Abstract
Trichoderma species are used commercially as biocontrol agents against a number of phytopathogenic fungi due to their mycoparasitic characterisitics. The mycoparasitic response is induced when Trichoderma specifically recognizes the presence of the host fungus and transduces the host-derived signals to their respective regulatory targets. We made deletion mutants of the tga3 gene of Trichoderma atroviride, which encodes a novel G protein α subunit that belongs to subgroup III of fungal Gα proteins. Δtga3 mutants had changes in vegetative growth, conidiation, and conidial germination and reduced intracellular cyclic AMP levels. These mutants were avirulent in direct confrontation assays with Rhizoctonia solani or Botrytis cinerea, and mycoparasitism-related infection structures were not formed. When induced with colloidal chitin or N-acetylglucosamine in liquid culture, the mutants had reduced extracellular chitinase activity even though the chitinase-encoding genes ech42 and nag1 were transcribed at a significantly higher rate than they were in the wild type. Addition of exogenous cyclic AMP did not suppress the altered phenotype or restore mycoparasitic overgrowth, although it did restore the ability to produce the infection structures. Thus, T. atroviride Tga3 has a general role in vegetative growth and can alter mycoparasitism-related characteristics, such as infection structure formation and chitinase gene expression.
Several fungi belonging to the genus Trichoderma can act as mycoparasites and are used commercially as biological control agents against plant-pathogenic fungi, such as Rhizoctonia solani, Botrytis cinerea, Sclerotium rolfsii, Sclerotinia sclerotiorum, and Pythium spp. (5, 6, 18). Mycoparasitic strains can penetrate and kill the host fungi. Mycoparasitism is accompanied by the secretion of antifungal metabolites, such as peptaibol antibiotics (32, 47), and morphological changes, such as coiling around the host and development of hooks and/or appressorium-like structures (11). Hydrolytic enzymes, such as chitinases, glucanases, and proteases, also are important for biocontrol activity, since they enable Trichoderma to degrade the host's cell wall and to utilize its cellular contents (20). Chitinase gene expression is induced by colloidal chitin, fungal cell walls, or the chitin monomer N-acetylglucosamine (29). In Trichoderma atroviride the N-acetylglucosaminidase (NAGase)-encoding gene, nag1, has a major impact on the induction by chitin of other chitinases (4). In mycoparasitic interactions between T. atroviride and R. solani, expression of ech42, which encodes endochitinase 42, is triggered by a low-molecular-weight diffusible factor released from the host prior to physical contact between the two fungi (10, 50). Lectins in the host's cell wall can induce coiling of the mycoparasite around the host (1, 24, 41).
Elucidation of the signaling pathways underlying mycoparasitism has only recently begun. Heterotrimeric G proteins are composed of α, β, and γ subunits. Gα subunits play pivotal roles in the recognition process, virulence, and virulence-dependent development in a number of plant-pathogenic fungi (3, 15, 26, 27, 31). Fungal Gα subunits are highly conserved and can be divided into three major subgroups (3). In T. atroviride, the subgroup I Gα protein Tga1 affects light-induced conidiation and mycoparasitism-related coiling (41). The corresponding protein in Trichoderma virens is involved in antagonism against S. rolfsii but not in antagonism against R. solani. ΔtgaA loss-of-function mutants sporulate and have coiling behavior similar to that of the wild type (37), suggesting that the subgroup I Gα subunits have different functions in these mycoparasites. The T. virens subgroup II G protein α subunit, TgaB, has no specific role in development or mycoparasitism (37). Fungal subgroup III Gα proteins also regulate morphological and developmental processes, such as germination, conidiation, and secondary metabolite production (28, 53), and they may increase intracellular cyclic AMP (cAMP) levels by stimulating adenylyl cyclase (28, 33, 39).
The objectives of this study were to isolate a subgroup III Gα subunit deletion mutant of T. atroviride and to determine its role in development and mycoparasitism. This report is the first report of the role of the Tga3-mediated G protein pathway in mycoparasitism-related properties, such as host recognition, chitinase expression, and secretion.
MATERIALS AND METHODS
Strains and culture conditions.
T. atroviride strain P1 (formerly Trichoderma harzianum ATCC 74058) was used for this study and was grown on potato dextrose agar (PDA) (Difco, Detroit, Mich.) at 28°C until sporulation. B. cinerea and R. solani were used as pathogens and were obtained from the collection of the Institute of Plant Pathology, Università degli Studi di Napoli Federico II, Naples, Italy. Escherichia coli JM 109 was the host for plasmid amplification.
T. atroviride was grown in liquid synthetic medium (SM) as described previously (4). For induction experiments, the fungus was precultivated for 36 h in SM containing 1% (wt/vol) glycerol as the carbon source, harvested by filtration, washed with sterile tap water, and transferred to fresh SM containing 1% (wt/vol) colloidal chitin or 1% (wt/vol) N-acetyl-β-d-glucosamine. To test the influence of exogenous cAMP (Sigma, St. Louis, Mo.), cAMP was added to PDA cooled to 48°C after autoclaving.
Cloning of tga3.
Two products (∼1,000 and ∼1,100 bp) were obtained by PCR amplification of genomic DNA from T. atroviride strain P1 with degenerate oligonucleotide primers based on conserved regions of Gα sequences (primers GαF [5′-GGNGCNGGNGARWSNGGNAA-3′] and GaR [5′-GTRTCNGTNGCNYDNGT-3′]). The products were subcloned into pGEM-T (Promega, Madison, Wis.) and sequenced. For screening of a genomic λ BlueStar library of T. atroviride P1, the tga3-containing PCR fragment was radioactively labeled with 32P by random priming and used as a probe. Sequencing of isolated clones was performed by using the following internal primers: Tga3intF1 (5′-CTGCTTCGAGAACGTTACCTC-3′; bp 881 to 901), Tga3intF2 (5′-CGATGGCATTTACGCGAAG-3′; bp 1632 to 1650), Tga3intF3 (5′-GTATTAACTTGCTGCGCTCATTG-3′; bp 2476 to 2498), Tga3intR1 (5′-GTATTCGCTCAGCGCAACAC-3′; bp 913 to 932), Tga3intR2 (5′-GACAATGGTCGACTTGCCAC-3′; bp 220 to 239), Tga3intR3 (5′-CAGGGCTGGCAATGATGAAAG-3′; bp −486 to −466), and Tga3intR4 (5′-CTGTGCCAGCCTAAAAGTGTGC-3′; bp −1287 to −1266).
