Abstract
Stenotrophomonas maltophilia is one of the most prevalent opportunistic bacteria causing nosocomial infections. It has become problematic because most of the isolates are resistant to multiple antibiotics, and therefore, development of phage therapy has attracted strong attention. In this study, eight S. maltophilia phages were isolated from clinical samples including patient specimens, catheter-related devices, and wastewater. These phages can be divided into four distinct groups based on host range and digestibility of the phage DNAs with different restriction endonucleases. One of them, designated φSMA5, was further characterized. Electron microscopy showed it resembled Myoviridae, with an isometric head (90 nm in diameter), a tail (90 nm long), a baseplate (25 nm wide), and short tail fibers. The φSMA5 double-stranded DNA, refractory to digestion by most restriction enzymes, was tested and estimated to be 250 kb by pulsed-field gel electrophoresis. This genome size is second to that of the largest phage, φKZ of Pseudomonas aeruginosa. In sodium dodecyl sulfate-polyacrylamide gel electrophoresis, 25 virion proteins were visualized. N-terminal sequencing of four of them suggested that each of them might have had its N terminus cleaved off. Among the 87 S. maltophilia strains collected in this study, only 61 were susceptible to φSMA5, indicating that more phages are needed toward a phage therapy strategy. Since literature search yielded no information about S. maltophilia phages, φSMA5 appears to be the first reported.
Stenotrophomonasmaltophilia, previously known as Xanthomonas maltophilia (37) and Pseudomonas maltophilia (22), is an aerobic, gram-negative bacillus widespread in a variety of environments. It has been found in the environment as a growth-promoting agent in the rhizospheres of plants, as well as in soil, water, sediment, sewage, frozen foods, and some other habitats (3, 8, 14, 20, 23, 25). More importantly, S. maltophilia is increasingly prevalent in hospitals as an opportunistic human pathogen causing nosocomial infections in immunocompromised individuals and involved in postoperative infections, the infections of urinary tracts and respiratory tracts, and other disease syndromes (15, 20, 34, 40). This organism was reported to be the second most frequently isolated nosocomial bacterium after Pseudomonas aeruginosa, and its infections have a growing clinical importance (43). The prevalence of S. maltophilia in Taiwan has also been increasing. For example, among the 6,092 bacterial strains isolated from the nosocomial infections of respiratory tracts in Taichung Veterans Hospital of Taiwan in the year 2003, 451 strains (7.4%) were identified as S. maltophilia. This bacterium ranked fifth among 86 causative bacterial species, after P. aeruginosa, Acinetobacter baumannii, Klebsiella pneumoniae, and Staphylococcus aureus (Clinical Microbiology Report, Taichung Veterans Hospital, Taichung, Taiwan, 2004). Furthermore, most of these isolates were resistant to multiple antibiotics, including imipenem (H.-C. Chang and S.-F. Weng, unpublished results), which is similar to the situations in other parts of the world (4, 11, 12, 13, 32) and in the closely related P. aeruginosa, in which resistance to multiple drugs, including imipenem, has been rapidly rising in recent years (26). Therefore, different approaches are needed for effective therapy of S. maltophilia infections. The development of phage therapy, a possible alternative treatment, has attracted strong attention.
Although S. maltophilia is important in both medical and environmental concerns, no research on bacteriophages infecting this organism has been documented. In this study, S. maltophilia strains collected in our laboratory were used as the indicator hosts for phage isolation. Eight phages were obtained from clinical samples, including patient specimens, wastewater, and catheter-related devices. They were divided into four distinct groups based on the host range and digestibility of the phage DNAs with different restriction endonucleases. One of them, designated φSMA5, was further characterized. This phage appears to be the first S. maltophilia bacteriophage to have been characterized.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study, including 87 strains of S. maltophilia recently collected, are listed in Table 1. The S. maltophilia strains used as the hosts for plaque assay and propagation of phages were T39 for φSMA2, φSMA5, and φSMA6; T36 for φSMA1, φSMA3, and φSMA4; and ATCC 13637 and T26 for φSMA7 and φSMA8, respectively. S. maltophilia strains were cultivated in tryptic soy broth (TSB) or TSB agar (Bacto) at 30°C, and the other bacteria were grown in Luria-Bertani broth or L agar at either 28°C for Xanthomonas strains or 37°C for Escherichia coli, Enterobacter cloacae, K. pneumoniae, P. aeruginosa, Proteus mirabilis, and Serratia marcescens. Ampicillin (50 μg/ml) was added when necessary. Bacterial growth was monitored turbidimetrically by measuring optical density at 600 nm (OD600). For S. maltophilia T39, an OD600 of 1.0 corresponded to 1.0 × 109 cells/ml.
