Abstract
In the past decade, there has been increasing interest in use of small molecules for immunomodulation. The affinity-based pull-down purification is an essential tool for target identification of small molecules and drug discovery. This study presents our recent efforts to investigate the cellular target(s) of Compound A, a small molecule with demonstrated immunomodulatory properties in human peripheral blood mononuclear cells (PBMCs). While we have previously observed the immunomodulatory activity of Compound A in PBMCs, the specific molecular targets underlying its effects remains elusive. To address this challenge, we synthesized a trifluoromethyl phenyl diazirine (TPD)-bearing trifunctional Probe 1 based on the chemical structure of Compound A, which could be used in a pull-down assay to efficiently bind to putative cellular targets via photoaffinity labelling. In this report, we utilized bovine serum albumin (BSA) as a model protein to establish a proof-of-concept in order to assess the suitability of Probe 1 for binding to an endogenous target. By the successful synthesis of Probe 1 and demonstrating the efficient binding of Probe 1 to BSA, we propose that this method can be used as a tool for further identification of potential protein targets of small molecules in living cells. Our findings provide a valuable starting point for further investigations into the molecular mechanisms underlying the immunomodulatory effects of Compound A.
Graphical abstract
1. Introduction
Immunotherapy has become a promising approach to treating cancer and other diseases by harnessing the power of the immune system. It works by stimulating or enhancing the body's natural defences to recognise and attack cancer cells or other harmful agents [1,2]. The identification of the cellular targets and their underlying mechanisms responsible for immunomodulatory activities is one of the obstacles in the development of effective immunotherapies [1,3].
In order to overcome this challenge, drug discovery researchers have turned to advanced chemical biology techniques, such as bio-orthogonal reactions and photoaffinity labelling (PAL), to elucidate the molecular mechanisms of immunomodulatory compounds [4,5]. By combining these two innovative techniques, a trifunctional probe molecule can serve as a powerful tool to access a biologically significant space or target. A trifunctional approach utilizes a small molecule containing three key components: an affinity warhead, a photo-crosslinker, and a bio-orthogonal handle [6]. The general process of this approach involves the pre-association between the molecular probe and the interacting target, exposure to a selected ultraviolet (UV) irradiation, and a bio-orthogonal reaction to introduce a desired reporter tag (Fig. 1A). This method allows the performance of a variety of biological assays, including affinity pull-down purification or related microscopic analyses [7,8].
Fig. 1.
A) The trifunctional approach. B) Schematic photoaffinity labeling with the photo-crosslinker trifluoromethyl phenyl diazirine. C) The proposed trifluoromethyl phenyl diazirine (TPD) probe molecule.
Photoaffinity labelling makes use of a photo-labile functionality, namely a photo-crosslinker, to construct a covalent bond with the target molecule when in close proximity [9]. This technique relies on the generation of reactive species, such as a carbene or radical, followed by bond insertion or cross-linking with the endogenous target (Fig. 1B). In comparison, methods that rely on non-covalent interactions have diminished success due to the extensive washing conditions required for the isolation of target–molecule complex. Photoaffinity labelling has the potential to overcome this downside and improves the likelihood of isolating the target–probe complex of interest [10]. Bio-orthogonal chemistry, on the other hand, has emerged as a powerful tool in the field of chemical biology [11,12]. This concept was brought up by the Bertozzi group and adapted widely to biological settings [13]. Among them, copper-catalyzed azide-alkyne cycloaddition (CuAAC) or later coined “click reaction”, was proposed and has been studied extensively by Sharpless and Meldal since the 2000s [[14], [15], [16]]. Bio-orthogonal reactions enable chemical reactions in living organisms with high yield, high rate, and good selectivity. This is often used toward the installation of a reporter tag or a fluorophore for further analyses.
With the help of click chemistry and PAL techniques, researchers are able to identify specific protein targets of immunomodulatory compounds and understand their mode of action, which can aid the development of more effective immunotherapies [17,18] These advanced chemical techniques have also allowed for the design and synthesis of novel immunomodulatory compounds with improved efficacy and specificity [[19], [20], [21]]. Small molecules have become a promising class of immunotherapy drugs. They have the potential to overcome some of the limitations of traditional immunotherapies, such as their high cost and adverse side effects [20,22]. They work by targeting specific molecules involved in the immune response, which can lead to more selective and effective treatments. Additionally, small molecule-based therapies have the advantages of oral administration, whose convenience increases patient compliance, as well as relatively rapid clearance, which diminishes the risk of a serious adverse immune response. As a result, they have become a prospective area of research in the field of immunotherapy [1,23].
Our team has recently published a report on Compound A (Fig. 1C), a small molecule with immunomodulatory properties inducing interleukin (IL-2 secretion) and T-cell proliferation in human PMBCs [24]. While we have observed these significant immunomodulatory activities of Compound A, the cellular target(s) underlying its effects have yet to be identified. Given the complex nature of studying the mixed cellular populations of BMCs, we sought to develop a procedure of a pull-down purification in combination with click chemistry and photoaffinity labelling. This experiment aims to demonstrate the efficient binding of Probe 1 to plausible cellular targets using a non-cellular protein model. Therefore, BSA was utilized as the model and interacting protein with Probe 1 to establish a proof-of-concept method of protein pull down. BSA was selected as the model protein for its desired character of binding to small, drug-like molecules [25]. The goal of this study is to demonstrate efficient labeling of BSA by Probe 1, which would validate the use of Probe 1 in cell lysates or live cells in order to isolate the unknown target(s) of Compound A.