Fungal transformation.
Agrobacterium-mediated transformation of T. atroviride was carried out as previously described (51). Briefly, after cocultivation of a conidial suspension (107 spores ml−1) for 24 h with an Agrobacterium tumefaciens strain containing the disruption construct pTSZ-Δtga3, in which the entire tga3 coding region was replaced by an hph (hygromycin B phosphotransferase-encoding) expression cassette (51), fungal transformants were selected on PDA containing 200 μg of hygromycin B per ml. Transformants were recovered and purified to mitotic stability by repeated transfer to selective medium and by two rounds of single-spore isolation.
DNA and RNA manipulations.
Genomic DNA was isolated as previously described (16); Southern hybridizations were carried out as described by Sambrook et al. (43). RNA was isolated as described by Chomczynski and Sacchi (7). Northern blotting of total RNA was performed by using BiodyneB nylon membranes (Pall Europe, Portsmouth, United Kingdom) and applying 20 μg of RNA. For hybridization, the radioactively labeled PCR product containing most of the tga3 structural gene described above was used. For analysis of chitinase gene transcription, a 1.9-kb SalI/XbaI fragment of nag1, a 1.0-kb PstI fragment of ech42, and a 1.9-kb KpnI fragment of act1 were used as probes.
Real-time reverse transcriptase PCR.
RNAs were incubated with DNase I (1 U/μg of RNA; Fermentas, St. Leon-Rot, Germany) according to the manufacturer's instructions to remove any remaining chromosomal DNA. First-strand cDNA synthesis was carried out with an oligo(dT)18 primer (0.5 μg/μl), a random hexamer primer (0.2 μg/μl), 0.45 μg of total RNA, and RevertAid H Minus Moloney murine leukemia virus reverse transcriptase (200 U/μl; Fermentas) according to the manufacturer's instructions. For amplification of an ∼60-bp fragment of the actin-encoding gene act1, real-time PCR was carried out with primers act1F (5′-TATGTGCAAGGCCGGTTTCG-3′) and act1R (5′-GGTCTTCCGACAATGGAGG-3′). A 370-bp nag1 fragment was amplified with primers nag1intF (5′-GCAAATGCGCTGTGGCCCATC-3′) and nag1intR (5′-GCTTGAAGGTGGTCGCGGTATC-3′).
Real-time PCR amplification was carried out with the iCycler real-time detection system (Bio-Rad, Hercules, Calif.) in a 50-μl mixture containing iQ SYBR Green Supermix (Bio-Rad), each primer at a concentration of 100 nM, 5 μl of sample, 10 nM fluorescein, and 1× SYBR Green. The act1 fragment was amplified with the following PCR program: initial denaturation for 90 s at 95°C, followed by 50 cycles consisting of 95°C for 15 s, 57°C for 20 s, and 72°C for 20 s. nag1 was amplified by 50 cycles of 95°C for 15 s, 61°C for 20 s, and 72°C for 20 s. Successful amplification was verified by determination of the melting temperature and by agarose gel electrophoresis. A nonamplification control and a control with RNA without reverse transcription to exclude DNA contaminants were performed in parallel for every assay. All determinations were performed three times with two different sample dilutions. The results were displayed in the PCR base line subtracted mode, where the software (Bio-Rad) determined the cycle number at which the reaction reached the threshold.
The efficiency (E) of the real-time PCR was calculated by amplification of two different dilutions of the samples by using the equation E = 101/tc (12), and the threshold cycles (tc) were amended for an optimum efficiency of 2. To compare different samples, the threshold cycle of the analysis with the primers for nag1 was corrected with a factor for the act1 amplification. The sample of wild-type T. atroviride P1 harvested 10 h after replacement with glucose (Gluc/10h sample) was assigned the factor 1 because it had the highest threshold cycle. As 3.32 cycles theoretically gives a 10-fold increase in the amount of PCR product, the difference between the threshold cycle of the wild-type Gluc/10h sample and the corrected threshold cycle of the other samples was divided by 3.32, yielding X. Inducibility (I) was calculated by using the equation I = 10X.
Protoplasting and determination of growth inhibition by SDS.
Protoplasts were prepared from mycelia grown for 15 h on PDA containing cellophane (Maildor, Gournay en Bray, France) as previously described (16) with lysing enzymes from T. harzianum (Glucanex; Sigma). The inhibitory effect of sodium dodecyl sulfate (SDS) was determined by measuring the radial growth rates on plates containing PDA supplemented with 0.01 or 0.02% SDS and comparing these growth rates to the growth rates on plates containing PDA without SDS. Hyphal elongation was measured twice a day, and the growth rate was determined.
Biocontrol assays.
Tests of inhibition of B. cinerea spore germination were performed as described previously (4). Inhibition by culture filtrates was adjusted for the biomass present in the culture. Plate confrontation assays with R. solani or B. cinerea as the host were performed as previously described (32, 50).
Enzyme assays.
Chitinase activity was determined in culture filtrates, in culture broth including mycelia, and in intracellular samples. Mycelium-bound activity was determined with medium containing mycelium, which was removed by centrifugation at 13,000 × g at 22°C for 5 min before spectrophotometric analysis. For determination of intracellular enzyme activities, harvested mycelium was ground in liquid N2 and suspended in 1.5 volumes of protease inhibitor cocktail containing 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin A, and 0.6 μM leupeptin hemisulfate (all obtained from Sigma). Cells were broken by sonication for 1 min at 4°C (cycle 30 with 40% power; Sonopuls Sonicator [Bandelin, Berlin, Germany]), and the extract was centrifuged for 20 min at 20,000 × g at 4°C. The supernatant was collected and stored at −20°C after addition of 10% glycerol.
Enzyme assays were performed as previously described (17) with p-nitrophenyl N-acetyl-β-d-glucosaminide as the substrate for determination of N-acetylglucosaminidase activity and with p-nitrophenyl β-d-N′,N"-triacetylchitotriose as the substrate for determination of endochitinase activity (all chemicals were obtained from Sigma). Enzyme activity was related to biomass by determination of the mycelial dry weight.
Measurement of intracellular cAMP levels.