TABLE 1.
Bacterial strains used in this study
| Strain(s) | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Escherichia coli | ||
| DH5-α | endA1 hsdR17(rk−mk+) supE44 thi-1 recA1 gyrA relA1φ80d lacZΔM15Δ(lacZYA-argF)U169 | 18 |
| E1 to E13 | Clinical isolates, Apr | L. T. Wua |
| Enterobacter cloacae | ||
| Ec33, Ec45, Ec90, Ec91, Ec104, Ec106, Ec114, Ec158, Ec169, Ec172, Ec173, Ec176 | Clinical isolates, Apr | L. T. Wu |
| Klebsiella pneumoniae | ||
| 10693 | ATCC type strain | ATCCb |
| Kp6, Kp13, Kp18, Kp55, Kp59, Kp60, Kp70, Kp98, Kp112 | Clinical isolates, Apr | L. T. Wu |
| Proteus mirabilis | ||
| Pm33, Pm36, Pm40, Pm51, Pm59, Pm69, Pm70, Pm72, Pm73, Pm84, Pm85 | Clinical isolates | L. T. Wu |
| Pseudomonas aeruginosa | ||
| 27853 | ATCC type strain | 33 |
| P16041, P16052, P16163, P21057, P21107, P21108 | Clinical isolates | L. T. Wu |
| Serratia marcescens | ||
| SmI2 to SmI4, SmI8, SmI15 to SmI18, SmI22, SmI24, SmI26, SmI27, SmI29, SmI30, SmI34, SmI36, SmI37 | Clinical isolates | L. T. Wu |
| SmII2, SmII3, SmII5, SmII7, SmII8, SmII10 to SmII13, SmII15 to SmII17, SmII21, SmII22, SmII25, SmII27 | Clinical isolates | L. T. Wu |
| Stenotrophomonas maltophilia | ||
| 13637 | ATCC type strain | 21 |
| 11901 | Isolated from soil, Apr | 19 |
| 15678 | Isolated from compost, Apr | BCRCc |
| T2, T4 to T6, T9 to T17, T19, T21 to T29, T33, T34, T36 to T48, T50 to T54, T57 to T69, T72 to T80, T83 to T85, T89 to T92, T94 to T104 | Clinical isolates, Apr | This study |
| Xanthomonas campestris pv. campestris | ||
| 33913 | ATCC type strain | 39 |
| P20H | Nonmucoid mutant, Apr | 42 |
| Xc6 | K. C. Tzengd | |
| Xc11 | Wild-type strain, Apr Rifr | 41 |
| Xc17 | Wild-type strain, Apr Rifr | 41 |
| Xc85 | Wild-type strain, Apr | K. C. Tzeng |
| Xc114 | Wild-type strain, Apr | K. C. Tzeng |
| X. axonopodis pv. citri | ||
| Xcc60 | Wild-type strain, Apr | K. C. Tzeng |
| X. axonopodis pv. dieffenbachiae | ||
| Xcd65 | Wild-type strain, Apr | K. C. Tzeng |
| X. axonopodis pv. glycines | ||
| Xcg69 | Wild-type strain, Apr | K. C. Tzeng |
| X. axonopodis pv. phaseoli | ||
| Xcp73 | Wild-type strain, Apr | K. C. Tzeng |
| X. axonopodis pv. vesicatoria | ||
| Xv2, Xv4, Xv7, Xv12 to Xv16, Xv18 to Xv25, Xv28 to Xv33, Xv35 to Xv43, Xv45 to Xv54, Xv56 to Xv59 | Wild-type strains, Apr | Y. H. Tsenge |
| X. oryzae pv. oryzae | ||
| Xo21 | Wild-type strain | K. C. Tzeng |
| Xo2a | Wild-type strain | Y. H. Tseng |
L. T. Wu, Institute of Medical Science, China Medical University, Taichung, Taiwan.
ATCC, American Type Culture Collection.
