Abstract
Antibodies (mAbs) and antibody fragments (Fabs) constitute one of the largest and most rapidly expanding groups of protein pharmaceuticals. In particular, antibody fragments have certain advantages over mAbs in some therapeutic settings. However, due to their greater chemical diversity, they are more challenging to purify for large-scale production using a standard purification platform. Besides, the removal of Fab-related byproducts poses a difficult purification challenge. Alternative Fab purification platforms could expedite their commercialization and reduce the cost and time invested. Accordingly, we employed a strong cation exchanger using a pH-based, highly linear gradient elution mode following Protein L affinity purification and developed a robust two-step purification platform for an antibody fragment. The optimized pH gradient elution conditions were determined on the basis of purity level, yield, and the abundance of Fab-related impurities, particularly free light chain. The purified Fab molecule Ranibizumab possessed a high degree of similarity to its originator Lucentis. The developed purification platform highly intensified the process and provided successful clearance of formulated Fab- and process-related impurities (∼98 %) with an overall process recovery of 50 % and, thus, might be a new option for Fab purification for both academic and industrial purposes.
Keywords: Antibody fragment, Pharmaceutical purification, Biosimilar production, Liquid chromatography, Mass spectrometry
Highlights
-
•
The removal of Fab-related impurities by pH gradient-based cation exchange chromatography.
-
•
An optimized two-step purification platform for Fab molecules.
-
•
Comparable profile of in-house purified Fab molecule with the originator.
1. Introduction
Fabs are antigen-binding fragments of a full-length antibody, which comprise the variable heavy domain (VH) and light domain (VL) in conjunction with the first domain of the constant chain (CH1) and the constant region of the light chain (CL) [1]. Fabs are capable of penetrating tissues and being cleared from circulation more quickly, making them useful for treating disorders [2]. However, Fabs are more challenging to purify than full-length antibodies as they are of diverse structure and typically lack an Fc region. Thus, establishing tailored and intricate purification methods takes a significant investment of time, money, and process development resources [3]. Fab purification platform options might help advance their commercialization and minimize the cost and time invested. Protein A-based affinity media are frequently used to capture full-length antibodies by recognizing the Fc region. However, Protein A resins are not particularly useful for Fab purification in many cases. Protein G and, notably Protein L are used as the affinity chromatography step for Fab purification [[3], [4], [5], [6]].
Due to their small size and lack of glycosylation, antibody fragments can be cost-effectively expressed in bacteria. Although the bacterial expression of Fab fragments is favorable, the formation of an active heterodimer has proven to be difficult [7]. Similar to other proteins with disulfide bonds, the synthesis of Fabs in the reducing environment of the bacterial cytoplasm is challenging [8]. The correct formation of disulfide bonds can be facilitated by targeting recombinant proteins to the periplasmic region of E. coli, nevertheless, the balance of light chain (LC) and heavy chain (HC) expression in the periplasm of E. coli is important to obtain functional Fab [9]. To produce soluble functional Fab, light chain (LC) and heavy chain (HC) antibody precursors must be properly produced and delivered into the periplasm, where they are assembled into a folded functional molecule. Crude E. coli homogenates and fermentation broth produce a wide variety of protein species; ideally, the formation of these fermentation by-products should be prevented during cell cultivation. However, current analytical techniques are limited to reliably distinguish and identify impurities in homogenates. Thus, these Fab-related impurities (residual light and heavy chain, intermediates, and aggregates) must be eliminated during the purification process to obtain a purified drug substance [10]. In cases where LC/HC balance cannot be reached or optimal conditions for periplasmic expression and isolation cannot be maintained, the co-capture of correctly folded Fab and free LC is one of the most critical challenges in the downstream process of Fab purification by Protein L. The separation of free LC from functional Fab requires a more precise strategy due to the small difference between their molecular weights and isoelectric points. Herein, we investigate the viability of using a pH-based gradient elution strategy in the subsequent cation-exchange chromatography (CEX) to remove Fab-related impurities following Protein L purification and offer an optimized two-step platform for Fab purification that is also suitable for pilot-scale production. For this, purification of a Fab therapeutic Ranibizumab was optimized and the characteristics of in-house purified Ranibizumab were compared to those of originator molecule Lucentis to assert the utility of the proposed purification platform.
2. Experimental section
2.1. Production and purification
Production of Ranibizumab: A single vector expressing both chains of recombinant Fab with a signal leader sequence (Heat-stable enterotoxin II – STII) for translocation into the periplasm was used for production. Ranibizumab was expressed in E. coli W3110 cells by inducing the expression for 16 h at 20 °C with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG). Following production, the cell pellet was resuspended in a homogenization buffer (100 mM Tris, 10 mM EDTA pH 7.4). The system was cooled to 4 °C before homogenization begins. PMSF was added to a final concentration of 1 mM. High-pressure homogenization was carried out in three passages, with a pressure of 800–850 bar in all stages using the Homolab 2.20 homogenizer system (FBF, Italy). Samples were centrifuged for 2 h at 23,800×g (Hitachi CR22 N Refrigerated Ultracentrifuge), and the homogenate supernatant containing the soluble product was filtered with a 0.22 μm sterile membrane filter.
Protein L affinity chromatography: Chromatographic separations were carried out on an AKTA Avant 25 chromatography system (Cytiva, USA) equipped with an integrated pH electrode, enabling continuous pH monitoring throughout the run. The pumps and data acquisition were controlled using Unicorn software (Cytiva, USA). Prepacked Protein L columns were equilibrated with 5 column volumes (CV) of starting buffer (20 mM sodium phosphate, pH 7.6). Following sample application, the column was washed with 6 C V of 20 mM sodium phosphate (pH 6.0) and then eluted with 10 C V of 100 mM glycine HCl (pH 2.5) using a step elution. Following elution, the mAb pool from Protein-L chromatography was subjected to viral inactivation by being held at a low pH (2.5–3.0) for 1 h at room temperature and adjusted to the desired pH for a further step of purification [[11], [12], [13]]. High-scale affinity purification experiments were carried out on AKTA Pilot 600 system using Axichrom 70/300 columns packed with Protein L resin (17,547,804, Cytiva, USA).
Cation exchange chromatography: An HiTrap SP-FF column was used for cation exchange chromatography (CEX). A flow rate of 1 mL/min was applied. 20 mM phosphate buffer (pH 5.0), was used as an equilibration and wash buffer. The column was equilibrated for at least two CV, and then eluates from the affinity chromatography were loaded. After a wash step of three CV, a salt-based elution was performed using a 20 mM sodium phosphate buffer containing 1 M NaCl, with varying pH values. Detailed elution profiles for different runs are described in the results section (Table 2). Additional regeneration with 0.1 M NaOH was performed regularly. High-scale affinity purification experiments were carried out on AKTA Pilot 600 chromatography system using Axichrom 50/300 columns packed with SP-FF resin (17,072,904, Cytiva, USA).
Table 2.