Mycelia from strains grown for 72 h on PDA plates covered with a cellophane membrane were ground in liquid N2 to a fine powder, weighed, and homogenized in 10 volumes of 0.1 M HCl. The samples were centrifuged at 600 × g at 22°C, and the cAMP content was measured with a Direct cAMP enzyme immunoassay kit (Sigma) used according to the manufacturer's instructions. cAMP levels were related to the protein concentrations (as measured by the Bio-Rad protein assay [Bio-Rad]) of the samples and were expressed as means ± standard deviations for three independent experiments.
Determination of cell wall chitin and glucan contents.
Triplicate cultures of each strain were grown for 36 h in potato dextrose broth (PDB) (Difco). Mycelia were harvested, pressed dry between sheets of filter paper, weighed, ground in liquid N2, and suspended in distilled water. Cells were broken by sonication for 1 min at 4°C (cycle 50 and 70% power; Sonopuls Sonicator), and the extract was centrifuged for 10 min at 13,000 × g at 4°C, washed four times with distilled water, and lyophilized.
Chitin content was determined as previously described (48) after the samples were hydrolyzed by boiling for 4 h with 4 M HCl. For glucan determination, the samples were hydrolyzed as described by Nemcovic and Farkas (38), and the amount of glucan was determined as described previously (35).
Nucleotide sequence accession number.
The nucleotide sequence of tga3 has been deposited in the GenBank database under accession number AF452097.
RESULTS
Cloning and characterization of tga3.
PCR amplification resulted in two products that were approximately 1,000 and 1,100 bp long, both of which correspond to Gα subunits based on amino acid sequences deduced from the sequenced PCR fragments. A 958-bp fragment encoded Tga1 (GenBank accession number AY036905) (41). A 1,189-bp fragment, designated tga3, encoded a protein that is 88% identical (9) to Neurospora crassa GNA-3, which belongs to subgroup III of fungal Gα subunits as classified by Bölker (3). The 1,189-bp PCR product was used to screen a genomic λ BlueStar library and to identify multiple clones carrying 10- to 15-kb inserts. Based on sequencing with internal primers Tga3intF1-F3 and Tga3intR1-R4, tga3 (GenBank accession number AF452097) consists of a predicted 1,470-bp open reading frame interrupted by five putative introns at positions conserved in other fungal Gα homologues. The amino acid sequence was 90% identical to the MAGA sequence (GenBank accession number AF011340) from Magnaporthe grisea and 88% identical to the GNA-3 sequence (GenBank accession number AF281862) from N. crassa. Most of the differences occurred in the N-terminal one-half of the protein. Tga3 was only 49% identical to T. atroviride Tga1. Based on Southern analyses performed with genomic DNA of T. atroviride digested with several restriction enzymes and with the 1,189-bp PCR product as the probe, tga3 is present in a single copy in the genome of T. atroviride (data not shown).
tga3 expression during confrontation between T. atroviride and R. solani.
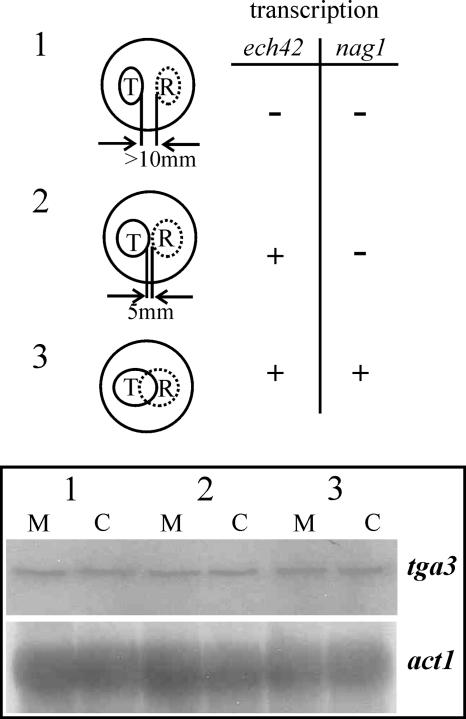
tga3 gene transcription was examined during confrontation of T. atroviride with R. solani by isolating total RNA from different stages during a plate confrontation assay (Fig. 1). Samples from stage 1 (the distance between Trichoderma and the host was >10 mm; neither ech42 nor nag1 was expressed), stage 2 (precontact; ech42 was expressed, but nag1 was not expressed), and stage 3 (direct contact; both nag1 and ech42 were expressed) (50), including controls in which T. atroviride was confronted with itself, were subjected to Northern analysis. tga3 was transcribed constitutively whether the host was present or not (Fig. 1), although detection of the tga3 transcript required a long exposure of the Northern blot, suggesting that tga3 was not highly expressed.
FIG. 1.
Transcription of tga3 during different stages of a direct confrontation of T. atroviride P1 with R. solani. A schematic representation of the different confrontation stages (stages 1, 2, and 3) of the mycoparasitic interaction is shown, and expression of the chitinase-encoding genes ech42 and nag1 is indicated by a plus sign or a minus sign. T, T. atroviride; R, R. solani. For Northern analysis of tga3 gene transcription, 20 μg of total RNA was hybridized with a ∼1.1-kb PCR fragment of the tga3 structural gene or a 1.9-kb KpnI fragment of the actin-encoding gene act1 as a probe. Lanes M, mycoparasitic interaction between Trichoderma and Rhizoctonia; lanes C, control, in which Trichoderma was confronted with itself.
Phenotype of tga3 disruptants.