BCRC, Bioresources Collection and Research Center, Hsinchu, Taiwan.
K. C. Tzeng, Department of Plant Pathology, National Chung Hsing University, Taichung, Taiwan.
Y. H. Tseng, Center for Research and Development, Chungtai Institute of Health Sciences and Technology, Taichung, Taiwan.
Spot test.
The strains to be tested were grown overnight either in TSB broth for S. maltophilia or in Luria-Bertani broth for the other bacteria. Three milliliters of the molten soft TSB or L agar (0.7%) was mixed with 100 μl of the cells, which was then overlaid on the surface of the solidified basal TSB or L agar (1.5%). Ten microliters (ca. 1.0 × 108 PFU/ml) of a phage suspension was spotted onto the plate, which was then incubated overnight. Bacterial sensitivity to a bacteriophage was established by bacterial lysis at the spot where the phage was deposited. According to the degrees of clarity, the spots were differentiated into three categories: clear, turbid, and no reaction.
Plaque assay.
Phage lysates were subjected to serial dilution with sterilized, deionized water. Then, 100 μl of a phage suspension and 100 μl of the cells from an overnight culture of S. maltophilia were mixed with 3 ml of the molten soft TSB agar and poured on the surface of a regular MRS agar plate. The numbers of the plaque were counted after incubating the plates overnight.
Isolation of bacteriophages.
The samples potentially containing bacteriophages, collected in China Medical University Hospital, were kindly donated by L.-T. Wu. They included 24 patient specimens (10 urine, 6 sputum, 4 pus, 2 pleural effusion, and 2 cerebral spinal fluid samples), 4 batches of catheter tip washings (obtained by soaking the used tips in a cylinder containing distilled water with shaking), and 6 wastewater samples from drainages. These samples were centrifuged (5,000 × g, 10 min at 4°C), and the supernatants were filter (0.45-μm-pore-size membrane) sterilized and checked for the presence of phages by depositing 10 μl on the double-layered plates containing the lawns of the S. maltophilia cells. The top agar, together with phages within the clearing zones, was picked and soaked in TSB. After appropriate dilution, the suspensions were plated for plaque formation. At least two more successive single-plaque isolations were performed to obtain pure cultures.
Phage adsorption.
Phage adsorption experiments were carried out as described in Foschino et al. (17) with some modifications. Cells of S. maltophilia T39 (0.6 U of OD600) were infected with a phage suspension to give a multiplicity of infection (MOI) of 0.0005 and incubated at 30°C. Aliquots (100 μl) were taken at 10-min intervals (up to 100 min) and diluted in 0.9 ml of cold TSB. Following centrifugation (12,000 × g, 5 min), the supernatants containing unadsorbed phages were diluted and titrated by the double-layered-agar plate technique. Adsorption was expressed as percent decrease of phage titer in the supernatant compared to that in controls.
One-step growth.
One-step growth experiments were performed as described in Pajunen et al. (31) with some modifications. Cells of S. maltophilia T39 (0.6 U of OD600) were harvested by centrifugation and resuspended in the fresh TSB (ca. 109 CFU/ml). Phage φSMA5 was added at an MOI of 0.0005 and allowed to adsorb for 5 min at room temperature. The mixture was centrifuged (12,000 × g, 10 min), and the pellets containing infected cells were suspended in 25 ml of TSB, followed by incubation at 30°C. Samples were taken at 10- or 20-min intervals (up to 4 h) and immediately diluted, and then titers were determined by the double-layered-agar plate method.
Purification of phage particles.
High-titer lysates of φSMA5 (400 ml, ca. 1.0 × 1010 PFU/ml) were centrifuged at 6,000 rpm for 20 min at 4°C (Sorvall RC 5C, GS3 rotor). The supernatants were passed through a 0.45-μm-pore-size membrane filter and then centrifuged at 8,400 rpm for 2 h at 4°C with the same rotor. The pellets were suspended in 1.0 ml of TE buffer (10 mM Tris-HCl, pH 7.0, containing 1.0 mM EDTA adjusted to pH 7.0) and loaded on the block gradient of CsCl, followed by ultracentrifugation at 25,000 rpm for 2 h at 4°C with the TH641 rotor in a Sorvall ultracentrifuge OTD Combi instrument. The banded phage particles were recovered, dialyzed against TE buffer, and then stored at 4°C until used.