Salt-based elution conditions for cation exchange chromatography.
| Cation Exchange Chromatography (HiTrap SP-FF) | ||||
|---|---|---|---|---|
| Buffer A |
20 mM sodium phosphate pH 5.0 |
|||
| Buffer B |
20 mM sodium phosphate, 1 M NaCl pH 6.0 |
|||
| Elution | Flow rate (mL/min) | Number of CV | Concentration B | |
| SE - 1 | Linear gradient | 0.6 | 16 | 50 % |
| SE - 2 | Linear gradient | 0.6 | 18 | 100 % |
| SE - 3 | Linear gradient | 0.6 | 30 | 50 % |
| SE - 4 | Step | 0.6 | 18 10 10 |
25 % 50 % 100 % |
| SE - 5 | Linear gradient | 0.6 | 20 6 5 |
25 % 50 % 100 % |
| SE - 6 | Linear gradient | 0.6 | 20 10 5 |
25 % 50 % 100 % |
2.2. Biophysical and biochemical characterization of ranibizumab
SDS-PAGE and Immunoblotting: Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) was conducted on 12 % polyacrylamide gels. Fab samples loaded (2 μg/lane) onto the gels were stained with Coomassie blue for detection. For immunoblotting, after protein samples were run on 12 % polyacrylamide gel, blotting was carried out for 80 min at 4 °C and 100 V by using 0.22 μm Nitrocellulose membrane (Bio-Rad) and wet transfer system (Bio-Rad). Membranes were incubated with 5 % non-fat dry milk in TBS-T (Bio-Rad). HRP-conjugated antibody binding was performed overnight at RT for 2 h with gentle agitation. After washing the membrane with TBS-T, the membrane was treated with Clarity™ ECL Western Blotting Substrate (Bio-Rad) and visualized via ChemiDoc Imaging System (Bio-Rad, Richmond, CA, USA).
Peptide Mapping: Purified Ranibizumab was firstly denatured with 8 M urea, and then subsequently reduced with 20 mM DTT (A2452, Biomatik) and alkylated with 40 mM iodoacetamide (I-1149, Sigma-Aldrich). Proteins were overnight digested by 1:20 (w/w) sample:trypsin (Promega) in 100 mM ammonium bicarbonate (pH 8.0) at 37 °C. Trypsin activity was stopped by formic acid. Identification of digested peptides via mass spectrometry was performed by using Acquity UPLC coupled with Waters Synapt G2-Si HDMS (Waters Corp). Mobile phase A was water, mobile phase B was acetonitrile (ACN), and mobile phase C was water containing 1 % (v/v) formic acid. Digested samples were loaded on BEH C18 RP (Waters, 130 Å, 1.7 μm, 2.1 mm × 100 mm) analytical column. Peptides separation was accomplished with a gradient of 1–42 % mobile phase B over 60 min. Mobile phase C and flow rate were kept constant at 10 % and 200 μL/min, respectively, during the run, and the column temperature was maintained at 65 °C. All analyses were performed in positive electrospray ionization and resolution mode. Glu-fibrinopeptide B was infused to calibrate time-of-flight analyzer from 50 to 2000 m/z. Data analysis was performed in UNIFI (Waters) and MassLynx (Waters).
Intact/Reduced Mass: 10 μL purified protein was injected onto an Acquity UPLC protein BEH C4 analytical column (Waters 300 Å, 1.7 μm, 2.1 mm × 50 mm) with a 30-min gradient. In reduced protein analysis, purified protein was incubated with 20 mM DTT (A2452, Biomatik) at 80 °C for 25 min. Reduced protein was also injected to the same column used in intact mass analysis. The column temperature was maintained at 80 °C. Data analysis was performed in UNIFI (Waters) and MassLynx (Waters).
Size exlusion chromatography: High molecular weight and low molecular weight variants of sample were separated using a pre-equilibrated Biosuite SEC analytical column (Waters, 250 Å, 5 μm, 7.8 mm × 300 mm) with 30-min 0.5-mL/min isocratic elution. Mobile phase was 200 mM potassium phosphate and 250 mM KCl (pH 6.2). The column temperature was 25 °C.
Capillary electrophoresis: Reduced and non-reduced capillary electrophoresis-sodium dodecyl sulfate (CE-SDS) analyses were performed with a capillary electrophoresis system (PA800 plus Pharmaceutical Analysis System; Sciex) equipped with photodiode array (PDA) detector. For analysis in reducing conditions, Fab was mixed with a 10 kDa internal standard (5 mg/mL), SDS-MW sample buffer (100 mM Tris-HCl, 1 % SDS, pH 9.0) (A10663, Beckman Coulter, Brea, CA, USA), and 2-mercaptoethanol (1:20 v/v) (M7154, Sigma-Aldrich) and then boiled at 100 °C for 3 min. For analysis in non-reducing conditions, 12.5 mM iodoacetamide (I-1149, Sigma-Aldrich) was used instead of 2-mercaptoethanol and the mixture was incubated at 70 °C for 10 min. The sample mixture was introduced onto a 50 μm I.D. bare-fused-silica capillary (338,451, Beckman Coulter) by applying voltage at −5 kV for 20 s. Separation voltage remained constant at −15 kV with an applied field strength of −497 V/cm 32 Karat software (SCIEX) was used to collect and process the data. For isoelectric focusing, PA 800 Plus Pharmaceutical Analysis System was utilized with the commercial 50 μm “Neutral Coated Capillary” and UV detector. Samples were mixed with 200 μL of 3.75 mM urea dissolved in commercial cIEF gel solution (200 μL), 12 μL of pharmalyte 3–10 (17-0456-01, Cytiva), 20 μL of 500 mM arginine (A5006, Sigma-Aldrich) as cathodic stabilizer, 2 μL of 200 mM iminodiacetic acid (220,000, Sigma-Aldrich) as anodic stabilizer, and pI markers (pH 10, pH 9.5 and pH 7.0; A58481, Beckman Coulter, Brea, CA, USA). Peaks on electropherogram for main and variant peaks are analyzed via Karat32 software. Isoelectric points and abundance percentages of main and variant peaks are reported.
Endotoxin levels: To detect Gram-negative bacterial endotoxins, Limulus Amebocyte Lysate (LAL) Chromogenic Endotoxin Quantitation Kit was used according to manufacturer's instructions (Thermo Scientific, #88282).
Ligand Leakage: Protein L concentrations leaking from the HiTrap Protein L (5 mL) column after 2-step purification were determined by ELISA following the manufacturer's instructions (Medicago, #10-0027-1).
Host cell proteins: To determine the presence of E. coli HCP impurities in products manufactured by recombinant expression, an immunoenzymetric assay kit was used according to manufacturer's instructions (Cygnus Technologies, #F410).
ELISA: Fab concentrations were determined by Anti-VEGF ELISA kit following the manufacturer's instructions (Alpha Diagnostic International, #200-880-LUG).
Native/Ion Mobility Mass Spectrometry: The buffer of the purified protein was exchanged for 100 mM ammonium acetate via 10 kDa-cutoff Amicon filters (Merck Millipore). Concentration of sample was adjusted to 10 μM. Sample was directly infused into electrospray ionization source (ESI) source.
Human Umbilical Vein Endothelial Cells (HUVEC) Proliferation Inhibition: Endothelial cell growth basal medium (CC-3121, Lonza, Switzerland) supplemented with EGM-2 supplements and growth factors without VEGF (CC-4176, Lonza, Switzerland) was used for the cultivation of HUVEC cells in a humidified atmosphere containing 95 % air and 5 % CO2 at 37 °C. 100 ng/mL Reference product (Lucentis), 100 ng/mL Fab product (Ranibizumab), and 100 ng/mL VEGF were applied to HUVEC (Lonza, #191027) cells (1x104 cells). Lucentis and in-house purified Ranibizumab were incubated with VEGF in a tube at room temperature for 90 min prior to application. The VEGF/protein mix was further applied to the cells. Inhibition of HUVEC proliferation was monitored in real-time with xCELLigence RTCA DP instrument.