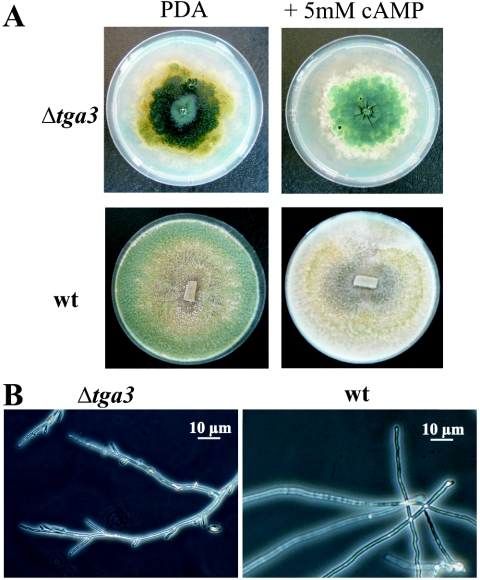
Sixty-two percent of the transformants had single-copy integration of the disruption cassette at the homologous tga3 gene locus. Three arbitrarily chosen disruptants, Δtga3-1, Δtga3-3, and Δtga3-5, were analyzed. The disruptants grew at ∼45% the rate of the wild type on PDA and formed little aerial mycelium. The mutants formed flat, glossy colonies that continuously produced dark green conidia (Fig. 2A) that were the normal size and shape. The Δtga3 strains also sporulated in the dark, suggesting that the Tga3 Gα subunit has a role in light-induced conidiation. The intracellular cAMP levels in the mutants (6.5 ± 1.3 pmol of cAMP/mg of protein) were less than one-half those found in the wild-type strain (15.9 ± 4.2 pmol/mg) or a transformant with an ectopic integration of the disruption cassette (13.5 ± 2.9 pmol/mg). Addition of exogenous cAMP (2, 5, and 10 mM) did not alter the slowly growing, hypersporulating phenotype of the mutants (Fig. 2A), although inclusion of 5 mM cAMP in the growth medium more than doubled the intracellular cAMP levels (35.5 ± 5.4 pmol of cAMP/mg of protein) in all three strains tested compared to the wild-type strain grown on PDA without cAMP.
FIG. 2.
Morphology of strain Δtga3-3 compared to the morphology of wild-type T. atroviride P1 (wt). (A) Colony morphology after growth on solid medium (PDA) for 7 days and effect of addition of 5 mM exogenous cAMP on the phenotype. (B) Microscopic examination of the T. atroviride wild type and Δtga3-3 mutant after growth in liquid culture (PDB) for 36 h at 28°C.
In liquid cultures in PDB or SM with 1% glycerol for 36 h, no conidiation occurred. The germination of conidia from the Δtga3 mutants was both reduced and delayed compared to the germination of conidia from wild-type strain P1. After 9 h of incubation at 28°C, ∼30% of the wild-type conidia, but only 10% of the Δtga3 conidia, had formed germ tubes, and after 11 h, 55% of the wild-type conidia and 33% of the Δtga3 conidia had germinated. After incubation for 36 h in liquid cultures, the Δtga3 mutants produced ∼30% less biomass than the comparable wild-type cultures produced. The mycelia of Δtga3 strains were more highly branched than those of the wild-type strain (Fig. 2B).
Mycoparasitic ability of Δtga3 mutants.
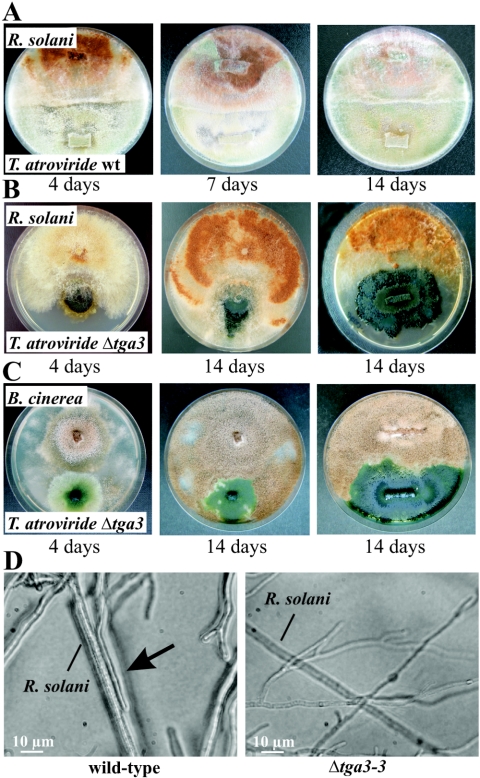
After 3 to 4 days of cocultivation of wild-type strain P1 with R. solani, P1 began to overgrow and lyse the host fungus (Fig. 3A), while the Δtga3 mutants were avirulent in similar cultures (Fig. 3B). Even when the mutants were grown until their mycelia covered about one-half of the plate before R. solani was inoculated on the opposite side of the petri dish, the Δtga3 strains were unable to parasitize the R. solani colonies (Fig. 3B). Thus, the Tga3 Gα subunit could have an important role in signal transduction during host recognition and/or the mycoparasitic response of T. atroviride. Similar results were obtained for the Δtga3 mutants when B. cinerea (Fig. 3C) or S. sclerotiorum (data not shown) was used in the confrontation assays. Typical morphological changes associated with mycoparasitism (34) (e.g., growth of the mycoparasite alongside the host's hyphae and attachment to these hyphae) were not observed in confrontations with the Δtga3 mutants (Fig. 3D). Addition of exogenous cAMP to the plate confrontation assays (PDA supplemented with 2, 5, or 10 mM cAMP) did not restore the mycoparasitic behavior (i.e., overgrowth and host lysis) of the Δtga3 strains, although cultures supplemented with 5 mM cAMP regained the ability to form mycoparasitism-related infection structures (data not shown). Culture filtrates of the Δtga3 mutants from cultures grown on N-acetylglucosamine for 10 h and on colloidal chitin for 48 h as sole carbon sources had 25 to 75% of the antifungal activity (inhibition of Botrytis spore germination) of the wild-type culture filtrates.
FIG. 3.
Biocontrol assays for analyzing mycoparasitic ability. (A to C) Plate confrontation assays of T. atroviride wild-type strain P1 (wt) (A) and the Δtga3-3 mutant with R. solani (B) and B. cinerea (C). In the plates on the right in panels B and C, the tga3-negative mutant was grown until it covered about one-half of the plate, and then the host fungi were inoculated. Pictures were taken 4, 7, and 14 days (A) or 4 and 14 days (B and C) after inoculation. (D) Microscopic examination of the mycoparasitic interaction between the T. atroviride wild type or the Δtga3-3 mutant and R. solani. Attachment of the wild type to the host's hyphae is indicated by the arrow.
Extracellular chitinase activities in Δtga3 strains.
In the wild-type strain T. atroviride P1, no chitinase production was observed during incubation on glucose or glycerol for 5 and 10 h (data not shown). When induced with N-acetylglucosamine, the wild type produced up to 300 U of NAGase per g (dry weight) for up to 10 h; after 12 h the NAGase activity began to decline again (Table 1). Upon incubation on colloidal chitin for 48 h, both NAGase activity (up to 1,900 U/g [dry weight]) and endochitinase activity (up to 205 U/g [dry weight]) also were induced in T. atroviride P1.
