Abstract
The nodulation of legumes has for more than a century been considered an exclusive capacity of a group of microorganisms commonly known as rhizobia and belonging to the α-Proteobacteria. However, in the last 3 years four nonrhizobial species, belonging to α and β subclasses of the Proteobacteria, have been described as legume-nodulating bacteria. In the present study, two fast-growing strains, LUP21 and LUP23, were isolated from nodules of Lupinus honoratus. The phylogenetic analysis based on the 16S and 23S rRNA gene sequences showed that the isolates belong to the genus Ochrobactrum. The strains were able to reinfect Lupinus plants. A plasmid profile analysis showed the presence of three plasmids. The nodD and nifH genes were located on these plasmids, and their sequences were obtained. These sequences showed a close resemblance to the nodD and nifH genes of rhizobial species, suggesting that the nodD and nifH genes carried by strain LUP21T were acquired by horizontal gene transfer. A polyphasic study including phenotypic, chemotaxonomic, and molecular features of the strains isolated in this study showed that they belong to a new species of the genus Ochrobactrum for which we propose the name Ochrobactrum lupini sp. nov. Strain LUP21T (LMG 20667T) is the type strain.
Plants from the family Leguminosae are usually capable of dinitrogen fixation because of their symbiotic interaction with nodulating bacteria belonging to the order Rhizobiales. Most bacteria that establish a symbiosis with legume plants, including some nonrhizobial species of Methylobacterium (16, 45) and Devosia (40, 41), belong to the α subclass of Proteobacteria, although some species from genera of the β subclass, such as Ralstonia and Burkholderia, can also nodulate legumes (6, 32, 53). In the last few years there has been an increasing amount of research focused on bacteria that nodulate stems or roots of tropical legume species. However, the identity of many of the endosymbionts of temperate legumes still remains unknown. The genus Lupinus groups up to 200 species of herbs and small shrubs, broadly distributed in the Mediterranean area and in the American continent, where they colonize very different environments. Despite the agronomic and ecological interest of Lupinus, this plant has been poorly studied with respect to its symbionts. Plants from the genus Lupinus are nodulated by fast- and slow-growing rhizobia; however, slow-growing rhizobia are more frequently isolated from this legume (3, 5, 21, 30). The data obtained from the small-subunit (SSU) rRNA gene indicate a very close relationship between some bradyrhizobia isolated from Lupinus and Bradyrhizobium japonicum (3, 12, 30). However, bacterial strains nodulating Lupinus plants have been poorly characterized thus far, and fast-growing species nodulating this legume have not been not officially described; nevertheless, in the past the species Rhizobium lupini was proposed (15) and was later abandoned (14).
During a study of rhizobia nodulating Lupinus plants in several geographical regions, we isolated two fast-growing strains from nodules of two Lupinus honoratus plants. According to the rRNA gene sequences, they were identified as members of the genus Ochrobactrum within the α2 subclass of Proteobacteria. This genus belongs to the family Brucellaceae and was first described by Holmes et al. in 1988 (11) with a single species, Ochrobactrum anthropi. Later, four more species were described from different origins: Ochrobactrum intermedium from clinical sources (54), Ochrobactrum grignonense and Ochrobactrum tritici from soil and the wheat rhizoplane (23), and Ochrobactrum gallinifaecis from chicken feces (17).
Two nonrhizobial species belonging to the α subclass of Proteobacteria have been described hitherto as the legume endosymbionts Devosia neptuniae (41) and Methylobacterium nodulans (16). Recently, a strain isolated from Acacia mangium (33) was reported to belong to the genus Ochrobactrum in the family Brucellaceae, although there are no data about the symbiotic genes carried by this strain, nor its species affiliation. Therefore, we sought here to examine the phylogenetic relationships of strains LUP21 and LUP23 and to detect and characterize the symbiotic genes that encode for nodulation and nitrogen fixation. Finally, a polyphasic study of the isolates was also performed to establish the taxonomic position of the new strains.
MATERIALS AND METHODS
Isolation from plant nodules.
Strains LUP21T and LUP23 were isolated from root nodules of L. honoratus growing in Argentina. Isolations were made according to the method of Vincent (56) with yeast mannitol agar (YMA) (4). The cultures used to inoculate Lupinus albus plants were purified from a single colony after 2 days of incubation at 28°C and cultivated on YMA medium.
Nodulation tests.
Surface-sterilized seeds of L. albus were germinated axenically in petri dishes. Seedlings were transferred to pots with sterile vermiculite and watered with nitrogen-free Rigaud and Puppo (38) nutrient solution. Five plants were inoculated with 1 ml each of a bacterial suspension of LUP21 or LUP23 containing 8 × 108 cells/ml. The inoculated plants were placed for 6 weeks in a plant growth chamber with mixed incandescent and fluorescent lighting (400 microeinsteins m−2 s−1; 400 to 700 nm), programmed for a 16-h photoperiod, day-night cycle, with a constant temperature in the range of 25 to 27°C and 50 to 60% relative humidity. The strain Bradyrhizobium sp. strain ISLU35, a strain nodulating Lupinus in the Canary Islands (12), was used as positive control. As a negative control, uninoculated L. albus plants watered with nitrogen-free Rigaud and Puppo solution were used. After 6 weeks, the nodules were counted, and the dry weight of the aerial part of the plants was determined. The data obtained were analyzed by one-way analysis of variance, with the mean values compared by using the Fisher protected least significant difference (LSD) analysis (P = 0.05).
Plasmid profile analysis.
The isolates were subjected to plasmid profile analysis according to the method of Plazinski et al. (37), except that electrophoresis was done at 2 V cm−1 for 90 min, followed by 3 V cm−1 for 60 min and finally at 6 V cm−1 for 4 h. The 175- and 205-kb plasmids of Sinorhizobium meliloti GR4 (48) were used as size markers. Plasmid DNA was capillary transferred to a nylon membrane according to the method of Southern (44) and immobilized by baking at 80°C for 2 h.
nodD and nifH detection.
Oligonucleotide primers were designed to amplify a conserved fragment of the nodD and nifH genes among members of the family Rhizobiaceae (40). The PCR-amplified fragments of nifH and nodD genes were sequenced. For use as probes, they were digoxigenin labeled with a DIG DNA Labeling Kit (Roche Diagnostics) according to the manufacturer's instructions. Hybridization was detected with the DIG Nucleic Acid Detection Kit (Boehringer Mannheim) with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium as substrates for alkaline phosphatase according to the manufacturer's instructions.
DNA extraction and sequence analysis.
The 16S rRNA, 23S rRNA, nodD, and nifH genes were amplified and sequenced as previously described (40, 52). The sequences obtained were compared to those from GenBank by using the BLAST program (1). Sequences were aligned by using the CLUSTAL X software (46). The distances were calculated according to Kimura's two-parameter method (18), and phylogenetic trees were inferred by using the neighbor-joining method (42). Bootstrap analysis was based on 1,000 resamplings. The MEGA2 package (19) was used for all analyses.
Phenotypic characterization.
