Abstract
Neurodegenerative diseases (ND) are heterogeneous disorders of the central nervous system that share a chronic and selective process of neuronal cell death. A computational approach to investigate shared genetic and specific loci was applied to 5 different ND: Amyotrophic lateral sclerosis (ALS), Alzheimer's disease (AD), Parkinson's disease (PD), Multiple sclerosis (MS), and Lewy body dementia (LBD). The datasets were analyzed separately, and then we compared the obtained results. For this purpose, we applied a genetic correlation analysis to genome-wide association datasets and revealed different genetic correlations with several human traits and diseases. In addition, a clumping analysis was carried out to identify SNPs genetically associated with each disease. We found 27 SNPs in AD, 6 SNPs in ALS, 10 SNPs in PD, 17 SNPs in MS, and 3 SNPs in LBD. Most of them are located in non-coding regions, with the exception of 5 SNPs on which a protein structure and stability prediction was performed to verify their impact on disease. Furthermore, an analysis of the differentially expressed miRNAs of the 5 examined pathologies was performed to reveal regulatory mechanisms that could involve genes associated with selected SNPs. In conclusion, the results obtained constitute an important step toward the discovery of diagnostic biomarkers and a better understanding of the diseases.
Keywords: Neurodegenerative diseases, Bioinformatics, GWAS, SNPs
Graphical Abstract
1. Introduction
Neurodegenerative diseases are chronic disorders affecting the central nervous system, causing a progressive loss of neuronal function [1]. These disorders manifest with a wide range of symptoms and signs, including problems with memory, movement, coordination, speech, and other cognitive and motor functions. They often have a degenerative and progressive course, with no effective treatments currently available to reverse their condition [1]. The mechanisms underlying the onset of neurodegenerative diseases are not always fully understood, but involve a multiplicity of factors [2]. These diseases are often characterized by the accumulation of specific proteins and anatomical fragility and exhibit several fundamental processes associated with progressive neuronal dysfunction and cell death [3]. Among these are proteotoxic stress, alterations of the ubiquitin-proteasome, autophagosome/lysosome systems, oxidative stress and neuroinflammation [3].
The most common and studied neurodegenerative diseases include Amyotrophic lateral sclerosis (ALS), Alzheimer's disease (AD), Parkinson's disease (PD), Multiple sclerosis (MS), and Lewy body dementia (LBD). Each of these diseases has unique clinical, neurobiological and pathological characteristics.
The similarities in pathological and molecular mechanisms among neurodegenerative diseases are also present at the genetic level. Previous studies applied Genome-wide association studies (GWAS) on pairs of neurodegenerative diseases to investigate genetic correlations, or overlap [4], [5]. However, the genetic variants responsible for these mechanisms are not yet clear.
ALS is a deadly neurodegenerative disease that affects approximately one person in 350, with an average age of onset between 50 and 60 years. Cases of juvenile ALS (before the age of 25) and early-onset ALS (before the age of 45) represent a small percentage (∼1% and ∼10%) of all patients [6]. Upper and lower motor neuron degeneration causes progressive paralysis, ultimately leading to respiratory failure within 3–5 years of disease onset [6]. ALS is a complex and heterogeneous disorder, with patients exhibiting variable clinical features and progression patterns [6]. Currently, the diagnostic criteria and available therapies are limited, since the complexity of ALS requires a multidisciplinary approach and the development of targeted therapies [6].
3 drugs were approved by the FDA. Riluzole, the first drug approved in 1995, acts mainly on glutamate presynaptic release reducing excitotoxic neuronal cell death [7]. It can decrease the progression of ALS, extended the survival time of ALS patients. However, the drug cannot reduce the damage to motor neurons [8].
Edaravone, approved by FDA in 2015, regulates oxidative stress process. However, some clinical trials showed contradictory results in America and Europe. Recent studies reported inconsistent benefits on survival in Italy and Germany, compared to America [9], [10], [11].
This difference may be due to genetic components based on ethnic origins.
AMX0035, third drug recently approved by FDA in 2022, acts on mitochondria and endoplasmic inhibitor decreasing neuronal death [12]. A phase 3 PHOENIX trial is occurring in the United States and Europe and it will offer more knowledge into safety and efficacy of AMX0035. NCT05021536 [13].
Recent multi-omics studies have highlighted genomic, transcriptomic, proteomic and metabolomic alterations in neuronal cells involved in ALS. At the genomic level, genome-wide association studies (GWAS) have been conducted and have identified numerous causal and susceptibility genes in ALS [14], [15]. These genes encode proteins involved in the cytoskeleton, mitochondrial metabolism, autophagy, proteostasis, and other cellular processes. In particular, the SOD1, EPHA4, KIFAP3 and UNC13A genes appear to influence disease progression and survival of ALS patients [14]. Furthermore, at the proteomic level, it is widely known that the accumulation of misfolded cytoplasmic proteins in degenerating motor neurons represents a hallmark of the disease [15]. Among the main proteins involved in this process, there is TDP-43, which translocates into the cytoplasm under stress conditions or when it presents mutations. TDP-43 aggregates are present in most cases of ALS, including in patients with pathogenic variants of C9ORF72 [15]. This mutation in C9orf72 causes nuclear loss and cytoplasmic aggregation of TDP-43. A recent study demonstrated a correlation between C9orf72 and components of Ran-GTPase cycle [16], involved in the regulation and activation of nucleocytoplasmic transport [17]. In vitro studies found that TDP-43 can create amyloid like fibrils that are typical of patient brains [18], [19], [20]. These aggregations can bind native TDP-43 in healthy cells and spread to the central nervous system suggesting a potential mechanism for the propagation of pathology [18], [19], [20].
AD is a progressive, incurable neurodegenerative disease characterized by a gradual decline in cognition, memory and thinking. It is the most common form of dementia, accounting for about 60–70% of total cases, usually with an onset in adulthood over the age of 65 [21], [22]. To date, the disease is estimated to affect more than 40 million people worldwide, with an exponential increase expected in the coming decades [21], [22]. Currently, the therapeutic approaches available for AD offer modest results in symptomatic improvement and a definitive curative therapy does not yet exist [23]. This limitation is due to a lack of comprehensive understanding of the cause and progression of AD [23]. Immunotherapy is considered the new frontier of the medicine and the most effective treatment of AD [24].
Monoclonal antibodies, aducanumab and lecanemab recently approved by FDA for the therapy of AD, are designed against Aβ plaques and oligomers [25].
Aducanumab was approved by FDA on 7 June 2021. However, post-approval studies will be completed in October 2023 as clinical trials have given controversial results [26].
Lecanemab was approved by FDA in January 2023. Given the adverse events associated with the therapy, longer trials are required to demonstrate the efficacy of the drug [27]. A study integrating transcriptomic, proteomic and epigenomic analyses in AD using postmortem human brain samples, revealed high expression of transcription- and chromatin-related genes, including histone acetyltransferase activity for H3K27ac and H3K9ac, and H3K122ac loss [28]. Interestingly, Gene Ontology analysis of genes targeted by H3K122ac revealed pathways associated with purine/guanosine triphosphate catabolic processes. In addition, the authors suggested a positive circuit in AD where H3K9ac gain at CREBBP grows CBP expression, thus activating histone acetylation in the epigenome [28].
Chung et al. investigated genomics and transcriptomics data and found a correlation between AD and SNPs in MGMT (O6-methylguanine-DNA methyltransferase), which is implicated in DNA damage repair process amongst among women lacking POE ε4 allele in sporadic AD [29]. A recent integrative multi-omics approach of genome-wide and transcriptomic studies identified several genes and genetic variants related to lipid metabolism [30]. In addition, the authors proposed a neuroprotective rôle of modulators of S1P metabolism in AD treatment [30].
PD is also a chronic and progressive neurodegenerative disease that affects the nervous system, in particular, it affects the nerve cells responsible for the production of dopamine in the brain. There is currently no specific cure for PD, but numerous studies have been conducted over the years to discover new therapies.
Among the last efficacy drugs approved by FDA in treating PD there are safinamide, istradefylline and pimavanserin [31]. Safinamide, approved in 2017 by FDA, is a derivative of benzylamino and MAO-B inhibitor, with different modes of action. [32] Istradefylline, is the first non-dopaminergic drug approved by FDA (2019), and acts on adenosine A2A receptor antagonist. [33]. Pimavanserin, approved in 2016 by FDA is an antagonist of 5-hydroxytryptamine (HT)2 A receptor [34]. Numerous GWASs have been conducted over the years, exceeding 24 analyzes since 2009, identifying a total of 78 associated loci [35]. A recent GWAS study used a fine-mapping approach to carefully examine 74 of these loci. 2 specific SNPs were identified: rs7294619, located at the LRRK2 locus, which affects LRRK2 expression via a specific microglial cell control element, and rs4771268, located at the MBNL2 locus, which affects a regulatory control of MBNL2 specifically in oligodendrocytes [36].
MS is a chronic disease involving the central nervous system and is characterized by inflammation, demyelination and neuronal degeneration [37].