TABLE 1.
Extracellular, mycelium-bound, and intracellular NAGase activities of N-acetylglucosamine-induced cultures of T. atroviride wild-type strain P1 and the Δtga3 mutant
| Sample | Induction time (h) | NAGase activity (U/g [dry wt])a
|
|
|---|---|---|---|
| Wild type | Δtga3 | ||
| Extracellular | 5 | 75 ± 7 | 16 ± 9 |
| 10 | 300 ± 70 | 21 ± 3 | |
| 12 | 270 ± 50 | 37 ± 8 | |
| Myceliumbound | 5 | 900 ± 400 A | 1,100 ± 500 C |
| 10 | 1,500 ± 400 A | 700 ± 300 C | |
| 12 | 700 ± 200 A | 700 ± 100 C | |
| Intracellular | 5 | 10,500 ± 300 B | 12,400 ± 300 D |
| 10 | 10,600 ± 100 B | 12,500 ± 300 D | |
| 12 | 11,400 ± 300 B | 12,800 ± 400 D | |
The values are means ± standard deviations of three independent experiments. The values followed by the same letter were pooled, and the data for the Δtga3 mutant were compared to the data for the wild type by using the Mann-Whitney U test. For the mycelium-bound samples two replicates of nine samples were examined, and the P value was >0.5. For the intracellular samples two replicates of nine samples were examined, and the P value was ≤0.001.
As with the wild type, chitinase production was not observed in the Δtga3 mutants when they were cultured on glucose or glycerol. The extracellular NAGase activities of the N-acetylglucosamine-induced cultures of the Δtga3 strains were 7 to 21% that of the wild-type strain (Table 1). In cultures induced with colloidal chitin, both the NAGase and endochitinase activities of the Δtga3 mutants were no more than 22 to 60% those of the wild type. These results suggest that Tga3 influences the production of extracellular endo- and exochitinases.
nag1 and ech42 transcription in Δtga3 mutants.
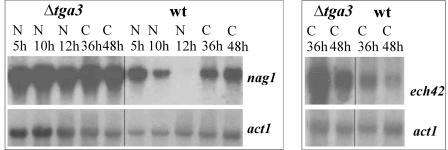
In wild-type strain P1, nag1 transcript formation was greatest 5 h after transfer to a medium containing N-acetylglucosamine, lower after 10 h, and undetectable by 12 h. Following transfer to colloidal chitin, nag1 and ech42 transcripts could be detected at both 36 and 48 h. At all times tested, the Δtga3 mutant produced significantly more nag1 transcript than the wild type produced when the organisms were induced with N-acetylglucosamine, and it produced both nag1 and ech42 transcripts when the organisms were induced with colloidal chitin (Fig. 4).
FIG. 4.
Analysis of transcription of the chitinase-encoding genes nag1 and ech42 in the T. atroviride wild type (wt) and the Δtga3-3 mutant strain following induction with N-acetylglucosamine or colloidal chitin: Northern analysis of nag1 gene transcription 5, 10, and 12 h after transfer to 1% N-acetylglucosamine (lanes N) and 36 and 48 h after transfer to 1% colloidal chitin (lanes C). A 1.9-kb SalI/XbaI fragment of nag1 was used as the probe. ech42 gene transcription was analyzed 36 and 48 h after transfer to colloidal chitin by using a 1.0-kb PstI fragment of ech42 as the probe. Hybridization with a 1.9-kb KpnI fragment of the actin-encoding gene act1 was used as a loading control.
The nag1 mRNA levels of selected samples were quantified by real-time reverse transcriptase PCR. nag1 mRNA was barely detectable in both the wild type and the Δtga3-3 mutant 10 h after transfer to glucose or glycerol. After induction with N-acetylglucosamine for 10 h, the nag1 transcript levels were at their highest values in both strains; the wild type showed ∼2,700-fold induction, and Δtga3 showed ∼3,500-fold induction. After 12 h, the nag1 transcript level for the wild-type strain had decreased to ∼600-fold inducibility, reflecting the results of the Northern blot analysis, while the nag1 transcript level for the Δtga3 strain had dropped only slightly to ∼2,650-fold inducibility. When organisms were induced with colloidal chitin, a similar pattern was observed; the Δtga3 strain again had elevated nag1 transcript levels at both 36 h (1.9-fold higher than the wild type) and 48 h (1.5-fold higher than the wild type) after induction.
Localization of N-acetylglucosaminidase activity in Δtga3 mutants.
Although the Δtga3 mutant had elevated nag1 and ech42 transcript levels, its extracellular chitinase activities in induced cultures were reduced. Similar levels of mycelium-bound NAGase activity were found in the wild type and the mutant, but intracellular NAGase activity was significantly higher in the Δtga3 mutant than in the wild-type control (Table 1).
Cell wall of the Δtga3 mutants.
We could not produce protoplasts from the Δtga3 strains even if we incubated them for up to 20 h (normal incubation time, 3 to 4 h) and added twice as much of the lysing enzymes. Resistance to protoplasting by cell wall lytic enzymes may be due to a change in cell wall polymer content, an increase in cross-linking, or a change in the type of cross-linking occurring in the cell wall. Any of these changes could affect cell wall porosity (21). Nevertheless, neither the cell wall chitin nor the glucan content of the Δtga3 mutants was significantly different from the content of the wild type or the transformant with ectopic integration of the disruption construct (data not shown). Growth inhibition by SDS often is used as an indicator of cell wall integrity (8, 21), but the reduction in hyphal growth by the Δtga3 mutants in the presence of 0.01 or 0.02% SDS was comparable to that of the control strains (data not shown).