Colony morphology was studied on yeast mannitol (56) and nutrient agars. Cells were Gram stained according to the method of Doetsch (8). For electron microscopy the cells were grown on nutrient agar for 2 days and then stained with 0.2% (vol/vol) uranyl acetate. Thin sections were examined at 80 kV with a Zeiss EM 209 transmission electron microscope. Physiological studies were performed as described in reference 55, and the API 50CH, API 20E, and API 20NE systems were used according to the manufacturer's instructions. Catalase and oxidase activity were tested as described previously (41). Susceptibility to various antibiotics was examined by using penicillin (10 U), ampicillin (2 μg), oxytetracycline (30 μg), neomycin (5 μg), cloxacillin (1 μg), erythromycin (2 μg), cefuroxime (30 μg), ciprofloxacin (5 μg), polymyxin B (300 IU), gentamicin (10 μg), and streptomycin (300 μg) disks (Becton Dickinson) with Antibiotic agar (Oxoid) as the basal medium.
Extraction and analysis of fatty acids.
For fatty acid methyl ester (FAME) analysis, strains were grown for 24 h at 28°C on TSA plates containing 30 g of Trypticase soy broth (BBL) supplemented with 15 g of Bacto agar (Difco) liter of distilled water−1. Cells were saponified, and fatty acids were methylated to FAMEs and extracted by using Sherlock Microbial Identification System version 3.0 (MIDI) (31, 29). FAMEs were separated on an Agilent 6890A series gas chromatograph with a 7683 autoinjector and an autosampler tray module (Agilent Technologies). Separation of FAMEs was achieved with a fused-silica capillary column (25 m by 0.2 mm) with cross-linked 5% phenylmethyl silicone (film thickness, 0.33 μm; HP Ultra2). Hydrogen served as carrier gas. Peak integration and identification were performed by using the Hewlett Packard Chemstation and Sherlock software.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of whole-cell proteins.
Strains were grown on nutrient agar (Oxoid CM3) at 28°C for 24 h. Whole-cell protein extracts were prepared and separated by electrophoresis by using small modifications of the procedure of Laemmli (22) as described previously (7).
TP-RAPD patterns.
Crude DNA (2 μl) was used as a template for obtaining two-primer-[random(ly) amplified polymorphic DNA] (TP-RAPD) patterns. PCR was performed by using an AmpliTaq Gold reagent kit (Perkin-Elmer Biosystems) according to the manufacturer's instructions. Primers 8F (5′-AGAGTTTGATCCTGGCTCAG-3′, Escherichia coli positions 8 to 27) and 1522R (5′-GGTTACCTTGTTACGACTT-3′, E. coli positions 1509 to 1491) were used at 2 μM final concentration (39). PCR conditions were as follows: preheating at 95°C for 9 min; followed by 35 cycles of denaturing at 95°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 2 min; with a final extension at 72°C for 7 min. The PCR products were separated by electrophoresis on a 1.5% agarose gel in TBE buffer (100 mM Tris, 83 mM boric acid, 1 mM EDTA [pH 8.5]) at 6 V cm−1, stained in a solution containing 0.5 μl of ethidium bromide ml−1, and photographed under UV light. Standard VI (Boehringer-Roche) was used as a size marker. Then, 3 μl of 6× loading solution (40% sucrose and 0.25% bromophenol blue) was added to each sample.
DNA base composition and DNA-DNA hybridization.
DNA was extracted by the procedure of Marmur (26). To determine the DNA base composition, DNA was degraded enzymatically into nucleosides (28). The resulting nucleoside mixtures were separated by high-performance liquid chromatography using a Waters Symmetry Shield C8 column at 37°C. The solvent was 0.02 M NH4H2PO4 (pH4.0) with 1.5% acetonitrile. Nonmethylated lambda phage DNA (Sigma) was used as the calibration reference. DNA-DNA hybridizations were performed by using a microplate method modified from Ezaki et al. (9) as described by Willems et al. (60), with hybridizations carried out at 45°C.
RESULTS AND DISCUSSION
Nodulation of Lupinus.
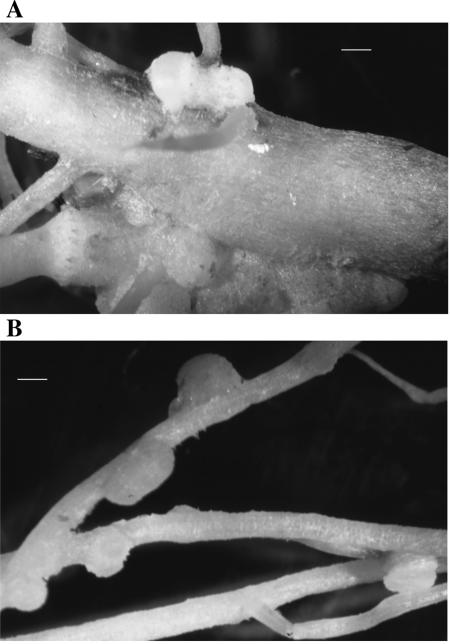
The fast-growing isolates LUP21T and LUP23 were able to nodulate Lupinus albus plants that had a lower number of nodules than those inoculated with strain ISLU35 used as a positive control. The strains LUP21T and LUP23 induced a mean of 8 nodules per plant, whereas Bradyrhizobium sp. strain ISLU35 used as control formed a mean of 14 nodules per plant (Table 1). This was a significant difference at P = 0.05 according to Fisher protected LSD analysis. The morphology of nodules induced by strains isolated in the present study (Fig. 1A) was different from that of nodules induced by strain ISLU35 (Fig. 1B). The nodules induced by the LUP strains were formed at the intersection of the main and secondary roots, whereas those induced by strain ISLU35 were formed along the secondary roots. At 6 weeks postinoculation, no significant differences were observed in the dry matter of plants inoculated with strain ISLU35 and the strains from the present study (Table 1). Significant differences were observed in the dry matter of plants inoculated with the strains from the present study and the negative controls, suggesting that strains LUP21T and LUP23 are able to fix nitrogen in symbiosis with L. albus.
TABLE 1.
Symbiotic characteristics of the strains from this study
| Treatment | Nodule location | No. of nodules/planta | Shoot dry matterb (g) |
|---|---|---|---|
| Control without N | None | 0 | 0.29 |
| O. lupini LUP21T | At the intersections of the main and secondary roots | 8 | 0.44 |
| O. lupini LUP23 | At the intersections of the main and secondary roots | 7 | 0.42 |
| Bradyrhizobium sp. strain ISLU35 | Along secondary roots | 14 | 0.41 |
The number of nodules per plant was significantly different between the LUP isolates and strain ISLU35 at P = 0.05 according to Fisher protected LSD analysis. Both were also significantly different from the control.
The shoot dry matter was not significantly different between the LUP isolates and strain ISLU35 at P = 0.05 according to Fisher protected LSD analysis. They were significantly different from the control.
FIG. 1.
Nodules induced by strain LUP21T (A) and Bradyrhizobium sp. strain ISLU35 (B) in L. albus roots. Bars, 0.2 cm.
nodD and nifH amplification and phylogenetic analysis of strain LUP21T.