Like other neurodegenerative diseases, it is a multifactorial disease due to complex interactions of genetic, environmental, infectious, and possibly vascular factors. From a genetic point of view, although MS is not considered a hereditary disease, genetic variations that increase the risk of developing it have been identified and, the most consistent data is the association between the disease and the HLA-DR15 and HLA alleles -DQ6 [38]. From an environmental point of view, decreased exposure to sunlight was associated with an increased risk, probably due to the reduced production of vitamin D [39]. There is no definitive curative therapy for MS. However, there are treatment approaches that can help manage symptoms, slow disease progression and improve the quality of life of MS patients. These treatments include immunomodulatory drugs that aim to reduce inflammation in the central nervous system, physical and rehabilitation therapies aimed at maintaining mobility and promoting general well-being. [40]. The newest drugs are Briumvi and Ponvory [41].
Briumvi, approved by FDA in 2022, is a glycoengineered anti-CD20 monoclonal antibody. Given the importance of B cells in MS, it acts via their depletion.
Ponvory, approved by FDA in 2021, is an iminothiazolidinone derivative. It binds to S1P1 receptor promoting its degradation [42].
A recent multi-omics approach combining methylation and gene expression identified more than 200 genetic regions that may influence susceptibility to MS. Notably, two independent genome segments (lincRNAs: RP11–326C3.13 and TNFSF14) were identified, that show significant associations with multiple genomic features across different levels of omics analyses [43].
LBD is a neurodegenerative disease that represents a form of dementia with characteristics similar to AD, with an earlier onset and also related to PD. Patients with LBD have a wide range of cognitive, motor, and autonomic symptoms [44]. The main feature of the disease is the presence of abnormal accumulations of a protein called alpha-synuclein, known as Lewy bodies, within brain cells. These accumulations form in specific areas of the nervous system and brain, damaging neuronal cells. Because of symptomatic similarities to other forms of dementia, LBD is often misdiagnosed or underdiagnosed [45]. In addition, it has variability both between patients and over time within the same individual, making it a complex disease to treat. Available treatments may aim to alleviate specific symptoms but may aggravate others, further complicating disease management [44].
To date, there are not drugs approved by FDA [46], [47].
The aim of our study is to investigate separately 5 neurodegenerative diseases, including ALS, AD, PD, MS, and LBD using a computational approach. To our knowledge there are few works that use the same procedures for different neurological pathologies in a single study, making the results more comparable [48]. Specifically, the genetic correlations between the different diseases and phenotypes derived from the UK Biobank (UKBB), affecting the lifestyle and medical history of the participants, will be explored. Subsequently, a clumping analysis will be performed to identify independent loci, i.e., SNPs present in the genome that are genetically associated with the specific diseases under analysis. The relevant SNPs in the coding regions of the genes will be subjected to analysis to evaluate how they influence the structural stability of the proteins. Finally, we will perform an analysis to identify differentially expressed microRNAs (miRNAs) obtained from miRNA expression profiles of patients affected by the 5 diseases studied. We will evaluate their possible interactions with genes deriving from the initial datasets or directly with the analyzed SNPs. Through this approach, our study aims to better understand the genetic and molecular mechanisms underlying neurodegenerative diseases. The results obtained could contribute to the discovery of diagnostic biomarkers, to the understanding of the pathogenic mechanisms of diseases and potentially open new perspectives for the development of targeted and personalized therapies.
2. Material and methods
2.1. Workflow
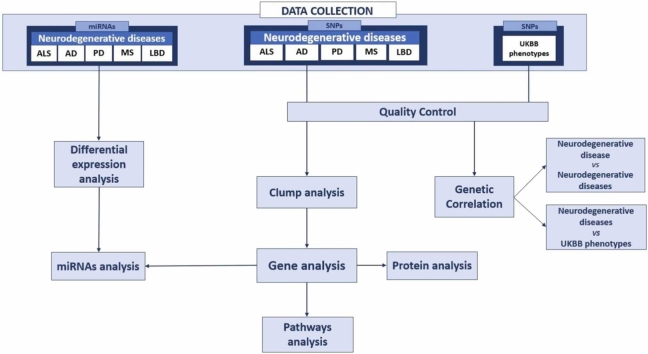
Fig. 1 briefly shows the various steps of our study. We analyzed GWAS of 5 neurological diseases ALS, AD, PD, MS, and LBD. After a quality control step, we performed a genetic correlation analyses between the neurological diseases and human traits derived by UKBB in order to observe the phenotypes most correlated with the diseases. A clumping analysis was carried out to identify SNPs implicated in each neurological disease. The genes present in the SNPs identified for each disease were analyzed and their biological processes were studied by functional annotation. Structural stability predictions analysis was performed on the SNPs in the coding regions of the genes to evaluate how they influence the structural stability of the protein. In the last step, we collected miRNA expression profiles of the 5 neurological diseases and we identified differentially expressed miRNAs for each disease. We studied their possible interactions with SNPs and genes deriving from the previous steps.
Fig. 1.
The figure shows the summary scheme of this study. Starting from 5 datasets of genome-wide association study (GWAS) profiles of patients with neurodegenerative diseases: Amyotrophic lateral sclerosis (ALS), Alzheimer's disease (AD), Parkinson's disease (PD), Multiple sclerosis (MS), and Lewy body dementia (LBD) the study was conducted using a multi-omics approach to analyze possible biomarkers and identify new or known drug targets.
2.2. Data collection
In this study we collected GWAS summary statistics and miRNA expression profiles from different public repositories.
GWAS summary statistics of 5 different neurodegenerative diseases: ALS, MS and LBD datasets were downloaded from the GWAS catalog (Accessed on May 2022; https://www.ebi.ac.uk/gwas/docs/about); AD dataset from the GWAS Atlas, (Accessed on May 2022; https://atlas.ctglab.nl/documentation) and finally the PD dataset from the IEU OpenGwas project (Accessed on May 2022; https://gwas.mrcieu.ac.uk/about/). The 5 datasets are all derived from European cohorts and have participants of both sexes.
ALS dataset consists of 152,268 individuals, including 29,612 cases and 122,656 controls. Summary statistics of AD contain 71,880 cases and 383,378 control. PD dataset includes 33674 cases and 449056 controls. MS cohorts is composed of 4888 cases and 10395. The number of cases was 2591 and 4027 for LBD.
GWAS phenotype samples were collected from the UK Biobank (UKBB), with a wide range of phenotypic information. The phenotypic data was not from the same patients of the GWAS with the neurodegenerative diseases. UKBB recruited approximately 500,000 participants aged 40–69 years, of both sexes, and from the United Kingdom, who provided informed consent and answered questions about their health, lifestyle, and sociodemographic aspects. All the summary statistics used in this study are downloaded from the United Kingdom Biobank database (Accessed on May 2022; http://www.nealelab.is/uk-biobank). The number of partecipants for each phenotype can be visualized in: Catalogues (ox.ac.uk).
Four different datasets of miRNAs related to the 5 neurodegenerative diseases under study were downloaded from Gene Expression Omnibus (Accessed on May 2022; https://www.ncbi.nlm.nih.gov/geo/). The datasets are GSE120584 (AD and LBD), GSE52917 (ALS), GSE16658 (PD) and GSE17846 (MS).
2.3. Quality control
The goal of this step is to eliminate low-quality data that may compromise the results of the analysis. Quality Control includes the removal of samples with low DNA quality, removal of markers with low genotyping rates, correction of population structure, and correction of any confounding effects. This minimizes false positives and false negatives, improving the accuracy of the analysis and the reliability of the results [49].
In particular, we considered for further analyses only phenotypes with SNP-heritability z > 4 for neurological diseases and UKB phenotypes. In addition, SNPs with a minor allele frequency (MAF) < 1% were removed.
2.4. Genetic correlation
To estimate the genetic correlation (GC) between two traits from GWAS summary statistics, we used the GitHub package LDSC (Accessed on May 2022; LD Score: https://github.com/bulik/ldsc). To perform LDSC analysis, it is necessary to use genetic data from a large number of individuals representative of a given population [50], [51]. We used as representative samples to calculate genetic linkage disequilibrium and apply LDSR analysis accurately, data of individuals of European ancestry from the 1000 Genomes Project [50], [51]. We tested genetic correlations between neurological diseases and UKBB traits with SNP-based heritability z score > 4 [50], [51]. We considered statistically significant genetic correlations as those obtaining a 5% False Discovery Rate (FDR) correction (FDR Q value < 0.05).
Two different correlations were calculated: i) among the five neurodegenerative diseases, ii) between each neurodegenerative disease and the phenotypes from the UKBB.
2.5. Clumping procedure
Clumping analysis was conducted for the 5 neurodegenerative disease datasets after quality control. Clumping analysis is able to select relevant SNPs for each neurological disease. The analysis was conducted using PLINK 1.9 [52]. We used the datasets individually as each dataset can be influenced by batch effects, namely experimental and data processing conditions can hide the biological effect of interest [53].
From clumping analysis, we obtained the SNPs independently associated with the neurodegenerative disease phenotypes. We considered only SNPs with a p-value < 5 × 10–8 and filtered out the others. In GWAS studies, a significance threshold of 5 × 10 − 8 is commonly used as a reference value of a genome wide association. This threshold is based on a Bonferroni correction for 1 million independent tests, which is a standard approximation for the number of independent SNPs in the human genome [54], [55].
EnsDb.Hsapiens.v79 and biomaRt were used to obtain the genes associated with each SNP. Once the genes containing SNPs were identified, DAVID functional annotation analyses were conducted (Accessed on 22 September 2023; https://david.ncifcrf.gov/summary.jsp).