Isolation and restriction enzyme digestion of the phage DNA.
The purified phages (1.0 × 1010 PFU/ml) were treated with 10 μg of RNase A and DNase I (37°C, 30 min). To the mixture, sodium dodecyl sulfate (SDS) was added at a final concentration of 1% and then the mixture was treated with 20 U of proteinase K at 65°C. After 1 h, an equal volume of phenol-chloroform (1:1) was added to remove the proteinaceous materials. The extraction was repeated twice, and the DNA was precipitated according to the standard procedures (35). Restriction enzyme digestions of the phage DNA were carried out following the instructions provided by the suppliers. Restriction endonucleases tested for digestibility of the φSMA5 DNA were AatII, AccI, AluI, ApaI, AseI, AvaI, BamHI, BanII, BfaI, BglII, BspDI, BssHII, BstYI, ClaI, DdeI, DraI, EcoRI, EcoRV, HaeII, HaeIII, HincII, HindIII, HinfI, HpaI, KpnI, MfeI, MluI, NaeI, NcoI, NdeI, NheI, NotI, NsiI, PstI, PvuI, PvuII, SacI, SacII, SalI, Sau3AI, ScaI, SmaI, SpeI, SphI, SspI, StuI, StyI, XbaI, XhoI, and XmnI.
PFGE.
The procedures described by Tseng et al. (38) were followed for pulsed-field gel electrophoresis (PFGE) in a CHEF-DR III apparatus from Bio-Rad (Richmond, Calif.), except that the purified φSMA5 particles (ca. 1.0 × 1010 PFU/ml) were used as the source for the preparation of intact genomic DNA.
Electron microscopy.
The phage suspension was dropped onto the 300-by 300-mesh grid, which had been treated by coating with one drop of 0.1% bacitracin. After 3 min, the phage particles were stained with 2% uranyl acetate for 10 s and then examined under a JEM-1200 EX II electron microscope (JEOL, Peabody, Mass.) at an operating voltage of 120 kV.
SDS-PAGE.
Structural proteins of φSMA5 were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Phage particles purified by ultracentrifugation were mixed with the sample buffer and then heated in a boiling water bath for 3 min, followed by separating the proteins in the gel (12%). Protein bands were visualized by staining the gels with Coomassie brilliant blue.
N-terminal amino acid sequencing of proteins.
Proteins from the φSMA5 particles separated in SDS-PAGE gels were transferred to a polyvinylidene difluoride membrane and stained with Coomassie brilliant blue. Membrane strips containing the protein bands were excised and subjected to Edman degradation for determining the N-terminal sequences, performed on a 477A sequencer (PE Applied Biosystems).
RESULTS AND DISCUSSION
Eight S. maltophilia bacteriophages were isolated.
Thirty-four hospital samples, including patients' specimens, catheter washings, and wastewater from drainages, were obtained. These samples were separately spot tested on the lawns of 10 S. maltophilia strains, including the three type strains and seven from our collections. Eight bacteriophages were thus isolated and designated φSMA1 to φSMA8, each of which was obtained from an independent sample collected at different times: φSMA1, φSMA3, φSMA5, φSMA7, and φSMA8 from sputum samples, φSMA2 and φSMA4 from pleural effusions, and φSMA6 from catheter tips. The results of the spot test showed that these phages could be divided into four groups based on the host susceptibility: φSMA2, φSMA5, and φSMA6 in group I; φSMA1, φSMA3, and φSMA4 in group II; and φSMA7 and φSMA8 in groups III and IV, respectively (Table 2).
TABLE 2.
