Abstract
Although copious qualitative information describes the members of the diverse microbial communities on Earth, statistical approaches for quantifying and comparing the numbers and compositions of lineages in communities are lacking. We present a method that addresses the challenge of assigning sequences to operational taxonomic units (OTUs) based on the genetic distances between sequences. We developed a computer program, DOTUR, which assigns sequences to OTUs by using either the furthest, average, or nearest neighbor algorithm for each distance level. DOTUR uses the frequency at which each OTU is observed to construct rarefaction and collector's curves for various measures of richness and diversity. We analyzed 16S rRNA gene libraries derived from Scottish and Amazonian soils and the Sargasso Sea with DOTUR, which assigned sequences to OTUs rapidly and reliably based on the genetic distances between sequences and identified previous inconsistencies and errors in assigning sequences to OTUs. An analysis of the two 16S rRNA gene libraries from soil demonstrated that they do not contain enough sequences to support a claim that they contain different numbers of bacterial lineages with statistical confidence (P > 0.05), nor do they contain enough sequences to provide a robust estimate of species richness when an OTU is defined as containing sequences that are no more than 3% different from each other. In contrast, the richness of OTUs at the 3% level in the Sargasso Sea collection began to plateau after the sampling of 690 sequences. We anticipate that an equivalent extent of sampling for soil would require sampling more than 10,000 sequences, almost 100 times the size of typical sequence collections obtained from soil.
An outstanding challenge in microbial ecology is to estimate species richness based on 16S rRNA gene sequences. The computational methods available to address this challenge are limited. Sequences are usually grouped as operational taxonomic units (OTUs) or phylotypes, both of which are defined by electrophoretic pattern (9, 12, 18) or DNA sequence (1, 21). Screening for unique 16S rRNA genes by electrophoretic pattern can be complicated either by sequences that are more than 3% different sharing the same pattern or by sequences with less than 3% difference having different patterns (9, 22). Nucleotide sequences provide a more precise analysis. Sequences with greater than 97% identity are typically assigned to the same species, those with >95% identity are typically assigned to the same genus, and those with >80% identity are typically assigned to the same phylum, although these distinctions are controversial (1, 2, 11, 13, 21, 24, 29). A genetic distance is approximately equal to the converse of the identity percentage. These cutoff values are simply a best fit of historical taxonomy with modern 16S rRNA gene sequencing, not a rigorously validated hierarchy.
There are few methods available to assign sequences to OTUs quickly based on sequence data (26). Investigators typically analyze a distance matrix manually for values that are less than the cutoff level. This approach is problematic when the distance relationships between three or more sequences are not transitive, which forces the creation of a decision rule that may not be consistently enforced. Furthermore, manually applying a decision rule to a large distance matrix can be too unwieldy, tedious, and time-consuming to be accurate.
To assign sequences quickly and accurately to OTUs, we developed DOTUR (Distance-Based OTU and Richness; the source code is available from the authors at http://www.plantpath.wisc.edu/fac/joh/dotur.html). A PHYLIP (http://evolution.genetics.washington.edu/phylip.html)-generateddistance matrix is used as an input file to DOTUR, which assigns sequences to OTUs for every possible distance. DOTUR then calculates values that are used to construct randomized rarefaction and collector's curves of observed OTUs, diversity indices, and richness estimators. In this paper, we demonstrate DOTUR's dexterity by analyzing and comparing 16S rRNA gene libraries constructed from soil and seawater, which have been studied with other methods in previous reports.
MATERIALS AND METHODS
Sequence assignment in DOTUR.
Three sequence assignment methods are available in DOTUR: nearest neighbor, furthest neighbor, and average neighbor. The nearest neighbor (i.e., single linkage) algorithm constructs a link when one object (an individual sequence or groups of sequences) is similar to any of the sequences in the object it is joining. The furthest neighbor (i.e., complete linkage) method is a more constrained criterion, which assigns a sequence to an object only if it is similar to all of the sequences in the group it is joining. The average neighbor method (i.e., unweighted pair-group method by using arithmetic averages) finds the two most similar entities and links them by averaging the differences between the entities being joined and all of the other entities. A more complete description of these methods is provided elsewhere (17), and a manually calculated example is shown in the manual on the DOTUR website.
Richness estimation in DOTUR.
