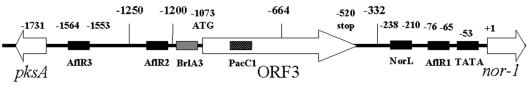
We focused on the expression of nor-1, a gene encoding an enzyme that catalyzes the conversion of the first stable aflatoxin biosynthesis intermediate, norsolorinic acid, to averantin (29, 34). The nor-1 promoter (Fig. 1) includes a consensus AflR binding site (AflR1; TCGNNNNNCGR) (14) located at positions −75 to −64 with respect to the primary nor-1 transcriptional start site (position 1). Additional upstream consensus AflR binding sites are located at positions −1213 (AflR2) and −1563 (AflR3) (14). Genes upstream from nor-1 include an open reading frame with an unknown function (ORF3; translational start at position −1073) and the divergently transcribed pksA gene (translational start at position −1731) (7, 30).
FIG. 1.
Schematic diagram of the A. parasiticus nor-1 promoter region. The numbers indicate the numbers of nucleotide residues upstream from the transcriptional start site. The positions of potential cis-acting sites are indicated, including the positions of AflR1, AflR2, and AflR3 (AflR binding sites), NorL, and TATA, as well as PacC1 and BrlA3, which were reported previously to be involved in pksA transcriptional regulation (14). The location of an open reading frame (ORF3) with an unknown function is also shown. The sizes of the nor-1 promoters used in this study also are indicated (332, 76, and 64 bp).
In previous studies, A. parasiticus nuclear extracts (13, 22) and AflR expressed in Escherichia coli (13) could not bind to nor-1 promoter fragments containing AflR1 in vitro, suggesting that AflR1 might not be functionally relevant. Directed mutations in AflR1 did not affect transcription of the divergently transcribed pksA gene in vivo (14), suggesting that AflR1 is not important for expression of pksA. By using the same mutagenesis approach, both AflR2 and AflR3 were shown to be necessary for pksA transcription, but the effects of AflR1, AflR2, and AflR3 mutations on nor-1 and ORF3 expression were not reported (14). Furthermore, alteration of specific BrlA and PacC cis-acting sites in the pksA-nor-1 intergenic region by directed mutagenesis significantly altered pksA transcription, indicating that AflR may not be the only transcriptional regulator for pksA (14).
Based on the previous data, we hypothesized that interaction of AflR with the AflR1 site plays an important role in regulation of nor-1 expression but that one or more additional transcription factors are required for formation of this protein-DNA complex.
The objectives of this study were (i) to clarify the role of AflR and AflR1 in expression of nor-1 in A. parasiticus and (ii) to identify additional cis-acting sites required for maximum nor-1 transcriptional activation. This study demonstrated conclusively that multiple transcription factors (including AflR and NorLbp) are required for maximum nor-1 promoter activity under our experimental conditions. Because the NorL site appears only once in the aflatoxin gene cluster, its association with the nor-1 promoter may have important implications for the overall regulatory scheme for the aflatoxin pathway.
MATERIALS AND METHODS
Bacterial and fungal strains.
E. coli DH5α F′ [φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rk− mk+) phoA supE44 λ− thi1 gyrA96 relA1] (Invitrogen, Carlsbad, Calif.) was used to amplify plasmid DNA by standard procedures (1). A. parasiticus SU-1 (= ATCC 56775) was the wild-type aflatoxin-producing strain used in this study. A. parasiticus AFS10 is an aflR gene disruption strain (5) and does not produce aflatoxin. A. parasiticus NR-1 (niaD) (18) was the recipient strain used for plasmid transformation.
Plasmid constructs.
Construction of plasmid pNANG-3 and pAPGUSNN-B has been described previously (10). To test the functionality of cis-acting sites, nor-1 promoter fragments were generated by PCR by using pAPGUSNN-B as the template and primers with NotI (3′) and PacI (5′) tails. Promoter fragments were ligated into the NotI and PacI sites in pNANG-3, resulting in the β-glucuronidase (GUS) reporter plasmid series designated pBNGn (where n indicates the size of the promoter in base pairs).
(i) pBNG332 and pBNG332TATAmut.
The polyadenylation site for ORF3, located 332 bp upstream from the transcriptional start site of nor-1, was used to define the 5′ end of the nor-1 promoter. This 332-bp promoter fragment was amplified by PCR with primers JL267 (5′-GTTAATTAAGTCGAGCGGACATGGCCACG-3′; PacI site underlined) and JL186 (5′-TCGCGGCCGCTAAGTGATCCATTCATTATGTC-3′; NotI site underlined). To generate pBNG332TATAmut, the TATA box was replaced by a PmeI site (5′-ATATATAG-3′ changed to 5′-GTTTAAAC-3′) in the context of the 332-bp nor-1 promoter. The 332-bp nor-1 promoter was divided into two PCR fragments (PacI/PmeI and NotI/PmeI) that were joined at the TATA box. The primers used for the PacI/PmeI fragment were JL267 and JL414 (5′-ATGTTTAAACTGGGATACGATCATGGGTC-3′; PmeI site underlined). The primers used for the NotI/PmeI fragment were JL186 and JL415 (5′-GGGTTTAAACGGCGGTGTGTTGGTCG-3′; PmeI site underlined). After digestion with the appropriate restriction endonucleases, a three-fragment ligation was performed with pNANG-3, the PacI/PmeI fragment, and the NotI/PmeI fragment. The nor-1 promoter in pBNG332 and pBNG332TATAmut was verified by nucleotide sequence analysis.
(ii) pBNG298, pBNG268, pBNG238, and pBNG210.