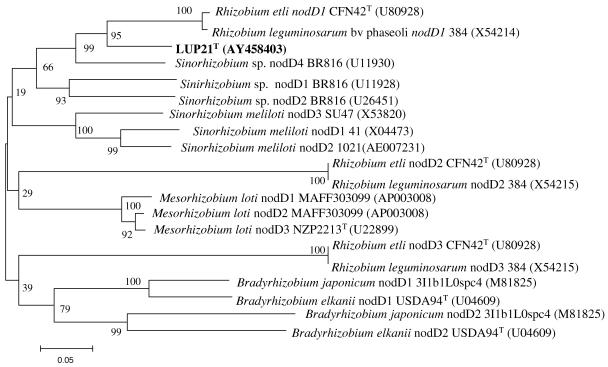
The nodD and NifH genes (accession numbers AY458403 and AY458402, respectively) of strain LUP21T were amplified and sequenced by using specific primers (40). The phylogenetic analysis of the sequences obtained is shown in Fig. 2 and 3. The nodD gene sequence of strain LUP21T showed 86.4% similarity with respect to the nodD gene sequence of Rhizobium etli CFN42T. The sequence of the nifH gene showed 99.6% similarity with several strains of Mesorhizobium ciceri reported to nodulate chickpea in Africa (25). These results differ from those obtained previously based on nifD sequences from Bradyrhizobium strains that showed a great degree of local divergence (36); nevertheless, additional studies including other rhizobial species from various geographical locations should be performed to establish definitive conclusions about the relation between the phylogeny of nif genes and the geographical origin of strains carrying these genes.
FIG. 2.
Comparative sequence analysis of nodD gene sequences from strain LUP21T and representative related strains from GenBank. The significance of each branch is indicated by a bootstrap value calculated for 1,000 subsets. The analysis was based on 376 nt. Bar, 5 nt substitutions per 100 nt.
FIG. 3.
Comparative sequence analysis of nifH gene sequences from strain LUP21T and representative related strains from GenBank. The significance of each branch is indicated by a bootstrap value for 1,000 subsets. The analysis was based on 438 nt. Bar, 5 nt substitutions per 100 nt.
Plasmid profile analysis and location of nifH and nodD genes.
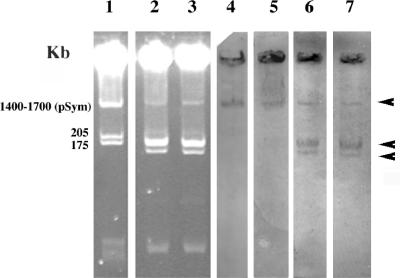
In fast-growing species nodulating legumes, the symbiotic genes encoding for nodulation and nitrogen fixation are located on plasmids. These symbiotic plasmids can be transferred in the rhizosphere conferring the ability to nodulate legumes to nonrhizobial species (32, 40, 45). Figure 4 shows the plasmid profile of strains LUP21T and LUP23 (lanes 2 and 3) carrying a megaplasmid of more than 1,700 kb (plasmid A) and two megaplasmids of ca. 200 kb (plasmid B) and 150 kb (plasmid C), respectively. The probes used allowed the detection of both nodD and nifH genes in the three plasmids carried by the strains isolated in the present study (Fig. 4, lanes 6 and 7).
FIG. 4.
Plasmid profile in a horizontal 0.7% agarose gel. Lanes: 1, S. meliloti GR4; 2, strain LUP21T; 3, LUP23. The results of hybridization (marked by arrows), using the nodD probe, for strain GR4 (lane 4) and strains LUP21T and LUP23 (lane 6) are also shown. The results of the hybridization using the nifH probe for strain GR4 (lane 5) and strains LUP21T and LUP23 (lane 7) are as indicated.
rRNA sequencing and analysis.
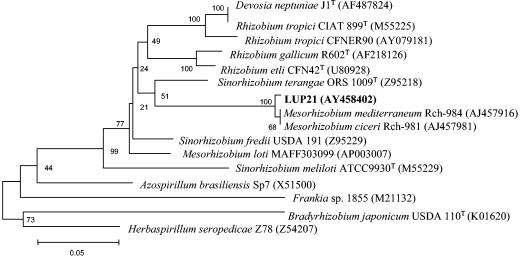
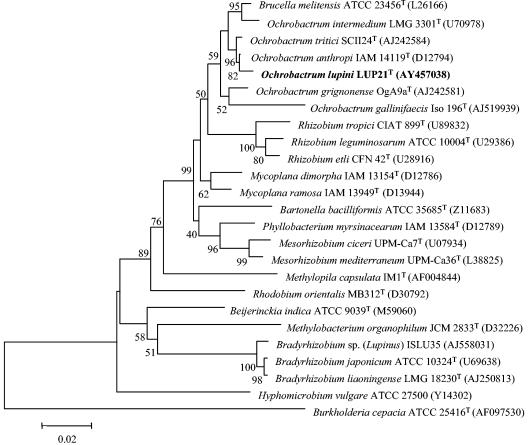
A single colony of each strain was used for all molecular analyses. The complete 16S rRNA sequences of strains LUP21T (accession number AY457038) and LUP23 were obtained before and after their reisolation from Lupinus nodules to check the purity of the strains. They were also compared to sequences from the public databases. The 16S rRNA sequence of strain LUP23 showed 100% identity with that of the strain LUP21T. Isolates LUP21T and LUP23 were tentatively identified as members of the genus Ochrobactrum after a comparison with the sequences deposited in the GenBank. This genus is located in the order Rhizobiales (α2 subclass of Proteobacteria), in the family Brucellaceae, and does not contain nitrogen fixing, legume-nodulating species (Fig. 5). The closest relatives found were O. anthropi with 99.5% sequence similarity, followed by O. tritici with 99.3% sequence similarity.
FIG. 5.
Comparative sequence analysis of 16S rRNA from strain LUP21T and representative related strains from GenBank. The significance of each branch is indicated by a bootstrap value for 1,000 subsets. The analysis was based on 1,481 nt. Bar, 2 nt substitutions per 100 nt.
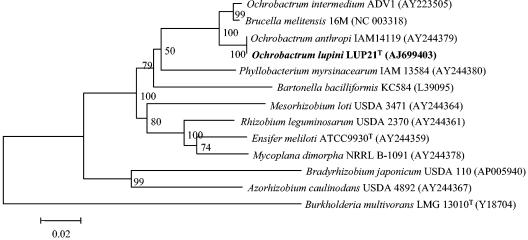
The results of 16S rRNA sequencing clearly showed that the strains from the present study do not belong to the rhizobial genera described until now. However, it was recently reported that fragments of 16S rRNA may be transferred among bacteria, including rhizobia, by lateral gene transfer, creating mosaic genes in which different segments have affinities to different relatives (51, 57), obscuring classification on the basis of this molecule. The 16S rRNA sequence of the strains isolated in the present study notably differs from all rhizobial species along the complete gene. For example, with respect to R. leguminosarum ATCC 10004T and B. japonicum LMG 6138T, variations are located along the complete 16S rRNA gene showing differences in 80 and 151 nucleotides (nt), respectively. These observations exclude the possibility of horizontal transfer of a 16S rRNA fragment from Ochrobactrum to the strains from the present study. Nevertheless, a partial sequence of the 23S rRNA gene (1,890 nt, corresponding to E. coli positions 208 to 2233 [10]) was obtained to confirm the results obtained by 16S rRNA sequencing. The sequences of strains LUP21T (accession number AJ699403) and LUP23 were identical. A comparison with the sequences held in the GenBank showed that the sequence obtained was 99.8% similarity to that of O. anthropi LMG 3331T. Figure 6 shows the phylogenetic location of strain LUP21T within the family Brucellaceae. This result confirms that the isolates from the present study belong to the genus Ochrobactrum and are distinct from rhizobial genera currently reported.
FIG. 6.
Comparative sequence analysis of 23S rRNA from strain LUP21T and representative related strains from GenBank. The significance of each branch is indicated by a bootstrap value for 1,000 subsets. The analysis was based on 1,890 nt. Bar, 2 nt substitutions per 100 nt.