2.6. Protein structure analysis
Starting from the SNPs obtained from the previous analysis, we focused on SNPs found in coding regions. Protein stability studies and analyzes were conducted following the MAVISp (Multi-layered Assessment of VarIants by Structure for Proteins) protocol [56]. MAVISp can be applied to single three-dimensional (3D) structures of proteins and their complexes (simple mode) or to an ensemble of conformations (ensemble mode) [56].
The framework is designed in a modular way so that we can apply to our studies all the modules or only a selection of them and each module relies on Python scripts, often supported by specific virtual environments [56]. For our case studies, we used the simple mode, since we focused on the static structure of each protein under investigation. The first step is collecting the data to analyze. Variants and information for each protein were retrieved using ClinVarMiner and Cancermuts [57], [58].
2.6.1. Structure Selection
We used the Structure Selection module of MAVISp to retrieve models from the AlphaFold2 database to use as initial structure for calculations of the effects of the variants on the protein structural stability [56] . Its primary objective is to address the challenge of predicting three-dimensional protein structures from protein sequences. The resulting protein predictions obtained are accompanied by a confidence metric. This confidence metric is applied for each protein residue and is called the predicted local distance difference test (pLDDT). The pLDDT is an indicator that assesses the level of consistency of the prediction with respect to an experimental context, based on the local distance difference test [59].
The model confidence is expressed on a scale ranging from 0 to 100. Confidence levels are categorized as follows: Very High (pLDDT > 90), Confident (90 > pLDDT > 70), Low (70 > pLDDT > 50), and Very Low (pLDDT < 50) (Accessed on May 2022; https://alphafold.ebi.ac.uk/).
For our analysis, we used a trimming strategy to obtain the best structural coverage and minimize the number of low confidence regions by going to trim and not considering protein residues with a confidence level less than 70 (pLDDT > 70).
2.6.2. Stability energy calculation
Following the trimming of proteins according to the predictive confidence level provided by AlphaFold, the next step involves calculating the change in folding free energy (∆∆G), which is used to describe the thermodynamic stability of a protein or molecule during the process of folding or conformational change between the mutated variants and the wild-type protein. For this purpose, two distinct software programs, MutateX and RosettaDDGpredictions, were employed [60], [61]. Missense mutations, which involve substitutions of amino acids, can significantly impact protein stability. Understanding the molecular effects of protein mutations is crucial for comprehending biological processes and facilitating interventions such as structure-based drug development or targeted mutagenesis [60], [61].
The MAVISp protocol employs a consensus approach that combines the outcomes from both methods. Mutations exhibiting a stability change greater than 3 kcal/mol are considered destabilizing at the cellular level. Variants with ∆∆G values ranging from − 2 kcal/mol to 2 kcal/mol are deemed neutral, while mutations with ∆∆G values below − 3 kcal/mol are classified as stabilizing according to both methods [62]. After conducting the analyses with MutateX and RosettaDDGprediction, the results obtained from both softwares were compared [60], [61].
2.7. miRNA analysis
Four miRNA datasets of the 5 neurodegenerative diseases were downloaded from Gene Expression Omnibus (GEO): GSE52917 (ALS), GSE120584 (AD and LBD), GSE16658 (PD) and GSE17846 (MS).
In the initial step, we selected the GEO datasets and specified the miRNA groups to be compared for each dataset, representing control and pathological cases. For the analysis, we utilized the force normalization option available in the Options tab of GEO2R, which applies quantile normalization to the expression data, ensuring that all selected samples exhibit an identical value distribution (Accessed on May 2022; https://www.ncbi.nlm.nih.gov/geo/geo2r/). GEO2R employs the Limma (linear models for microarray data) package to perform the statistical analysis and calculate fold changes and p-values for each gene. By utilizing this tool, we were able to sort and filter the results based on statistical significance, thereby identifying miRNAs that were significantly upregulated or downregulated between control and pathological cases. The statistical significance of the results was determined considering FDR < 0.05 for all datasets. A log fold change (logFC) threshold of 1 and − 1 was used to identify upregulated and down-regulated miRNAs. For the LBD dataset we used |logFC| > 0.5 as no differentially expressed miRNAs were obtained with the logFC > 1.
After identifying the differentially expressed miRNAs for each of the five neurodegenerative diseases, we used the miRSystem to conduct further analyses [44]. This database integrates seven well-known miRNA target gene prediction programs, including DIANA, miRanda, miRBridge, PicTar, PITA, rna22, and TargetScan [63]. Results are obtained using prediction algorithms that assess the complementarity between the miRNA and the binding site within the target gene. To ensure greater confidence in predicted target genes, we only included those that had been validated and had a minimum count of 3. This threshold was established to provide a good confidence margin in predictive analysis. Next, we perofmed an intersection between the target genes identified for each miRNA and the genes obtained from the clumping analysis and by this examination of the overlap between the two sets of genes, we identified the genes common to the two analyses.
3. Results
3.1. Quality control
We downloaded from the GWAS Catalog, GWAS Atlas and IEU OpenGwas project databases the summary statistics of the five neurodegenerative diseases. All five datasets are found to have a Z-score greater than 4. Table 1 shows the number of SNPs for each neurodegenerative disease before and after quality control. Regarding UKB, 957 traits passed quality control.
Table 1.
Total number of SNPs before and after quality control for each neurodegenerative disease. Amyotrophic lateral sclerosis (ALS), Alzheimer's disease (AD), Parkinson's disease (PD), Multiple sclerosis (MS), and Lewy body dementia (LBD).
| Diseases | Pre-quality control | Post-quality control |
|---|---|---|
| ALS | 10461755 | 7054478 |
| AD | 13367299 | 9736043 |
| PD | 17891936 | 8571928 |
| MS | 7968107 | 7915405 |
| LBD | 7843595 | 7513830 |
3.2. Genetic correlation
Genetic correlation was performed using LD score method. LD score of a SNP is the sum of LD r2 calculated with all other SNPs [64].
First, we calculated genetic correlations between neurodegenerative diseases. The obtained results that achieved an FDR < 0.05 are shown in Table 2.
Table 2.
Genetic correlation with False Discovery Rate (FDR) and standard deviation (sd) among neurodegenerative diseases. Amyotrophic lateral sclerosis (ALS), Alzheimer's disease (AD), Parkinson's disease (PD), Multiple sclerosis (MS), and Lewy body dementia (LBD).
| Diseases | Genetic correlation (sd) | FDR |
|---|---|---|
| ALS Vs AD | -0.289 (0.103) | 0.025 |
| ALS Vs LBD | 0.440 (0.194) | 0.047 |
| ALS Vs PD | 0.156 (0.064) | 0.037 |
| PD Vs AD | -0.208 (0.078) | 0.025 |
| PD Vs LBD | 0.612 (0.176) | 0.005 |
Only some diseases were correlated to each other. It can be noted that MS has no significant genetic correlation with the other diseases.
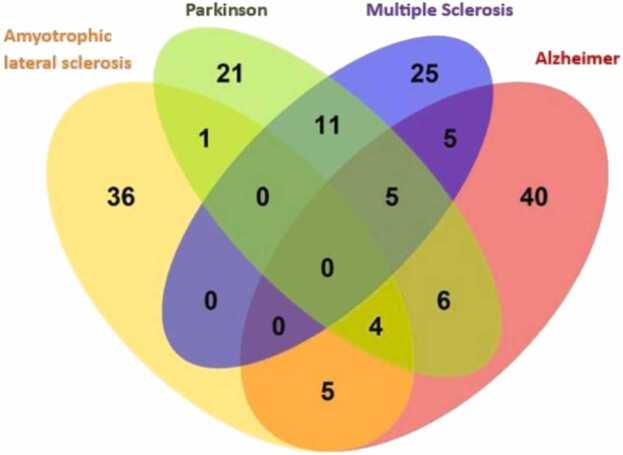
In addition, we calculated the genetic correlation for each disease with 957 phenotypes downloaded from the UKBB, that passed quality control. These analyses are useful to understand whether diseases are influenced by a range of habits and lifestyle characteristics. We found: 46 significant phenotypes correlate with ALS, 65 with AD, 48 with PD, and 46 with MS. In contrast, no significant genetic correlations between phenotypes and LBD were found. Fig. 2 shows Venn diagram between phenotypes and neurological diseases.
Fig. 2.
The number of common and non-common phenotypes present in the statistically significant correlations among the 4 diseases.
By examining the Venn diagram, it is evident that there are no common phenotypes with significant correlations among all neurodegenerative diseases. However, there are 4 common phenotypes among ALS, AD and PD, and 5 common phenotypes among AD, MS and PD.
Nine phenotypes are correlataed with ALS and AD. Among them the most relevant are: "Dried fruit intake" "Duration screen displayed", "FI3 word interpolation", "Fluid intelligence score", "Qualifications", "Time spent using computer" and "Time spent watching television (TV)".
Five phenotypes exhibit correlations in both ALS and PD. Among them: "Age started wearing glasses or contact lenses" "Dried fruit intake" "Qualifications" and "Time spent watching television (TV)".
There are no phenotypes that show correlations in both ALS and MS. Fifteen phenotypes demonstrate correlations in both AD and PD. The most relevant to the study are: namely "Age at first live birth", "Age completed full-time education", "Age first had sexual intercourse", "Attendance or disability or mobility allowance", "Average total household income before tax", "Body mass index (BMI)", "Cereal type Muesli", "Dried fruit intake", "Leg fat", "Qualifications", and "Smoking status current". Interesting to note the correlation between BMI and leg fat. Ten phenotypes show correlations in both AD and MS. Among them: "Body mass index (BMI)", "Frequency of tiredness or lethargy in the last 2 weeks", "Leg fat", "Long-standing illness disability or infirmity", "Other serious medical condition or disability diagnosed by doctor", and "Taking other prescription medications".