2. Methodology
2.1. Synthesis of TPD-containing trifunctional probes
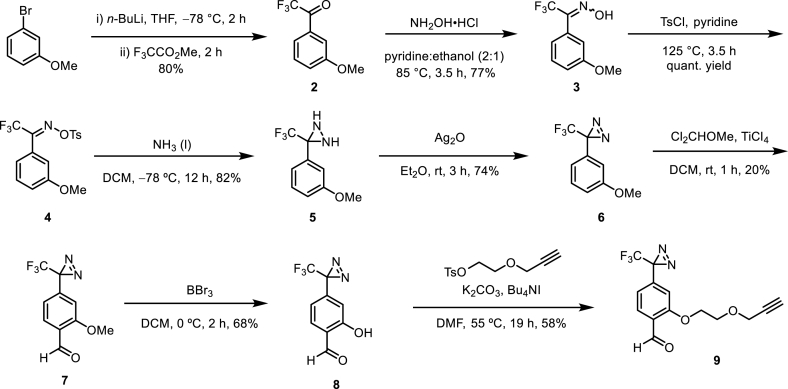
Taking into consideration of the chemical structure of Compound A and the efficacy between different photo-crosslinkers, we identified the TPD-containing trifunctional Probe 1 as our target compound (Fig. 1D). We initially examined the recently published method by Kumar and Manetch (23), but ultimately opted to follow the more established route reported by Mayer and Maier [26]. Beginning from 3-bromoanisole, reaction with n-BuLi and methyl trifluoroacetate afforded aryl trifluoromethyl ketone 2. Subsequent reaction with hydroxylamine hydrochloride yielded oxime 3, which could be readily converted to tosyloxime 4. Diaziridination with liquid ammonia followed by oxidation using silver (II) oxide afforded diazirine 6. The formyl group para to the diazirine was installed via a Friedel–Crafts alkylation using dichloromethyl methyl ether. Finally, the alkyne handle was installed through a sequential demethylation and SN2 reaction with the tosylated alkyne to furnish the desired TPD 9. (Scheme 1).
Scheme 1.
The synthesis of the of the TPD–aldehyde building block 9.
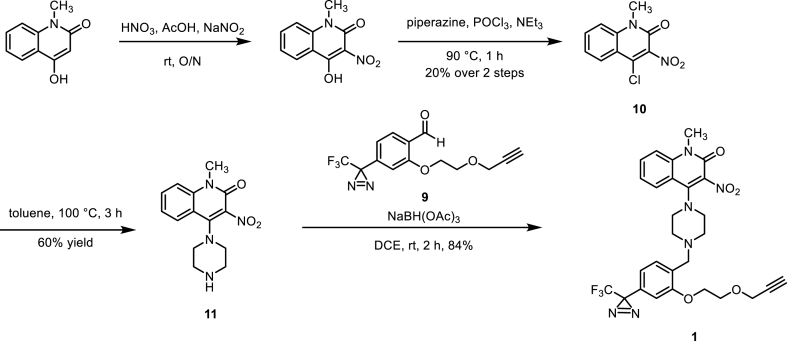
On the other hand, the synthesis of the affinity component started from the commercially available 4-hydroxy-1-methylquinolin-2(1H)-one. The enol form of 4-hydroxy-1-methylquinolin-2(1H)-one underwent a nitration using nitric acid. Next, substitution of the hydroxy group with a chlorine at the beta-carbonyl position was achieved by using phosphorus(V) oxychloride (POCl3). The affinity warhead was then synthesized through an SNAr reaction between intermediate 10 and piperazine. Finally, reductive amination with TPD 9 and the affinity warhead 11 allowed for the construction of TPD-containing trifunctional Probe 1 in good yield (Scheme 2).
Scheme 2.
The synthesis of the affinity component 11 and the final Probe 1.
2.2. Assessment of TPD stability with different reducing agents
Before performing the click reaction of the TPD-containing trifunctional Probe 1 with a protein, we sought to examine the stability of the TPD functionality under the selected reaction conditions and its compatibility with different reducing agents. To elaborate, the synthesized TPD Probe 1 was mixed properly with the chosen CuAAC reagents, including copper catalyst, ligand, and reducing reagents. Two different reducing reagents, sodium ascorbate [15,27] and tris(2-carboxyethyl)phosphine (TCEP) [28]; [29], were chosen to compare their compatibility with the TPD Probe 1.
2.2.1. Experimental for the stability test of the TPD trifunctional probe 1
-
1.
The stock solutions of the TPD Probe 1 in DMSO (1 mM), CuSO4·5H2O in water (20 mM), sodium ascorbate in water (200 mM), TCEP·HCl in NaOH solution (200 mM), and TBTA in DMSO (10 mM) were prepared prior to the experiment.
-
2.
To one chromatography vial was added sequentially with the stock solutions of Probe 1 (20 μL), CuSO4·5H2O (25 μL), TCEP·HCl (25 μL), TBTA (25 μL), and water (400 μL). To another new chromatography vial was added with the stock solutions of Probe 1 (20 μL), CuSO4·5H2O (25 μL), sodium ascorbate (25 μL), TBTA (25 μL), and water (400 μL).
-
3.
The vials were vortexed and sealed properly with plastic caps.
-
4.
The vials were covered with aluminum foil and kept in the dark for an hour.
-
5.
After 1 h, the vials were directly submitted to LC-MS for analysis.
2.2.2. The selection of reducing agent for the click reaction condition
-
1.
The stock solution of biotin–(PEG)3–azide in DMSO (2 mM) were prepared.
-
2.
To one chromatography vial was added sequentially with the stock solutions of Probe 1 (20 μL), water (375 μL), biotin–(PEG)3–azide (20 μL), CuSO4·5H2O (25 μL), Tris(benzyltriazolylmethyl)amine (TBTA) (25 μL), and sodium ascorbate (25 μL). To another new chromatography vial was added with the stock solutions of Probe 1 (20 μL), water (375 μL), azide (20 μL), CuSO4·5H2O (25 μL), TBTA (25 μL), and TCEP·HCl (25 μL).