To generate a nor-1 promoter deletion series, different upstream PCR primers with PacI sites (underlined sequences) were used with the same downstream PCR primer (JL186 with a NotI site). The upstream primers used were JL411 (5′-CCTTAATTAAACTGCTATGGTGACCTATTG-3′) for pBNG298, JL412 (5′-CATTAATTAACCACATAGGCTACTCAAAAT-3′) for pBNG268, JL413 (5′-GGTTAATTAAAGATCTCTGCTATTAAGTCGG-3′) for pBNG238, and JL302 (5′-CCCTTAATTAATAGCGTGCTGGATGCGCGAA-3′) for pBNG210. The nor-1 promoters in this deletion series were verified by nucleotide sequence analysis.
(iii) pBNGNorLmut.
To generate pBNGNorLmut, a 28-bp region (positions −210 to −238) was replaced (changed from 5′-AGATCTCTGCTATTAAGTCGGTGATTAG-3′ to 5′-GTATAAGAAGTTTGTGATGGGATTCGTC-3′) in the context of the 332-bp nor-1 promoter. The 332-bp nor-1 promoter was divided into two PCR fragments (PacI/210 and NotI/238) that were joined at position −224. The primers used for the PacI/210 fragment were JL267 and JL613 (5′-CAAACTTCTTATACGCTCATGTCAATTTTGAG-3′). The primers used for the NotI/238 fragment were JL186 and JL612 (5′-TGATGGGATTCGTCCGTGCTGGATGCGC-3′). JL612 and JL613 do not have restriction endonuclease sites. After digestion with the appropriate restriction endonucleases, a three-fragment ligation was performed with pNANG-3, the PacI/210 fragment, and the NotI/238 fragment. The nor-1 promoter in pBNG332NorLmut was verified by nucleotide sequence analysis.
(iv) pAFLRMBP.
The pMAL protein fusion and purification system (New England Biolabs, Beverly, Mass.) was used for AflR production by using methods described previously for Ver-1 (20). A DNA fragment encompassing the entire AlfR open reading frame was generated by PCR by using an aflR cDNA fragment and PCR primers 5′-TTTCTAGAGTTGACCATATCTCCCCCC-3′ and 5′-CCAAGCTTTCATTCTCGATGCAGGTAAT-3′. The resulting product was cloned in frame into the XbaI and HindIII sites of expression vector pMALc2. Expression and purification of the AflR::MBP fusion protein (approximately 94 kDa) from E. coli were performed according to the manufacturer' s instructions (New England Biolabs). Approximately 0.4 μg of fusion protein was included in electrophoretic mobility shift assay (EMSA) reaction mixtures as described below. A 40-fold excess of purified maltose binding protein (MBP) (16 μg) generated by using the same methods that were used for AflR::MBP was included in control reaction mixtures.
Generation and analysis of A. parasiticus transformants.
Transformation of A. parasiticus protoplasts was performed as described by Horng et al. (18). Two to four micrograms of DNA and approximately 107 protoplasts resulted in approximately 100 transformants. The site of plasmid integration was determined by PCR analysis (10) and was confirmed by Southern analysis. With the PCR assay, a 2.0-kb amplicon was diagnostic for 3′ nor-1 integration. Genomic DNA was purified from A. parasiticus cultures shaken for 48 h in 100 ml of YES liquid medium (2% yeast extract, 6% sucrose; pH 5.8) at 29°C with five 6-mm-diameter glass beads (28) and was subjected to Southern hybridization analysis by using standard procedures (1). A 2.5-μg portion of genomic DNA was digested with ScaI and probed with a 900-bp ClaI fragment isolated from the nor-1 terminator region of pAPGUSNN-B (10). Digestion of DNA from the recipient strain, NR-1, with ScaI generated a 3.0-kb restriction fragment, while a 3′ integrant (nor-1 terminator) resulted in 3.7- and 4.0-kb restriction fragments.
GUS reporter assays.
As an indirect measure of nor-1 promoter function, GUS reporter activity (19) was measured in a liquid culture and a solid culture essentially as reported previously (10). The fluorescence of protein from the liquid culture was analyzed by using a Cytofluor II fluorimeter (Biosearch Co., Bedford, Mass.). The excitation and emission wavelengths were 360 and 460 nm, respectively. A standard curve for 4-methylumbelliferone (10 to 600 nM) in GUS lysis buffer also was generated, and GUS activity was expressed in picomoles of 4-methylumbelliferone produced per minute per milligram of protein.
EMSA. (i) Protein extraction.
Cell protein extracts were generated by using modifications of the procedures of Peters and Caddick (25) and Peres-Esteban et al. (24). Conidiospores (1 × 107) were inoculated into 1-liter flasks containing 500 ml of appropriate growth medium and 10 glass beads. The cultures were incubated at 29°C for 48 h with shaking (150 rpm). Mycelia were collected by filtration by using Miracloth (Calbiochem, San Diego, Calif.), washed with sterile cold water, frozen with liquid nitrogen, and ground with a mortar and pestle. The ground mycelia were transferred to a 125-ml flask containing 5 ml of lysis buffer (25 mM HEPES-KOH [pH 7.5], 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 ml of fungal protease inhibitor cocktail [Sigma, St. Louis, Mo.] per 20 g of mycelia) and stirred on ice for 15 min. Saturated ammonium sulfate was slowly added to a final concentration of 10%, and the mixture was stirred for 15 min on ice and allowed to stand on ice for 15 min. Cellular debris was pelleted by centrifugation at 36,500 × g for 30 min at 4°C. The concentration of ammonium sulfate in the supernatant was raised from 10 to 70% by adding solid ammonium sulfate slowly over 90 min with gentle stirring on ice. Cellular proteins were pelleted by centrifugation (10,000 × g for 20 min at 4°C), resuspended in dialysis buffer (15% glycerol, 15 mM HEPES-KOH [pH 7.9], 100 mM KCl, 1 mM EDTA, 2 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 1 ml of fungal protease inhibitor cocktail [Sigma] per 20 g of mycelia), and dialyzed twice for 2 h each time (10,000-molecular-weight-cutoff dialysis membrane) in 2 liters of dialysis buffer. The protein concentration was determined by using the Bio-Rad reagent (Bio-Rad, Hercules, Calif.). The protein solution was divided into aliquots and stored at −80°C.