Formulation: The formulation and concentration of Ranibizumab were carried out using an automated TFF system called Sartoflow Smart Small-Scale Benchtop TFF (Sartorius, Germany). Sartocon Slice 200 ultrafiltration cassette with a 10 kDa pore size (Sartorius, Germany) was used. The ultrafiltration cassette and the TFF system were sequentially rinsed with distilled water and formulation buffer. Subsequently, the filtration process was performed during the product cycle and the product was exchanged with the formulation buffer (10 mM histidine HCl, 292 mM trehalose, 0.01 % polysorbate 20, pH 5.5).
3. Results and discussion
An analytical characterization and similarity testing plan was designed to evaluate and compare the structure, biological function, and heterogeneity of our in-house purified Fab molecule Ranibizumab to those of the originator Lucentis using cutting-edge techniques. Where applicable, orthogonal methods were used to assess product attributes. Methods used for the analysis and similarity test between the purified Fab and reference product were classified according to the relevant analytical field (Table 1). A well-established platform process that exploits unique Fab molecule characteristics has been adopted to develop a purification strategy.
Table 1.
Analytical characterization plan for the Fab molecule.
| Category | Assays or Analytical Techniques |
|---|---|
| Primary structure | Primary amino acid sequence and profile by peptide mapping Isoelectric point and profile by cIEF Extinction coefficient by UV spectroscopy |
| Mass | Molecular mass of intact protein by intact protein MS Molecular mass of reduced HC and LC by LC-MS |
| Product-related substances and impurities | Size variants by CE-SDS Charge variants by cIEF |
| Process-related impurities | HCP-ELISA Endotoxin levels by LAL test Ligand leakage analysis by Protein L ELISA |
| Particles and aggregates | Multimers and aggregates by SEC-UPLC |
| Higher-order structure | Secondary structure by Far-UV CD Tertiary structure by native MS and IMMS |
| Biological activity | VEGF binding by ELISA Potency; inhibition of proliferation of HUVEC cells |
3.1. Characterization of Fab-related impurities following protein L chromatography
Given that Fab can be attached specifically to resins functionalized with the surface-binding Protein L, which has a high affinity for the kappa light chain, a prepacked HiTrap Protein L column was used for the capture of Fab molecules from crude E. coli samples. W3110 cells expressing recombinant Fab with a signal leader sequence for translocation into the periplasm were used for production. First, clarified E. coli homogenates containing Fab and Fab-related impurities were injected onto the Protein L column. Due to the affinity of Protein L to the kappa chain of immunoglobulin, both Fab assembly and free light chain (LC) are capable of binding to the Protein L column and are commonly co-eluted by gradient elution at an acidic pH, especially in higher protein loads. Supp Figure A1A shows the purification of Fab on a 1 mL HiTrap Protein L column by the application of a linear gradient over 10 column volumes (CV) from the pH range of 7.0–7.6 to 3.0–2.5. To assess the optimal binding and elution conditions for the Fab molecule, flow-through and elution fractions were applied to SDS-PAGE and analyzed by western blotting. According to the chromatograms and blots, we determined the optimal capture buffer to be pH 7.6 and 2.97 mS/cm and the optimal elution buffer to be pH 2.5. SDS-PAGE and Western blot analyses also confirmed that the peak contained the Fab molecule (Supp Figure A1C). Further, optimal flow rate conditions for binding and elution were analyzed based on the plan displayed in Supp Figure A2. According to the chromatograms, optimum binding and elution flow rates were determined as 1.0 and 0.8 mL/min, respectively.
Following the optimization of efficient capture of Fab molecules from E. coli homogenates (Fig. 1A), the secondary structure, oligomerization status, and abundance of free LC were analyzed. The molecular weight distribution of the Fab molecule was estimated using size-exclusion chromatography (SEC-UPLC) and capillary gel electrophoresis (CE-SDS). CE-SDS analysis under non-reducing conditions represented the presence of product-related impurities. Free light chain was the major impurity after Protein L chromatography (9.68 %). Of all protein species, 9.68 % were light chain and 4.48 % were heavy chain, yielding an 83.5 % final purity (Fig. 1B). However, it should be noted that the feed material quality varied from study to study due to the large number of experiments, thereby changing the abundance of product-related impurities. SEC-UPLC profile of the Fab molecule demonstrated low molecular weight substances as well as trace amounts of oligomeric fraction of the protein (Fig. 1C). The Fab molecule used in this study had a molecular weight of around 48 kDa, as determined by intact mass analysis (Fig. 1D and E). Based on the analyses, we demonstrated that HiTrap Protein L chromatography can be utilized as an effective capture step for Fab purification; however, a second chromatography step is required to remove product-related contaminants, especially co-captured free LC of the Fab molecule.
Fig. 1.
Purification and characterization of the Fab molecule. A) Chromatogram of Protein L run on a HiTrap Protein L column using 50 mL homogenate sample containing Fab, gradient pH 7.6 to pH 2.5 over 10 CV. B) Electropherogram of Protein L eluate showing the purity and the percentage of Fab-related impurities C) Analysis of aggregates by SEC-UPLC D) Deconvoluted mass spectra for intact mass E) Mass spectrum of Fab molecules for intact mass. HMW: High molecular weight impurities, LMW: Low molecular weight impurities.
3.2. Development of CEX chromatography for the removal of Fab-related impurities
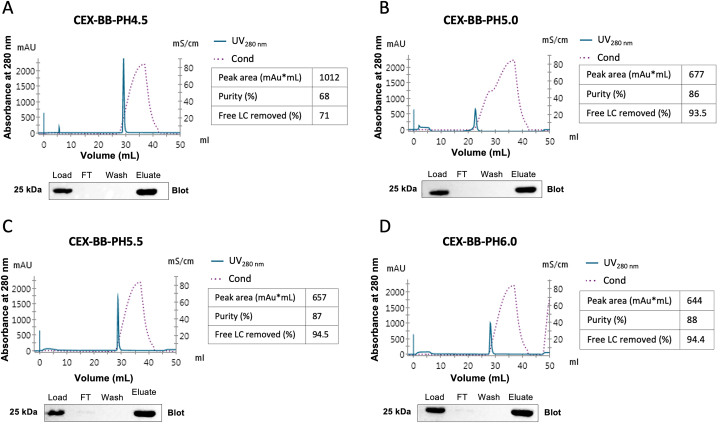
Following Protein L chromatography, all eluted fragments were then used as feed material and separated using a strong cation exchanger. The pH of the feed material was adjusted to the binding pH in CEX runs. Four pH conditions ranging from 4.5 to 6.0 were tested for the optimum binding of the Fab molecule to Cytiva HiTrap SP-FF cation exchanger column. Elution was carried out using a salt-based linear gradient, 20 mM sodium phosphate buffer (pH 6.0) containing 1 M NaCl. Eluted fractions were collected and analyzed by CE-SDS. Even though Fab was the predominant protein species in the eluates, CE-SDS data revealed the presence of product-related impurities (Fig. 2). According to UV spectroscopy measurements, immunoblots, and CE-SDS analyses, optimum binding for the Fab molecule with a relatively high percentage of purity was achieved at pH 5.0. In parallel, Fab fragment binding has been previously found most successful with multimodal resins at pH 5.0, which is far below the isoelectric point range of the target Fab molecules [14]. For each run, free LC removed (%) has been calculated from the ratio of the initial percentage of LC following Protein L chromatography and the final percentage of LC after the CEX run, since the abundance of product-related impurities in the samples used for the runs varied due to the variations in the upstream process. When binding was performed at pH 5.0, Fab accounted for 86 % of the proteins, while 5.56 % of all protein species were free LC (CEX-BB-pH5.0). This condition led to a considerably higher percentage of free LC removal, resulting in a greater purity of functional Fab when compared to the condition in which binding occurred at pH 4.5. Nevertheless, the efficacy of salt-based elution in cation-exchange chromatography to remove free LC was insufficient in these conditions.