-
3.
The vials were vortexed and sealed properly with plastic caps.
-
4.
The vials were covered with aluminum foil and kept in the dark for an hour.
-
5.
After 1 h and 3 h, 100 μL of sample was taken out each time and submitted for LC-MS analyses to know the stability of the synthesized Compound A (data not shown).
2.3. Optimization of irradiation time for photoaffinity labelling
After the establishment of the CuAAC condition, we sought to determine the optimal time for the photoaffinity labelling experiment. Methanol was used as a model substrate to measure the consumption of the generated carbene under UV irradiation [26]. In this experiment, the TPD-containing Probe 1 was dissolved in methanol in glass vials before exposed to UV light, while one sample was kept in the dark as the negative control. The samples were irradiated over different time periods, including 30, 60, 90, and 120 min, followed by LC-MS analysis.
2.3.1. The optimization of photoaffinity labelling reaction
-
1.
A stock solution of Probe 1 in methanol (1 mM) was prepared.
-
2.
The prepared stock solution (a volume of 100 μL each) was added to five 5 mL glass vials.
-
3.
The reaction solutions were purged with N2 and sealed with a plastic cap and parafilm.
-
4.
One vial was wrapped with aluminum foil and kept in the dark to serve as the negative control. The other four vials were placed 3 cm underneath a UV lamp that was pre-warmed for 30 min.
-
5.
After 30, 60, 90, and 120 min, one vial was removed from the light each time and submitted for LC-MS analysis immediately.
2.4. Measuring IL-2 production in human PBMCs
The concentration of IL-2 was quantified by ELISA kits (R&D, Cat. No. DY202) using PBMC samples from healthy human participants. PBMCs were obtained by leukapheresis from healthy human controls, thawed, and cultured in RPMI 1640 with 10 % FBS (Sigma-Aldrich, USA) and 1 % penicillin/streptomycin (Sigma-Aldrich, USA). PBMCs (2 × 105 cells) were seeded in a 96-well U-bottomed plate and stimulated with 100 ng/mL Staphylococcal Enterotoxin B (SEB) (Sigma-Aldrich, USA) for 72 h with or without probe 1. The research was approved by the University of Alberta's Institutional Review Board (IRB #Pro00046064). All participants provided written informed consent prior to participating in the study [24,30].
2.5. BSA labelling experiments
With the optimized reaction conditions in hand, we designed a proof-of-concept BSA labelling experiment utilizing the synthesized Probe 1 (the stepwise method is listed below). In this labelling experiment, the CuAAC was performed first. The formation of the cycloaddition product 12 and the consumption of the TPD Probe 1 were monitored by LC-MS. Next, the reaction mixture containing the cycloaddition product was treated directly to a solution of BSA in PBS buffer without further purification. The sample was placed in the dark for incubation, followed by 1 h of UV irradiation to enable the formation of the covalent interaction between BSA and the cycloaddition product 12. Then, a C18 reverse-phase column was used to remove unreacted cycloaddition product 12 and salt presented in the buffer (Fig. 2). Finally, the irradiated sample was analyzed by matrix-assisted laser desorption/ionization (MALDI) (Fig. 3).
Fig. 2.
The workflow of the proof-of-concept BSA labelling experiment.
Fig. 3.
The MALDI result of the first photoaffinity labelling experiment.
2.5.1. The BSA labelling protocol
-
1.
The stock solutions of the TPD Probe 1 in DMSO (3 mM), biotin–(PEG)3–azide in DMSO (12 mM), CuSO4·5H2O in water (20 mM), sodium ascorbate in water (200 mM), and TBTA in DMSO (100 mM) were prepared prior to the experiment.
-
2.
In 5 mL glass vials was added sequentially with the stock solutions of Probe 1 (20 μL), water (375 μL), azide (20 μL), CuSO4·5H2O (25 μL), TBTA (25 μL), and sodium ascorbate (25 μL).
-
3.
The reaction was vortexed, sealed with a plastic cap, and wrapped with aluminum foil overnight.
-
4.
The reaction progress was accessed by LC-MS after 24 h. LC-MS indicated that the diazirine starting material almost was consumed after 24 h, and the signal corresponding to the triazole product with correct molecular weight was observed.
-
5.
Assuming that the click reaction went to completion, 350 μL of the reaction mixture was added to 250 μL of BSA solution (1 mg/mL in PBS).
-
6.
The sample was vortexed for three times and kept under room temperature in the dark for 20 min to ensure proper mixture of BSA and the compound.
-
7.
The sample was placed 3 cm underneath a UV lamp and irradiated for an hour.
-
8.
The irradiated sample was desalted with a C18 column according to the procedure provided by the manufacturer.
-
9.
The purified protein sample was subjected to MALDI analyses.
2.6. Pull down assay using BSA and probe 1
-
1)
The stock solutions of the TPD Probe 1 in DMSO (2.125 mM), biotin–(PEG)3–azide in DMSO (4 mM), CuSO4·5H2O in PBS (2 mM), sodium ascorbate in PBS (66.66 mM), and TBTA in DMSO (3.33 mM), were prepared prior to the experiment. For this experiment involving pull down assay with streptavidin, the reducing agents were dissolved in PBS.
-
2)
Preparation of the Probe 1–BSA reaction mix: 15.9 μg (15) of BSA was dissolved in 19.4 μL of PBS in an Eppendorf tube and then 1.1 μL of Probe 1 (stock concentration of 2.125 mM in DMSO) was added to the mix. The ratio of BSA to Probe 1 was 1:10. Note: The ratio of BSA to Probe 1 is variable depending upon the nature of Probe 1 that needs to be optimized.
-
3)
Incubation of the reaction mix: Incubated the above mixture at 37 °C in a 5 % CO2 incubator for 6 h (the incubation time was based on the time of onset of immunostimulatory effect of Compound A in PBMCs).