(ii) Probe generation.
DNA probes were end labeled with [γ32P]ATP by using Ready-to-Go kinase and following the manufacturer' s instructions (Amersham, Piscataway, N.J.). The oligonucleotide probes and competitors used in EMSA for the NorL site were 206/244 (positions −244 to −205: ATGAGCAGATCTCTGCTATTAAGTCGGTGATTAGCGTGCT) and 206/244mut (positions −244 to −205: ATGAGCGTATAAGAAGTTTGTGATGGGATTCGTCCGTGCT) (the underlined residues represent the region from position −210 to position −238 designated NorL, and the residues in boldface type are part of a 14-base sequence with dyad symmetry). The probes used for analysis of the AflR site in the nor-1 and ver-1 promoter regions were nor-1 AflR1 (5′-ACCGCCCAACTCGGCCAGCGACCAACACAC-3′; AflR1 in boldface type) and ver-1 AflR (5′-TCAGATATTCGGTCTCCGAGGAAAGAT-3′; AflR in boldface type).
(iii) EMSA.
EMSA and competition EMSA were performed as previously described (1). Binding reactions were performed at 30°C for 15 min by using the following components: 2 μg of nonspecific competitor DNA (polydeoxyinosinic-deoxycytidylic acid), 7.5 mg of bovine serum albumin, 20 fmol of labeled probe, and 32 μg of protein extract or 400 ng of the AflR::MBP fusion. When competitors were used, a 250-fold molar excess (5,000 fmol) of competitor was added to the binding reaction mixture. All components of the binding reaction mixture (including the competitor) were added to the tubes prior to addition of the protein extract.
RESULTS
In vitro AflR binding to AflR1 in the A. parasiticus nor-1 promoter.
Recombinant AflR::MBP (full-length AflR fused to MBP and expressed in E. coli) did not form a stable complex with an oligonucleotide containing the nor-1 AflR1 site (Fig. 2) but did form a stable complex with an oligonucleotide containing the consensus AflR binding site in the ver-1 promoter (Fig. 2). This complex was not detected when no protein or only MBP was added in the EMSA. These data are consistent with data from previous in vitro studies (13) that suggested that A. parasiticus AflR has little or no affinity for AflR1 in nor-1 but has high affinity for the AflR binding site in several A. parasiticus aflatoxin gene promoters, including the ver-1 promoter.
FIG. 2.
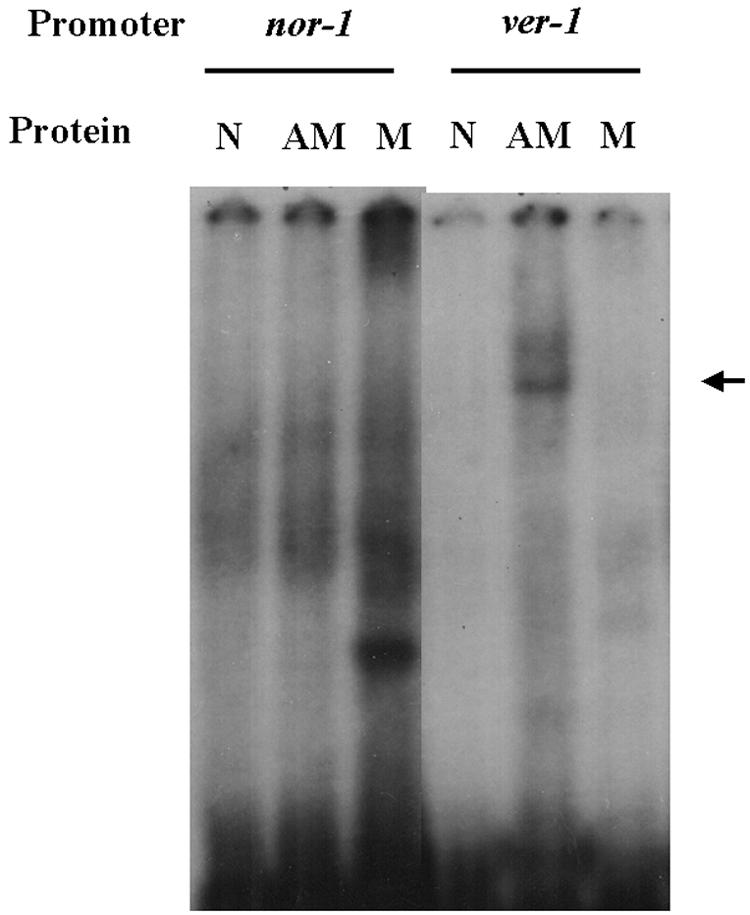
EMSA of AflR1 sites in nor-1 and ver-1 promoters. Twenty femtomoles of radiolabeled oligonucleotides carrying AflR1 from the nor-1 and ver-1 promoters was subjected to EMSA in the presence of an AflR::MBP (AM) fusion protein (0.4 μg per reaction mixture), MBP (M) (as a control; 16 μg per reaction mixture), or no protein (N) (see Materials and Methods). The arrow indicates the location of the specific AflR::MBP/ver-1 oligonucleotide complex.