DISCUSSION
In filamentous fungi, α subunits of heterotrimeric G proteins, which are activated upon ligand binding to a corresponding receptor, play crucial roles in regulating functions such as asexual development, sexual reproduction, and pathogenicity, in which fungi sense the presence of mating partners by pheromones and pathogenic fungi respond to signals from their hosts (22, 25, 26, 28, 31, 39, 44, 52). We identified and characterized a novel G protein α subunit-encoding gene, tga3, of the mycoparasitic fungus T. atroviride, which groups with subgroup III of fungal Gα proteins. Thus, Trichoderma spp. have members of each Gα protein subgroup, as previously shown for, e.g., N. crassa and M. grisea (28, 31, 45). The tga3 gene is transcribed constitutively at a low level during different growth stages and irrespective of the presence of a host fungus. Deleting the T. atroviride subgroup I Gα protein-encoding gene tga1 did not alter tga3 transcript levels (Zeilinger, unpublished data), which is consistent with results obtained for N. crassa, where deleting any of the three Gα subunit-encoding genes did not affect the expression of the other G proteins (28).
For generating Δtga3 knockout mutants, the hph (hygromycin B phosphotransferase-encoding) gene was employed as a selection marker. This marker gene is frequently used for transformation of T. atroviride (34, 41, 50, 51), and its gene product so far has not been reported to interfere with growth or the mycoparasitic ability of the fungus. This is consistent with our results for an ectopic transformant used as a control in selected experiments, in which it behaved like the wild type.
Δtga3 mutants grow slowly with nearly no aerial hyphae, and they continuously produce conidia when they are cultured on solid media, a phenotype similar to that reported for an N. crassa Δgna-3 mutant (28). However the Neurospora mutant can form conidia in submerged culture. Conidia from Δtga3 mutants also have reduced and delayed germination, a characteristic similar to a characteristic observed for the gasC mutants of Penicillium marneffei (53).
Mutants of some subgroup III fungal Gα proteins, including Cryphonectria parasitica CPG-2 (14), N. crassa GNA-3 (28), Ustilago maydis Gpa3 (39), and Podospora anserina MOD-D (33), can be phenotypically suppressed by exogenous cAMP. The finding that the Δtga3 mutant phenotype was not altered by the addition of exogenous cAMP suggests that these properties are mediated through a cAMP-independent pathway. Nevertheless, intracellular cAMP levels were significantly reduced in Δtga3 mutants, suggesting that Tga3 can increase the internal cAMP level.
In Trichoderma, as in other filamentous fungi, asexual sporulation is activated by light (2, 13, 42, 49), and no conidia are produced when the fungus is grown in the dark. When grown in the dark, Δtga3 mutants sporulated in the same manner as when they were cultured in the light, like tga1 antisense mutants (41). Thus, both Gα proteins (Tga1 and Tga3) act as negative regulators of conidiation, perhaps by sharing downstream signaling components. Overlapping roles for Gα subunit proteins also are known in N. crassa, in which both GNA-1 and GNA-3 regulate conidiation (25, 28), and in P. marneffei, in which GasA- and GasC-mediated signaling blocks conidiation and regulates production of a red pigment (52, 53).
Tga3 also has a major impact on mycoparasitism, as Δtga3 mutants were avirulent in plate confrontation assays with R. solani, B. cinerea (Fig. 3), and S. sclerotiorum (data not shown). The antifungal activity of chitinolytic enzymes and their role during mycoparasitism are well established (29, 40, 46). While Δtga3 strains had reduced extracellular endochitinase and N-acetylglucosaminidase levels, nag1 and ech42 gene transcription and intracellular chitinase levels were increased in the mutants. Compared to Aspergillus nidulans, in which the subgroup I Gα subunit FadA and the Gβ subunit SfaD regulate cell wall porosity and chitin content (8), cell wall composition was not altered in Δtga3 mutants. Thus, the observed accumulation of chitinolytic enzymes inside the cell and the retention of these enzymes in the cell wall may be due to Tga3-mediated posttranscriptional regulation of chitinase gene expression and its influence on chitinase secretion.
When chitinase-induced culture supernatants were used in in vitro Botrytis spore germination assays, the antifungal activity of the Δtga3 mutants was reduced compared to the activity of the wild type and roughly reflected the extracellular chitinase activity. These experiments showed that the Δtga3 mutants can still produce extracellular chitinases and suggested that the complete loss of mycoparasitism in direct confrontation assays is caused by other defects. One hypothesis is that the Δtga3 strains no longer recognize the host. This hypothesis is strongly supported by the lack of mycoparasitism-related infection structures (e.g., growth alongside, attaching to, or coiling around the host's hyphae) (11, 34) in dual cultures of the Δtga3 mutants with a host fungus. In U. maydis, deleting the subgroup III Gα protein Gpa-3 rendered the fungus avirulent. The resulting mutants also were mating deficient, as they could not respond to the pheromone signal (39). Another hypothesis for the loss of mycoparasitism by the Δtga3 mutants is that hyphal polarity could be changed due to the altered, hyperbranching phenotype, resulting in an inability to form the typical mycoparasitism-accompanying morphological changes.
Recognition of the host fungus by Trichoderma seems to play an important role in triggering the mycoparasitic response (1, 36, 37). The host-derived signals identified so far are diffusible, low-molecular-weight compounds released from the host's cell wall by the action of Trichoderma-secreted chitinases, which function as precontact inducers of antagonistic genes in the mycoparasite and as elicitors of plant defense mechanisms (19, 30, 50). Since most of the chitinase activity is retained inside the cells of Δtga3 strains, one hypothesis for the loss of host recognition could be a reduction in or a complete lack of this inducer, resulting in the failure of precontact induction of mycoparasitism-relevant genes. In addition, lectins present on the host's cell wall may be involved in host recognition, as they induce mycoparasitic coiling upon direct contact (23, 24). When different lectins were used to coat nylon fibers for biomimetic experiments, the lectins promoted attachment to and coiling around the fibers, regardless of their sugar moiety, suggesting that lectins were not the specificity-determining factor in host recognition (41). Tga1 antisense transformants could not overgrow R. solani and exhibited decreased coiling around nylon fibers when they were tested in biomimetic assays (41), similar to Δtga3 mutants which showed a loss of mycoparasitism-related infection structure formation upon direct contact with a living host fungus.
In summary, the data strongly support the hypothesis that the loss of mycoparasitism of Δtga3 mutants is a combination of indirect effects (e.g., reduced growth, a defect in chitinase secretion) and direct effects (e.g., loss of infection structure formation). Furthermore, Tga3 directly influences the regulation of mycoparasitism-related chitinase gene transcription, as the mutants had enhanced nag1 and ech42 mRNA levels.