DOTUR was designed to calculate various diversity indices and richness estimators. Diversity indices and richness estimators are useful to compare the relative complexity of two or more communities and to estimate the completeness of sampling of a community. Once DOTUR assigns sequences to OTUs, it performs a random sampling without replacement procedure. The probability of drawing a representative from an OTU is the number of times the OTU was observed divided by the total number of sequences in the library. For each randomization, DOTUR calculates the Shannon-Weaver and Simpson diversity indices (19), the abundance-based coverage estimator (ACE) (5, 6), and the bias-corrected Chao1 (4), interpolated jackknife (3), and bootstrap (28) richness estimators with a 95% confidence interval (CI), when applicable, as a function of sampling effort. Sample calculations are provided in the manual on the DOTUR website.
If the sequences are inputted in the order in which they were obtained, DOTUR can construct the actual collector's curve for each estimate by plotting the CI against the sequencing effort to determine how many sequences are required to obtain a desired level of precision for the estimate. Input and output data for all of the sequence collections analyzed in this paper are available at the DOTUR website.
RESULTS
Validation and evaluation of sequence assignments.
To test DOTUR, we analyzed the clone library constructed from improved Scottish soil (21) (Table 1; Fig. 1). The sequences were aligned by using ClustalW (ftp://ftp.ebi.ac.uk/pub/software/unix/clustalw/), and we constructed a Jukes-Cantor corrected distance matrix (10) by using the DNADIST program from PHYLIP. We applied the nearest neighbor, average neighbor, and furthest neighbor assignment algorithms implemented in DOTUR and observed 114, 115, and 116 OTUs, respectively, for each of the assignment algorithms (Table 1). The frequency distributions observed by using the furthest neighbor algorithm at a distance of 3% were identical to the distribution described by McCaig et al. (21), indicating that DOTUR makes appropriate assignments.
TABLE 1.
Comparison of various techniques to determine the frequency distribution and richness estimates for 16S rRNA gene collections from Scottish and Amazonian soilsa
| Source of 16S rRNA gene library | Analysis method | Total no. of se- quences | No. of unique OTUs | No. of OTUs with nx sequencesb
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| n1 | n2 | n3 | n4 | n5 | n6 | ||||
| Scottish soil | McCaig | 137 | 114 | 98 | 12 | 2 | 1 | 1 | 0 |
| Hughes | 137 | 113 | 96 | 13 | 2 | 1 | 1 | 0 | |
| FastGroup | 137 | 131 | 127 | 3 | 0 | 1 | 0 | 0 | |
| NN | 137 | 112 | 96 | 12 | 2 | 0 | 1 | 1 | |
| AN | 137 | 113 | 97 | 12 | 2 | 0 | 2 | 0 | |
| FN | 137 | 114 | 98 | 12 | 2 | 1 | 1 | 0 | |
| Amazonian soil | FN | 98 | 84 | 75 | 6 | 1 | 2 | 0 | 0 |
OTUs were defined by using a distance level of 3%. The frequency distribution of the improved Scottish soil 16S rRNA gene library of McCaig et al. (21) was determined from rank abundance data from McCaig et al. (21) and Hughes et al. (14), from output of the FastGroup program of Seguritan and Rowher (26), and by using the three assignment algorithms implemented in DOTUR. For comparison, the Amazonian soil library (2) was analyzed by using the furthest neighbor algorithm in DOTUR. NN, nearest neighbor assignment algorithm; AN, average neighbor assignment algorithm; FN, furthest neighbor assignment algorithm.
n1, no. of singletons; n2, no. of doubletons; etc.
FIG. 1.
Rarefaction curves (A) and lineage-through-time plot (B) from DOTUR analysis using furthest neighbor assignment algorithm with unimproved Scottish soil 16S rRNA gene library for various distance levels. Error bars represent the 95% CI.
There have been previous attempts to assign sequences from this data set to OTUs. The authors of the Scottish soil study identified 114 at the 3% distance level (Table 1; 21), whereas 113 have also been reported (14). These analyses illustrate how DOTUR can provide better accuracy than manual counts of sequences, and this level of accuracy will become even more important as the size of clone libraries increases. We also used the FastGroup program (26), which selects a reference sequence to which every other sequence is compared. If the query sequence is within a designated percentage similarity, it joins the group. This method is similar to the nearest neighbor method with the exception that FastGroup compares each query sequence to a single reference sequence instead of to all of the sequences in the OTU. This analysis yielded 131 OTUs at the 3% distance level. The problem with this approach is that results can be skewed depending on which sequence is selected by the program to be the reference. In this case, it appears that the assignment was perhaps overly conservative.