AflR1 and nor-1 promoter activity in A. parasiticus.
Transformants carrying nor-1::GUS reporter plasmids with 332-, 76-, and 64-bp nor-1 promoter fragments (AflR1 located between positions −76 and −64) were constructed. Transformants carrying nor-1:GUS with a substitution mutation in AflR1 (TCGGCCAGCGA changed to AGTTTAAACAG) in the context of the 332-bp promoter also were produced.
A. parasiticus NR-1 transformants were screened with a PCR assay to identify clones with a single copy of the reporter plasmid at the 3′ end of the nor-1 gene. Single-copy integration at 3′ nor-1 was subsequently confirmed by Southern blot analysis (10). This location is known to be important for correct regulation of nor-1 promoter activity (10). The percentage of transformants with the correct integration ranged from 2 to 10% depending on the experiment (data not shown).
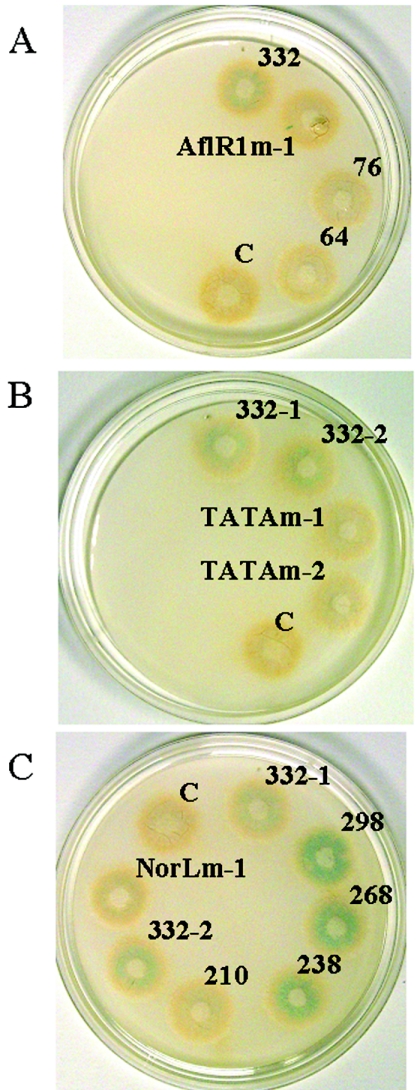
GUS activity, as an indirect indicator of nor-1 promoter activity, was measured in duplicate for two confirmed 3′ nor-1 integrants for each reporter construct in liquid shake cultures (glucose minimal salts medium, 48 h) and on solid growth medium (YES medium, 46 h) (26). In the liquid medium, two independent isolates carrying the wild-type 332-bp promoter fragment (isolates 332-1 and 332-2; wild-type AflR1) had easily measurable GUS activity (5.4 pmol/min/mg), while two independent isolates carrying the 332-bp AflRmut promoter fragment with an AflR1 substitution in the context of the 332 promoter (AflR1m-1 and AflR1m-2) and the recipient strain NR-1 (control) had no detectable GUS activity. In agreement with these data, on solid medium, 332-1 and 332-2 showed clearly detectable GUS activity (blue color in colonies), while AflR1m-1 and AflR1m-2 and NR-1 (control) displayed no detectable GUS activity (Fig. 3A). Of particular interest, shorter promoter fragments either with AflR1 (76 bp) or without AflR1 (64 bp) showed no detectable GUS activity (22). These data confirmed that AflR1 is necessary but not sufficient for nor-1 transcriptional activation in A. parasiticus and suggested that there is an important cis-acting site between positions −76 and −332.
FIG. 3.
Deletion and substitution analysis of the nor-1 promoter. A GUS activity analysis was performed with two independent single-copy 3′ integrants from each nor-1::GUS reporter construct grown on YES solid medium as described in Materials and Methods. Representative colonies are shown as follows: in panel A, isolate 1 carrying the wild-type 332-bp promoter fragment (332), deletion mutants 76 and 64, isolate 1 carrying the 332-bp fragment with an AflR1 substitution mutation (AflRm-1), and the control strain NR-1 (C) (no plasmid integrated into the genome); in panel B, isolates 1 and 2 carrying the wild-type 332-bp promoter fragment (332-1 and 332-2), isolates 1 and 2 carrying the 332-bp fragment with a TATA substitution mutation (TATAm-1 and TATAm-2), and the same control strain (C) as in panel A; and in panel C, isolates 1 and 2 carrying the wild-type 332-bp promoter fragment (332-1 and 332-2), deletion mutants 298, 268, 238, and 210, isolate 1 carrying the 332-bp fragment with a NorL substitution mutation (NorLm-1), and the same control strain (C) as in panel A.
Activity of the TATA box in the nor-1 promoter of A. parasiticus.
Two independent fungal isolates carrying the wild-type 332-bp nor-1 promoter fragment and two independent isolates carrying the 332-bp nor-1 promoter with a TATA substitution mutation (332TATAmut) in nor-1::GUS constructs were tested for GUS activity in liquid medium and on solid YES growth medium. Isolates with the TATA substitution mutation (isolates TATAm-1 and TATAm-2) and the recipient strain NR-1 (control) had no detectable GUS activity either in liquid medium or on solid growth medium (Fig. 3B), while isolates with the wild-type 332-bp nor-1 promoter (isolates 332-1 and 332-2) had easily detectable activity in liquid GMS medium (5.4 pmol/min/mg) and on solid YES medium (Fig. 3B). These data confirmed that the TATA box is required (under the conditions tested) for nor-1 transcriptional activation in vivo.