Lastly, 16 phenotypes exhibit correlations in both PD and MS. Specifically, to note are: "Arm fat", "Body mass index (BMI)" "C reactive protein (quantile)", "Leg fat", "Mouth or teeth dental problems", "Waist circumference", "Weight" and "Whole body fat mass". Many of these phenotypes appear to involve characteristics related to body mass. mos.
Table Supplementary 1 shows the main phenotypes associated with the neurological diseases.
3.3. Gene selection
Through clump analysis, we successfully identified SNPs that showed independent associations with one or more of the five neurodegenerative diseases studied. Then, genes containing the statistically significant SNPs obtained were identified.
We identified 27 genes in AD, 6 genes in ALS, 10 genes in PD, 3 genes in LBD, and 18 genes in MS (Table Supplementary 2). There are no independently associated SNP-carrying genes shared among all five diseases (Fig. 3).
Fig. 3.
Upset plot shows the intersection of genes found in Lewy body dementia (LBD), Amyotrophic lateral sclerosis (ALS), Parkinson's disease (PD), Multiple sclerosis (MS) and Alzheimer's disease (AD).
Clump's analysis showed that only 5 SNPs were located within the coding regions of specific genes (KIF5A, CFAP410, PILRA, CYP2R1, and TMEM175), while most of the identified SNPs were located in the intronic regions or regions following (downstream) or preceding (upstream) the coding sequences of the genes. Although these regions do not directly encode the protein sequence, the presence of a SNP in these areas can be of critical importance. This is because these regions may contain regulatory elements that play a crucial role in protein synthesis. Therefore, variations present in these noncoding regions can also influence gene expression and have significant functional implications.
3.4. Functional annotation
Knowing the functional annotation of genes containing the SNPs under analysis can offer greater biological insights and help generate directly testable hypotheses. We reported the results in the Table Supplementary 3.
Regarding ALS-related genes, among others there are biological processes focused on the antiport (CLCN3, SLC9A8) and differentiation (SLC9A8 and UNC13A).
Focusing on the genes related to AD, one of the biological processes obtained is the host-virus interaction (CD2AP, BIN1, CR1, PILRA). The identified genes have also significant roles in the endocytosis (BIN1, PICALM, and SORL1) and complement pathway (CLU and CR1).
Most of the biological processes associated with PD are involved in apoptosis (BAG3) and endocytosis (HIP1R).
The biological processes involved in MS are implicated among others in transcription (ERG, L3MBTL3, MAZ, AND GFI1) and immunity (CD86, IL2RA and HLA-DRB1).
Finally, regarding the biological processes of LBD identified by our analysis it is possible to observe a significant relevance in the cholesterol metabolism (APOE), and differentiation (BIN1).
3.5. Protein structure analysis
Although most GWAS identify SNPs associated with diseases in non-coding regions, those SNPs in protein-coding regions can influence the protein sequence [65], [66]. We conducted an analysis to reveal whether the presence of SNP associated with neurological diseases, which causes a missense mutation, can cause conformational changes in the corresponding protein structure.
3.5.1. Protein description
The proteins analyzed are: KIF5A, CFAP410, PILRA, CYP2R1, and TMEM175 (Table 3).
Table 3.
SNPs of the 5 neurodegenerative disease datasets present in coding regions. In the table for each SNP are defined: the associated genes, the associated neurodegenerative disease, the Uniprot name of the protein encoded by the associated gene and the missense mutation caused by the SNP (wild type amino acid, position, mutated amino acid).
| SNPs (rsID) | Associated gene | Associated Illnesses | Uniprot Entry | Mutations |
|---|---|---|---|---|
| rs113247976 | KIF5A | Amyotrophic lateral sclerosis | Q12840 | P986L P986R |
| rs75087725 | CFAP410 | Amyotrophic lateral sclerosis | O43822 | V58L |
| rs1859788 | PILRA | Alzheimer's disease | Q9UKJ1 | R78G |
| rs202122669 | CYP2R1 | Parkinson's disease | Q6VVX0 | P36L |
| rs34311866 | TMEM175 | Parkinson's disease | Q9BSA9 | M393T |
The Uniprot code for the KIF5A protein is Q12840. A SNP identified in our analysis is SNP rs113247976, which leads to P986L and P986R mutations. This SNP is considered one of the potential causes of ALS [67]. The Uniprot code for the CFAP410 protein is O43822. SNP identified in the above analysis is rs113247976, which leads to the V58L mutation and is considered one of the possible causes of ALS. The Uniprot code for the PILRalpha protein is Q9UKJ1. SNP identified is SNP rs1859788, which leads to the R78G mutation and is considered one of the potential causes of AD. The Uniprot code for the CYP2R1 protein is Q6VVX0. SNP identified is SNP rs202122669, which leads to the P36L mutation and is associated with PD. The Uniprot code for the TMEM175 protein is Q9BSA9. SNP identified is SNP rs34311866, resulting in the M393T mutation, and it is considered one of the possible causes of PD.
We analyzed 4 out of 5 proteins through the simple mode steps of the MAVISp protocol [56]. As the fifth protein (i.e., TMEM175), is a transmembrane protein and all of its domains are located in an intermembrane zone, the pipeline for its analysis has yet to be optimized to deal with transmembrane proteins and no results are available at the moment.
3.5.2. Retrieval of variants and information
Using the ClinVarMiner and CancerMuts tools, we conducted an analysis to retrieve variants and information related to each protein. Specifically, the variants of interest under study found on ClinVarMiner were identified, which included KIF5A and CYP2R1 mutations. However, regarding PILRA and CFAP410, the mutations of interest were not available or investigated on ClinVarMiner, so they were manually entered.
Then, using CancerMuts, we downloaded and retrieved all relevant information, including REVEL pathogenicity scores and other annotations regarding the protein context in which these mutations are found.
Regarding KIF5A (P986L and P986R), the acquired data reveal the presence of the mutation within the amino acid region from 986 to 990 aa, which harbors an interaction motif with USP7 (Ubiquitin Specific Protease 7), which has been identified and documented in the ELM database. In addition, the data indicate that the region affected by the mutation exhibits features of the disorder.
In the case of CYP2R1 (P36L), the region that is affected by the mutation is characterized as a region of conformational disorder. In addition, by analyzing the information collected, it was found that at the location of the mutation, specifically in amino acid 36, a cleavage site is present. Using the ELM database, an anchor motif for Mitogen-Activated Protein Kinase (MAPK) was present at the mutation location. Also, between amino acids 34 and 36, there is a retention or recovery signal containing two arginine residues in the protein, which is involved in protein retention or recovery within specific cellular compartments.
Information was obtained regarding the mutation identified in PILRA (R78G) indicating its location within a structurally ordered region. Lastly, for CFAP410 (V58L), no further information was obtained during the analysis.
3.5.3. Structure Selection
Protein structures were identified using the AlphaFold2 model. The protein predictions obtained are accompanied by a calculated confidence metric applied to each protein residue. The stability of the protein structure is assessed by the confidence level represented by the pLDDT value. Each confidence level is associated with a specific color, as highlighted in the figures of the four proteins under study (Fig. 4, Fig. 5, Fig. 6, Fig. 7). Regions colored blue indicate a high confidence level (pLDDT > 90), while regions colored blue indicate good confidence (90 > pLDDT > 70). Regions colored yellow indicate low confidence (70 > pLDDT > 50), while regions colored orange indicate very low confidence (pLDDT < 50). In addition, the amino acid of interest that is subject to mutation was highlighted with a red-colored ball. Each graphic representation below illustrates the three-dimensional model of the proteins under study, which is essential for determining the regions to be eliminated in order to retain only the portion of the protein structure with a high degree of reliability for subsequent analysis.
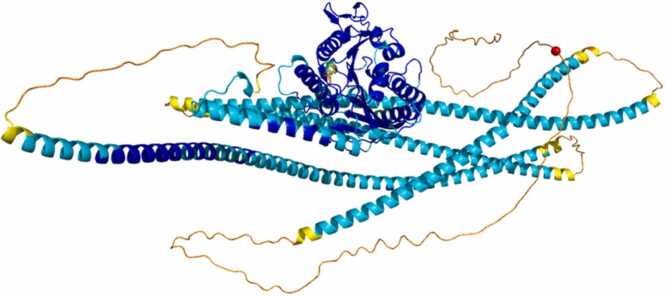
Fig. 4.
This figure represents the structural prediction of the KIF5A protein, where the different colors present correspond to the associated confidence level. The red sphere represents the amino acid Proline at position 986 involved in the mutation (P986L/P986R).
Fig. 5.
This figure represents the structural prediction of the CFAP410 protein, where the different colors present correspond to the associated confidence level. The red sphere represents the amino acid Valine at position 58 involved in the mutation (V58L).