Very recently a strain classified in the genus Ochrobactrum (Ochrobactrum sp. strain DASA 35030) was reported as an endosymbiont of Acacia mangium; however, its taxonomic status within the genus was not fully established (33). When the deposited 16S rRNA sequence of strain DASA 35030 with 1,151 nt was compared to the sequence of LUP21T, the similarity found was to be 93.1%.
Phenotypic characterization.
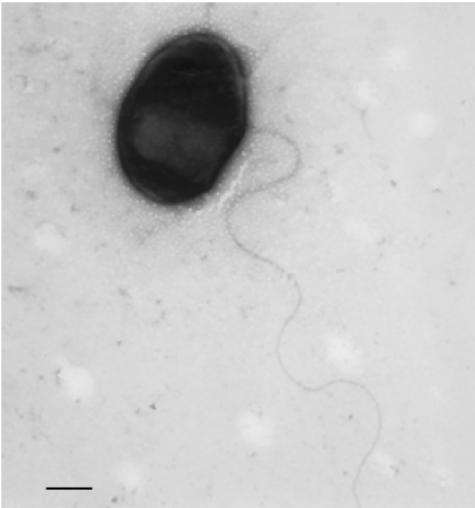
Strains LUP21T and LUP23 were gram-negative short-rods and motile by a polar flagellum (1.4 to 1.5 μm by 0.2 to 0.4 μm). Figure 7 shows the cell morphology of isolate LUP21T as observed by transmission electronic microscope. Cells form white mucoid colonies on YMA medium. The generation times were 3 to 4 h in YMB, and both strains were catalase and oxidase positive.
FIG. 7.
Transmission electron micrograph of strain LUP21T grown in nutrient agar for 48 h. Bar, 0.7 μm.
The phenotypic characteristics of strains LUP21T and LUP23 were compared to those of other Ochrobactrum species and various representatives from rhizobial genera. Differentiating physiological characteristics are presented in Table 2. According to the data, the strains from the present study clearly differ from those representing the genera Rhizobium, Mesorhizobium, and Bradyrhizobium. Isolates LUP21T and LUP23 showed a growth rate similar to that of other Ochrobactrum species, which is faster than rhizobial strains. LUP21T and LUP23 differed from their phylogenetic closest neighbors, O. anthropi and O. tritici, in nitrate reduction and d-turanose and d-arabitol assimilation (which were negative for strains LUP21T and LUP23) and in esculin hydrolysis and melibiose and citrate assimilation (which were positive for these strains). In addition to these differences, the strains from the present study also differed from O. anthropi in resistance to polymyxin B, which was positive for strains LUP21T and LUP23, and from O. tritici in their ability to assimilate mannose and l-arabinose. The differences with respect to the remaining Ochrobactrum species and representative strains of Bradyrhizobium, Mesorhizobium, and Rhizobium are recorded in Table 2.
TABLE 2.
Differentiating physiological characteristicsa
| Parameter | Physiological characteristics of straina:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | |
| Reference | This study | This study | 11 | 23 | 23 | 54 | 17 | 20, 61, 62 | 20, 62 | 34, 35 | 55 | 43, 63 | 27, 63 |
| Type of flagellation | Polar | Polar | Subpolar or peritrichous | Subpolar or peritrichous | Subpolar or peritrichous | Polar | Nonmotile | Polar or subpolar | ND | ND | ND | ND | Subpolar or peritrichous |
| Incubation time at 28°C in YMA or TSA | 12-24 h | 12-24 h | 12-24 h | 12-24 h | 12-24 h | 12-24 h | 12-24 h | 4-7 days | 6-10 days | 3-5 days | 4-5 days | 2-3 days | 1-2 days |
| Growth at 40°C | w | w | v | + | + | ND | ND | − | − | + | + | − | + |
| Growth at pH 10 | + | + | ND | − | − | ND | ND | − | − | + | − | − | − |
| Growth in 2% NaCl | + | + | ND | ND | ND | ND | ND | − | − | + | + | − | + |
| β-Galactosidase | − | − | − | − | − | − | − | − | −c | − | − | + | + |
| Urease | + | + | + | − | + | + | ND | ND | ND | ND | ND | + | + |
| Nitrate reduction | − | − | + | + | + | + | ND | + | ND | ND | ND | ND | + |
| Arginine dehydrolase | − | − | ND | − | − | − | ND | ND | ND | ND | ND | ND | −f |
| Esculin | + | + | − | +b | − | v | − | v | − | ND | ND | ND | +f |
| Assimilation of: | |||||||||||||
| l-Arabinose | + | + | + | + | − | − | + | + | + | + | + | + | +f |
| Adipate | − | + | v | − | − | − | − | ND | ND | ND | ND | ND | +f |
| Citrate (24h) | + | + | − | − | − | + | − | v | v | ND | ND | −e | Vh |
| Phenyl-acetate | + | − | − | − | − | − | − | ND | ND | ND | ND | ND | −f |
| N-Acetylglucosamine | + | + | + | −b | + | +b | − | ND | ND | + | + | ND | +f |
| d-Mannose | + | + | + | + | − | v | + | + | + | + | + | + | +f,g |
| Maltose | + | + | + | − | + | − | − | + | − | + | + | +g | + |
| Rhamnose | + | + | + | + | + | −b | − | + | v | + | + | ND | ND |
| Mannitol | + | + | + | − | +c | − | ND | + | v | + | + | + | + |
| d-Turanose | − | − | + | + | + | ND | ND | + | v | + | + | ND | ND |
| Melibiose | + | + | − | + | − | − | − | v | − | + | + | ND | ND |
| d-Arabitol | − | − | + | + | + | ND | ND | + | v | + | + | +i | vi |
| Antibiotic resistance | |||||||||||||
| Chloramphenicol | S-I | S-I | R | S-I | I | R | ND | I | I | R | S | Sj | Rk |
| Polymyxin B (300 IU) | R | R | S | R | S | v | ND | Rd | ND | R | S | Sg | Rf,g |
| G+C content (mol%) | 56.8 | 57 | 56-59 | 58 | 59 | 57-59 | ND | 61-65 | 60-64 | 63-64 | 63-64 | 59-64 | 59-64 |
Strains: 1, O. lupini LUP21T (LMG 22726T); 2, O. lupini LUP23 (LMG 22727); 3, O. anthropi; 4, O. grignonense; 5, O. tritici; 6, O. intermedium; 7, O. gallinifaecis; 8, B. japonicum; 9, B. liaoningense; 10, M. ciceri; 11, M. mediterraneum; 12, Rhizobium etli; 13, R. tropici. +, Positive; −, negative; v, variable; w, weak; ND, no data; S, sensitive; R, resistant; I, intermediate.
Kämpfer et al. (17).
This study.
Yao et al. (62).
Zurdo-Piñeiro et al. (64).
van Berkum et al. (49).
Amarger et al. (2).
Wei et al. (59).
Vincent (56).
Fatty acid analysis.