Role of NorL in nor-1 transcriptional activity in A. parasiticus.
Deletion analysis of the nor-1 promoter described above suggested the presence of an important cis-acting site localized between positions −332 and −76. To localize this potential site, the GUS activities of two independent fungal isolates carrying nor-1::GUS constructs with 332-, 298-, 268-, 238-, and 210-bp promoter fragments were measured in liquid medium and on solid growth medium. Isolates carrying nor-1::GUS constructs with the 332-, 298-, 268-, and 238-bp promoter fragments all had easily detectable GUS activity in liquid medium (ranging from 1 to 13 pmol/min/mg) and on solid medium (Fig. 3C). The isolates with the 210-bp promoter (with a wild-type AflR1 site) had no detectable activity in either medium (Fig. 3C), supporting the hypothesis that there is an important cis-acting site located between positions −210 and −238.
To verify the significance of the region from position −210 to position −238 (designated NorL) in vivo, two independent isolates carrying a nor-1::GUS construct with a NorL substitution mutation in the context of the 332-bp nor-1 promoter fragment (NorLmut) were generated. In liquid medium, a NorL substitution mutation (isolates NorLm-1 and NorLm-2) resulted in a 2.5-fold reduction in GUS activity relative to the wild-type 332-bp promoter fragment (5.4 ± 2.2 pmol/min/mg) and a more-than-threefold reduction relative to the 238-bp promoter fragment (7.2 ± 5.8 pmol/min/mg). The variation between duplicate samples for the two independent isolates was relatively large in liquid medium. However, the data did demonstrate a clear trend, which was confirmed by GUS activity data derived from the same isolates grown on solid medium (Fig. 3C). Deletion or substitution of NorL in isolates NorLm-1 and NorLm-2 consistently and reproducibly reduced the GUS activity relative to the activities observed with the 332-, 298-, 268-, and 238-bp promoter fragments on YES solid medium. When a fluorescence assay was performed with extracts prepared from the same isolates grown on solid YES medium for 72 h, replacement of NorL reduced GUS activity approximately sixfold in NorLm-1 (isolate #124A) and approximately twofold in NorLm-2 (isolate #85) compared to the 332-bp wild-type nor-1 promoter fragment (7 pmol/min/mg) (isolate #10A).
In previous studies, exogenous cyclic AMP (cAMP) (5 mM) had a strong positive regulatory effect on nor-1 promoter activity in A. parasiticus grown in liquid medium and on solid medium. This effect was mediated at least in part by a novel cis-acting site designated CRE-1 (26, 27). We found that the relative ability of the nor-1 promoter to respond to exogenous cAMP was not affected in either NorLm-1 or NorLm-2, which was consistent with the observation that the cAMP response is mediated by CRE-1 (27). Thus, while a functional NorL site is not sufficient for nor-1 transcriptional activation, it is necessary for maximum nor-1 transcriptional activation under a variety of growth conditions.
DNA-protein complexes with NorL.
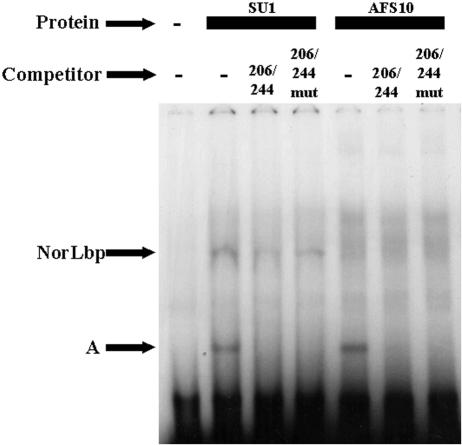
To detect DNA binding proteins that interact with NorL, we performed EMSA with a radiolabeled, 40-bp, wild-type oligonucleotide probe (206/244) or a mutant derivative (206/244mut) covering the region from position −210 to position −238 (NorL) and cell extracts from strains SU-1 (wild type) and AFS10 (AflR knockout) grown under aflatoxin-inducing conditions. Extracts from each strain were prepared on two independent occasions and gave similar EMSA results. The 206/244mut oligonucleotide had the same substitution in the region from position −210 to position −238 as the nor-1::GUS reporter construct 332NorLmut. The substitution changed 21 of 28 nucleotides while maintaining the G+C content.
With the wild-type 206/244 oligonucleotide probe, two shifted complexes carrying DNA binding proteins designated NorLbp and protein A were identified with an SU-1 protein extract (Fig. 4). A 250-fold excess of unlabeled wild-type probe 206/244 effectively competed with the NorLbp/DNA complex and the protein A/DNA complex, suggesting that both complexes represent specific DNA-protein interactions. However, a 250-fold excess of unlabeled probe 206/244mut was not an effective competitor for the NorLbp/DNA complex but prevented protein A/DNA complex formation. These data confirmed that NorLbp participated in a specific complex whose formation was mediated by the 28-bp region designated NorL. NorL substitution mutations resulted in two- to sixfold reductions in the nor-1 promoter activity complex, supporting this conclusion. The NorLbp/DNA complex was not observed with the wild-type 206/244 oligonucleotide probe and AFS10 protein extracts (Fig. 4) or with the 206/244mut oligonucleotide probe and either SU-1 or AFS10 protein extracts (data not shown). We concluded that a specific DNA/protein (NorLbp) complex depends on NorL for formation and requires AflR either directly or indirectly for maximum binding capacity. We did not identify the number of proteins in the NorLbp/DNA complex.