When we compared the data produced by DOTUR to those expected based on rarefaction theory, manual calculations, and output from EstimateS (http://viceroy.eeb.uconn.edu/estimates), the results were similar. DOTUR was considerably faster than EstimateS (data not shown). Furthermore, DOTUR performed the sequencing assignment procedure while EstimateS could not. As DOTUR calculates the various richness and diversity parameters for each distance level, it produces separate files that can be used to generate lineage-through-time plots, which describe how many OTUs are present for various evolutionary distances (Fig. 1B). This type of analysis has been described elsewhere (20, 23).
Application of DOTUR to the Amazonian soil clone library.
Hughes et al. (14) asked the provocative question, “Are microbes too diverse to count?” The most widely cited paper to support an answer of yes is that of Borneman and Triplett (2), who sampled 98 bacterial 16S rRNA gene from two Amazonian rainforest soils and concluded that the 98 bacterial sequences were unique. Using DOTUR, we identified two pairs of identical sequences (GenBank accession numbers U686617 and U68641 and U68620 and U68618) in the pooled Amazonian data set. Instead of 98 singletons, there were actually 94 singletons and 2 doubletons when the definition of an OTU was uniqueness.
Performing the DOTUR analysis with the pooled Amazonian clone library, we found that the frequency distribution of sequences in OTUs was comparable to that in the improved Scottish clone library at other distances as well (Table 1) (Fig. 2). The rarefaction curve generated from the improved Scottish sequences, when uniqueness determined entry into an OTU, indicated that there was a 95% chance that if only 98 sequences had been sampled then the authors would have identified between 93 and 97 OTUs. When a distance smaller than 3% defined entry into an OTU, the CI after 98 sequences was between 80 and 89 OTUs. The number of OTUs observed from the Amazonian library was 94 when the OTU definition was uniqueness and 84 when the OTU definition was set at 3%. Therefore, it is impossible to conclude with confidence that there is a difference in the richness observed between the “exotic” east Amazonian soil and the “common” Scottish agricultural soil, because the number of observed OTUs from the Amazonian soil falls within the 95% CI of the Scottish soil after the sampling of 98 sequences. However, it is possible that with further sequencing, differences in richness between the two libraries would have been observed. Although the two samples may have the same level of richness, it is possible that the libraries contained a different composition of 16S rRNA genes.
FIG. 2.
Rarefaction curves (A) and lineage-through-time plot (B) from DOTUR analysis using the furthest neighbor assignment algorithm with the Amazonian soil 16S rRNA gene library for various distance levels. Error bars represent the 95% CI.
Application of DOTUR to the Sargasso Sea metagenome sequence.
Recently, Venter et al. (31) published an extensive sequencing project that consisted of nearly 2 million sequencing reads and a total of 1.7 Gbp from uncultured organisms in a composite 1.5-m3 sample from the Sargasso Sea. We obtained each of the sequence readings from the GenBank FTPserver (ftp://ftp.ncbi.nih.gov/pub/TraceDB/environmental_sequence/) and screened each one for a universal 16S rRNA oligonucleotide (8) and a modified universal RNA polymerase gene (rpoB) oligonucleotide (16). For each gene, we extracted the 300 bp surrounding the probe sequences so that each sequence started and ended at approximately the same location within the gene and maximized the number of gene fragments in the final gene library. There were 690 partial 16S rRNA gene fragments and 507 partial rpoB fragments in our final sequence collections.
We applied DOTUR to the two gene fragment collections as described above (Fig. 3 and 4). We identified 114 16S rRNA species and 304 rpoB species by using the 6% difference definition of species of Venter et al. (31) for protein coding sequences. A DOTUR analysis showed that when we varied the rpoB species definition between 19 and 21% difference, the 95% CI for the final richness estimate based on the two genes overlapped. By rarefaction, we found that the number of observed rpoB OTUs fell within the 95% CI of the 16S rRNA species fragment rarefaction curve (between 90 and 104 species when 507 sequences were sampled) when the rpoB species definition was between 22 and 23% difference and the 16S rRNA species definition was 3% difference. By using the same method, if we assumed that a 6% difference is appropriate for defining a species by rpoB sequences, all members of a species would need to have identical 16S rRNA sequences in order for the rpoB and 16S rRNA richness estimates to have overlapping 95% CIs.
FIG. 3.