Spot testa of phages φSMA1 to φSMA8 on 10 strains of S. maltophilia
| Phage | T8 | T26 | T36 | T54 | T62 | T64 | T71 | ATCC 13637 | BCRC 11901 | BCRC 15678 |
|---|---|---|---|---|---|---|---|---|---|---|
| φSMA1 | − | − | +++ | − | − | − | − | − | − | − |
| φSMA2 | − | +++ | +++ | +++ | +++ | − | − | ++ | ++ | +++ |
| φSMA3 | − | − | ++ | − | − | − | − | − | − | − |
| φSMA4 | − | − | +++ | − | − | − | − | − | − | − |
| φSMA5 | − | +++ | +++ | +++ | ++ | − | − | ++ | ++ | +++ |
| φSMA6 | − | +++ | +++ | +++ | ++ | − | − | ++ | ++ | +++ |
| φSMA7 | − | − | − | − | − | − | − | ++ | ++ | − |
| φSMA8 | − | +++ | − | − | − | − | − | − | − | − |
−, no reaction; ++, turbid; +++, clear.
Propagation and adsorption of φSMA5.
Phage φSMA5 formed clear plaques, which were about 1 mm in diameter, on the lawns of S. maltophilia T39. Infection of the T39 liquid cultures at early exponential phase (OD600 at 0.6) with φSMA5 caused a rapid lysis of the cells. With an MOI of 1.0, the infected cultures ceased to grow at 2.5 h postinfection and then became clear at about 6.0 h. With an MOI of 10, the cultures were cleared at 3 h, and a φSMA5 lysate of 5.0 × 1010 PFU/ml could be obtained. The adsorption rate of φSMA5 on strainT39 is shown in Fig. 1. Approximately 70% of the φSMA5 was adsorbed to the host cells after 50 min, rising slowly to 80% at 100 min postinfection.
FIG. 1.
Adsorption curve of bacteriophage φSMA5 on S. maltophilia T39.
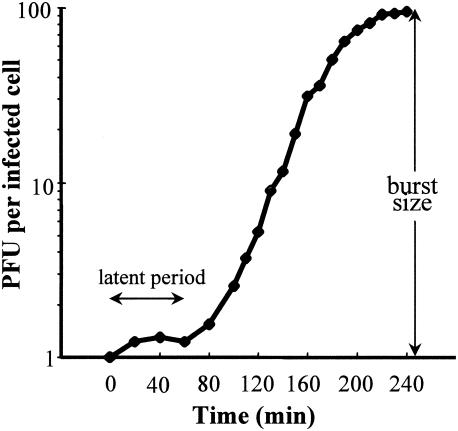
Burst size and latent period of φSMA5.
The one-step growth curve of φSMA5 on S. maltophilia T39 was determined (Fig. 2). The latent period, defined as the time interval between the adsorption (not including 15 to 20 min of pretreatment) and the beginning of the first burst, was about 80 min. The average burst size was about 95 PFU/cell, calculated as the ratio of the final count of liberated phage particles to the initial count of infected bacterial cells during the latent period.
FIG. 2.
One-step growth curve of bacteriophage φSMA5 on S. maltophilia T39. Shown are the PFU per infected cell in cultures at different time points. Each data point is a mean from four experiments.
φSMA5 was stable for 16 months at 4°C.
To test for phage stability, crude lysates of φSMA5 (ca. 5.0 × 1010 PFU/ml) were stored at 4°C and the titers were assayed at intervals of 10 days. After 16 months, no significant loss in infectivity was observed.
φSMA5 has a buoyant density between 1.36 and 1.40 g/cm3 and is morphologically similar to the members of family Myoviridae.
During the course of phage purification, the φSMA5 lysates (total of about 2.0 × 1012 PFU) were subjected to ultracentrifugation in the block gradient of CsCl representing 1.26, 1.30, 1.36, 1.40, and 1.45 g/cm3. The phage particles were found to band on top of the 1.40-g/ml block, suggesting that φSMA5 has a buoyant density between 1.36 and 1.40 g/cm3.
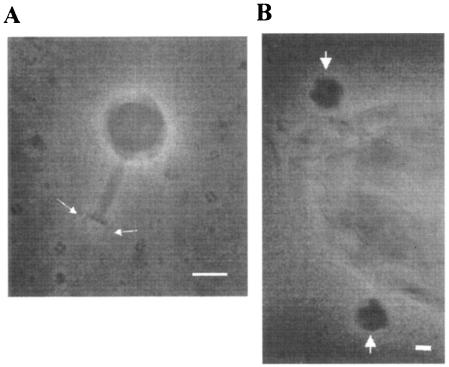
To reveal the morphology of φSMA5, the purified phage was examined by electron microscopy. As shown in Fig. 3, it had an isometric head (ca. 90 by 90 nm), a visible collar, and a relatively short contractile tail (90 nm long) with a terminal plate (25 nm wide) and short tail fibers, which were characteristics of the family Myoviridae or Bradley's group A1 (1, 2).