FIG. 4.
EMSA of NorL/NorLbp DNA-protein complex in the A. parasiticus nor-1 promoter. Twenty femtomoles of radiolabeled oligonucleotide 206/244 was used as a probe with 32 μg of protein extract from SU-1 or AFS10 grown under aflatoxin-inducing conditions. EMSA competitors (206/244 and 206/244mut) were used in a 250-fold excess. The positions of DNA-protein complexes containing NorLbp and protein A are indicated by arrows.
DISCUSSION
Experiments with AflR null mutants and AflR-inducible expression strains along with AflR-DNA binding studies demonstrated that AflR is required for aflatoxin biosynthesis and the expression of specific aflatoxin pathway genes. For example, transcriptional activation of the middle pathway gene avnA occurs through a single consensus cis-acting site (4). An additional potential AflR binding site (TCGNNNNNCGR) in the avnA promoter, when mutated, did not alter avnA transcription in vivo, and AflR protein expressed in E. coli was unable to bind to this nonfunctional binding site in vitro (4). The stcU promoter has three consensus AflR binding sites within 800 bp of the translational start site (15). Mutational analysis of the three AflR binding sites revealed that the most distal site, at position −762, had no effect on stcU transcription, while AflR consensus binding sites at positions −81 and −168 both appeared to be functional in vivo (15). The activities of the two functional AflR binding sites were not additive, as stcU promoters with either the position −81 site or the position −168 site were indistinguishable from strains that had both AflR binding sites (15).
In contrast, EMSA with A. parasiticus AflR expressed in E. coli and fungal protein extracts suggested that A. parasiticus AflR either does not bind AflR1 in the nor-1 promoter or has a much lower affinity for AflR1 than for the consensus AflR binding sites in several other aflatoxin biosynthetic promoters (13;this study), casting doubt on the functional significance of AflR1. We tested this hypothesis directly by analyzing the effects of substitution mutations on nor-1 promoter function using nor-1::GUS reporter constructs. Substitution of AflR1 in the 332-bp A. parasiticus nor-1 promoter resulted in nondetectable levels of GUS activity, confirming that AflR1 is necessary for nor-1 transcriptional activation in A. parasiticus under the growth conditions utilized in this study.
The case for AflR being the sole regulator of all aflatoxin biosynthesis structural genes is not as strong. For example, we found that a TATA box and a CRE-1 site (27) play important roles in regulation of nor-1 promoter activity. AflJ also has been putatively assigned a role as a transcriptional coactivator (8) and has been reported to interact directly with AflR (9). Furthermore, studies with the pksA promoter (14) provided evidence that both the PacC (pH sensing) and BrlA (sporulation) regulatory proteins can alter pksA transcriptional regulation through cis-acting sites in the pksA-nor-1 intergenic region (Fig. 1). However, in preliminary experiments, deletion of the PacC and BrlA consensus cis-acting sites did not affect nor-1 transcriptional regulation in A. parasiticus under the conditions tested (22). These data suggest either that nor-1 does not require the PacC and BrlA sites for transcriptional activation or that these sites are not required under our specific growth conditions.
In the present study, analysis of A. parasiticus isolates carrying deletions in the nor-1 promoter resulted in the important conclusion that although AflR is necessary for nor-1 expression, it is clearly not sufficient for maximum expression. The data also demonstrated that at least one additional cis-acting site(s) located between positions −332 and −76 contributes to nor-1 transcriptional activation. There are no additional consensus AflR binding sites (TCGNNNNNCGR) in this promoter region. Therefore, one or more unknown transcriptional activators presumably bind in this promoter region and influence nor-1 transcriptional activation in A. parasiticus. Based on deletion analysis, there is a cis-acting site, NorL, in the region from position −210 to position −238 of the nor-1 promoter. Substitution of NorL reduced nor-1 promoter activity two- to sixfold in liquid growth medium and on solid growth medium, confirming the biological significance of this site. Mutation of NorL did not influence the relative ability of the nor-1 promoter to respond to cAMP, which is consistent with the role of CRE-1 in mediating the cAMP regulatory effect (27). EMSA identified two specific complexes containing DNA binding proteins, designated NorLbp and protein A, that formed when a 40-bp oligonucleotide probe carrying the NorL site was used; both complexes were effectively competed with wild-type probe, but only the protein A/DNA complex was competed with a 40-bp mutant oligonucleotide probe sequence. The presence of the protein A/DNA complex and the absence of the NorLbp/DNA complex as determined by EMSA when the AFS10 cell extract (aflR knockout) was used suggested that formation of the protein A/DNA complex is independent of aflR expression, while formation of the NorLbp/DNA complex depends on aflR.
The wild-type 40-base oligonucleotide probe used in EMSA contained a 14-bp sequence that is a candidate for the NorL cis-acting site: 5′-AGCAGATCTCTGCT-3′ (the nucleotides in boldface type display dyad symmetry). Based on the location of this site in the NorL region, we propose that NorLbp may be a heterodimer that forms around the 14-bp NorL site. Protein A (AflR independent and part of a faster-migrating complex) could be one protein in the pair that binds to either NorL half-site, 5′-AGCAGA-3′. Consistent with this hypothesis, the NorLmut oligonucleotide, which retains four of six bases in this 5′ half-site (AGCGTA; retained bases are indicated by boldface type), can compete with wild-type oligonucleotide for protein A/DNA complex formation. The putative NorL site is present only in the nor-1 promoter and not in the promoters of other genes in the aflatoxin pathway. Thus, the regulation of nor-1 could be unique, which would not be totally unexpected given the function of the Nor-1 protein in the first committed step of the aflatoxin biosynthetic pathway. In preliminary experiments, deletion of a 50-bp fragment containing AflR2 (located immediately upstream of ORF3) greatly reduced nor-1 promoter activity (22). These results could be explained if ORF3 encodes the AflR-dependent component of NorLbp.