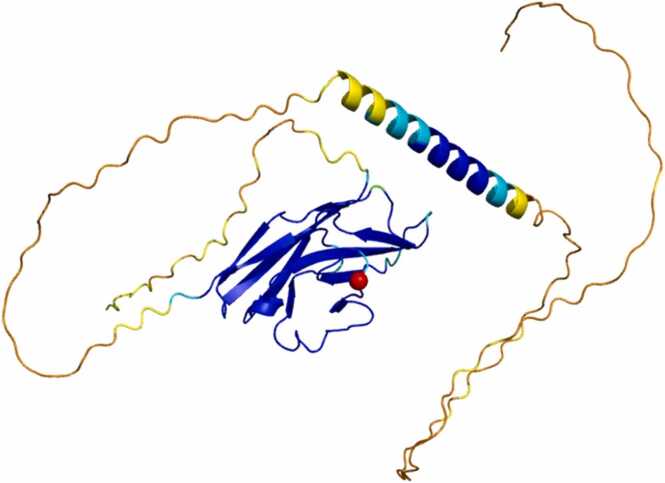
Fig. 6.
This figure represents the structural prediction of the PILRA protein, where the different colors present correspond to the associated confidence level. The red sphere represents the amino acid Arginine at position 78 involved in the mutation (R78G).
Fig. 7.
This figure represents the structural prediction of the CYP2R1 protein, where the different colors present correspond to the associated confidence level. The red sphere represents the amino acid Proline at position 36 involved in the mutation (P36L).
As evident from the figure, the mutated amino acid in KIF5A, specifically proline 986, is located in a region characterized by a low structural confidence level. For the purpose of subsequent analysis, this low-confidence region would be removed, thus disrupting the study of the mutation in question and preventing further detailed investigation.
Regarding the CFAP410 protein, the mutation of interest is located at the level of the amino acid Valine at position 58. The image clearly shows that the amino acid is located in a highly reliable and high-confidence region. Accordingly, the protein was trimmed taking into account only the portion between amino acid 1 and amino acid 147 for subsequent analysis.
In PILRA, the mutation of interest is located at the level of the amino acid Arginine at position 78. Again, as in the previous case, the amino acid is located in a highly reliable region with a high confidence level. We took into account for subsequent analysis the portion between amino acid 32 and amino acid 153.
In relation to the last protein under study, CYP2R1, the mutated amino acid is proline at position 36. It is located within a region characterized by high reliability. In this circumstance, it was decided not to perform truncation of the protein, as this approach would not result in a significant loss of precision and would allow complete analysis of the protein.
3.5.4. Stability Energy Calculation
After selecting the protein structure cut, we proceeded to calculate the change of folding free energy (∆∆G) by comparing the mutated variants with the wild-type protein. To perform this calculation, we used both MutateX and RosettaDDGpredictions software. Next, we focused on mutations caused by the SNPs in the three different proteins with high-confidence predictive structure, which are CFAP410, PILRA and CYP2R1.
Focusing on the analysis of the results obtained through MutateX, we initially generated, using the ddg2density tool, a density plot representing all the calculated differences in ∆∆G. This density plot is the first step in the analysis, as it allows us to determine the scale of the ∆∆G values used in creating the heatmap. By plotting the density plot for all three proteins (Fig. 8), we observed that the majority of the ∆∆G values vary approximately from about − 3 to about 10.
Fig. 8.
Plots a, b and c represent the calculated density and individual energy values identified under the x-axis. Graph a represents the density relative to CFAP410, graph b represents the density relative to PILRA, and graph c represents the density relative to CYP2R1.
Next, we used the ddg2heatmap tool to generate a heatmap of the calculated ∆∆G mutation values. The wild-type amino acids are represented on the y-axis and the mutated amino acids on the x-axis. We used as value bounds those previously obtained from the density map, and values outside this range were fitted to the bounds of the range itself. These out-of-range values should simply be interpreted as "very destabilizing", with a reddish colour, or as "very stabilizing", with a bluish colour. The heatmap colors range on a blue to red color scale with green, yellow and orange in the central area representing different energy stability scores. Below we report the heatmaps of the proteins only in the area of the respective amino acids of interest. A cut-off value of ∆∆G > 3 kcal/mol was considered for destabilizing mutations.
As regards the CFAP410 protein, in the heatmap (Fig. 9), it is highlighted that the substitution of the amino acid Valine in position 58 with Leucine does not produce significant variations in the free energy according to MutateX prediction. This mutation is represented by the blue color in the heat map and results in a highly stabilizing mutation. Indeed, the value of ΔΔG for this mutation is − 1.70 kcal/mol. This result shows that the mutation of valine to leucine does not impact the structural stability of the protein, as there is no significant change in stability. In the context of this mutation, the two amino acids are essentially identical as they belong to the apolar amino acid category and have very similar side chains. Consequently, it is reasonable to expect that this mutation would not result in significant changes in the structure or stability of the protein.
Fig. 9.
The figure represents a heat map of the CFAP410 protein, in which the amino acids from amino acid 38 to amino acid 74 are displayed. Within this sequence, the amino acid 58 of interest, corresponding to valine, is specifically identified.
Focusing on the PILRA protein, within the thermographic representation (Fig. 10), it can be seen that the replacement of the amino acid Arginine at position 78 with Glycine does not induce significant changes in free energy. It should be considered that however that in MutateX analyses mutations at Glycine are not reliable because the backbone is not relaxed. This mutation is also represented by blue color in the heat map, indicating a highly stabilizing mutation. The value of ΔΔG for this mutation is 0.10 kcal/mol. It is of relevance to point out that glycine represents the smallest amino acid from a structural point of view. This implies that its presence at position 78, replacing arginine, would hardly cause spatial occupancy that could result in destabilizing changes. This result suggests that the mutation of arginine to glycine does not affect the structural stability of the protein, as no significant alterations in stability are found.
Fig. 10.
The figure represents a heat map of the PILRA protein, in which the amino acids from amino acid 63 to amino acid 93 are displayed. Within this sequence, the amino acid 78 of interest, corresponding to valine, is specifically identified.
Finally, focusing on the CYP2R1 protein, it is evident from the heat map (Fig. 11) that also in this situation the alteration of the mutation, in this case, proline in position 36 with leucine, does not determine a destabilization of the free energy. This, like the previous one, is also a highly sensitive case as MutateX struggles even with the amino acid proline as they are not reliable since the backbone is not relaxed. Again, the color associated with this mutation corresponds to a shade of blue, indicating a highly stabilizing effect. In fact, the ΔΔG value for this mutation is 0.83 kcal/mol, slightly higher than the mutations previously analyzed in the other proteins, but still confirming a very stabilizing mutation.
Fig. 11.
The figure represents a heat map of the CYP2R1 protein, in which the amino acids from amino acid 1 to amino acid 52 are displayed. Within this sequence, the amino acid 36 of interest, corresponding to valine, is specifically identified.
Analyzing the results obtained through RosettaDDGpredictions, a remarkable similarity with the values calculated through MutateX is observed. In the case of the V58L mutation in CFAP410, the value of ΔΔG obtained is − 1.95 kcal/mol, differing only by 0.25 from the result obtained by MutateX. For the R78G mutation in PILRA, the value of ΔG obtained is − 0.45 kcal/mol, differing by 0.55 from the result obtained with MutateX. Finally, for the P36L mutation in CYP2R1, the value of ΔΔG obtained is 1.10 kcal/mol, a difference of 0.27 from the result obtained with MutateX.
By comparing the results obtained with MutateX and those obtained with RosettaDDGprediction, it is possible to state that all three mutations analyzed do not cause significant changes in the protein conformation. Consequently, it can be inferred that these mutations are not structurally involved as a possible cause of neurodegenerative diseases.
3.6. miRNA analysis
In this step, we conducted an analysis on the differentially expressed miRNAs. Differential expressed miRNAs were reported in Table Supplementary 4. Regarding the ALS dataset, 7 up-regulated miRNAs, and 4 down-regulated miRNAs were identified. The differential expression analysis in AD revealed 2 up-regulated miRNAs and no down-regulated miRNAs.
In contrast, no up-regulated miRNAs and 8 down-regulated miRNAs were identified in the PD. Related to MS, 30 up-regulated miRNAs and 7 down-regulated miRNAs were identified.
Finally, we found 5 up-regulated miRNAs and 5 down-regulated miRNAs for LBD dataset.
3.7. miRNA-gene interaction
After identifying the differentially expressed miRNAs for each of the 5 diseases, we used the miRSystem software to find the interactions occurring between these miRNAs and the genes we previously identified. These genes carry SNPs, which were statistically associated with one of the diseases under study by the previous clumping analysis.
The idea is that the presence of SNPs in an oncogene or a tumor suppressor (targets of miRNA) might influence gene regulation and change the capability to bind the miRNAs [68], [69].
Only interactions were found in PD and MS and listed in the Table 4.
Table 4.