The two G protein α subunits of T. atroviride have overlapping roles in conidiation and mycoparasitism, so it is necessary to identify the specific receptors and the signaling pathways used to transmit the host-derived signals to downstream effectors in order to understand the complex biochemical processes that result in fungal biocontrol.
Acknowledgments
This work was supported by grants from the Austrian Programme for Advanced Research and Technology (APART 10764) of the Austrian Academy of Sciences and from Fonds zur Förderung Wissenschaftlicher Forschung (grant FWF P15483). Part of the work was performed within the scope of the Marie Curie training site GEMCAT (contract HPMT-CT-2001-00243) supported by the European Community.
We thank C. P. Kubicek for helpful discussions and for critically reviewing the manuscript.
REFERENCES
- 1.Barak, R., Y. Elad, D. Mirelman, and I. Chet. 1985. Lectins: a possible basis for specific recognition in the interaction of Trichoderma and Sclerotium rolfsii. Phytopathology 75:458-462. [Google Scholar]
- 2.Betina, V. 1995. Photoinduced conidiation in Trichoderma viride. Folia Microbiol. 40:219-224. [Google Scholar]
- 3.Bölker, M. 1998. Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet. Biol. 25:143-156. [DOI] [PubMed] [Google Scholar]
- 4.Brunner, K., C. K. Peterbauer, R. L. Mach, M. Lorito, S. Zeilinger, and C. P. Kubicek. 2003. The Nag1 N-acetylglucosaminidase of Trichoderma atroviride is essential for chitinase induction by chitin and of major relevance to biocontrol. Curr. Genet. 43:289-295. [DOI] [PubMed] [Google Scholar]
- 5.Chet, I. 1987. Trichoderma—application, mode of action and potential as a biocontrol agent of soilborne plant pathogenic fungi, p. 137-160. In I. Chet (ed.), Innovative approaches to plant disease control. John Wiley & Sons, Inc., New York, N.Y.
- 6.Chet, I., N. Benhamou, and S. Haran. 1998. Mycoparasitism and lytic enzymes, p. 153-171. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2. Enzymes, biological control and commercial application. Taylor and Francis Ltd., London, United Kingdom.
- 7.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 8.Coca, M. A., B. Damsz, D.-J. Yun, P. M. Hasegawa, R. A. Bressan, and M. L. Narasimhan. 2000. Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J. 22:61-69. [DOI] [PubMed] [Google Scholar]
- 9.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortès, C., A. Gutiérrez, V. Olmedo, J. Inbar, I. Chet, and A. Herrera-Estrella. 1998. The expression of genes involved in parasitism by Trichoderma harzianum is triggered by a diffusible factor. Mol. Gen. Genet. 260:218-225. [DOI] [PubMed] [Google Scholar]
- 11.Elad, Y., I. Chet, P. Boyle, and Y. Henis. 1983. Parasitism by Trichoderma spp. on Rhizoctonia solani and Sclerotium rolfsii—scanning electron microscopy and fluorescence microscopy. Phytopathology 73:85-88. [Google Scholar]
- 12.Freeman, W. M., S. J. Walker, and K. E. Vrana. 1999. Quantitative RT-PCR: pitfalls and potential. BioTechniques 26:112-122, 124-125. [DOI] [PubMed] [Google Scholar]
- 13.Froehlich, A. C., Y. Liu, J. J. Loros, and J. C. Dunlap. 2002. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297:815-819. [DOI] [PubMed] [Google Scholar]
- 14.Gao, S., and D. L. Nuss. 1996. Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc. Natl. Acad. Sci. USA 93:14122-14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gronover, C. S., D. Kasulke, P. Tudzynski, and B. Tudzynski. 2001. The role of G protein alpha subunits in the infection process of the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 14:1293-1302. [DOI] [PubMed] [Google Scholar]
- 16.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaff. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18:71-76. [DOI] [PubMed] [Google Scholar]
- 17.Harman, G. E., C. K. Hayes, M. Lorito, R. M. Broadway, A. Di Pietro, and A. Tronsmo. 1993. Chitinolytic enzymes of Trichoderma harzianum: purification of chitobiosidase and endochitinase. Phytopathology 83:313-318. [Google Scholar]
- 18.Harman, G. E., and T. Björkman. 1998. Potential and existing uses of Trichoderma and Gliocladium for plant disease control and plant growth enhancement, p. 229-265. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2. Enzymes, biological control and commercial application. Taylor and Francis Ltd., London, United Kingdom.
- 19.Harman, G. E., C. R. Howell, A. Viterbo, I. Chet, and M. Lorito. 2004. Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2:43-56. [DOI] [PubMed] [Google Scholar]
- 20.Hjeljord, L., and A. Tronsmo. 1998. Trichoderma and Gliocladium in biological control: an overview, p. 129-151. In G. E. Harman and C. P. Kubicek (ed.), Trichoderma and Gliocladium, vol. 2. Enzymes, biological control and commercial application. Taylor and Francis Ltd., London, United Kingdom.