The cellular fatty acid patterns of strains LUP21T and LUP23 and other Ochrobactrum and rhizobial strains are recorded in Table 3. The profile is dominated for all Ochrobactrum strains by C18:1ω7c which makes up 55 to 70% of the fatty acid methyl esters. Also present in all Ochrobactrum strains tested, in quantities of more than 2% are the straight chain fatty acids C16:0 and C18:0, the branched chain C18:1 2OH and the cyclic fatty acid C19:0 cyclo ω8c. The hydroxy fatty acid C18:1 2OH, present (at least 2%) in all Ochrobactrum strains tested, is not present in significant amounts in the genera Bradyrhizobium, Mesorhizobium, Sinorhziobium, or Rhizobium (13, 47). The overall fatty acid profiles of the new isolates support their inclusion in the genus Ochrobactrum.
TABLE 3.
Fatty acid profile of strains LUP21T and LUP23 compared to those of the remaining species from the genus Ochrobactrum and to those of the representative rhizobial species
| Fatty acid | Fatty acid profile compared to straina:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1b | 2b | 3b | 4b | 5b | 6b | 7c | 8d | 9d | 10d | 11d | 12d | 13d | |
| 10:0 3OH | 0.3 | ||||||||||||
| 11:0 ISO 3OH | 1.5 | ||||||||||||
| 12:0 2OH | 0.5 | 0.2 | |||||||||||
| 12:0 3OH | 0.8 | 0.5 | <0.1 | 0.3 | 0.1 | ||||||||
| 13:0 iso 3OH | 0.2 | ||||||||||||
| 14:0 | 0.2 | 0.7 | <0.1 | 0.4 | |||||||||
| 15:0 2OH | 0.7 | ||||||||||||
| 15:0 iso 3OH | 3.4 | ||||||||||||
| 16:0 | 4.2 | 5.5 | 7.2 | 2.7 | 3.9 | 2.5 | 8.9 | 12.0 | 14.8 | 13.4 | 10.3 | 2.5 | 7.7 |
| 16:0 3OH | 0.7 | 0.4 | 1.2 | 3.9 | |||||||||
| 16:1 ω5c | 1.1 | 0.9 | |||||||||||
| 17:0 | 1.6 | 1.1 | 1.6 | 1.6 | 0.9 | 2.3 | 0.2 | 0.8 | 1.3 | 1.7 | 0.3 | <0.1 | |
| 17:0 iso | 4.2 | 4.2 | 0.9 | ||||||||||
| 17:0 iso 3OH | 0.3 | ||||||||||||
| 17:0 cyclo | 0.8 | 0.9 | 0.7 | ||||||||||
| 17:1ωw6c | 1.4 | 1.2 | 0.4 | 1.1 | 0.2 | 0.7 | |||||||
| 17:1ω8c | 0.2 | 0.5 | 1 | 0.2 | 0.5 | <0.1 | |||||||
| 18:0 | 3.0 | 2.6 | 5.2 | 5.4 | 3.5 | 3.2 | 3.7 | 0.6 | 0.4 | 3.3 | 4.2 | 8.7 | 2.9 |
| 18:0 3OH | 1.0 | 0.8 | 0.5 | 1.9 | |||||||||
| 18:1 2OH | 6.2 | 13.4 | 2.4 | 2.6 | 12.9 | 3.9 | 1.5 | 0.8 | |||||
| 18:1ω7c | 70.8 | 59.5 | 62.8 | 68.3 | 68.6 | 55.8 | 28.8 | ||||||
| 11 methyl 18:1ω7c | 1.9 | 11.9 | 5.6 | 1.5 | 0.6 | ||||||||
| 19:0 10 methyl | 0.7 | 0.8 | 0.2 | 0.1 | <0.1 | ||||||||
| 19:0 cyclo ω8c | 4.3 | 8.3 | 14.5 | 16.4 | 6.3 | 30.5 | 47.2 | 1.2 | 5.0 | 37.3 | 33.4 | 10.2 | 49.3 |
| 20:1ω9t | 0.1 | 0.5 | 1 | ||||||||||
| 20:1ω7c | 0.4 | ||||||||||||
| 20:2ω6,9c | 0.1 | 1 | 0.8 | 0.4 | |||||||||
| 20:3ω6,9,12c | 2 | 1.8 | |||||||||||
| Summed feature 1f | 0.5 | 0.2 | |||||||||||
| Summed feature 2g | 1.9 | 1.4 | 1.6 | 0.6 | 6.0 | 2.3 | |||||||
| Summed feature 3h | 2.3 | 3.2 | 2.1 | 1.5 | 1.4 | 0.8 | 3.7 | 1.1 | 0.4 | 0.3 | 0.2 | 0.2 | |
| Summed feature 7i | 81.2 | 75.5 | 24.7 | 35.8 | 66.2 | 24.6 | |||||||
| Unknown 11.799 | 0.4 | ||||||||||||
| Unknown 18.814 | 1.2 | ||||||||||||
Strains: 1, O. lupini LUP21T (LMG 22726T); 2, O. lupini LUP23 (LMG 22727); 3, O. anthropi LMG 3331T; 4, O. grignonense LMG 18954T; 5, O. tritici LMG 18957T; 6; O. intermedium LMG 3301T; 7, O. gallinifaecis DSM 15295T; 8, B. japonicum; 9, B. liaoningense; 10, M. ciceri; 11; M. mediterraneum; 12, R. etli; 13, R. tropici.
Data from this study.
Kämpfer et al. (17).
Tighe et al. (47).
Fatty acid with values lower than 0.1% are not shown.
Summed feature 1: 13:0 OH/15:1 iso I/15:1 iso H.
Summed feature 2: 12:0 unknown aldehyde with ECL 10.928/16:1 iso I/14:0 3OH.
Summed feature 3: 16:1 ω7c/15:0 iso 2OH.
Summed feature 7: 18:1 ω7cis/ω9trans/ω12trans, 18:1ω7cis/ω9cis/ω12trans.
SDS-PAGE of whole-cell proteins.
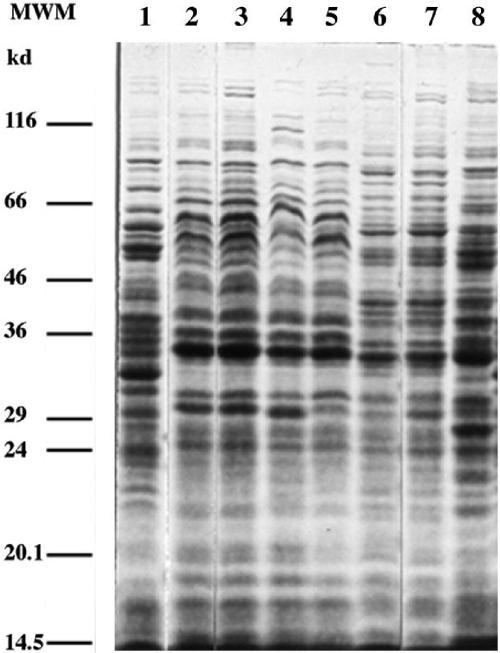
The protein profiles (Fig. 8) for strains LUP21T and LUP23 were nearly identical, indicating that both strains most probably belong to the same species. Their protein profile was similar to that of O. anthropi LMG 3331T but had some differences in the higher-molecular-weight part of the profile. It had some more differences with the protein pattern of O. tritici LMG 18957T and was clearly different from that of O. intermedium LMG 3301T and O. grignonense LMG 18954T. These results are in line with the fact that, also according to 16S rRNA sequence similarities, O. anthropi (and O. tritici) are the nearest neighbors of the Lupinus isolates.
FIG. 8.