-
4)
UV illumination: At the end of the incubation period, the mixture in an Eppendorf tube was irradiated with UV illumination for 1 h at room temperature using a UV lamp at 350 nm (Spectroline, ENF-240C, US).
-
5)
Initiation of the click chemistry reaction: Subsequently, 1.2 μL biotin–(PEG)3–azide (>2x of Probe 1), 1.5 μL CuSO4, 4.5 μL TBTA and 2.25 μL (total = 9.45 μL) sodium ascorbate were added sequentially to a total volume of 30 μL. Note: The ratio of CuSO4: TBTA: sodium ascorbate was 1:5:10. The above click reagents were added to the sample and incubated at RT for 1.3 h in dark
-
6)
Addition of prewashed streptavidin to the above reaction mixture. For our experiment, 5.6 μL (3 μg of biotin–conjugated BSA) of the after–click reaction mix (AC) was mixed in 90 μL of prewashed Streptavidin–Agarose bead slurry (Catalogue #16–126; Upstate cell signaling solutions, USA).
-
7)
Incubation at 4 °C: The above reaction mixture was incubated at 4 °C in a three-dimensional rotating mixer (Diamed, Canada) for 1 h and was subsequently centrifuged at 1000 g for 5 min at RT. The supernatant was aspirated and stored at 4 °C until loading.
-
8)
Wash of the beads: The washing buffer was prepared separately by mixing Tris (151.42 mg) with NaCl (0.5825 g) and 2.5 mL of 10 % SDS all in 22.5 mL of water. 1 mL washing buffer was added to each sample and incubated for 5 min at room temperature on a rotor. Centrifuged 1000 g for 5 min at room temperature for three times, repeated the above three steps thrice. The supernatant was aspirated and discarded.
-
9)
Release of proteins from beads: 30 μL of 4x Laemmli sample buffer (from Bio-Rad; cat #160747, premixed with β-mercaptoethanol), was added to the sample, incubated at 95 °C for 5 min to facilitate the release of beads from the protein. The samples were centrifuged at high speed (13000 g) for 1 min at room temperature and stored at −20 °C until loading.
-
10)
Western blotting: In a control pull-down experiment, the different amounts of BSA alone, the after–click sample (3 μg of AC) along with the ladder (LD) in comparison to Probe 12 were separated by 2.5 % sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Additionally, the supernatant following the step 5 (Sup) was also separated. The gel was electrophoresed at 200 V for 1 h, and proteins were transferred onto nitrocellulose membrane (Bio-Rad, catalogue # 1620115, Germany), The transfer was then run at 20 V for 1.20 min on a 1.5 mm gel, ensuring proper transfer. The membranes were blocked with blocking buffer (5 % milk in TBST) for 1 h at room temperature on a rocking platform (VWR, USA). Subsequently, the blots were incubated overnight at 4 °C with streptavidin IRDYE 680LT (Licor; catalogue # 926–68031, USA) in 5 % milk at 1:1000. The blot was washed three times with TBST for 10 min and imaged using an IR dye labelling instrument using Studio Lite software (LI-COR, Biosciences).
3. Results
3.1. Synthesis of TPD-containing trifunctional probes
As the click chemistry has been recently emerging as a modern tool to identify the cellular targets, in order to exploit this approach, we attempted to synthesize TPD trifunctional Probe 1 based on the structure of Compound A.
3.1.1. Compound data
3.1.1.1. 2-(2-(Prop-2-yn-1-yloxy)ethoxy)-4-(3-(trifluoromethyl)-3H-diazirin-3-yl)benzaldehyde (9)
The compound was synthesized according to a previously reported literature procedure [26], employing 2-hydroxy-4-(3-(trifluoromethyl)-3H-diazirin-3-yl)benzaldehyde (0.75 mmol, 0.17 g) the tosylated alkyne (0.84 mmol, 0.21 g), tetrabutylammonium iodide (Bu4NI) (0.083 mmol, 31 mg), K2CO3 (1.3 mmol, 0.18 mg), and DMF (18 mL) to give 9 (0.14 g, yield 58 %) as a pale yellow solid. Rf 0.25 (1:6 petroleum ether: diethyl ether); 1H NMR (500 MHz, CDCl3) δ 10.49 (s, 1H), 7.85 (d, J = 8.0 Hz, 1H), 6.85 (d, J = 8.0 Hz, 1H), 6.75 (s, 1H), 4.33–4.21 (m, 4H), 3.99–3.94 (m, 2H), 2.48 (t, J = 2.4 Hz, 1H); 13C {1H} NMR (125 MHz, CDCl3) δ 188.8, 160.9, 136.6, 128.9, 125.7, 121.8 (q, 1JC–F = 274 Hz, CF3), 119.0, 110.9, 79.1, 75.1, 68.4, 67.8, 58.6, 28.6 (q, 2JC–F = 41 Hz, CCF3); HRMS (ESI, [M+Na]+) calcd for C14H11F3N2NaO3 335.0614, found: m/z 335.0606. The TPD-containing compounds are stored in the dark to avoid light-mediated decomposition.