The table presents a list of identified Up-regulated or Down-regulated miRNAs related to the genes they interact with for Parkinson's and Multiple Sclerosis. In addition, the last column provides the rsID name of the SNPs present within the target gene.
| Diseases | miRNA | miRNA Up/Down-regulated | Gene-target | SNP-target |
|---|---|---|---|---|
| Parkinson | hsa-miR-19b hsa-miR-19a |
Down-regulated Down-regulated |
SYT11 | rs35749011 |
| Multiple Sclerosis | hsa-let-7b hsa-let-7c hsa-let-7d hsa-let-7e hsa-let-7 g |
Up-regulated Up-regulated Up-regulated Up-regulated Up-regulated |
CD86 | rs2681424 |
| hsa-miR-613 hsa-miR-130b hsa-miR-625 hsa-miR-499–5p |
Up-regulated Up-regulated Up-regulated Down-regulated |
ERG | rs2836425 | |
| hsa-miR-132 hsa-miR-20b hsa-miR-212 |
Up-regulated Up-regulated Down-regulated |
L3MBTL3 | rs4364506 | |
| hsa-miR-145 | Up-regulated | CLEC16A | rs6498168 | |
| hsa-miR-499–5p | Down-regulated | EVI5 | rs6689470 | |
| hsa-miR-223 | Up-regulated | RGS1 | rs7535818 |
The results highlight the potential role of miRNAs in regulating gene expression of identified target genes. This interaction between miRNAs and genes may have important functional implications and play a crucial role in the regulation of critical biological processes. In addition, such interactions may contribute to disease onset, providing a deeper understanding of the molecular mechanisms involved.
4. Discussion
In the present study, we used GWAS from UKBB to explore 5 neurodegenerative diseases liability in the context of different human traits and diseases. Genetic correlation analysis showed that ALS shows a negative correlation with AD and a positive correlation with LBD and PD. PD correlates with AD, and LBD.
In addition, the 5 neurodegenerative diseases are correlated with traits associated to different domains. ALS correlates with different phenotypic groups belonging to mental health and well-being, activity, health and physical conditions, social and recreational habits, fluid intelligence, education and qualifications, sexuality, and diet. AD correlates with traits belonging to diseases and medical conditions, fluid intelligence, education and qualifications, anthropometric measurements, employment status, activity, diet, housing and lifestyle, and sexuality. PD genetic correlations were related to: anthropometric measurements, diseases and medical conditions, diet, biological and clinical indicators, education and qualifications, sexuality, and housing and lifestyle. MS genetic correlations were related to different phenotypic groups: anthropometric measurements, biological and clinical indicators, diseases and medical conditions, and lifestyle.
Through a clumping analysis we investigated the genetic overlap existing in the 5 neurological diseases. It revealed that 3 genes, namely HLA-DRB1, TMEM175, and BIN1, exhibit a shared association with two diseases. The gene HLA-DRB1, a component of the human Major Histocompatibility Complex (MHC) Class II [70], harbors two distinct SNPs, namely rs3104373 associated with MS and rs6931277 associated with AD. Similarly, the TMEM175 gene, encoding the Transmembrane Protein 175 [71], contains two distinct SNPs, namely rs34311866 associated with PD and rs6599388 associated with LBD. Likewise, the BIN1 gene, known as Bridging Integrator 1, encodes a protein involved in diverse cellular functions, including intracellular vesicle trafficking and membrane formation [72]. This gene also encompasses two distinct SNPs, namely rs4663105 associated with AD and rs6733839 associated with LBD. The latter is a crucial process for the transmission of signals between nerve cells (neurons) in synapses. It involves the formation, anchoring, fusion and recycling of synaptic vesicles that contain neurotransmitters, the chemical molecules responsible for communication between neurons [73].
Notably, in our analysis we did not find APOE in AD dataset while we found it in LBD dataset. Apolipoprotein E (APOE4) is the common known genetic risk factor for AD, but only 7% of cases are due to APOE4, suggesting the role of other genetic factors with higher frequency in AD pathogenesi [74], [75].
Interestingly, only 5 SNPs identified in our study were located within the coding regions of specific genes (KIF5A, CFAP410, PILRA, CYP2R1, and TMEM175). In the next step, we evaluated if SNPs can alter the structural stability and the intermolecular interactions of the proteins, modifying the protein activity and influencing the possible formation of neurodegenerative diseases [65], [66]. As MAVISp protocol has yet to be optimized to deal with transmembrane proteins, no analysis for TMEM175 was performed.
After the protein structure cut, we calculated ∆∆G by comparing the mutated variants with the wild-type protein. This analysis showed that mutations in the 3 proteins do not impact the structural stability of the proteins and these mutations could not be structurally involved as a possible cause of neurodegenerative diseases.
Finally, we evaluated a potential implication in the regulation of genes containing SNPs derived from clumping analyis, by miRNAs.
Differential expression analysis generated a list of differentially expressed miRNAs for the 5 neurodegenerative diseases compared with control samples.
We investigated the interactions between differentially expressed miRNAs and the genes that carry SNPs obtained with clumping analysis. Only interactions in PD and MS were found. We can note that, in PD, the interaction of the down-regulated miR-19a and miR-19b with the SYT11 gene could play a role in the pathogenetic process. Down-regulation of these two miRNAs suggests reduced activity in PD, with possible consequences for biological processes involved in disease formation. Similarly, in the context of MS, the interaction between up-regulated and down-regulated miRNAs (Table 4) and the CD86, ERG, L3MBTL3, CLEC16A, EVI5, and RGS1 genes could represent a significant advance in understanding the complex dynamics of the disease. This interaction suggests that altered expression of these miRNAs may influence biological processes related to MS, opening new perspectives for understanding the disease.
Our study has some limitations to consider. Like the majority of GWAS study, we explored genetic data derived from individuals of European ancestry due to the lack of big cohorts from other ancestry groups. Therefore, further studies should be performed to confirm our results and demonstrate the generalizability of the associations obtained. In addition, the genetic correlation analysis could be influenced by the number of partecipants for each phenotype of UKBB data.
Another limitation of our study is related to the mapping of SNPs to genes by proximity. Indeed, it has been revealed that SNPs may sometimes involve distant genes [76], [77].
Our in silico analysis found relevant genes associated with SNPs identified, that could be also regulated by differentially expressed miRNAs. but, a limitation of this study is the lack of a large cohort of matched miRNA-mRNA expression data for each disease to perform a transcriptional analysis to support the influence of miRNAs on genes.
Last, miRNA-expression profiles for each neurological disease are derived by different body fluids (serum, whole blood, PBMSs) as public datasets from the same tissue are not easily available. This could lead to conflicting results due different miRNA expression levels based on the different tissues. In addition, some of the original publications related to miRNA datasets indicated that there was blood contamination in the serum, and because of that the upregulated miRNAs could be unreliable [78].
Funding
EP group has been supported by Danmarks Grundforskningsfond (DNRF125) and NovoNordisk Fonden Bioscience and Basic Biomedicine (NNF20OC0065262). Part of the calculations have been supported by a EuroHPC Benchmark Access Grant (EHPC-BEN-2023B02-010) on Discoverer. The work is also supported by A PhD Fellowship from the Danish Data Science Academy (DDSA) to M.A.
CRediT authorship contribution statement
Conceptualization, C.C. and F.M.; methodology, F.M., S.D., M.U., and M.A.; formal analysis, F.M., S.D., M.U., and M.A. writing—original draft preparation, C.C. and F.M.; Supervision, I.C., D.P., E.P.,P.G., and C.C. All authors have read and agreed to the published version of the manuscript.
All authors have read and agreed to the published version of the manuscript.
Declaration of Competing Interest
None Declared.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.10.031.
Appendix A. Supplementary material
Supplementary material Table 1: Genetic correlations related to different phenotypic groups in ALS, AD, PD and MS.
.
Supplementary material Table 2: SNPs that obtained independent associations with the 5 neurodegenerative diseases.
.
Supplementary material Table 3: Functional annotations of genes containing the SNPs associated with the 5 neurodegenerative diseases.
.
Supplementary material Table 4: The table shows differentially expressed miRNAs associated with the neurodegenerative diseases: ALS, AD, PD, MS, and LBD. The first column shows the miRNAs, the second column shows the corresponding FDR values and the third column shows the logFC values.
.