Fig. 2.
Selecting optimal binding conditions for the Fab in cation-exchange chromatography (CEX). Chromatograms of CEX runs on a HiTrap SP-FF column using 1 mL eluate obtained from Protein L step. Binding with A) pH 4.5 B) pH 5.0C) pH 5.5 D) pH 6.0 with a linear gradient to 100 % Buffer B over 10 CV. Immunoblots of peaks and purity ratios obtained by CE-SDS were given. Full, non-adjusted blots are represented in Supplementary File Fig. 2.
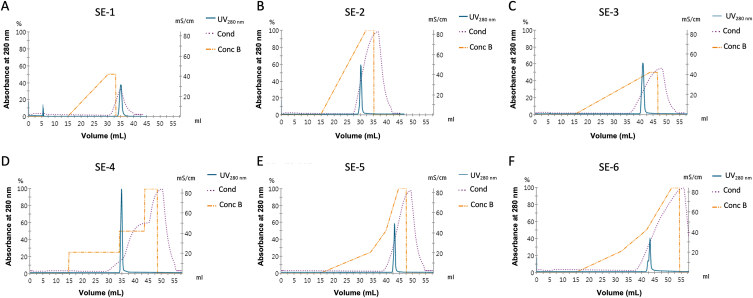
To improve the resolution, different salt-based elution settings were screened at a flow rate of 0.6 mL/min, and fractions were monitored by 12 % polyacrylamide gel and immunoblotting. The salt-based elution parameters for CEX runs are represented in Table 2. According to the chromatograms, the highest resolution was obtained by a gradual increase in the salt concentration in SE-6 representing a shoulder peak (Fig. 3). Nonetheless, the resolution of this method remained insufficient to completely get rid of traces of Fab-related impurities.
Fig. 3.
Salt elution profiles of ion-exchange chromatography using SP-FF column with different elution conditions at a flow rate of 0.6 mL/min. A) 50 % concentration B with linear gradient over 16 C V B) 100 % concentration B with linear gradient over 18 C V C) 50 % concentration B with linear gradient over 30 C V D) 25 %, 50 % and 100 % concentration B with step gradient over 18, 10 and 10 C V, respectively E) 25 %, 50 % and 100 % concentration B with linear gradient over 20, 6 and 5 C V, respectively E) 25 %, 50 % and 100 % concentration B with linear gradient over 20, 10 and 5 CV, respectively. SE: Salt elution, Cond: Conductivity, Conc B: Concentration B (20 mM sodium phosphate, 1 M NaCl pH 6.0).
We further employed cation-exchange chromatography (CEX) coupled with a pH gradient elution for the efficient removal of Fab-related impurities. The advantage of adopting a pH gradient as opposed to a typical salt gradient is that the collected fractions with low ionic strength contain extremely low salt concentrations, thereby avoiding the need for desalting. In addition, because the separation is based on relative variations in the pI of the protein, the resolution can be much higher than with salt gradient elution. Notably, the pH gradient-based CEX approach has been utilized and developed to assess the charge heterogeneity of mAbs for analytical purposes in many studies [[15], [16], [17], [18]]. In a purification train, it could provide a significant reduction in labor and equipment needs. For this, a HiTrap SP-FF column (particle size, ∼90 μm) was subjected to a pH gradient at a flow rate of 0.6 mL/min following a 5-CV equilibration in a pH 5.0 buffer. A linear pH gradient of 30 CV ranging from 5.0 to 7.0–8.5 was applied to achieve the elution and separation of Fab and Fab-related impurities. Over the course of the experiments, fractions of each peak were collected, and the pH of each fraction was monitored online. CEX runs yielded two well-separated peaks, indicating a distinct charge difference (Fig. 4A–C). In our case, Fab light chain has a pI of 6.34, whereas intact Fab has a pI of 7.97. According to the chromatograms, the highest resolution was achieved by the application of a linear gradient from pH 5.0 to pH 8.5 (PHE-8.5) (Fig. 4C). The presence of the Fab molecule in the fractions was confirmed by immunoblotting. In all polyacrylamide gels, the second fraction (FR 2) displayed a double band formation corresponding to the light and heavy chains of the Fab molecule, whereas the first fraction (FR 1) represented only one band with a slightly shorter kDa compared to the bands of the FR 2, indicating that the majority of the fully folded Fab molecule exists in the FR 2. To confirm this, fractions of the three experimental conditions were analyzed by capillary electrophoresis (CE-SDS). The presence of Fab-related impurities in the fractions was compared to the reference Fab molecule Lucentis produced by the originator for further fraction selection. FR 1 included a high percentage of light chain as a Fab-related impurity, whereas FR 2 was confirmed as the correctly processed Fab with a molecular mass of 48 kDa. As expected, the higher pI of the Fab assembly resulted in higher retention compared to the LC throughout the pH gradient. As pH increased, LC became more negative, leading to an earlier release from the column. The percentages of intact Fab and Fab-related impurities in the fractions were represented in Table 3 for each run. Of all experiments, PHE-8.5 displayed the highest purity in the last fraction obtained from the CEX run (Table 3). After CEX runs with salt elution, to move onto the next process, such as anion exchange chromatography (AEX) or multimodal chromatography (e.g., MMX), a pre-treatment is required to lower the conductivity. Since sample dilution requires a large amount of buffer, single-pass ultrafiltration/diafiltration (UF/DF) can be used to concentrate and dilute the target protein, thereby minimizing the amount of buffer required for pre-treatment [19]. In contrast, chromatography steps employing pH-based elution strategy yield salt-free eluates, avoiding the need for an UF/DF step between the chromatography runs. Consequently, our two-step strategy not only offered significant removal of product-related impurities, but also allowed for the easier application of a further chromatography step if necessary.
Fig. 4.
Elution profiles of ion-exchange chromatography using SP-FF column with a linear gradient of pH 7.0–8.5 in the presence of 20 mM sodium phosphate at a flow rate of 0.6 mL/min, and fractions screened by 12 % polyacrylamide gel and immunoblotting. A) Purity of fractions of PHE-7.0 CEX run eluted by Buffer B with pH 7.0 B) Purity of fractions of PHE-8.0 CEX run eluted by Buffer B with pH 8.0C) Purity of fractions of PHE-8.5 CEX run eluted by Buffer B with pH 8.5. Capillary electrophoresis electropherograms of fractions obtained from cation exchange chromatography (CEX) runs coupled to pH elution were displayed. The presence of Fab-related impurities in the fractions were compared to Lucentis produced by the originator for further fraction selection. PHE: pH elution, FR: fraction, FT: flow-through, LC: light chain, LLC: light-light chain dimer. Full, non-adjusted blots and gels are represented in Supplementary File Fig. 4.