SDS-PAGE protein profiles of O. grignonense LMG 18954T (lane 1), O. lupini LUP21T (lane 2), LUP23 (lane 3), O. anthropi 3331T (lane 4), O. anthropi LMG 371 (lane 5), O. tritici LMG 18957T (lane 6), O. tritici MG 18958 (lane 7), and O. intermedium LMG 3301T (lane 8). Molecular weight markers (MWM): lysozyme, 14.5 kDa; trypsin inhibitor, 20.1 kDa; trypsinogen, 24 kDa; carbonic anhydrase, 29 kDa; GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 36 kDa; egg albumin, 45 kDa; bovine albumin, 66 kDa; β-galactosidase, 116 kDa.
TP-RAPD patterns.
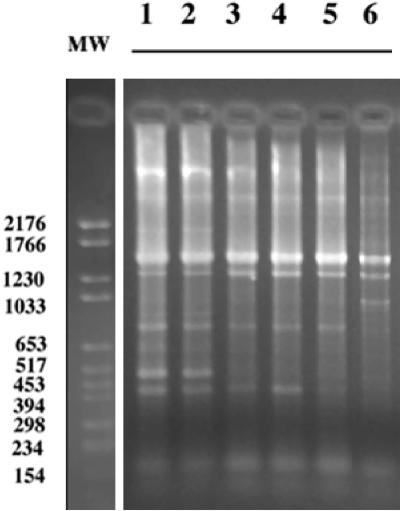
In a previous study we showed that TP-RAPD patterns allow the differentiation among rhizobial species (39) and that, within a species, the TP-RAPD pattern is identical for different strains. Figure 9 shows the TP-RAPD patterns of the strains isolated in the present study compared to those of O. anthropi and O. tritici. As expected, strains LUP21T and LUP23 showed the same pattern (Fig. 9, lanes 1 and 2), which differed from the patterns of O. anthropi (lanes 3, 4, and 5) and O. tritici (lane 6). Nevertheless, the profiles of LUP21T and LUP23 were closer to that of O. anthropi since only one difference between these profiles was found.
FIG. 9.
TP-RAPD profiles of O. lupini LUP21T (lane 1), O. lupini LUP23 (lane 2), O. anthropi LMG 3331T (lane 3), O. anthropi LMG 371T (lane 4), O. anthropi LMG 33 (lane 5), O. tritici LMG 18957T (lane 6). MW, Standard VI of Boehringer-Roche.
DNA-DNA hybridization.
The results of DNA-DNA hybridization were 95% between strain LUP21T and LUP23. Hybridization of these strains with labeled DNA of O. anthropi LMG 3331T resulted in 68 and 65% reassociation, respectively. The second strain, O. anthropi LMG 371 confirmed this observation, with 65% hybridization with LUP21T and LUP23. Both O. anthropi strains showed 86% hybridization. In light of the recommendation of a threshold value of 70% DNA-DNA similarity for definition of a species (58), these results indicate that strains LUP21T and LUP23 can be regarded as a separate genospecies, distinct from O. anthropi, although closely related.
In conclusion, the 16S rRNA and 23S rRNA sequences demonstrate that the isolates from Lupinus described here are phylogenetically unrelated to the rhizobia and represent a new species of the family Brucellaceae. To our knowledge, this is the first report of a member of this family with the ability to nodulate the legume Lupinus. The high similarity of the nodD and nifH sequences of strain LUP21T to the corresponding genes of rhizobial species suggests that this strain may have acquired these symbiotic genes by horizontal transfer from rhizobia. This new finding of a nonrhizobial species nodulating legumes is in line with recent reports of other new species outside the rhizobia, in the genera Devosia (40, 41) Methylobacterium (16, 45), Ralstonia (6), Blastobacter (50), Ochrobactrum (33), and Burkholderia (32, 53). It demonstrates the need for more studies on the diversity of bacteria nodulating legumes from both tropical and temperate soils.
On the other hand, the genotypic, phenotypic, and chemotaxonomic data presented here support the classification of strains LUP21T and LUP23 as new species of the genus Ochrobactrum for which the name O. lupini sp. nov. is proposed with isolate LUP21T representing the type strain. The strains were deposited in the LMG culture collection as LMG 22726T and LMG 22727, respectively.
Description of Ochrobactrum lupini sp. nov.
Ochrobactrum lupini (lu.pi′ni. N.L. masc. lupinus legume, N.L. gen. nov. lupini of the legume, referring to the isolation source of this microorganism, nodules of L. albus).
Cells are nonmotile, non-spore-forming, gram-negative rods. Good growth occurs on yeast-mannitol agar and nutrient agar at 25 to 30°C. Colonies on these media are white to beige mucoid with entire edges and have a diameter of between 2 and 3 mm. The two strains are oxidase and catalase positive. The fatty acid profiles were mainly composed of C18:1ω7c (50 to 70%), C16:0 (4 to 5%), and C18:0 (2 to 3%). Carbon source utilization includes l-arabinose, citrate, erythritol, d-fucose, l-fucose, gluconate, 2-ketogluconate, 5-ketogluconate, glucose, N-acetylglucosamine, lactose, d-mannose, maltose, mannitol, rhamnose, melibiose, and l-xylose. The following carbon sources are not assimilated: amygdalin, d-arabinose, cellobiose, ducitol, d-fructose, galactose, glycerol, glycogen, inositol, inulin, melezitose, α-methyl-d-glucoside, α-methyl-d-mannoside, d-raffinose, salicin, sorbitol, l-sorbose, trehalose, and turanose. Both strains hydrolyze esculin and urease. Various physiological differences between the two strains and other Ochrobactrum species are given in Table 2. The G+C contents of strains LUP21T and LUP23 were 56.8 and 57 mol%, respectively. The strains nodulate L. honoratus and L. albus. The type strain is LUP21T (LMG 22726T).
Acknowledgments
This study was financially supported by the MCYT (Spanish Government) grants to E.M.-M. and E.V. A.W. is grateful to the Fund for Scientific Research-Flanders for a postdoctoral fellowship.
We thank J. Gónzalez and M. Ortíz-Aranda for help with the electron microscopy preparations and R. Coopman and D. Dewettinck for excellent technical assistance.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amarger, N., V. Macheret, and G. Laguerre, G. 1997. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int. J. Syst. Bacteriol. 47:996-1006. [DOI] [PubMed] [Google Scholar]
- 3.Barrera, L. L., M. E. Trujillo, M. Goodfellow, F. J. García, I. Hernández-Lucas, G. Dávila, P. van Berkum, and E. Martínez-Romero. 1997. Biodiversity of Bradyrhizobia nodulating Lupinus spp. Int. J. Syst. Bacteriol. 47:1086-1091. [DOI] [PubMed] [Google Scholar]
- 4.Bergersen, F. J. 1961. The growth of Rhizobium in synthetic media. Australian J. Biol. 14:349-360. [Google Scholar]
- 5.Bottomley, P., H. H. Cheng, and S. R. Strain. 1994. Genetic structure and symbiotic characteristics of a Bradyrhizobium population recoverd from a pasture soil. Appl. Environ. Microbiol. 60:1754-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, W. M., S. Laevens, T. M. Lee, T. Coenye, P. de Vos, M. Mergeay, and P. Vandamme. P. 2001. Ralstonia taiwanensis sp. nov. isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 51:1729-1735. [DOI] [PubMed] [Google Scholar]
- 7.de Lajudie, P., A. Willems, B. Pot, D. Dewettinck, G. Maestrojuan, M. Neyra, M. D. Collins, B. Dreyfus, K. Kersters, and M. Gillis. 1994. Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int. J. Syst. Bacteriol. 44:715-733. [Google Scholar]
- 8.Doetsch, R. N. 1981. Determinative methods of light microscopy, p. 21-33. In P. Gerdhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G.B (ed.), Phillips manual of methods for general bacteriology. American Society for Microbiology, Washington, D.C.