Deletion analysis of the nor-1 promoter also suggested the presence of a negative regulator in the region from position −298 to position −332. Deletion of this region consistently increased GUS activity driven by the nor-1 promoter both in liquid growth medium and on solid growth medium. Although this region was not the focus of the present study, we will conduct a more thorough analysis of this region to identify this additional putative transcription factor binding site.
The variation in nor-1 promoter activity in A. parasiticus was greater in liquid shake cultures than in cultures grown on plates. GUS activity on solid medium was more consistent and reproducible for independent isolates on the same plate and for plates in the same experiment. Although differences in promoter activity could be due to a change in medium composition, we think that differences in the growth environment are more important. In submerged liquid shake cultures, asexual sporulation is not induced, and the organism grows in pellets that are very different from the fungal colonies observed on plant material. Growth on a solid surface more closely mimics the natural growth environment, where the fungus grows as a mycelial mat and sporulates asexually. A more detailed analysis of aflatoxin promoter function during growth in liquid medium and on solid medium is needed to resolve this issue.
A potential problem with many aflatoxin promoter studies is that the integration site for the reporter constructs is not in the aflatoxin gene cluster (e.g., trpC for stcU::GUS [15] and niaD for avnA::GUS [4]). If ver-1::GUS was integrated at niaD, then the promoter activity was >500-fold less than if it was integrated at ver-1 (20), even though the timing of transcription was the same. A similar effect has been reported for the pyrG locus (10). In the present study, the A. parasiticus nor-1::GUS reporter constructs were integrated at the nor-1 locus, which is critical to the normal timing and level of reporter transcription in a pattern similar to the wild-type nor-1 gene pattern (10).
Our objective in this study was to determine if the AflR1 site in the nor-1 promoter was functional or if other transcription factors played a role in nor-1 regulation. Using EMSA (in vitro) and functional mutation analysis (in vivo) of the nor-1 promoter, we clearly demonstrated that AflR1 is required but not sufficient for nor-1 transcriptional activation in A. parasiticus. We also showed that a TATA box and a unique and novel cis-acting site (NorL; located between residues −210 and −238) in the A. parasiticus nor-1 promoter are necessary for maximum transcriptional activation in vivo. Finally, a specific NorL-dependent DNA-protein (NorLbp) complex was identified that relies on a functional AflR, either directly or indirectly, for maximum binding activity. These data strongly suggest that regulation of nor-1 is important in the overall regulatory scheme for aflatoxin synthesis, in keeping with the important role of Nor-1 in catalyzing the synthesis of the first stable pathway intermediate.
Acknowledgments
This work was funded by the Michigan Agricultural Experiment Station, the National Food Safety and Toxicology Center, and a grant from NIH (CA RO1-52003-14).
REFERENCES
- 1.Ausubel, F. M., R. Brent, E. Kingston, D. D. Moore, J. G. Seidman, J. A Smith, and K. Struhl (ed.). 2004. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 2.Bennett, J. W., and K. E. Papa. 1988. The aflatoxigenic Aspergillus spp. Adv. Plant Pathol. 6:263-280. [Google Scholar]
- 3.Bhatnagar, D., T. E Cleveland, and P. J. Cotty. 1994. Mycological aspects of aflatoxin formation, p. 327-346. In D. L. Eaton and J. D. Groopman (ed.), The toxicology of aflatoxins. Academic Press, San Diego, Calif.
- 4.Cary, J. W., B. G Montalbano, and K. C. Ehrlich. 2000. Promoter elements involved in the expression of the Aspergillus parasiticus aflatoxin biosynthesis pathway gene avnA. Biochim. Biophys. Acta 1491:7-12. [DOI] [PubMed] [Google Scholar]
- 5.Cary, J. W., J. M. Dyer, K. C. Ehrlich, M. S. Wright, S. H. Liang, and J. E. Linz. 2002. Molecular and functional characterization of a second copy of the aflatoxin regulatory gene, aflR-2 from Aspergillus parasiticus. Biochem. Biophys. Acta 1576:316-323. [DOI] [PubMed] [Google Scholar]
- 6.Chang, P. K., J. W. Cary, D. Bhatnager, T. E. Cleveland, J. W. Bennett, J. E. Linz, C. P. Woloshuk, and G. A. Payne. 1993. Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:3273-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, P. K., J. W. Cary, J. Yu, D. Bhatnagar, and T. E. Cleveland. 1995. The Aspergillus parasiticus polyketide synthase gene pksA, homolog of Aspergillus nidulans wA, is required for aflatoxin biosynthesis. Mol. Gen. Genet. 248:270-277. [DOI] [PubMed] [Google Scholar]
- 8.Chang, P. K., J. W. Bennett, and P. J. Cotty. 2001. Association of aflatoxin biosynthesis and sclerotial development in Aspergillus parasiticus. Mycopathologia 153:41-48. [DOI] [PubMed] [Google Scholar]
- 9.Chang, P. K., and J. Yu. 2002. Characterization of a partial duplication of the aflatoxin gene cluster in Aspergillus parasiticus ATCC 56775. Appl. Microbiol. Biotechnol. 58:632-636. [DOI] [PubMed] [Google Scholar]
- 10.Chiou, C. H., M. J. Miller, D. L. Wilson, and J. E. Linz. 2002. Chromosomal location plays a role in regulation of aflatoxin gene expression in Aspergillus parasiticus. Appl. Environ. Microbiol. 68:306-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dvorackova, I. 1990. Aflatoxin and human health. CRC Press, Boca Raton, Fla.