Table 3.
of Fab-related impurities and intact Fab after each purification step in different fractions.
| Post-Protein L |
Post-CEX |
|||
|---|---|---|---|---|
| (%) | FR 1 (%) | FR 2 (%) | ||
| PHE-7.0 | Light chain (LC) | 10.5 | 16.1 | 0.84 |
| Heavy chain (HC) | 2.96 | 5.49 | 0.44 | |
| Light-light chain (LLC) | 1.05 | 0.67 | 0.54 | |
| Intact Fab | 72.5 | 72.1 | 90.8 | |
| PHE-8.0 | Light chain (LC) | 17.5 | 33.7 | 3.8 |
| Heavy chain (HC) | 6.50 | 11.5 | 0.40 | |
| Light-light chain (LLC) | 1.09 | 0.59 | 0.50 | |
| Intact Fab | 68.7 | 49.2 | 91.4 | |
| PHE-8.5 | Light chain (LC) | 17.5 | 33.6 | 1.48 |
| Heavy chain (HC) | 6.50 | 15.2 | 0.71 | |
| Light-light chain (LLC) | 1.09 | 0.72 | 0.12 | |
| Intact Fab | 68.7 | 48.5 | 95.9 | |
After the separation ability of the pH-based purification train employing a strong cation-exchanger was established, subsequent experiments were conducted with HiTrap SP-FF column. To test the batch-to-batch reproducibility of the resolution obtained by cation exchange chromatography, three CEX chromatography experiments using three different purification batches using the same feed material were conducted (Fig. 5A). CEX chromatograms yielded three partitioned peaks in the elution. The last fractions obtained from CEX elution were analyzed by CE-SDS in duplicates. According to the electrophoretic profile of the Fab molecule, overall purity was determined as 99.1 ± 0.15 (Fig. 5B).
Fig. 5.
Three batches for the Fab purification by Protein L followed by cation-exchange chromatography coupled with the pH elution. A) CEX chromatograms of different batches. Last fractions containing Fab are indicated by blue arrows B) Last fractions of each CEX runs analyzed by capillary electrophoresis as duplicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Three different batches were performed and purified using the established two-step purification strategy (n = 3; triplicates). The concentration of the starting feed material and final purified product of each run was determined by an antigen quantitative ELISA and overall Fab recovery was calculated. The resulting Fab product had a purity of >98 % and an overall process recovery of ∼50 % after two-step purification (Table 4). In a previous study, a purification strategy comprising two consecutive multimodal chromatography steps was shown to be effective in the purification of Fab molecules, with a final purity of 99.5 % and an overall process recovery of 32.55 % [20]. Similarly, multimodal resins have been reported to be superior to pure cation exchange resins, as they are more salt-tolerant and bind the Fabs more effectively. Nevertheless, the same study showed that isolating Fabs using Protein A and Protein L affinity resins and a final ultrafiltration/diafiltration resulted in the highest yield of 50 % [14].
Table 4.
Fab concentrations determined by an antigen quantitative ELISA. Overall yields and recovery were calculated as a result of 2-step purification from three different productions.
| Production Batch | Purification Batch | Purification Step | Fraction | Fab Concentration (mg/mL) | Volume (mL) | Fab Amount (mg) | Total Process Recovery (%) | Average | STDEV | RSD |
|---|---|---|---|---|---|---|---|---|---|---|
| Production Batch 1 | Purification Batch 1 | First Step (AC) | Load | 0.163 | 48.234 | 7.863 | 50.38 | 49.59 | 1.53 | 3.09 |
| Second Step (CEX) | Elution | 0.990 | 4.000 | 3.961 | ||||||
| Purification Batch 2 | First Step (AC) | Load | 0.206 | 47.056 | 9.702 | 50.56 | ||||
| Second Step (CEX) | Elution | 1.226 | 4.001 | 4.905 | ||||||
| Purification Batch 3 | First Step (AC) | Load | 0.217 | 48.307 | 10.469 | 47.82 | ||||
| Second Step (CEX) | Elution | 1.252 | 4.000 | 5.006 | ||||||
| Production Batch 2 | Purification Batch 1 | First Step (AC) | Load | 0.164 | 48.170 | 7.914 | 51.95 | 50.27 | 1.63 | 3.24 |
| Second Step (CEX) | Elution | 1.370 | 3.001 | 4.112 | ||||||
| Purification Batch 2 | First Step (AC) | Load | 0.187 | 48.372 | 9.039 | 48.7 | ||||
| Second Step (CEX) | Elution | 1.046 | 4.208 | 4.402 | ||||||
| Purification Batch 3 | First Step (AC) | Load | 0.174 | 48.875 | 8.516 | 50.16 | ||||
| Second Step (CEX) | Elution | 1.067 | 4.002 | 4.272 | ||||||
| Production Batch 3 | Purification Batch 1 | First Step (AC) | Load | 0.204 | 48.285 | 9.845 | 47.55 | 48.11 | 0.93 | 1.93 |
| Second Step (CEX) | Elution | 1.872 | 2.500 | 4.681 | ||||||
| Purification Batch 2 | First Step (AC) | Load | 0.229 | 48.415 | 11.082 | 49.18 | ||||
| Second Step (CEX) | Elution | 1.362 | 4.001 | 5.450 | ||||||
| Purification Batch 3 | First Step (AC) | Load | 0.236 | 49.916 | 11.795 | 47.59 | ||||
| Second Step (CEX) | Elution | 1.403 | 4.000 | 5.613 |
The final Fab product was taken into the formulation buffer and analyzed with regard to process-related impurities, including protein L leakage, host cell protein, and endotoxin abundance (Table 5). Protein L leakage, HCP and endotoxin concentrations for the products obtained after both steps were below the limit that is specified in the literature [4]. Optimized conditions for laboratory-scale two-step purification approach are represented in Table 6.
Table 5.
Protein L ligand leakage, host cell proteins and endotoxin levels determined by commercial kits after two-step purification.
| Purification Step | Fraction | Protein L Concentration (ng/mL) | Accepted Range |
|---|---|---|---|
| First Step (AC) | Load | – | Below 6.0 ng/mL |
| Elution | 5.36 | ||
| Second Step (CEX) | Elution | 3.76 | |
| Purification Step | Fraction | HCP Concentration (ng/mL) | |
| First Step (AC) | Load | 1.67 x 106 | Below 100 ng/mL |
| Elution | 661 | ||
| Second Step (CEX) | Elution | 16.8 | |
| Purification Step | Fraction | Endotoxin Concentration (EU/mL) | |
| First Step (AC) | Load | 1788 | Below 0.5 EU/mL |
| Elution | 0.065 | ||
| Second Step (CEX) | Elution | 0.031 |
Table 6.
Optimized laboratory-scale two-step chromatography setup for Fab purification, along with tabulated details of the buffers, flow rate(s), number of CVs used for the purification of Fab molecule.
| Affinity Chromatography (HiTrap Protein L Column) | |||
|---|---|---|---|
| Stage | Content | Flow rate (mL/min) | Number of CV |
| Equilibration | 20 mM sodium phosphate pH 7.6 | 1.0 | 5 |
| Load | pH adjusted to 7.6 | 0.8 | Variable |
| Wash | 20 mM sodium phosphate, pH 6.0 | 1.0 | 6 |
| Elution | 100 mM glycine-HCl, pH 2.5 | 1.0 | 10 |
| CIP | 0.1 M NaOH | 5 | |
| Storage |
20 % ethanol |
– |
– |
| Cation Exchange Chromatography (HiTrap SP-FF Column) | |||
| Stage | Content | Flow rate (mL/min) | Number of CV |
| Equilibration | 20 mM sodium phosphate pH 5.0 | 1.0 | 5 |
| Load | pH adjusted to 5.0 | 0.8 | Variable |
| Wash | Same as equilibration buffer | 1.0 | 6 |
| Elution (linear gradient) | 20 mM sodium phosphate pH 8.5 | 1.0 | 30 |
| CIP | 1 M NaOH | 5 | |
| Storage | 20 % ethanol | – | – |
3.3. Comparison of the final product with the originator molecule Lucentis
Analytical comparability between the originator and the biosimilar is vital to the development of biosimilar drugs [21]. In this regard, a side-by-side comparison was made between the final product Ranibizumab and the originator molecule Lucentis, on the basis of the critical quality attributes (CQA). Primary structure related to amino acid sequence and post-translational modifications; higher order structure; product-related substances; and purity and impurity, including size and charge variants, were all compared.