- 9.Ezaki, T., Y. Hashimoto, and E. Yabuuchi. 1989. Fluorometric deoxyribonucleic acid -deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39:224-229. [Google Scholar]
- 10.Guttel, R. R., N. Larsen, and C. R. Woese. 1994. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol. Rev. 58:1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes, B., M. Popoff, M. Kiredjian, and K. Kersters. 1988. Ochrobactrum anthropi gen. nov., sp. nov. from human clinic specimens and previously known as group Vd. Int. J. Syst. Bacteriol. 38:406-416. [Google Scholar]
- 12.Jarabo-Lorenzo, A., R. Pérez-Galdona, J. Donate-Correa, R. Rivas, E. Velázquez, M. Hernández, F. Temprano, E. Martínez-Molina, T. Ruiz-Argüeso, and M. León-Barrios. 2003. Genetic diversity of bradyrhizobial populations from diverse geographic origins that nodulate Lupinus spp. and Ornithopus spp. Syst. Appl. Microbiol. 26:611-623. [DOI] [PubMed] [Google Scholar]
- 13.Jarvis, B. D. W., S. Sivakumaran, S. W. Tighe, and M. Gillis. 1996. Identification of Agrobacterium and Rhizobium species based on cellular fatty acid composition. Plant Soil 184:143-158. [Google Scholar]
- 14.Jordan, D. C. 1984. Family III. Rhizobiaceae, p. 235-255. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. The Williams & Wilkins Co., Baltimore, Md.
- 15.Jordan, D. C., and O. N. Allen. 1974. Family III. Rhizobiaceae, p. 261-264. In R. E. Buchanan and N. E. Gibbons (ed.), Bergey's manual of determinative bacteriology, 8th ed. The Williams & Wilkins Co., Baltimore, Md.
- 16.Jourand, P., E. Giraud, G. Bena, A. Sy, A. Willems, M. Gillis, B. Dreyfus, and P. de Lajudie. 2004. Methylobacterium nodulans sp. nov., for a group of aerobic, factultatively methylotrophic, legume root-nodule forming and nitrogen-fixing bacteria. Int. J. Syst. Evol. Microbiol. 54:2269-2273. [DOI] [PubMed] [Google Scholar]
- 17.Kämpfer, P., S. Buczolits, A. Albrecht, H. J. Bussse, and E. Stackebrandt. 2003. Towards a standarized format for the description of a novel species (of an established genus): Ochrobactrum gallinifaecis sp. nov. Int. J. Syst. Evol. Microbiol. 53:893-896. [DOI] [PubMed] [Google Scholar]
- 18.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, S., K. Tamura, K., I. B. Jakobsen, and M. Nei. 2001. Molecular evolutionary genetics analysis software. Arizona State University, Tempe, Ariz.
- 20.Kuykendall, L. D., M. A. Roy, J. J. O'Neill, and T. E. Devine. 1988. Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int. J. Syst. Bacteriol. 38:358-361. [Google Scholar]
- 21.Kuykendall, L. D., B. Saxena, T. E. Devine, and S. E. Udell. 1991. Genetic diversity in Bradyrhizobium japonicum Jordan 1982 and a proposal for Bradyrhizobium elkanii sp. nov. Can. J. Microbiol. 38:501-505. [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lebuhn, M., W. Achouak, M. Schloter, O. Berge, H. Meier, M. Barakat, A. Hartmann, and T. Heulin. 2000. Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int. J. Syst. Evol. Microbiol. 50:2207-2223. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, W., R. Roselló-Mora, R. Aznar, S. Klugbauer, S. Springs, K. Reetz, C. Beimfohr, E. Brockmann, G. Kirchhof, S. Dorn, M. Bachleitner, N. Klugbauer, N. Springer, D. Lane, R. Nietupsky, M. Weizenegger, and K. H. Schleifer. 1995. Comparative sequence analysis of 23S rRNA from Proteobacteria. Syst. Appl. Microbiol. 18:164-188. [Google Scholar]
- 25.MaÂtallah, J., E. B. Berraho, S. Muñoz, J. Sanjuán, and C. Lluch. 2002. Phenotypic and molecular characterization of chickpea rhizobia isolated from different areas of Morocco. J. Appl. Microbiol. 93:531-540. [DOI] [PubMed] [Google Scholar]
- 26.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 27.Martínez-Romero, E., L. Segovia, F. M. Mercante, A. A. Franco, P. Graham, P., and M. A. Pardo. 1991. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int. J. Syst. Bacteriol. 41:417-426. [DOI] [PubMed] [Google Scholar]
- 28.Mesbah, M., U. Premachandran, and W. B. Whitman. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159-167. [Google Scholar]
- 29.MIDI. 1999. Sherlock microbial identification system operating manual, version 3. MIDI, Inc., Newark, Del.
- 30.Miller, M. S., and I. L. Pepper. 1988. Survival of a fast-growing strain of lupin rhizobia in Sonoran desert soils. Soil. Biol. Biochem. 20:323-327. [Google Scholar]
- 31.Miller, L. T. 1982. A single derivatization method for bacterial fatty acid methyl esters including hydroxy acids. J. Clin. Microbiol. 16. 16:584-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moulin, L., A. Munive, B. Dreyfus, and C. Boivin-Masson. 2001. Nodulation of legumes by members of β-subclass of Proteobacteria. Nature 411:948-950. [DOI] [PubMed] [Google Scholar]
- 33.Ngom, A., Y. Nakagawa, H. Sawada, J. Tsukahara, S. Wakabayashi, T. Uchiumi, A. Nuntagij, S. Kotepong, A. Suzuki, S. Higashi, and M. Abe. 2004. A novel symbiotic nitrogen-fixing member of the Ochrobactrum clade isolated from root nodules of Acacia mangium. J. Gen. Appl. Microbiol. 50:17-27. [DOI] [PubMed] [Google Scholar]
- 34.Nour, S. M., M. P. Fernández, P. Normand, and J. C. Cleyet-Marel. 1994. Rhizobium ciceri sp. nov., consisting of strains that nodulate chickpeas (Cicer arietinum, L.). Int. J. Syst. Bacteriol. 44:511-522. [DOI] [PubMed] [Google Scholar]
- 35.Nour, S. M., J. C. Cleyet-Marel, P. Normand, P., and M. P. Fernández. 1995. Genomic heterogeneity of strains nodulating chickpeas (Cicer arietinum, L.) and description of Rhizobium mediterraneum sp. nov. Int. J. Syst. Bacteriol. 45:640-648. [DOI] [PubMed] [Google Scholar]
- 36.Parker, M. A., B. Lafay, J. J. Burdon, and P. van Berkum. 2002. Conflicting phylogeographic patterns in rRNA and nifD indicate regionally restricted gene transfer in Bradyrhizobium. Microbiology 148:2557-2565. [DOI] [PubMed] [Google Scholar]
- 37.Plazinski, J., Y. H. Chen, and B. G. Rolfe. 1985. General method for the identification of plasmid species in fast-growing soil microorganisms. Appl. Environ. Microbiol. 48:1001-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigaud, J., and A. Puppo. 1975. Indole-3-acetic catabolism by soybean bacteroids. J. Gen. Microbiol. 8:223-228. [Google Scholar]