3.1.1.2. 4-Chloro-1-methyl-3-nitroquinolin-2(1H)-one (10)
In a 25 mL round bottom flask, a catalytic amount of NaNO2 was added to a stirring solution of 4-hydroxy-1-methylquinolin-2(1H)-one (2.86 mmol, 500 mg) in glacial acetic acid (5.0 mL) and concentrated nitric acid (500 μL). The flask was capped with a septum, and a needle was applied to serve as a vent. The reaction was kept under room temperature and stirred for 16 h. The reaction mixture was filtered, and the solid was dried in an oven for 30 min to provide a yellow solid (425 mg, yield 68 %) and used in the next step without further purification. Next, to a new, oven dried round bottom flask equipped with a stir bar and a rubber septum, was charged with the nitro intermediate (1.11 mmol, 245 mg). The system was sealed and purged with nitrogen. Then, phosphorus oxychloride (2.0 mL) and triethylamine (0.1 mL) were added to the flask. The reaction mixture was heated to 90 °C and stirred for 2 h and before cooled down to room temperature. The reaction mixture was added with ice-cold water (20 mL) and extracted with ethyl acetate (20 mL). The organic layer was separated, dried over MgSO4, filtered, and concentrated under reduced pressure. The crude mixture was purified by flash column chromatography, employing 1:1 ethyl acetate: hexanes as eluent to give 10 (130 mg, yield 20 %): Rf 0.49 (1:1 ethyl acetate:hexanes); IR (cast film): 3049, 2922, 2852, 1730, 1664, 1597, 1538, 1457, 1354, 1191, 1088, 767, 756 cm−1; 1H NMR (500 MHz, CDCl3) δ 8.13 (dd, J = 8.0, 1.5 Hz, 1H), 7.81–7.78 (m, 1H), 7.51–7.43 (m, 2H), 3.80 (s, 3H). 13C {1H} NMR (125 MHz, CDCl3) δ 153.8, 138.7, 135.9, 134.1, 127.6, 124.2, 116.8, 114.9, 30.5, one of the carbons signal was not observed likely due to spectral overlap; HRMS (ESI, [M+H]+) calcd for C10H7ClN2O3 239.0218; found: m/z 239.0218.
3.1.1.3. 1-Methyl-3-nitro-4-(piperazin-1-yl)quinolin-2(1H)-one (11)
To a 25 mL round bottom flask equipped with a magnetic stir bar was added 10 (1.26 mmol, 300 mg), piperazine (3.78 mmol, 325 mg) and toluene (2 mL). The flask was capped with a septum, and a needle was applied to serve as a vent. The reaction was heated to 100 °C and stirred for 3 h. The reaction progress was monitored by thin layer chromatography and LC-MS. Upon reaction completion, the reaction mixture was diluted with ethyl acetate (20 mL) and washed with saturated aqueous Na2CO3 (10 mL) and H2O (10 mL). The organic layer was dried over MgSO4, filtered, and concentrated under reduced pressure. The crude was purified by flash column chromatography, employing 1:9 methanol:ethyl acetate solution as eluent to afford 11 as light yellow sticky oil (216 mg, yield 60 %). Rf 0.19 (1:4 methanol:ethyl acetate); IR (cast film): 3333, 3053, 2951, 2916, 1849, 1650, 1613, 1593, 1527, 1460, 1398, 1327, 1189, 1084, 1025, 763 cm−1; 1H NMR (500 MHz, CDCl3) δ 7.96 (dd, J = 8.5, 1.5 Hz, 1H), 7.65 (ddd, J = 8.5, 7.0, 1.5 Hz, 1H), 7.39 (d, J = 8.0 Hz, 1H), 7.31 (ddd, J = 8.0, 7.0, 1.5 Hz, 1H), 3.71 (s, 3H), 3.25–3.19 (m, 4H), 3.11–3.05 (m, 4H), 1.70 (br s, 1H); 13C {1H} NMR (125 MHz, CDCl3) δ 156.2, 149.2, 139.7, 134.9, 132.6, 126.6, 122.7, 117.5, 115.1, 51.7, 46.1, 30.0.; HRMS (ESI, [M+H]+) calcd for C14H17N4O3 289.1295; found: m/z 289.1295.
3.1.1.4. The TPD trifunctional probe 1
An oven dried round bottom flask equipped with a stir bar and a rubber septum was added 9 (0.16 mmol, 51 mg), 11 (0.16 mmol, 48 mg), NaBH(OAc)3 (0.32 mmol, 68 mg), and DCE (1.5 mL). The reaction was stirred at room temperature for 16 h. After the reaction went to completion, the solvent was removed under reduced pressure. The reaction residue was re-dissolved in CH2Cl2 (10 mL), and the organic layer was washed with H2O (10 mL x 2) and brine (10 mL). The organic layer was separated and dried over MgSO4, filtered, and the solvent removed under reduced pressure. The crude mixture was purified by flash column chromatography, employing 9:1 ethyl acetate:methanol as eluent to afford 1 (79 mg, yield 85 %) as a yellow solid: Rf 0.22 (1:1 ethyl acetate:hexane); 1H NMR (500 MHz, CDCl3) δ 7.94 (d, J = 7.5 Hz, 1H), 7.66–7.63 (m, 1H), 7.42 (d, J = 7.5 Hz, 1H), 7.39 (d, J = 8.5 Hz, 1H), 7.30 (t, J = 7.5 Hz, 1H), 6.80 (d, J = 7.5 Hz, 1H), 6.65 (s, 1H), 4.28 (d, J = 2.5 Hz, 2H), 4.19–4.14 (m, 2H), 3.96–3.90 (m, 2H), 3.70 (s, 3H), 3.67 (s, 2H), 3.28 (app. t, J = 4.5 Hz, 4H), 2.70 (br s, 4H), 2.47 (t, J = 2.5 Hz, 1H). 13C {1H} NMR (125 MHz, CDCl3) δ 157.1, 156.1, 149.0, 139.7, 134.8, 132.5, 130.8, 129.1, 128.4, 126.7, 122.6, 122.1(q, 1JC–F = 274 Hz), 119.1, 117.4, 115.1, 109.8, 79.4, 74.9, 68.1, 68.0, 58.6, 55.7, 53.0, 50.5, 30.0, 28.5 (q, 2JC–F = 40 Hz); HRMS (ESI, [M+H]+) calcd for C28H28F3N6O5 585.2068; found: m/z 585.2070. The TPD-containing compounds are stored in the dark to avoid light-mediated decomposition.