- 21.Hong, Z., P. Mann, N. H. Brown, L. E. Tran, K. J. Shaw, R. S. Hare, and B. DiDomenico. 1994. Cloning and characterization of KNR4, a yeast gene involved in (1,3)-β-glucan synthesis. Mol. Cell. Biol. 14:1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horwitz, B. A., A. Sharon, S. Lu, V. Ritter, T. M. Sandrock, O. C. Yoder, and B. G. Turgeon. 1999. A G protein α subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet. Biol. 26:19-32. [DOI] [PubMed] [Google Scholar]
- 23.Inbar, J., and I. Chet. 1992. Biomimetics of fungal cell-cell recognition by use of lectin-coated nylon fibers. J. Bacteriol. 174:1055-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inbar, J., and I. Chet. 1994. A newly isolated lectin from the plant pathogenic fungus Sclerotium rolfsii: purification, characterization and its role in mycoparasitism. Microbiology 140:651-657. [DOI] [PubMed] [Google Scholar]
- 25.Ivey, F. D., P. N. Hodge, G. E. Turner, and K. A. Borkovich. 1996. The Gαi homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7:1283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain, S., K. Akiyama, K. Mae, T. Ohguchi, and R. Takata. 2002. Targeted gene disruption of a G protein α subunit gene results in reduced pathogenicity in Fusarium oxysporum. Curr. Genet. 41:407-413. [DOI] [PubMed] [Google Scholar]
- 27.Kasahara, S., and D. L. Nuss. 1997. Targeted gene disruption of a fungal G-protein α subunit gene results in increased vegetative growth but reduced virulence. Mol. Plant-Microbe Interact. 10:984-993. [DOI] [PubMed] [Google Scholar]
- 28.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the Gα protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubicek, C. P., R. L. Mach, C. K. Peterbauer, and M. Lorito. 2001. Trichoderma: from genes to biocontrol. J. Plant Pathol. 83:11-23. [Google Scholar]
- 30.Kullnig, C., R. L. Mach, M. Lorito, and C. P. Kubicek. 2000. Enzyme diffusion from Trichoderma atroviride (= T. harzianum P1) to Rhizoctonia solani is a prerequisite for triggering of Trichoderma ech42 gene expression before mycoparasitic contact. Appl. Environ. Microbiol. 66:2232-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, S., and R. A. Dean. 1997. G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol. Plant-Microbe Interact. 10:1075-1086. [DOI] [PubMed] [Google Scholar]
- 32.Lorito, M., S. L. Woo, M. D'Ambrosio, G. E. Harman, C. K. Hayes, C. P. Kubicek, and F. Scala. 1996. Synergistic action between cell wall degrading enzymes and membrane affecting compounds. Mol. Plant-Microbe Interact. 9:206-213. [Google Scholar]
- 33.Loubradou, G., J. Bégueret, and B. Turcq. 1999. MOD-D, a Gα subunit of the fungus Podospora anserina, is involved in both regulation of development and vegetative incompatibility. Genetics 152:519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, Z., R. Tombolini, S. Woo, S. Zeilinger, M. Lorito, and J. K. Jansson. 2004. In vivo study of Trichoderma-pathogen interactions using constitutive and inducible green fluorescent protein reporter systems. Appl. Environ. Microbiol. 70:3073-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, G. A. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 36.Mukherjee, P. K., J. Latha, R. Hadar, and B. A. Horwitz. 2003. TmkA, a mitogen-activated protein kinase of Trichoderma virens, is involved in biocontrol properties and repression of conidiation in the dark. Eukaryot. Cell 2:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukherjee, P. K., J. Latha, R. Hadar, and B. A. Horwitz. 2004. Role of two G protein alpha subunits, TgaA and TgaB, in the antagonism of plant pathogens by Trichoderma virens. Appl. Environ. Microbiol. 70:542-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nemcovic, M., and V. Farkas. 2001. Cell-wall composition and polysaccharide synthase activity changes following photoinduction in Trichoderma viride. Acta Biol. Hung. 52:281-288. [DOI] [PubMed] [Google Scholar]
- 39.Regenfelder, E., T. Spellig, A. Hartmann, S. Lauenstein, M. Bölker, and R. Kahmann. 1997. G proteins in Ustilago maydis: transmission of multiple signals. EMBO J. 16:1934-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rey, M., J. Delgado-Jarana, and T. Benitez. 2001. Improved antifungal activity of a mutant of Trichoderma harzianum CECT2413 which produces more extracellular proteins. Appl. Microbiol. Biotechnol. 55:604-608. [DOI] [PubMed] [Google Scholar]
- 41.Rocha-Ramirez, V., C. Omero, I. Chet, B. A. Horwitz, and A. Herrera-Estrella. 2002. Trichoderma atroviride G-protein α-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot. Cell 1:594-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roncal, T., and U. Ugalde. 2003. Conidiation induction in Penicillium. Res. Microbiol. 154:539-546. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Truesdell, G. M., Z. Yang, and M. B. Dickman. 2000. A Gα subunit gene from the phytopathogenic fungus Colletotrichum trifolii is required for conidial germination. Physiol. Mol. Plant Pathol. 54:131-140. [Google Scholar]
- 45.Turner, G. E., and K. A. Borkovich. 1993. Identification of a G-protein α subunit from Neurospora crassa that is a member of the Gi family. J. Biol. Chem. 268:14805-14811. [PubMed] [Google Scholar]
- 46.Viterbo, A., O. Ramot, L. Chemin, and I. Chet. 2002. Significance of lytic enzymes from Trichoderma spp. in the biocontrol of fungal plant pathogens. Antonie Leeuwenhoek 81:549-556. [DOI] [PubMed] [Google Scholar]
- 47.Wiest, A., D. Grzegorski, B.-W. Xu, C. Goulard, S. Rebuffat, D. J. Ebbole, B. Bodo, and C. Kenerley. 2002. Identification of peptaibols from Trichoderma virens and cloning of a peptaibol synthetase. J. Biol. Chem. 277:20862-20868. [DOI] [PubMed] [Google Scholar]
- 48.Yabe, T., T. Yamada-Okabe, S. Kasahara, Y. Furuichi, T. Nakajima, E. Ichishima, M. Arisawa, and H. Yamada-Okabe. 1996. HRK1 encodes a cell surface protein that regulates both cell wall β-glucan synthesis and budding pattern in the yeast Saccharomyces cerevisiae. J. Bacteriol. 178:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yager, L. N., H. O. Lee, D. L. Nagle, and J. E. Zimmerman. 1998. Analysis of fluG mutations that effect light-dependent conidiation in Aspergillus nidulans. Genetics 149:1777-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeilinger, S., C. Galhaup, K. Payer, S. L. Woo, R. L. Mach, C. Fekete, M. Lorito, and C. P. Kubicek. 1999. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 26:131-140. [DOI] [PubMed] [Google Scholar]
- 51.Zeilinger, S. 2004. Gene disruption in Trichoderma atroviride via Agrobacterium-mediated transformation. Curr. Genet. 45:54-60. [DOI] [PubMed] [Google Scholar]
- 52.Zuber, S., M. J. Haynes, and A. Andrianopoulos. 2002. G-protein signaling mediates asexual development at 25°C but has no effect on yeast-like growth at 37°C in the dimorphic fungus Penicillium marneffei. Eukaryot. Cell 1:440-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuber, S., M. J. Haynes, and A. Andrianopoulos. 2003. The G-protein α-subunit GasC plays a major role in germination in the dimorphic fungus Penicillium marneffei. Genetics 164:487-499. [DOI] [PMC free article] [PubMed] [Google Scholar]