FIG. 3.
Transmission electron micrographs of phage φSMA5. (A) φSMA5 consists of an elongated isometric head of 90 by 90 nm and a complex tail of 90 by 15 nm. One baseplate and two tail fibers (indicated with arrows) are visible on most phage particles. (B) Shown is a part of the S. maltophilia T39 cell with φSMA5 particles adsorbed (arrows). Scale bars, 50 nm.
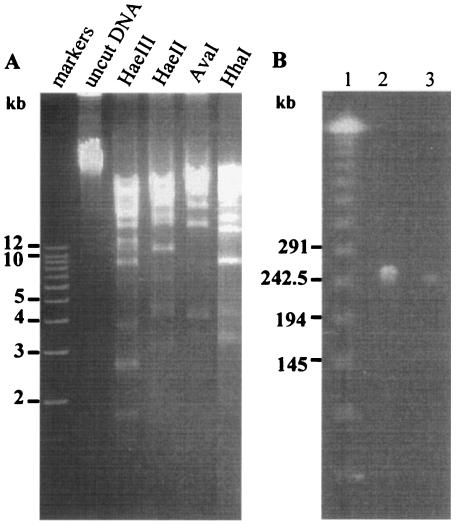
φSMA5 DNA is refractory to the digestion by many restriction endonucleases.
To test for digestibility of the φSMA5 DNA, 50 type II restriction endonucleases (listed in Materials and Methods) were used in this study. It was found that the DNA was refractory to the digestion by most of the restriction enzymes tested. It could be consistently cut only by AvaI, HaeII, HaeIII, or HhaI. Because the fragments generated were mostly larger than 12 kb, only the smaller fragments were resolved in 0.8% agarose gel (Fig. 4A). These restriction enzymes, AvaI, HaeII, HaeIII, and HhaI, have the sites consisting of sequences C↓PyCGPuG, PuGCGC↓Py, GG↓CC, and GCG↓C, respectively. Notably, both HaeIII and HhaI sites contain purely G and C, whereas the AvaI and HaeII sites are rich in GC, with both cleavages occurring between a C and a Py which can also be a C. Phage genome resistance to restriction digestion is common, and several possible explanations have been proposed (5, 30). First, phage genomes may adapt under the selection of widely occurring restriction modification systems and lose restriction sites during evolution. Second, phage genomes may encode methyltransferase enzymes that can modify the bases in one or more of the restriction sites. Third, phage genomes may contain unusual bases. For examples, the presence of 5-hydroxymethyl deoxycytosine in T4 and replacing of deoxycytosine with 5-methyl deoxycytosine in the Xanthomones oryzae phage Xp12 render the DNAs refractory to most restriction enzymes (5, 24). The first two possibilities are unlikely cases in φSMA5 because they can only lead to the resistance to just one or a few, but not to a great number of, enzymes concomitantly. Should this prediction be correct, the findings that G and C are favored for the restriction sites suggest that the φSMA5 genome may contain unusual bases analogous to A and/or T.
FIG. 4.
(A) Agarose gel electrophoresis of the DNA digests from the φSMA5 genome. The restriction enzymes used are labeled above the lanes. (B) PFGE of the φSMA5 genomic DNA. Lanes: 1, size markers; 2, DNA from 1.0 × 109 PFU; 3, DNA from 1.0 × 108 PFU.
In parallel experiments with the four competent enzymes, the DNAs from φSMA2 and φSMA6 were found to give the same restriction patterns as those from the φSMA5 DNA (data not shown). These results and the data of host range tests suggested that φSMA2, φSMA5, and φSMA6 are closely related or even the same. Since they were isolated from different niches of the same hospital, these data also suggest the prevalence and success of this phage in the environment.
The φSMA5 genome is about 250 kb in size.