Data Availability
GWAS summary statistics of ALS, MS and LBD were downloaded from the GWAS catalog (Accessed on May 2022; https://www.ebi.ac.uk/gwas/docs/about); AD dataset from the GWAS Atlas, (Accessed on May 2022; https://atlas.ctglab.nl/documentation) and finally the PD dataset from the IEU OpenGwas project (Accessed on May 2022; https://gwas.mrcieu.ac.uk/about/). Summary statistics related to human traits are available from the United Kingdom Biobank database (http://www.nealelab.is/uk-biobank). Gene expression profiles are available from Gene Expression Omnibus (Home - GEO - NCBI (nih.gov)). The data with the MAVISp assessment on the protein-coding variant are reported in the MAVISp database:https://services.healthtech.dtu.dk/services/MAVISp-1.0/
References
- 1.Gitler A.D., Dhillon P., Shorter J. Neurodegenerative disease: models, mechanisms, and a new hope. Dis Model Mech. 2017;10(5):499–502. doi: 10.1242/dmm.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morello G., Salomone S., D'Agata V., Conforti F.L., Cavallaro S. From multi-omics approaches to precision medicine in amyotrophic lateral sclerosis. Front Neurosci. 2020;14 doi: 10.3389/fnins.2020.577755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dugger B.N., Dickson D.W. Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol. 2017;9(7):a028035. doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellou E., Stevenson-Hoare J., Escott-Price V. Polygenic risk and pleiotropy in neurodegenerative diseases. Neurobiol Dis. 2020;142 doi: 10.1016/j.nbd.2020.104953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anttila V., Bulik-Sullivan B., Finucane H.K., et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360(6395) doi: 10.1126/science.aap8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Es M.A., Hardiman O., Chio A., Al-Chalabi A., Pasterkamp R.J., Veldink J.H., et al. Amyotrophic lateral sclerosis. Lancet. 2017;390(10107):2084–2098. doi: 10.1016/S0140-6736(17)31287-4. [DOI] [PubMed] [Google Scholar]
- 7.Jiang J., Wang Y., Deng M. New developments and opportunities in drugs being trialed for amyotrophic lateral sclerosis from 2020 to 2022. Front Pharm. 2022;13 doi: 10.3389/fphar.2022.1054006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang T., Al Khleifat A., Meurgey J.H., Jones A., Leigh P.N., Bensimon G., et al. Stage at which riluzole treatment prolongs survival in patients with amyotrophic lateral sclerosis: a retrospective analysis of data from a dose-ranging study. Lancet Neurol. 2018;17(5):416–422. doi: 10.1016/S1474-4422(18)30054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lunetta C., Moglia C., Lizio A., Caponnetto C., Dubbioso R., Giannini F., et al. The Italian multicenter experience with edaravone in amyotrophic lateral sclerosis. J Neurol. 2020;267(11):3258–3267. doi: 10.1007/s00415-020-09993-z. [DOI] [PubMed] [Google Scholar]
- 10.Witzel S., Maier A., Steinbach R., Grosskreutz J., Koch J.C., Sarikidi A., et al. Safety and effectiveness of long-term intravenous administration of edaravone for treatment of patients with amyotrophic lateral sclerosis. JAMA Neurol. 2022;79(2):121–130. doi: 10.1001/jamaneurol.2021.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brooks B.R., Berry J.D., Ciepielewska M., Liu Y., Zambrano G.S., Zhang J., et al. Intravenous edaravone treatment in ALS and survival: An exploratory, retrospective, administrative claims analysis. EClinicalMedicine. 2022 4;52 doi: 10.1016/j.eclinm.2022.101590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGinley L. Washington post; Washington, DC: 2022. FDA approves first ALS drug in 5 years after pleas from patients.https://www.washingtonpost.com/health/2022/09/29/als-drug-fda-approval/ (Available at) [Google Scholar]
- 13.Amylyx . U.S. National Library of Medicine; Betheseda, MD: 2021. Phase III Trial of AMX0035 for amyotrophic lateral sclerosis treatment.https://ClinicalTrials.gov/show/NCT05021536 ([Internet] Available at) [Google Scholar]
- 14.Landers J.E., Melki J., Meininger V., Glass J.D., van den Berg L.H., van Es M.A., et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2009;106(22):9004–9009. doi: 10.1073/pnas.0812937106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baloh R.H. TDP-43: the relationship between protein aggregation and neurodegeneration in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. FEBS J. 2011;278(19):3539–3549. doi: 10.1111/j.1742-4658.2011.08256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGoldrick P., Lau A., You Z., Durcan T.M., Robertson J. Loss of C9orf72 perturbs the Ran-GTPase gradient and nucleocytoplasmic transport, generating compositionally diverse Importin β-1 granules. Cell Rep. 2023;42(3) doi: 10.1016/j.celrep.2023.112134. [DOI] [PubMed] [Google Scholar]
- 17.McGoldrick P., Robertson J. Unraveling the impact of disrupted nucleocytoplasmic transport systems in C9orf72-associated ALS. Front Cell Neurosci. 2023;17 doi: 10.3389/fncel.2023.1247297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding X., Xiang Z., Qin C., Chen Y., Tian H., Meng L., et al. Spreading of TDP-43 pathology via pyramidal tract induces ALS-like phenotypes in TDP-43 transgenic mice. Acta Neuropathol Commun. 2021;9(1):15. doi: 10.1186/s40478-020-01112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smethurst P., Risse E., Tyzack G.E., Mitchell J.S., Taha D.M., Chen Y.R., et al. Distinct responses of neurons and astrocytes to TDP-43 proteinopathy in amyotrophic lateral sclerosis. Brain. 2020 1;143(2):430–440. doi: 10.1093/brain/awz419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H.J., Mitchell J.C. Mechanisms of TDP-43 proteinopathy onset and propagation. Int J Mol Sci. 2021;22(11):6004. doi: 10.3390/ijms22116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane C.A., Hardy J., Schott J.M. Alzheimer's disease. Eur J Neurol. 2018;25(1):59–70. doi: 10.1111/ene.13439. [DOI] [PubMed] [Google Scholar]
- 22.Brookmeyer R., Gray S., Kawas C. Projections of Alzheimer's disease in the United States and the public health impact of delaying disease onset. Am J Public Health. 1998;88(9):1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Esquerda-Canals G., Montoliu-Gaya L., Güell-Bosch J., Villegas S. Mouse models of Alzheimer's disease. J Alzheimers Dis. 2017;57(4):1171–1183. doi: 10.3233/JAD-170045. [DOI] [PubMed] [Google Scholar]
- 24.Golde T.E. Disease-modifying therapies for Alzheimer's disease: more questions than answers. Neurotherapeutics. 2022;19(1):209–227. doi: 10.1007/s13311-022-01201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buccellato F.R., D'Anca M., Tartaglia G.M., Del Fabbro M., Scarpini E., Galimberti D. Treatment of Alzheimer's disease: beyond symptomatic therapies. Int J Mol Sci. 2023;24(18):13900. doi: 10.3390/ijms241813900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cummings J., Zhou Y., Lee G., Zhong K., Fonseca J., Cheng F. Alzheimer's disease drug development pipeline: 2023. Alzheimers Dement (N Y) 2023 25;9(2) doi: 10.1002/trc2.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dyck C.H., Swanson C.J., Aisen P., Bateman R.J., Chen C., Gee M., et al. Lecanemab in Early Alzheimer's Disease. N Engl J Med. 2023;388(1):9–21. doi: 10.1056/NEJMoa2212948]. [DOI] [PubMed] [Google Scholar]
- 28.Nativio R., Lan Y., Donahue G., Sidoli S., Berson A., Srinivasan A.R., et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer's disease. Nat Genet. 2020;52(10):1024–1035. doi: 10.1038/s41588-020-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung J., Das A., Sun X., Sobreira D.R., Leung Y.Y., Igartua C., et al. Genome-wide association and multi-omics studies identify MGMT as a novel risk gene for Alzheimer's disease among women. Alzheimers Dement. 2022 doi: 10.1002/alz.12719. 10.1002/alz.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baloni P., Arnold M., Buitrago L., Nho K., Moreno H., Huynh K., et al. Multi-Omic analyses characterize the ceramide/sphingomyelin pathway as a therapeutic target in Alzheimer's disease. Commun Biol. 2022;5(1) doi: 10.1038/s42003-022-04011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivanandy P., Leey T.C., Xiang T.C., Ling T.C., Wey Han S.A., Semilan S.L.A., et al. Systematic review on parkinson's disease medications, emphasizing on three recently approved drugs to control Parkinson's symptoms. Int J Environ Res Public Health. 2021;19(1):364. doi: 10.3390/ijerph19010364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bette S., Shpiner D.S., Singer C., Moore H. Safinamide in the management of patients with Parkinson's disease not stabilized on levodopa: a review of the current clinical evidence. Ther Clin Risk Manag. 2018;14:1737–1745. doi: 10.2147/TCRM.S139545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J.F., Cunha R.A. The belated US FDA approval of the adenosine A2A receptor antagonist istradefylline for treatment of Parkinson's disease. Purinergic Signal. 2020;16(2):167–174. doi: 10.1007/s11302-020-09694-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno G.M., Gandhi R., Lessig S.L., Wright B., Litvan I., Nahab F.B. Mortality in patients with Parkinson disease psychosis receiving pimavanserin and quetiapine. Neurology. 2018;91(17):797–799. doi: 10.1212/WNL.0000000000006396. [DOI] [PubMed] [Google Scholar]
- 35.Schilder B.M., Raj T. Fine-mapping of Parkinson's disease susceptibility loci identifies putative causal variants. Hum Mol Genet. 2022;31(6):888–900. doi: 10.1093/hmg/ddab294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schilder B.M., Navarro E., Raj T. Multi-omic insights into Parkinson's Disease: From genetic associations to functional mechanisms. Neurobiol Dis. 2022;163 doi: 10.1016/j.nbd.2021.105580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margoni M., Preziosa P., Filippi M., Rocca M.A. Anti-CD20 therapies for multiple sclerosis: current status and future perspectives. J Neurol. 2022;269(3):1316–1334. doi: 10.1007/s00415-021-10744-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Compston A., Coles A. Multiple sclerosis. Lancet. 2008;372(9648):1502–1517. doi: 10.1016/S0140-6736(08)61620-7. [DOI] [PubMed] [Google Scholar]