3.2. Assessment of stability of TPD probe 1 and the efficiency of the bio-orthogonal reaction with different reducing agents
Following the synthesis of the compounds, we first looked at the stability of TPD probe molecule 1 in the presence of the selected CuAAC reaction conditions. Probe 1 was mixed with the chosen CuAAC reagents without the addition of the azide reaction counterpart and kept in the dark. Two different reducing reagents, sodium ascorbate and TCEP, were chosen to assess their compatibility with the TPD Probe 1. The TPD Probe 1 remained intact under the designated CuAAC conditions and showed no sign of decomposition in LC-MS analysis (Data not shown).
Next, the CuAAC reactions were carried out using TPD Probe 1 with two different reducing agents. Two reactions were performed in parallel, with TCEP or sodium ascorbate as the respective reducing agent and biotin-(PEG)3-azide. The formation of the cycloaddition product 12 was only seen in the reaction using sodium ascorbate as the reducing agent. The reaction involved using TCEP as the reducing agent did not give the cycloaddition product 12, likely due to the reduction of biotin-(PEG)3-azide to the corresponding amine via the Staudinger reduction. Based on the observations, the reaction condition for CuAAC was selected using sodium ascorbate as the reducing agent.
3.3. Optimization of irradiation time for photoaffinity labelling
Following the establishment of the reaction condition for the CuAAC, we attempted to determine the optimal time required for the photoaffinity labelling experiment. Following the dissolution of Probe 1 in methanol, the formation of the methanol adduct was observed after 30 min of UV irradiation with significantly more complex by-products when the irradiation exceeded 60 min. Therefore, 60 min was selected as the optimal time for the subsequent photoaffinity labelling experiments in this study.
3.4. BSA labelling experiments
To our delight, the designed experimental workflow gave successful result of the labelling experiment. In the attempt, we detected an obvious increase in the molecular weight of the labelled BSA (Fig. 3). Also, the peak broadening strongly indicates the formation of a heterogenous mixture of labelling products due to the reaction at more than one site, and some degree of reaction with more than one molecule of the cycloaddition product 12. The successful result from this labelling experiment demonstrated that the synthesized TPD Probe 1 is a powerful tool to probe a biologically relevant space.
3.5. Probe 1 enhances the production of IL-2 from human PBMCs
To verify the immunomodulatory properties of Probe 1 on various immune cells, we exposed PBMCs derived from healthy donors to the Probe 1 at different concentrations up to 10 μM. Our findings indicate that the treatment of Probe 1 with PBMCs in the presence of SEB for a duration of 72 h resulted in a higher secretion of IL-2 in a concentration-dependent manner compared to PBMCs treated with SEB alone. The observed effect was statistically significant at a concentration of 3 μM and 10 μM of Probe 1, as depicted in Fig. 5A.
Fig. 5.
A) The structure of the complex P3 (BSA + Probe 12). B) Concentration-dependent increases in IL-2 production from human PBMCs incubated with SEB for 72 h. Statistical analysis was performed using a one-way ANOVA, with Greenhouse–Geisser correction, followed by Tukey's multiple comparisons test, with individual variance computed for each comparison. (Mean compared to cells treated with SEB only), C) Western blot representing the pulldown assay. The blots were visualized using IRDye® 680RD Streptavidin. The signal present in the probe lane confirms the enrichment of biotinylated BSA (AC; lane 4) and P3 (lane 6). Lanes 1–3 correspond to the different amounts of BSA (1, 2 and 4 μg/mL). D) A blot showing the enriched biotinylated BSA in the AC (lane 2) and the supernatant (lane 3). Molecular weight marker units are displayed in kilodaltons (kDa) in the ladder (LD).
3.6. Photoaffinity labelling with BSA and probe 1
To initiate the click chemistry reaction involving Probe 1 and BSA, Probe 1 was added to BSA and incubated for 6 h before being exposed to a UV lamp for 1 h. At this stage, we anticipate the formation of a crosslink between the alkyne site of Probe 1 and BSA. The mixture was then incubated for 1.5 h, which formed the click chemistry (Step 1–4 of Fig. 4). To verify the efficacy of the click chemistry reaction, the supernatant from the click reaction (AC) was compared to the synthesized Probe 12 + BSA (P3) (Fig. 5B). Different amounts of BSA (1, 2 and 4 μg), the after-click sample (3 μg of AC) and P3 (Probe 12 + BSA) along with the ladder (LD) were separated by SDS-PAGE (Fig. 5C). Notably, a single clear band higher than 63 kDa was noted both in AC and P3 lanes indicating the successful incorporation of cross-linkage and completion of click reaction between the Probe 1 and BSA. However, the absence of any notable band in the lane loaded with BSA (with the click reaction reagents in the absence of the Probe 1) show the functional utility of the Probe 1 in the click reaction. Interestingly, the presence of a single clear band in P3 lane (from BSA labelling experiment in Section 2.5) corresponding to the band in the AC lane (from BSA labelling experiment in Section 2.7, step 4) further confirms the photo-crosslinking and click chemistry (Step 1–4) in these reactions.
Fig. 4.
The workflow of pulldown photoaffinity labelling with BSA and Probe 1. Steps 1–9 indicating the schematic pathway followed for incubation of BSA and Probe 1, photo-crosslinking, click chemistry, pull-down separation, and Western blot detection.
To prove the efficiency of pull down, 3 μg of the reaction mixture after the click reaction (from step 4, Fig. 4) was separated using streptavidin agarose beads, allowing streptavidin to capture the biotinylated complex (Probe 1 + BSA). After SDS-PAGE separation, a faint band was detected in the sup lane (lane 3, Fig. 5D) aligning well to the distinct dark band in AC lane (lane 2, Fig. 5D) correspondingly higher than 63 kDa of the ladder. The covalently attached probe results in a slight increase in molecular weight correspond to the added size of the Probe 1. This suggests that the dark band confirms the enrichment of biotinylated BSA in the AC compared to the supernatant. This finding indicates that the biotinylated complex was predominantly captured by streptavidin agarose beads and the pull-down assay can be successfully applied to utilize the Probe 1 to efficiently bind to a protein.