- 12.Ehrlich, K. C., J. W Cary, and B. G. Montalbano. 1999. Characterization of the promoter for the gene encoding the aflatoxin biosynthetic pathway regulatory protein AflR. Biochim. Biophys. Acta 1444:412-417. [DOI] [PubMed] [Google Scholar]
- 13.Ehrlich, K. C., B. J. Montalbano, and J. W. Cary. 1999. Binding of the C6-zinc cluster protein, AflR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 230:249-257. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich, K. C., B. J. Montalbano, J. W. Cary, and P. J. Cotty. 2002. Promoter elements in the aflatoxin pathway polyketide synthase gene. Biochim. Biophys. Acta 1576:171-175. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes, M., N. P. Keller, and T. H. Adams. 1998. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 28:1355-1365. [DOI] [PubMed] [Google Scholar]
- 16.Flaherty, J. E., and G. A. Payne. 1997. Overexpression of aflR leads to upregulation of pathway gene transcription and increased aflatoxin production in Aspergillus flavus. Appl. Environ. Microbiol. 63:3995-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, A. J., and C. P. Wild. 1994. Epidemiology of aflatoxin-related disease, p. 233-258. In D. L. Eaton and J. D. Groopman (ed.), The toxicology of aflatoxins. Academic Press, San Diego, Calif.
- 18.Horng, J. S., P. K. Chang, J. J. Pestka, and J. E. Linz. 1990. Development of a homologous transformation system for Aspergillus parasiticus with the gene encoding nitrate reductase. Mol. Gen. Genet. 224:294-296. [DOI] [PubMed] [Google Scholar]
- 19.Jefferson, R. A. 1987. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5:387-405. [Google Scholar]
- 20.Liang, S. H., T. S. Wu, R. Lee, F. S. Chu, and J. E. Linz. 1997. Analysis of mechanisms regulating expression of the ver-1 gene, involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 63:1058-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLean, M., and M. F. Dutton. 1995. Cellular interactions and metabolism of aflatoxin: an update. Pharmacol. Ther. 65:163-192. [DOI] [PubMed] [Google Scholar]
- 22.Miller, M. J. 2003. Transcriptional regulation of the Aspergillus parasiticus aflatoxin biosynthetic pathway gene nor-1. Ph.D. dissertation. Michigan State University, East Lansing.
- 23.Payne, G. A., G. J. Nystrom, D. Bhatnagar, T. E. Cleveland, and C. P. Woloshuk. 1993. Cloning of the afl-2 gene involved in aflatoxin biosynthesis from Aspergillus flavus. Appl. Environ. Microbiol. 59:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Esteban, B., M. Orejas, E. Gomez-Pardo, and M. A. Penalva. 1993. Molecular characterization of a fungal secondary metabolism promoter: transcription of the Aspergillus nidulans isopenicillin N synthase gene is modulated by upstream negative elements. Mol. Microbiol. 9:881-895. [DOI] [PubMed] [Google Scholar]
- 25.Peters, D. G., and M. X. Caddick. 1994. Direct analysis of native chimeric GATA specific DNA binding proteins from Aspergillus nidulans. Nucleic Acids Res. 22:5164-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roze, L. V., R. M. Beaudry, N. P. Keller, and J. E. Linz. 2004. Regulation of aflatoxin synthesis by FadA/cAMP/protein kinase A signaling in Aspergillus parasiticus. Mycopathologia 158:219-232. [DOI] [PubMed] [Google Scholar]
- 27.Roze, L. V., M. J. Miller, M. Rarick, N. Mahanti, and J. E. Linz,. 2004. A novel cAMP response element, CRE1, modulates expression of nor-1 in Aspergillus parasiticus. J. Biol. Chem. 26:27428-27439. [DOI] [PubMed] [Google Scholar]
- 28.Skory, C. D., P. K. Chang, and J. E. Linz. 1993. Regulated expression of the nor-1 and ver-1 genes associated with aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:1642-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trail, F., P. K. Chang, J. W. Cary, and J. E. Linz. 1994. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins in Aspergillus parasiticus. Appl. Environ. Microbiol. 60:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trail, F., N. Mahanti, M. Rarick, R. Mehigh, S. H. Liang, R. Zhou, and J. E. Linz. 1995. Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl. Environ Microbiol. 61:2665-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson, D. M., and G. A. Payne. 1994. Factors affecting Aspergillus flavus group infection and aflatoxin contamination of crops, p. 309-346. In D. L. Eaton and J. D. Groopman (ed.), The toxicology of aflatoxins. Academic Press, San Diego, Calif.
- 32.Woloshuk, C. P., K. R. Foutz, J. F. Brewer, D. Bhatnagar, T. E. Cleveland, and G. A. Payne. 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, J. H., R. A. Butchko, M. Fernandes, N. P. Keller, T. J. Leonard, and T. H. Adams. 1996. Conservation of structure and function of the aflatoxin regulatory gene aflR from Aspergillus nidulans and A. flavus. Curr. Genet. 29:549-555. [DOI] [PubMed] [Google Scholar]
- 34.Zhou, R., and J. E. Linz. 1999. Enzymatic function of the Nor-1 protein in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 65:5639-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]