Peptide mapping is employed to examine differences in protein sequences and PTMs between the Ranibizumab and the originator Fab molecule (Lucentis). Fig. 6 shows a mirror plot representation of the LC-MS peptide maps of Ranibizumab and Lucentis given as total ion chromatograms (TICs) (Fig. 6A). The MS intensity is displayed along with the retention time. Both molecules exhibited a similar peptide map profile after overnight tryptic digestion. Intact mass analysis was used to assess the molecular masses of the intact antibody and single chain following DTT reduction. Fig. 6B and C show the deconvoluted MS spectra of both intact and reduced forms of both the originator and in-house purified Fab, respectively. Both Fabs showed multiple peaks at the intact protein level (Fig. 6B). The same deconvoluted masses were identified for the light chains (MH+, 23432 Da) and heavy chains (MH+, 24956 Da) of both Fab molecules (Fig. 6C).
Fig. 6.
Similarity assessment for MS profile of Ranibizumab and Lucentis by mirror plots A) Comparison of LC-MS peptide maps from overnight tryptic digests of the originator (Lucentis) and Fab molecules (Ranibizumab) B) Comparison for deconvoluted mass spectrum of Fab and the originator C) Comparison for reduced mass analysis for light and heavy chains.
In protein-based biopharmaceuticals, measuring mass variants, particularly aggregates, is pivotal due to their potential immunogenicity [22]. Antibodies undergo charge heterogeneity changes as a result of modifications, altering their product features like stability and binding activity. Thus, detailed monitoring of these modifications, which can affect antibody characteristics is a critical requirement defined by regulatory agencies [23]. According to the CQAs, the presence of particles, aggregates, and charge variants were quantified. SEC-UPLC was employed as the standard separation technique for the quantification of protein oligomers where 99.93 % were found to be folded Fab monomers of all protein species (Fig. 7A). Electrophoretic mobility profile of the originator and in-house purified Fab molecule displayed a high degree of similarity and purity (Fig. 7B). Charge variant analysis carried out by cIEF yielded similar acidic variants of Fab to the originator molecule and indicated that the pI values of the main peak (8.08) and acidic variant peaks (7.87) were identical (Fig. 7C).
Fig. 7.
Similarity assessment for size and charge variants, and aggregates of Fab and the originator A) Analysis of the particles and aggregates by SEC-UPLC B) Electrophoretic mobility of Fab and the originator for size variant analysis C) cIEF for charge variant analysis of Fab and the originator.
Circular dichroism reflects the alterations in the secondary structure of proteins with far-UV scanning in the wavelength range 190–250 nm [24]. The obtained Far UV-CD spectrum of Fab molecule showed a high degree of similarity to the originator with an absorption minimum at 218 nm and a maximum at approximately 200 nm (Fig. 8A), revealing predominantly a β-sheet-content protein. Melting profiles of both Fab assembly and the originator showed a comparable shift in ellipticity at 218 nm with increasing temperature, having Tm value of 84 ± 0.5 °C (Fig. 8B). Further, native mass spectrometry was utilized to assess the structural heterogeneity of the Fab molecule. Four different reference products were compared with two different Ranibizumab productions. The results showed the same charge estimates (+16, +15, +14) and mass/charge ratios for the Lucentis and Ranibizumab in their native-like states (Fig. 8C). Next, ion-mobility mass spectrometry (IMMS) was employed to evaluate multimeric and conformational variants for the Fab molecules under equilibrium conditions in comparison with the originator. IMMS is a technique based on the propulsion of proteins moving in a drift tube filled with inert gas in their natural state, resulting from different amounts of collisions with the gas depending on their collision cross section. With this technique, the conformational landscape of proteins with the same mass can be detected precisely, considering the time the proteins spend moving in the drift tube. When +15 (most dominant) charge state distribution of each sample was analyzed using IMMS approach, in-house purified Fab molecules have exactly the same mobility time (10.36 msec, 10.47 msec) as the reference products (Fig. 8D).
Fig. 8.
Similarity assessment for the higher-order structure of Fab and the originator A) Comparison for secondary structure profiles of Fab and the originator B) CD (218 nm) temperature scan of the Fab and the originator molecule C) Comparison for native MS spectrum D) Comparison for mobilogram profiles of Fab and the originator.
The concentrations of the Fab molecules from two-step purification were determined by an antigen quantitative ELISA method. 100 ng/mL reference products from three different lots, 100 ng/mL Fab products from five different purification batches, and 100 ng/mL VEGF were applied to HUVEC cells (1x104 cells). The effects of purified Ranibizumab from 3 different productions on the proliferation of HUVEC cells are shown in Fig. 9, in comparison with the reference products. It was observed that the in-house purified Ranibizumab of three consecutive productions were equivalent to three reference products (Lucentis) at the 24th and 48th hours of incubation.
Fig. 9.
Comparison of HUVEC proliferation inhibition of Ranibizumab and the reference molecule (Lucentis). Three different batches were purified by the developed two-step purification strategy. The inhibition ability of in-house purified Fab molecules (100 ng/mL) for HUVEC proliferation was tested by xCELLigence and compared to the reference molecule (100 ng/mL) and VEGF treated cells (100 ng/mL) at A) 24th and B) 48th hours. Ranibizumab and Lucentis products were analyzed and compared to the control (VEGF+) by an ordinary one-way ANOVA with GraphPad Prism 8.0.1 (**p < 0.01, ***p < 0.001, ****p < 0.0001). No VEGF control was used for testing the efficiency of VEGF treatment.
3.4. Pilot-scale purification of Ranibizumab using a two-step purification strategy
To confirm the performance of two-step purification of Ranibizumab for pilot-scale production, we intend to scale-up the process from 50 mL to 5 of L clarified feed containing approximately 1 g of Ranibizumab using the AKTA Pilot 600 chromatography system and Axichrom columns packed with Protein L and SP-FF resins. Chromatograms obtained from the purifications are depicted in Fig. 10. Protein L chromatography yielded a sharp single peak containing Fab molecule (Fig. 10A), whereas CEX run yielded two very well-separated peaks in the elution phase (Fig. 10B). Of these two peaks, second one with a higher retention contained highly pure Ranibizumab Fab assembly, as in the lab-scale optimization experiments.
Fig. 10.
Two-step pilot-scale purification of Ranibizumab A) Chromatogram of Protein L run on a Axichrom column (70/300) packed with Protein L resin using 5 L clarified feed containing 1 mg of Ranibizumab and a pH gradient from 7.6 to pH 2.5 over 10 C V. B) Chromatogram of CEX run on a Axichrom column (30/300) packed with SP-FF resin using the eluate obtained from capture step and a linear pH gradient from 5.0 to pH 8.5 over 30 CV.