4. Discussion
Compound A has been reported to have potential immunomodulatory effects through unidentified cellular targets [24]. Identification of a drug's target is a crucial step in understanding its mechanism of action and developing more targeted and effective therapies [31]. However, this task is challenging and requires specialized protocols and tools. As part of our efforts and ongoing interest to develop a reliable and effective method for identifying target proteins in complex biological systems for our compound (Compound A), we employed a pull-down protocol as a proof-of-concept.
To achieve this goal, we first designed a trifunctional approach utilizing a small molecule with an affinity warhead derived from Compound A, a photo-crosslinker, and a bio-orthogonal handle (Probe 1), The stability of the molecular probe under common CuAAC conditions was confirmed and optimization experiments were performed to determine the ideal irradiation time to facilitate photoaffinity labelling. The synthesized Probe 1 showed capability in inducing IL-2 secretion as an indication of its immunomodulatory effect, similar to Compound A [24]. To test the efficiency of Probe 1 in binding to its target protein, we employed BSA as a single model protein. BSA is frequently used as a model in experimental conditions for studying drug-protein interactions due to its well-characterized properties [32,33]. For the purpose of optimizing our protocol for target identification of Compound A, we used the pull-down assay with Probe 1 and BSA as a proof of concept for the sequential processes of photoaffinity, click chemistry, and separation of Probe 1 with BSA. The presence of a clear, strong signal in the pull-down sample lane and a weak signal in the supernatant lane provides compelling evidence of the efficacy of our optimized pull-down protocol (Fig. 5D). Subsequently, we also conducted a BSA labelling experiment using Probe 12 as a comparison to validate the pulldown, where an intense band of BSA complexed with Probe 12 (P3) was corresponding to the signal of the pull down (Fig. 5C). Taken together, our results suggest that the structural modification of the parent compound with a TPD-containing trifunctional group(s) offers us a promising method for cross–linking the small molecule with the plausible proteins in the cell. Through this proof-of-concept study, we were able to establish the reliability and effectiveness of our pull-down protocol for identifying the target proteins. This protocol with photoaffinity labeling can be further applied to other biological systems to identify target proteins in cellular processes. This established protocol with Probe 1 could be used effectively in future studies to identify the cellular targets responsible for the immunomodulatory effect of Compound A.
5. Conclusion
In the present study, we have attempted to synthesize a TPD trifunctional Probe 1 of Compound A and utilized this probe to develop a protocol for a pull-down assay using BSA as a model protein. Our report demonstrates that we were successful in labeling and separating the BSA–Probe complex. The successful synthesis and application of this probe can pave the way for researchers to employ new labeling strategies for their small molecules in various biological applications.
Data availability
Data will be made available on request.
CRediT authorship contribution statement
Yasser Tabana: Writing – original draft, Methodology, Investigation, Conceptualization. Chih-Hsuan Lin: Writing – original draft, Methodology, Investigation, Data curation, Conceptualization. Dinesh Babu: Writing – review & editing, Project administration, Methodology, Data curation. Ramanaguru Siva‐Piragasam: Resources, Methodology. Ashley A. Ponich: Methodology, Investigation. Tae Chul Moon: Supervision, Project administration. Arno G. Siraki: Writing – review & editing, Project administration, Investigation, Conceptualization. Shokrollah Elahi: Writing – review & editing, Supervision, Conceptualization. Richard Fahlman: Writing – review & editing, Supervision, Conceptualization. Frederick G. West: Writing – review & editing, Supervision, Project administration, Investigation, Conceptualization. Khaled Barakat: Writing – review & editing, Resources, Project administration, Investigation, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was partly supported by a (CIHR) project grant, and an Alberta Cancer Foundation grant (to KB). The first author (YT) would like to thank Alberta Innovates for providing the Postdoctoral Recruitment Fellowship.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e21408.
Contributor Information
Frederick G. West, Email: fwest@ualberta.ca.