- 39.Rivas, R., E. Velázquez, A. Valverde, P. F. Mateos, and E. Martínez-Molina. 2001. A two primers random amplified polymorphic DNA procedure to obtain polymerase chain reaction fingerprints of bacterial species. Electrophoresis 22:1086-1089. [DOI] [PubMed] [Google Scholar]
- 40.Rivas, R., E. Velázquez, A. Willems, N. Vizcaino, N. S. Subba-Rao, P. F. Mateos, M. Gillis, F. B. Dazzo, and E. Martínez-Molina, E. 2002. A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (L.f.) druce. Appl. Environ. Microbiol. 68:5217-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rivas, R., A. Willems, N. S. Subba-Rao, P. F. Mateos, F. B. Dazzo, E. Martínez-Molina, M. Gillis, and E. Velázquez. 2003. Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst. Appl. Microbiol. 26:47-53. [DOI] [PubMed] [Google Scholar]
- 42.Saitou, N., and M. Nei. 1987. A neighbor-joining method: a new method for reconstructing phylogenetics trees. Mol. Biol. Evol. 44:406-425. [DOI] [PubMed] [Google Scholar]
- 43.Segovia, L., J. P. W. Young, and E. Martínez-Romero. 1993. Reclassification of American Rhizobium leguminosarum biovar phaseoli type I strains as Rhizobium etli sp. nov. Int. J. Syst. Bacteriol. 43:374-377. [DOI] [PubMed] [Google Scholar]
- 44.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 45.Sy, A., E. Giraud, P. Jourand, N. García, A. Willems, P. de Lajudie, Y. Prin, M. Neyra, M. Gillis, C. Boivin-Masson, and B. Dreyfus. 2001. Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 183:214-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1987. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tighe, S. W., P. de Lajudie, K. Dipietro, K. Lindström, G. Nick, and B. D. W. Jarvis. 2000. Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesothizobium, Rhizobium, and Sinorhizobium species using the Sherlock Microbial Identification Systems. Int. J. Syst. Evol. Microbiol. 50:787-801. [DOI] [PubMed] [Google Scholar]
- 48.Toro, N., and J. Olivares. 1986. Characterization of a large plasmid of Rhizobium meliloti involved in enhancing nodulation. Mol. Gen. Genet. 202:331-335. [Google Scholar]
- 49.van Berkum, P., D. Beyene, G. Bao, T. A. Campbell, and B. D. Eardly. 1998. Rhizobium mongolense sp. nov. is one of three genotypes identified which nodulate and form nitrogen-fixing symbioses with Medicago ruthenica [(L.) Ledebour]. Int. J. Syst. Bacteriol. 48:13-22. [DOI] [PubMed] [Google Scholar]
- 50.van Berkum, P., and B. D. Eardly. 2002. The aquatic budding bacterium Blastobacter denitrificans is a nitrogen-fixing symbiont of Aeschynomene indica. Appl. Environ. Microbiol. 68:1132-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Berkum, P., Z. Terefework, L. Paulin, S. Suomalaien, K. Lindström, and B. D. Eardly. 2003. Discordant phylogenies within the rrn loci of Rhizobia. J. Bacteriol. 185:2988-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Camp, G., S. Chapelle, and R. De Wachter. 1993. Amplification and sequencing of variable regions in bacterial 23S rRNA genes with conserved primer sequences. Curr. Microbiol. 27:147-151. [DOI] [PubMed] [Google Scholar]
- 53.Vandamme, P., J. Goris, W. M., Chen, P. de Vos, and A. Willems. 2003. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov. nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25:507-512. [DOI] [PubMed] [Google Scholar]
- 54.Velasco, J., C. Romero, I. López-Goñi, J. Leiva, R. Díaz, and I. Moriyón. 1998. Evaluation of the relatedness of Brucella spp. and Ochrobactrum anthropi and description of Ochrobactrum intermedium sp. nov. a new species with a closer relationship to Brucella spp. Int. J. Syst. Bacteriol. 48:759-768. [DOI] [PubMed] [Google Scholar]
- 55.Velázquez, E., J. M. Igual, A. Willems, M. P. Fernández, E. Muñoz, P. F. Mateos, A. Abril, N. Toro, P. Normand, E. Cervantes, M. Gillis, and E. Martínez-Molina. 2001. Description of Mesorhizobium chacoense sp. nov. that nodulates Prosopis alba in the Chaco Arido region (Argentina). Int. J. Syst. Evol. Microbiol. 51:1011-1021. [DOI] [PubMed] [Google Scholar]
- 56.Vincent, J. M. 1970. The cultivation, isolation and maintenance of rhizobia, p. 1-13. In J. M. Vincent (ed.), A manual for the practical study of root nodule. Blackwell Scientific Publications, Oxford, England.
- 57.Wang, Y., and Z. Zhang. 2000. Comparative sequence analyses reveal frequent occurrence of short segments containing an abnormally high number of non-random base variations in bacterial rRNA genes. Microbiology 146:2845-2854. [DOI] [PubMed] [Google Scholar]
- 58.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. L. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 59.Wei, G. H., E. T. Wang, Z. Y. Tan, M. E. Zu, and W. X. Chen. 2002. Rhizobium indigoferae sp. nov. and Sinorhizobium kummerowiae sp. nov., respectively, isolated from Indigofera spp. and Kummerowia stipulacea. Int. J. Syst. Evol. Microbiol. 52:2231-2239. [DOI] [PubMed] [Google Scholar]
- 60.Willems, A., F. Doignon-Bourcier, J. Goris, R. Coopman, P. De Lajudie, and M. Gillis. 2001. DNA-DNA hybridization study of Bradyrhizobium strains. Int. J. Syst. Evol. Microbiol. 51:1315-1322. [DOI] [PubMed] [Google Scholar]
- 61.Xu, L. M., C. Ge, Z. Cui, J. Li, and H. Fan. 1995. Bradyrhizobium liaoningense sp. nov., isolated from the root nodules of soybeans. Int. J. Syst. Bacteriol. 45:706-711. [DOI] [PubMed] [Google Scholar]
- 62.Yao, Z. Y., F. L. Kan, E. T. Wang, and W. X. Chen. 2002. Characterization of rhizobia that nodulate legume species within the genus Lespedeza and description of Bradyrhizobium yuanmingense sp. nov. Int. J. Syst. Evol. Microbiol. 52:2219-2230. [DOI] [PubMed] [Google Scholar]
- 63.Young, J. M., L. D. Kuykendall, E. Martínez-Romero, A. Kerr, and H. Sawada. 2001. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola, and R. vitis. Int. J. Syst. Evol. Microbiol. 51:89-103. [DOI] [PubMed] [Google Scholar]
- 64.Zurdo-Piñeiro, J. L., E. Velázquez, M. J. Lorite, G. Brelles-Mariño, E. C. Schröder, E. J. Bedmar, P. F. Mateos, and E. Martínez-Molina. 2004. Identification of fast-growing rhizobia nodulating tropical legumes from Puerto Rico as Rhizobium gallicum and Rhizobium tropici. Syst. Appl. Microbiol. 27:469-477. [DOI] [PubMed] [Google Scholar]