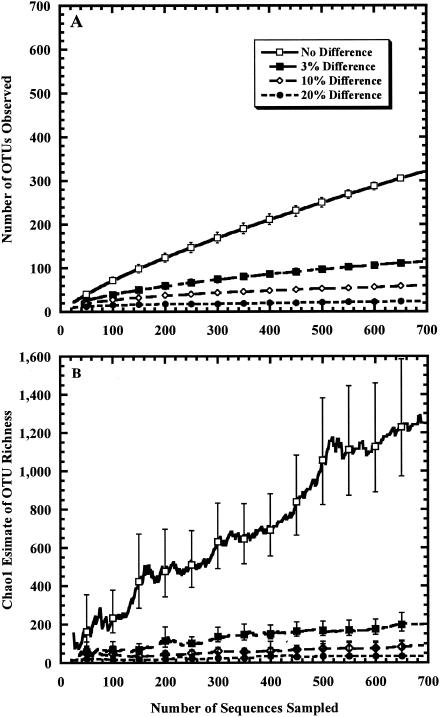
Rarefaction curve (A) and Chao1 richness estimate collector's curve (B) using partial 16S rRNA gene sequences from the Sargasso Sea metagenomic sequence. Error bars represent the 95% CI.
FIG. 4.
Rarefaction curve (A) and Chao1 richness estimate collector's curve (B) using partial rpoB sequences from the Sargasso Sea metagenomic sequence. Error bars represent the 95% CI.
Collector's curves for evaluating sampling progress.
Nonparametric richness estimators, such as ACE, Chao1, bootstrap, and jackknife, enable researchers to use observed frequencies of each OTU to estimate the richness of organisms in a community without having to sample each organism. We liken richness estimation to completing a sample-based census of a population, where the goal is to determine the total number of people living in a country without having to account for every individual in the population. Since it is not possible to know a priori what the true richness of any community is, we must decide upon a criterion for determining the minimum number of sequences required to obtain an accurate statistical census.
Earlier in this report, we used rarefaction curves to compare the relative richness between two communities (Scottish versus Amazonian soil) and to compare appropriate cutoff valves for other phylogenetic anchors for measuring richness (16S rRNA versus rpoB). However, if instead of measuring relative richness (OTUs observed) we are interested in the estimated richness (OTUs expected) and in determining the number of sequences necessary to obtain a measure of richness, then it is necessary to use a nonrandomized collector's curve. This analysis assumes that the probability of drawing any sequence is independent of the sequence that was drawn before it and that we do not know the probability of drawing each sequence. The goal of determining the richness within a clone library is to determine the probability of drawing a sequence that will change the estimate. When that probability converges to zero, then there is a high probability that the estimate is accurate and that continued sampling beyond that point will increase the confidence and precision of the estimate.
Because a rarefaction curve is the average of a large number of randomized collector's curves, the ability to measure the probability of drawing a sequence that will change the richness estimate is lost. A rarefaction curve of the Chao1 richness estimate creates a smooth curve whose final value is the final estimated value. Therefore, the shape of the rarefaction curve will change as the terminal estimate changes. Since the curve is smooth, the ability to gauge the probability that the estimate will change with additional sequences is lost when a rarefaction curve is used, but the overall shape of the collector's curve does not change. As new sequences are sampled, the preceding data points in a collector's curve stay the same, but the terminal estimate changes.
Previous studies that used Chao1 estimate rarefaction curves have suggested that when a nonparametric richness estimator rarefaction curve levels off, the value to which the curve converges is a reasonable estimate of the true richness (14). An analysis of the soil clone library derived from the improved Scottish soil suggested that the authors were confident that the true species richness was 467, with a 95% confidence interval between 333 and 681 OTUs after 137 sequences were sampled. They noted that the Chao1 richness estimate rarefaction curve began to level off after about 70 sequences (14). However, when we assumed that the sequences in the Scottish soil clone library were sampled in the order of their GenBank accession number and used DOTUR to calculate the Chao1 richness estimator, it was clear that the estimate continues to grow with additional sampling and that the estimate is sensitive to the addition of sequences (data not shown). Furthermore, when we analyzed the range of the 95% CI as a function of sampling effort by using the collector's curve, there was a modest positive correlation with sequencing effort (R2 = 0.37) so that the estimate's uncertainty increases with additional sampling. These results indicate that the 95% CI of between 333 and 681 OTUs is most likely too low. These results were masked by using a Chao1 estimator rarefaction curve.