Table 7 depicts the results of testing quality attributes for the two-step purification process, including protein L capture, cation exchange, and a final buffer exchange step performed via the Sartoflow Smart Small-Scale TFF system using a cellulose-based membrane. The data confirmed that, Fabs were successfully produced with high purity with a two-step purification strategy. The results for obtaining minimal impurities were deemed satisfactory when compared to the originator molecule Lucentis (Table 7). Further, the concentrations were measured by ELISA (Table 8). The resulting Fab product had a purity of ∼98 % and a total process recovery of ∼%51. To evaluate the comparative biological activity of the purified Ranibizumab, HUVEC proliferation induced by VEGF was inhibited by Ranibizumab and Lucentis. Fig. 11 shows the relative VEGF-dependent proliferation levels in HUVEC cells in the presence and absence of Ranibizumab and Lucentis. The results suggested that our purified Ranibizumab is comparable to Lucentis in neutralizing VEGF and in full bioactivity.
Table 7.
The analyses of mass, size and charge variants, aggregates, purity and biological activity of formulated Ranibizumab.
| Ranibizumab | Lucentis | ||
|---|---|---|---|
| Intact Mass (Da) | 48380 | 48380.199 ± 0.089 | |
| Reduced Mass | |||
| Light chain (Da) | 23432.5 ± 0.5 | 23432.48 ± 0.098 | |
| Heavy chain (Da) | 24956 | 24956.55 ± 0.049 | |
| CE-SDS | |||
| Light chain (%) | 0.965 ± 0.5 | ||
| Heavy chain (%) | 0.885 ± 0.065 | ||
| Intact Protein (%) | 97.95 ± 0.15 | 98.48 ± 0.16 | |
| SEC-HPLC | |||
| Main Peak (%) | 99.75 ± 19 | 99.640.34 | |
| IEX-HPLC | |||
| Acidic variant (%) | 0.14 ± 0.08 | ||
| Basic variant (%) | 0.765 ± 0.625 | ||
| Main peak (%) | 99.1 ± 0.7 | 99.57 ± 0.04 | |
| HUVEC relative potency (%) | 97.8 ± 0.7 | 98.47 ± 0.64 | |
| HCP level (ppm) | 0.07 ± 0.02 | 0.09 ± 0.02 | |
| Endotoxin Concentration (EU/mL) | <0.05 | <0.05 | |
Table 8.
Fab concentrations determined by an antigen quantitative ELISA. Overall yields and recovery were calculated as a result of two-step purification from two different batches.
| Two-step Purification | Fraction | Fab Amount (mg) | Recovery (%) | Total Process Recovery (%) |
|---|---|---|---|---|
| Purification | Load | 1077.2 ± 58.8 | 71.485 ± 3.475 | 51.2 ± 0.56 |
| Final | 772.189 ± 79.5 | |||
| TFF | Load | 770.149 ± 79.4 | 71.89 ± 2.67 | |
| Final | 551.89 ± 36.2 |
Fig. 11.
Comparison of HUVEC proliferation inhibition of Ranibizumab and the reference molecule (Lucentis). Two different batches were purified by the developed two-step purification strategy. The inhibition ability of in-house purified Fab molecules (100 ng/mL) for HUVEC proliferation was tested by xCELLigence and compared to the reference molecule (100 ng/mL) and VEGF treated cells (100 ng/mL) at A) 24th and B) 48th hours. Ranibizumab and Lucentis were analyzed and compared to the control (VEGF+) by an ordinary one-way ANOVA with GraphPad Prism 8.0.1 (**p < 0.01, ***p < 0.001, ****p < 0.0001). No VEGF control was used for testing the efficiency of VEGF treatment.
Here, we established a two-step purification strategy for Fab molecules and showed that Fab produced using our platform process is equivalent to the originator molecule produced in E. coli. Employing Protein L in combination with a strong cation-exchanger column using a pH based gradient elution mode provided a high purity and good yield for Fab purification. The entire process was carried out using the optimized conditions defined in the purification steps (Table 9). It should be noted, however, that various aspects of the upstream process can affect the downstream steps, including the state of cells, process parameters, feeding strategy, process robustness, inefficient translocation, and incomplete signal sequence processing. Variations in the upstream process can have a dramatic impact on the operations and performance of the downstream process. Therefore, both upstream and downstream processes need to be explored as an integrated system in order to build a productive and economically viable process. Besides, the protocol is developed for a single molecule, and each new candidate of this structural class could require the same effort.
Table 9.
Conditions for optimized pilot-scale two-step chromatography setup for Fab purification, along with tabulated details of the buffers, flow rate(s), number of CVs used for the purification of Fab molecule.
| Affinity Chromatography (AxiChrom 70/300 packed with Protein L resin) | |||
|---|---|---|---|
| Stage | Content | Flow rate (mL/min) | Number of CV |
| Equilibration | 20 mM sodium phosphate pH 7.6 | 230 | 5 |
| Load | Not adjusted | 100 | Variable |
| Wash | 20 mM sodium phosphate, pH 6.0 | 230 | 6 |
| Elution | 100 mM glycine-HCl, pH 2.5 | 230 | 10 |
| CIP | 0.1 M NaOH | 100 | 3 |
| Storage |
20 % ethanol |
230 |
3 |
| Cation Exchange Chromatography (AxiChrom 50/300 packed with SP-FF resin) | |||
| Stage | Content | Flow rate (mL/min) | Number of CV |
| Equilibration | 20 mM sodium phosphate pH 5.0 | 100 | 5 |
| Load | pH adjusted to 5.0 | 65 | Variable |
| Wash | 20 mM sodium phosphate 5.0 | 100 | 3 |
| Elution (linear gradient) | 20 mM sodium phosphate pH 8.5 | 100 | 30 |
| CIP | 0.5 M NaOH | 65 | 3 |
| Storage | 20 % ethanol | 100 | 3 |
4. Conclusions
Alternatives for Fab purification platforms could significantly improve commercialization of Fab molecules and reduce investment costs and time, thereby enabling more Fabs to reach the clinic and consequently more patients. For the straightforward purification of Fabs and Fab-related molecules, we propose a downstream process utilizing an affinity purification with a Protein L column followed by the use of a strong cation exchanger with the application of a linear pH gradient. Herein, it has been indicated that a linear pH gradient is superior to a salt gradient for a better separation of Fab-related impurities and intact Fab molecules. Besides, a two-step strategy is possible for the purification of Fab molecules, which reduces the cost and time of the downstream process. Overall, our in-house purified Fab molecule showed high similarity to the originator molecule. The established purification platform significantly intensified the process and enabled successful clearance of Fab- and process-related impurities (>95 %) with an overall product yield of 50 %, suggesting that it could be a new option for Fab purification for both academic and industrial applications.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
CRediT authorship contribution statement
Ozge Tatli: Conceptualization, Investigation, Methodology, Writing – original draft. Yagmur Oz: Data curation, Investigation, Methodology, Writing – review & editing. Baran Dingiloglu: Data curation, Investigation, Methodology, Writing – review & editing. Duygu Yalcinkaya: Data curation, Investigation, Methodology, Writing – review & editing. Ezgi Basturk: Data curation, Investigation, Methodology, Writing – review & editing. Melis Korkmaz: Data curation, Investigation, Methodology, Writing – review & editing. Latif Akbulut: Data curation, Methodology, Writing – review & editing. Derya Hatipoglu: Data curation, Methodology, Writing – review & editing. Cansin Kirmacoglu: Data curation, Methodology, Writing – review & editing. Buse Akgun: Data curation, Methodology. Kubra Turk: Data curation, Methodology. Orkun Pinar: Investigation, Methodology. Berna Sariyar Akbulut: Investigation, Methodology. Zeynep Atabay: Funding acquisition, Visualization. Eda Tahir Turanli: Investigation, Methodology, Writing – review & editing. Dilek Kazan: Funding acquisition, Investigation, Methodology, Writing – review & editing. Gizem Dinler Doganay: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study is supported by the Scientific & Technological Research Council of Türkiye (TUBITAK) 1007 program [grant number: 115G078)].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21001.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
figs1.
figs2.