Khaled Barakat, Email: kbarakat@ualberta.ca.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Tabana Y., Moon T.C., Siraki A., et al. Reversing T-cell exhaustion in immunotherapy: a review on current approaches and limitations. Expert Opin. Ther. Targets. 2021;25:347–363. doi: 10.1080/14728222.2021.1937123. [DOI] [PubMed] [Google Scholar]
- 2.Chen R., Ganesan A., Okoye I., et al. Medicinal research reviews; 2019. Targeting B7‐1 in Immunotherapy. [DOI] [PubMed] [Google Scholar]
- 3.Wang D.-R., Wu X.-L., Sun Y.-L. Therapeutic targets and biomarkers of tumor immunotherapy: response versus non-response. Signal Transduct. Targeted Ther. 2022;7:331. doi: 10.1038/s41392-022-01136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha J., Park H., Park J., et al. Recent advances in identifying protein targets in drug discovery. Cell Chem. Biol. 2021;28:394–423. doi: 10.1016/j.chembiol.2020.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z., Jiang Z., Chen N., et al. Target discovery of ebselen with a biotinylated probe. Chem. Commun. 2018;54:9506–9509. doi: 10.1039/c8cc04258f. [DOI] [PubMed] [Google Scholar]
- 6.Lapinsky D.J. Tandem photoaffinity labeling–bioorthogonal conjugation in medicinal chemistry. Bioorg. Med. Chem. 2012;20:6237–6247. doi: 10.1016/j.bmc.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 7.Zhao G., Li Z., Zhang R., et al. Tetrazine bioorthogonal chemistry derived in vivo imaging. Front. Mol. Biosci. 2022;9 doi: 10.3389/fmolb.2022.1055823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim R.K., Lin Q. Bioorthogonal chemistry: a covalent strategy for the study of biological systems. Sci. China Chem. 2010;53:61–70. doi: 10.1007/s11426-010-0020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassan M.M., Olaoye O.O. Recent advances in chemical biology using benzophenones and diazirines as radical precursors. Molecules. 2020;25:2285. doi: 10.3390/molecules25102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherratt A.R., Nasheri N., McKay C.S., et al. A new chemical probe for phosphatidylinositol kinase activity. Chembiochem. 2014;15:1253–1256. doi: 10.1002/cbic.201402155. [DOI] [PubMed] [Google Scholar]
- 11.Bertozzi C.R. A decade of bioorthogonal chemistry. Acc. Chem. Res. 2011;44:651–653. doi: 10.1021/ar200193f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scinto S.L., Bilodeau D.A., Hincapie R., et al. Bioorthogonal chemistry. Nature Rev. Method. Primers. 2021;1:30. doi: 10.1038/s43586-021-00028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hang H.C., Yu C., Kato D.L., et al. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. USA. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tornøe C.W., Christensen C., Meldal M. Peptidotriazoles on solid phase:[1, 2, 3]-triazoles by regiospecific copper (I)-catalyzed 1, 3-dipolar cycloadditions of terminal alkynes to azides. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 15.Rostovtsev V.V., Green L.G., Fokin V.V., et al. A stepwise huisgen cycloaddition process: copper (I)‐catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. 2002;114:2708–2711. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 16.Lal S., Rzepa H., Díez-González S. ACS Catal. 2014;4:2274−–2287. [Google Scholar]; (b) Worrell B.T., Malik J.A., Fokin V.V. Science. 2013;340:457–460. doi: 10.1126/science.1229506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauwaet T., Miyamoto Y., Ihara S., et al. Click chemistry-facilitated comprehensive identification of proteins adducted by antimicrobial 5-nitroimidazoles for discovery of alternative drug targets against giardiasis. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., Dong T., Zhou Y., et al. Exploring the binding proteins of glycolipids with bifunctional chemical probes. Angew. Chem. Int. Ed. 2016;55:14330–14334. doi: 10.1002/anie.201608827. [DOI] [PubMed] [Google Scholar]
- 19.Lee S., Nam Y., Koo J.Y., et al. A small molecule binding HMGB1 and HMGB2 inhibits microglia-mediated neuroinflammation. Nat. Chem. Biol. 2014;10:1055–1060. doi: 10.1038/nchembio.1669. [DOI] [PubMed] [Google Scholar]
- 20.Adams J.L., Smothers J., Srinivasan R., et al. Big opportunities for small molecules in immuno-oncology. Nat. Rev. Drug Discov. 2015;14:603–622. doi: 10.1038/nrd4596. [DOI] [PubMed] [Google Scholar]
- 21.Smith E., Collins I. Photoaffinity labeling in target-and binding-site identification. Future Med. Chem. 2015;7:159–183. doi: 10.4155/fmc.14.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Y., Yang Z., Cheng K., et al. Small molecule-based immunomodulators for cancer therapy. Acta Pharm. Sin. B. 2022 doi: 10.1016/j.apsb.2022.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Zanden S.Y., Luimstra J.J., Neefjes J., et al. Opportunities for small molecules in cancer immunotherapy. Trends Immunol. 2020;41:493–511. doi: 10.1016/j.it.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Ganesan A., Ahmed M., Okoye I., et al. Comprehensive in vitro characterization of PD-L1 small molecule inhibitors. Sci. Rep. 2019;9:1–19. doi: 10.1038/s41598-019-48826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang H., Huang L., Zhao D., et al. Interaction mechanism of flavonoids on bovine serum albumin: insights from molecular property-binding affinity relationship. Spectrochim. Acta Mol. Biomol. Spectrosc. 2020;239 doi: 10.1016/j.saa.2020.118519. [DOI] [PubMed] [Google Scholar]
- 26.Mayer T., Maier M.E. 2007. Design and Synthesis of a Tag‐free Chemical Probe for Photoaffinity Labeling. [Google Scholar]
- 27.Tron G.C., Pirali T., Billington R.A., et al. Click chemistry reactions in medicinal chemistry: applications of the 1, 3‐dipolar cycloaddition between azides and alkynes. Med. Res. Rev. 2008;28:278–308. doi: 10.1002/med.20107. [DOI] [PubMed] [Google Scholar]
- 28.Pujari S.S., Seela F. Parallel stranded DNA stabilized with internal sugar cross-links: synthesis and click ligation of oligonucleotides containing 2′-propargylated isoguanosine. J. Org. Chem. 2013;78:8545–8561. doi: 10.1021/jo4012706. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q., Chan T.R., Hilgraf R., et al. Bioconjugation by copper (I)-catalyzed azide-alkyne [3+ 2] cycloaddition. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 30.Shahbaz S., Okoye I., Blevins G., et al. Elevated ATP via enhanced miRNA-30b, 30c, and 30e downregulates the expression of CD73 in CD8+ T cells of HIV-infected individuals. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenone M., Dančík V., Wagner B.K., et al. Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol. 2013;9:232–240. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawar S.K., Jaldappagari S. Interaction of repaglinide with bovine serum albumin: spectroscopic and molecular docking approaches. J. pharm.analys. 2019;9:274–283. doi: 10.1016/j.jpha.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abdulateef S., Raypah M.E., Omar A., et al. Rapid synthesis of bovine serum albumin-conjugated gold nanoparticles using pulsed laser ablation and their anticancer activity on hela cells. Arab. J. Chem. 2023;16 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.