We constructed collector's curves of the Chao1 estimator to study the 16S rRNA fragment collection from the Sargasso Sea to evaluate the completeness of sampling with a 3% difference definition of a species. The Chao1 estimator predicted a minimum of 198 species (95% CI, between 187 and 211) by using the 16S rRNA gene fragment collection and 187 species (95% CI, 161 to 233) by using the rpoB sequence collection and an OTU definition of 20% difference (Fig. 3B and 4B). At the species level, there were no instantaneous 5% changes after the 230th 16S rRNA gene fragment was sampled (Fig. 3B). The species richness at this point in the sampling was 95 species (95% CI, between 78 and 130), which was 48% of the richness obtained after 690 sequences were sampled. Although the estimated variability was substantially decreased when more sequences were added to the collection, the richness estimate was significantly lower than that observed after the 690th sequence was sampled. To improve the accuracy of the estimate, we selected a smaller instantaneous change criterion of 2.5%. We did not find any instantaneous changes greater than 2.5% after the 662nd sequence was sampled; however, only 28 sequences were sampled after that point, making it difficult to judge the robustness of the estimate. The species richness after the 662nd sequence was added was 194 species, which was not significantly different from the final estimate of 198 species (95% CI, between 163 and 258). An alternative method of reducing the negative bias was to hypothetically cease sampling after a total of 430 sequences, which is 200 sequences more than the number required to reach the last instantaneous change of greater than 5%. The richness estimate after the addition of the 430th sequence was 155 species, or 78% of the richness, after all the sequences were collected.
A final approach we considered was to relax the definition of an OTU to the phylum level or 20% difference. We did not identify any instantaneous changes in the Chao1 estimate of 34 phyla after sampling the 486th sequence. This result gives strong confidence in the estimate that the 95% CI (between 24 and 111) surrounding the estimate of 34 OTUs contains the true richness when defining an OTU at 20% distance. The estimate's lack of precision is due to the number of singleton OTUs (n1 = 7) relative to doubleton OTUs (n2 = 1). As the number of singleton OTUs decreases, the precision of the estimate will improve.
DISCUSSION
DOTUR assigns sequences rapidly and systematically to OTUs by using all possible distances. In both clone libraries that we analyzed, DOTUR assigned sequences to OTUs more accurately and consistently than had previous methods. DOTUR also assists in assessing the completeness of a sequencing effort and the reliability of richness estimates.
Analysis with DOTUR indicates that it is not possible to state with confidence that the richness in the Amazonian library differs from that in the improved Scottish soil library. However, in spite of the relative dearth of sequences in each of these libraries compared to the estimated species richness in 1 g of soil, which is expected to be in the thousands of species (30), further sequencing might indicate a difference in richness between the libraries. In addition, the application of methods described elsewhere may demonstrate that while the richness between these two libraries is similar, their phylogenetic composition is different (20, 25, 27). Finally, a connection between species richness or community composition with ecological mechanisms remains to be determined. It is possible that two communities could have considerably different membership yet conduct similar biological processes.
The inclusion of the Sargasso Sea metagenome sequence has provided an interesting application of DOTUR for describing richness, comparing species definitions used for genes with phylogenetic information, and evaluating the level of sampling necessary to have confidence in an estimate. Venter et al. (31) found 143 different 16S rRNA species and 428 different rpoB species, and we found 114 and 303 different 16S rRNA and rpoB species, respectively. This difference may be explained by the fact that they restricted their analysis to those sequences that overlapped by at least 40 bp, while we required all sequences to overlap by the same 300 bp. Regardless of this difference in method, they predicted a minimum number of species of near 1,000 species by using rpoB sequences and we predicted a richness of 1,040 species by using their 6% difference species definition, suggesting that the methods produce comparable results.
Since DOTUR compares multiple OTU definitions simultaneously, we were able to compare various species level definitions by using 16S rRNA and rpoB gene sequences. Assuming that the 3% difference in 16S rRNA sequence is a valid definition of a species, a protein coding sequence species definition would then be near 20%. We found similar results with protein coding sequences other than rpoB that have been used as phylogenetic anchors (data not shown). A value of 20% is more consistent than 6% with previous definitions of species using protein coding sequences. For example, a difference of 30% in DNA-DNA hybridization analysis is used to differentiate between species. Our use of DOTUR accounts for differences in the rate of evolution for these two genes. One potential concern is that any estimates made by using 16S rRNA sequences is inflated, since bacteria are known to have multiple copies of this gene in their genome. Although it is predicted that most slow-growing bacteria that dominate the environment have, on average, close to 1 copy per genome (15), multiple copies from a single genome would have to be more than 3% different to have an effect on our analysis. If intragenomic variability was greater than 3%, the number of 16S rRNA OTUs would decrease, resulting in an even lower species definition for protein coding sequences. Any distance level that is selected to differentiate species will be arbitrary and consequently controversial, but it will serve as a useful benchmark for future analyses.