References
- 1.Holliger P., Hudson P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005;23:1126–1136. doi: 10.1038/NBT1142. [DOI] [PubMed] [Google Scholar]
- 2.Chames P., van Regenmortel M., Weiss E., Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br. J. Pharmacol. 2009;157:220–233. doi: 10.1111/J.1476-5381.2009.00190.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta S.K., Shukla P. vol. 43. 2016. pp. 31–42. (Microbial Platform Technology for Recombinant Antibody Fragment Production: A Review). 10.3109/1040841X.2016.1150959. [DOI] [PubMed] [Google Scholar]
- 4.Rodrigo G., Gruvegård M., van Alstine J.M. Antibody fragments and their purification by protein L affinity chromatography. Antibodies. 2015;4:259–277. doi: 10.3390/ANTIB4030259. 4 (2015) 259–277. [DOI] [Google Scholar]
- 5.Chen X., Wang Y., Li Y. Protein L chromatography: a useful tool for monitoring/separating homodimers during the purification of IgG-like asymmetric bispecific antibodies. Protein Expr. Purif. 2020;175 doi: 10.1016/J.PEP.2020.105711. [DOI] [PubMed] [Google Scholar]
- 6.Chen S.W., Tan D., Yang Y.S., Zhang W. Investigation of the effect of salt additives in Protein L affinity chromatography for the purification of tandem single-chain variable fragment bispecific antibodies. mAbs. 2020;12 doi: 10.1080/19420862.2020.1718440/SUPPL_FILE/KMAB_A_1718440_SM5218.DOCX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plückthun A. Antibody Engineering: sdvances from the pse of Escherichia coli expression systems. Bio/Technology. 1991;9:6. doi: 10.1038/nbt0691-545. 9 (1991) 545–551. [DOI] [PubMed] [Google Scholar]
- 8.Venturi M., Seifert C., Hunte C. High level production of functional antibody fab fragments in an oxidizing bacterial cytoplasm. J. Mol. Biol. 2002;315:1–8. doi: 10.1006/JMBI.2001.5221. [DOI] [PubMed] [Google Scholar]
- 9.Humphreys D.P. The Periplasm; 2014. Periplasmic Expression of Antibody Fragments; pp. 361–388. [DOI] [Google Scholar]
- 10.Quehenberger J., Wurm D., Kopp J., Pflügl S., Patil R.S., Anupa A., Gupta J.A., Rathore A.S. Challenges in expression and purification of functional fab fragments in E. coli: current dtrategies and Perspectives. Fermentation. 2022;8:175. doi: 10.3390/FERMENTATION8040175. 8 (2022) 175. [DOI] [Google Scholar]
- 11.Junter G.A., Lebrun L. Polysaccharide-based chromatographic adsorbents for virus purification and viral clearance. Biochemistry. 2020;10:291–312. doi: 10.1016/J.JPHA.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brorson K., Krejci S., Lee K., Hamilton E., Stein K., Xu Y. Bracketed generic inactivation of rodent retroviruses by low pH treatment for monoclonal antibodies and recombinant proteins. Biotechnol. Bioeng. 2003;82:321–329. doi: 10.1002/BIT.10574. [DOI] [PubMed] [Google Scholar]
- 13.Miesegaes G.R., Lute S., Strauss D.M., Read E.K., Venkiteshwaran A., Kreuzman A., Shah R., Shamlou P., Chen D., Brorson K. Monoclonal antibody capture and viral clearance by cation exchange chromatography. Biotechnol. Bioeng. 2012;109:2048–2058. doi: 10.1002/BIT.24480. [DOI] [PubMed] [Google Scholar]
- 14.Nascimento A., Pinto I.F., Chu V., Aires-Barros M.R., Conde J.P., Azevedo A.M. Studies on the purification of antibody fragments. Sep. Purif. Technol. 2018;195:388–397. doi: 10.1016/J.SEPPUR.2017.12.033. [DOI] [Google Scholar]
- 15.Fekete S., Beck A., Fekete J., Guillarme D. Method development for the separation of monoclonal antibody charge variants in cation exchange chromatography, Part II: pH gradient approach. J. Pharm. Biomed. Anal. 2015;102:282–289. doi: 10.1016/J.JPBA.2014.09.032. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L., Patapoff T., Farnan D., Zhang B. Improving pH gradient cation-exchange chromatography of monoclonal antibodies by controlling ionic strength. J. Chromatogr. A. 2013;1272:56–64. doi: 10.1016/J.CHROMA.2012.11.060. [DOI] [PubMed] [Google Scholar]
- 17.Füssl F., Cook K., Scheffler K., Farrell A., Mittermayr S. J. Bones, charge variant analysis of monoclonal antibodies using Direct coupled pH gradient cation exchange chromatography to high-resolution native mass spectrometry. Anal. Chem. 2018;90:4669–4676. doi: 10.1021/ACS.ANALCHEM.7B05241/SUPPL_FILE/AC7B05241_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- 18.Rea J.C., Moreno G.T., Lou Y., Farnan D. Validation of a pH gradient-based ion-exchange chromatography method for high-resolution monoclonal antibody charge variant separations. J. Pharm. Biomed. Anal. 2011;54:317–323. doi: 10.1016/J.JPBA.2010.08.030. [DOI] [PubMed] [Google Scholar]
- 19.Rathore A.S., Shirke A. Recent developments in membrane-based separations in biotechnology processes: review. Prep. Biochem. Biotechnol. 2011;41:398–421. doi: 10.1080/10826068.2011.613976. [DOI] [PubMed] [Google Scholar]
- 20.US20200095277A1 - A process for the purification of recombinant antibody fragments - Google Patents, (n.d.). https://patents.google.com/patent/US20200095277A1/en (accessed February 2, 2023).
- 21.Ramanan S., Grampp G. Drift, evolution, and divergence in biologics and biosimilars manufacturing. BioDrugs. 2014;28:363–372. doi: 10.1007/S40259-014-0088-Z. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg A.S. Effects of protein aggregates: an Immunologic perspective. AAPS J. 2006;8 doi: 10.1208/AAPSJ080359/METRICS. E501–E507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khawli L.A., Goswami S., Hutchinson R., Kwong Z.W., Yang J., Wang X., Yao Z., Sreedhara A., Cano T., Tesar D., Nijem I., Allison D.E., Wong P.Y., Kao Y.H., Quan C., Joshi A., Harris R.J., Motchnik P. Charge variants in IgG1: isolation, characterization, in vitro binding properties and pharmacokinetics in rats. mAbs. 2010;2:613–624. doi: 10.4161/MABS.2.6.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Provencher S.W., Glöckner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/BI00504A006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.