- 39.Ascherio A., Munger K.L. Environmental risk factors for multiple sclerosis. Part II: noninfectious factors. Ann Neurol. 2007;61(6):504–513. doi: 10.1002/ana.21141. [DOI] [PubMed] [Google Scholar]
- 40.La Mantia L., Di Pietrantonj C., Rovaris M., Rigon G., Frau S., Berardo F., et al. Comparative efficacy of interferon β versus glatiramer acetate for relapsing-remitting multiple sclerosis. J Neurol Neurosurg Psychiatry. 2015;86(9):1016–1020. doi: 10.1136/jnnp-2014-309243. [DOI] [PubMed] [Google Scholar]
- 41.Lee A. Ublituximab: first approval. Drugs. 2023;83(5):455–459. doi: 10.1007/s40265-023-01854-z. [DOI] [PubMed] [Google Scholar]
- 42.Alnaif A., Oiler I., D'Souza M.S. Ponesimod: an oral second-generation selective sphingosine 1-phosphate receptor modulator for the treatment of multiple sclerosis. Ann Pharm. 2023;57(8):956–965. doi: 10.1177/10600280221140480. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y., Cuellar-Partida G., Simpson Yap S., Lin X., Claflin S., Burdon K., et al. Utilising multi-large omics data to elucidate biological mechanisms within multiple sclerosis genetic susceptibility loci. Mult Scler. 2021;27(14):2141–2149. doi: 10.1177/13524585211004422. [DOI] [PubMed] [Google Scholar]
- 44.Taylor J.P., McKeith I.G., Burn D.J., Boeve B.F., Weintraub D., Bamford C., et al. New evidence on the management of Lewy body dementia. Lancet Neurol. 2020;19(2):157–169. doi: 10.1016/S1474-4422(19)30153-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanford A.M. Lewy body dementia. Clin Geriatr Med. 2018;34(4):603–615. doi: 10.1016/j.cger.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 46.MacDonald S., Shah A.S., Tousi B. Current therapies and drug development pipeline in lewy body dementia: an update. Drugs Aging. 2022;39(7):505–522. doi: 10.1007/s40266-022-00939-w. [DOI] [PubMed] [Google Scholar]
- 47.Sabbagh M.N., Taylor A., Galasko D., Galvin J.E., Goldman J.G., Leverenz J.B., et al. Listening session with the US Food and drug administration, lewy body dementia association, and an expert panel. Alzheimers Dement (N Y) 2023;9(1) doi: 10.1002/trc2.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wainberg M., Andrews S.J., Tripathy S.J. Shared genetic risk loci between Alzheimer’s disease and related dementias, Parkinson’s disease, and amyotrophic lateral sclerosis. Alz Res Ther. 2023;15:113. doi: 10.1186/s13195-023-01244-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurie C.C., Doheny K.F., Mirel D.B., Pugh E.W., Bierut L.J., Bhangale T., et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34(6):591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marees A.T., de Kluiver H., Stringer S., Vorspan F., Curis E., Marie-Claire C., et al. A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res. 2018;27(2) doi: 10.1002/mpr.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lazar C., Meganck S., Taminau J., Steenhoff D., Coletta A., Molter C., et al. Batch effect removal methods for microarray gene expression data integration: a survey. Brief Bioinform. 2013;14(4):469–490. doi: 10.1093/bib/bbs037. [DOI] [PubMed] [Google Scholar]
- 54.Pe'er I., Yelensky R., Altshuler D., Daly M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 55.Clarke G.M., Anderson C.A., Pettersson F.H., Cardon L.R., Morris A.P., Zondervan K.T. Basic statistical analysis in genetic case-control studies. Nat Protoc. 2011;6(2):121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnaudi M. , Beltrame L. , Degn K. , Utichi M. , Pettenella A. , Scrima S. , et al. MAVISp: Multi-layered Assessment of VarIants by Structure for proteins bioRxiv 2022.10.22.513328; doi: https://doi.org/10.1101/2022.10.22.513328.
- 57.Henrie A., Hemphill S.E., Ruiz-Schultz N., Cushman B., DiStefano M.T., Azzariti D., et al. ClinVar miner: demonstrating utility of a web-based tool for viewing and filtering ClinVar data. Hum Mutat. 2018;39(8):1051–1060. doi: 10.1002/humu.23555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiberti M., Di Leo L., Vistesen M.V., Kuhre R.S., Cecconi F., De Zio D., et al. The Cancermuts software package for the prioritization of missense cancer variants: a case study of AMBRA1 in melanoma. Cell Death Dis. 2022;13(10):872. doi: 10.1038/s41419-022-05318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., et al. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596(7873):590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tiberti M., Terkelsen T., Degn K., Beltrame L., Cremers T.C., da Piedade I., et al. MutateX: an automated pipeline for in silico saturation mutagenesis of protein structures and structural ensembles. Brief Bioinform. 2022;23(3):bbac074. doi: 10.1093/bib/bbac074. [DOI] [PubMed] [Google Scholar]
- 61.Sora V., Laspiur A.O., Degn K., Arnaudi M., Utichi M., Beltrame L., et al. RosettaDDGPrediction for high-throughput mutational scans: From stability to binding. Protein Sci. 2023;32(1) doi: 10.1002/pro.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arnaudi M. , Beltrame L. , Degn K. , Utichi M. , Pettenella A. , Scrima S. , et al. MAVISp: Multi-layered Assessment of VarIants by Structure for proteins bioRxiv 2022.10.22.513328; doi: https://doi.org/10.1101/2022.10.22.513328.
- 63.Lu T.P., Lee C.Y., Tsai M.H., Chiu Y.C., Hsiao C.K., Lai L.C., et al. miRSystem: an integrated system for characterizing enriched functions and pathways of microRNA targets. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ni G., Moser G., Schizophrenia Working Group of the Psychiatric Genomics Consortium, Wray N.R., Lee S.H. Estimation of genetic correlation via linkage disequilibrium score regression and genomic restricted maximum likelihood. Am J Hum Genet. 2018;102(6):1185–1194. doi: 10.1016/j.ajhg.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smigielski E.M., Sirotkin K., Ward M., Sherry S.T. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28(1):352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brenner D., Yilmaz R., Müller K., Grehl T., Petri S., Meyer T., et al. Hot-spot KIF5A mutations cause familial ALS. Brain. 2018;141(3):688–697. doi: 10.1093/brain/awx370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng H., Song F., Zhang L., Yang D., Ji P., Wang Y., et al. Genetic variants at the miR-124 binding site on the cytoskeleton-organizing IQGAP1 gene confer differential predisposition to breast cancer. Int J Oncol. 2011;38(4):1153–1161. doi: 10.3892/ijo.2011.940]. [DOI] [PubMed] [Google Scholar]
- 69.Chhichholiya Y., Suryan A.K., Suman P., Munshi A., Singh S. SNPs in miRNAs and target sequences: role in cancer and diabetes. Front Genet. 2021;12 doi: 10.3389/fgene.2021.793523]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.https://www.genecards.org/cgi-bin/carddisp.pl?gene=HLA-DRB1.
- 71.https://www.genecards.org/cgi-bin/carddisp.pl?gene=TMEM175&keywords=TMEM17.
- 72.https://www.genecards.org/cgi-bin/carddisp.pl?gene=BIN1.
- 73.Thiel G. Synapsin I, synapsin II, and synaptophysin: marker proteins of synaptic vesicles. Brain Pathol. 1993;3(1):87–95. doi: 10.1111/j.1750-3639.1993.tb00729.x. [DOI] [PubMed] [Google Scholar]
- 74.Livingston G., Sommerlad A., Orgeta V., Costafreda S.G., Huntley J., Ames D., et al. Dementia prevention, intervention, and care. Lancet. 2017;390(10113):2673–2734. doi: 10.1016/S0140-6736(17)31363-6. [DOI] [PubMed] [Google Scholar]
- 75.Stocker H., Perna L., Weigl K., Möllers T., Schöttker B., Thomsen H., et al. Prediction of clinical diagnosis of Alzheimer's disease, vascular, mixed, and all-cause dementia by a polygenic risk score and APOE status in a community-based cohort prospectively followed over 17 years. Mol Psychiatry. 2021;26(10):5812–5822. doi: 10.1038/s41380-020-0764-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Visel A., Rubin E.M., Pennacchio L.A. Genomic views of distant-acting enhancers. Nature. 2009;461(7261):199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brodie A., Azaria J.R., Ofran Y. How far from the SNP may the causative genes be? Nucleic Acids Res. 2016;44(13):6046–6054. doi: 10.1093/nar/gkw500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freischmidt A., Müller K., Zondler L., Weydt P., Volk A.E., Božič A.L., et al. Serum microRNAs in patients with genetic amyotrophic lateral sclerosis and pre-manifest mutation carriers. Brain. 2014;137(Pt 11):2938–2950. doi: 10.1093/brain/awu249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Table 1: Genetic correlations related to different phenotypic groups in ALS, AD, PD and MS.
Supplementary material Table 2: SNPs that obtained independent associations with the 5 neurodegenerative diseases.
Supplementary material Table 3: Functional annotations of genes containing the SNPs associated with the 5 neurodegenerative diseases.
Supplementary material Table 4: The table shows differentially expressed miRNAs associated with the neurodegenerative diseases: ALS, AD, PD, MS, and LBD. The first column shows the miRNAs, the second column shows the corresponding FDR values and the third column shows the logFC values.
Data Availability Statement
GWAS summary statistics of ALS, MS and LBD were downloaded from the GWAS catalog (Accessed on May 2022; https://www.ebi.ac.uk/gwas/docs/about); AD dataset from the GWAS Atlas, (Accessed on May 2022; https://atlas.ctglab.nl/documentation) and finally the PD dataset from the IEU OpenGwas project (Accessed on May 2022; https://gwas.mrcieu.ac.uk/about/). Summary statistics related to human traits are available from the United Kingdom Biobank database (http://www.nealelab.is/uk-biobank). Gene expression profiles are available from Gene Expression Omnibus (Home - GEO - NCBI (nih.gov)). The data with the MAVISp assessment on the protein-coding variant are reported in the MAVISp database:https://services.healthtech.dtu.dk/services/MAVISp-1.0/