When the number of different OTUs observed is less than twice the square root of the total richness, the Chao1 richness estimator is strongly correlated with the sequencing effort. If we assume that there are roughly 4,000 species OTUs in a gram of soil (30) and 150 in a milliliter of seawater (7), then at least 125 and 17 different OTUs, respectively, would need to be sampled before the correlation between richness and sequencing effort begins to decrease. However, we do not know how many sequences are required to reach the condition in which there is no correlation between sequencing effort and richness. In soil samples, we demonstrated that 137 sequences were insufficient to estimate richness reliably when distances of 3% were used to define an OTU. Using the Sargasso Sea samples, which is thought to contain one-tenth the richness of soil (7), we found that a total of 690 sequences was almost sufficient to obtain an accurate estimate of species richness and was sufficient to estimate richness when a 20% difference was used to define an OTU. It is likely that at least 10,000 sequences would be necessary to approach an estimate of the true species richness in soil. To evaluate sampling progress, we suggest tracking the richness estimation collector's curve and sampling until there are no instantaneous 2.5% changes in richness over 300 sequences.
When the Scottish and Amazonian soil sequences were reported, sequencing was quite expensive and laborious. With present technology, which is both less expensive and largely automated, we have the opportunity to generate and sequence large 16S rRNA gene libraries that may be sufficient in size to provide accurate estimates and comparisons of richness even in species-rich environments, such as soil.
Acknowledgments
A USDA postdoctoral fellowship in Soil Biology to P.D.S., the NSF Microbial Observatory program (MCB-0132085), the Howard Hughes Medical Institute, and the University of Wisconsin—Madison College of Agricultural and Life Sciences provided funding for this project.
REFERENCES
- 1.Bond, P. L., P. Hugenholtz, J. Keller, and L. L. Blackall. 1995. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl. Environ. Microbiol. 61:1910-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burnham, K. P., and W. S. Overton. 1979. Robust estimation of population size when capture probabilities vary among animals. Ecology 60:927-936. [Google Scholar]
- 4.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 5.Chao, A., and S. M. Lee. 1992. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 87:210-217. [Google Scholar]
- 6.Chao, A., M. C. Ma, and M. C. K. Yang. 1993. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 80:193-201. [Google Scholar]
- 7.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durbin, R., S. R. Eddy, A. Krogh, and G. Mitchison. 1998. Biological sequence analysis: probabilistic models of proteins and nucleic acids. Cambridge University Press, Cambridge, United Kingdom.
- 11.Everett, K. D. E., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 12.Felske, A., H. Rheims, A. Wolterink, E. Stackebrandt, and A. D. L. Akkermans. 1997. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology 143:2983-2989. [DOI] [PubMed] [Google Scholar]
- 13.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko, K. S., H. K. Lee, M. Y. Park, M. S. Park, K. H. Lee, S. Y. Woo, Y. J. Yun, and Y. H. Kook. 2002. Population genetic structure of Legionella pneumophila inferred from RNA polymerase gene (rpoB) and DotA gene (dotA) sequences. J. Bacteriol. 184:2123-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legendre, P., and L. Legendre. 1998. Numerical Ecology. Elsevier, New York, N.Y.
- 18.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, N.J.
- 20.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moyer, C. L., J. M. Tiedje, F. C. Dobbs, and D. M. Karl. 1996. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl. Environ. Microbiol. 62:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nee, S., R. M. May, and P. H. Harvey. 1994. The reconstructed evolutionary process. Philos. Trans. R. Soc. B 344:305-311. [DOI] [PubMed] [Google Scholar]
- 24.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 25.Schloss, P. D., B. R. Larget, and J. Handelsman. 2003. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seguritan, V., and F. Rohwer. 2001. FastGroup: a program to dereplicate libraries of 16S rDNA sequences. BMC Bioinformatics 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, E. P., and G. van Belle. 1984. Nonparametric estimation of species richness. Biometrics 40:119-129. [Google Scholar]
- 29.Stackebrandt, E., and B. M. Goebel. 1994. A place for DNA-DNA reassociation and 16S rRNA sequence-analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 30.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]