Because the φSMA5 genome was large, the AvaI, HaeII, HaeIII, and HhaI digests displayed many large and multiplex bands in agarose gel after electrophoretic separations, rendering it difficult to estimate the genome size by summing up the restriction fragment sizes. Therefore, PFGE was performed to mobilize the uncut phage DNA, and the genome size of φSMA5 thus estimated was about 250 kb (Fig. 4B).
Several giant bacteriophages, all belonging to family Myoviridae, are known. Phage φKZ of Pseudomonas aeruginosa, being the largest, has a long contractile tail and an icosahedral capsid 120 nm in diameter and possesses a linear double-stranded DNA genome of 280,334 bp (27). The broad-host-range vibriophage KVP40, with a head 140 nm long and 70 nm wide, has a linear, circularly permuted chromosomal DNA of 244,835 bp (29). The Aeromonas hydrophila phage Aeh1, with a head 134.4 nm long and 89.0 nm wide, has a double-stranded DNA genome of 233,234 bp (10). Compared to these phages, φSMA5, with a genome of about 250 kb, is among the largest reported so far.
Many bacteriophage genomes have been sequenced, and it is known that they contain great numbers of nonessential genes, most of which have no known functions. In T4, only 62 genes are essential and about 130 of the potential genes (about 300 open reading frames [ORFs] on the 168,903-bp genome) remain uncharacterized (28). About 80% of the 306 φKZ ORFs and 65% of the 386 KVP40 ORFs have no known functions (27, 29). By analogy, φSMA5 with a giant genome must have many genes that are not essential for the phage life cycle and may represent a good source of useful genes.
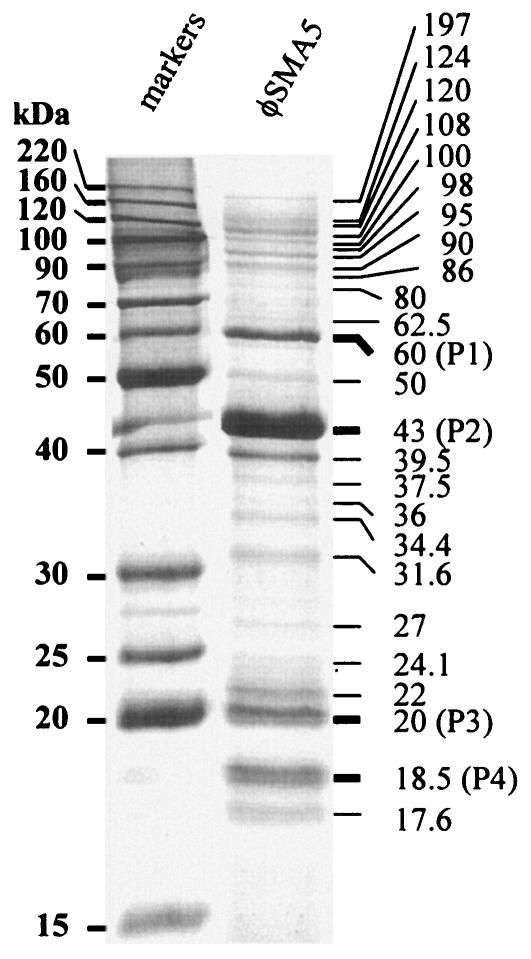
The φSMA5 virion consists of at least 25 proteins.
To further characterize φSMA5, the structural protein composition was analyzed by SDS-PAGE (12% polyacrylamide gel). At least 25 protein bands were visualized in the gel (Fig. 5), and the 43-kDa virion component was the most abundant protein of the φSMA5 particle. Four bands with molecular masses of 60, 43, 20, and 18.5 kDa were recovered from the gel and subjected to sequence determination of the N-terminal 10 amino acid residues (Table 3). None of these N-terminal sequences started with a methionine, suggesting the removal of the N-terminal residue(s) from the nascent polypeptides. Interestingly, two of the mature φSMA5 proteins had an Ala and the other two had a Gly at their N termini, suggesting that proteolysis may occur between X and Ala or X and Gly, where X represents an unknown residue and remains to be revealed after gene sequencing.
FIG. 5.
SDS-PAGE of the φSMA5 virion proteins. About 5.0 × 108 PFU of purified phage particles was boiled in cracking buffer (total of 20 μl) and loaded onto the well. Shown are the 25 bands with estimated molecular masses. Bands P1 to P4 were recovered from the gel and subjected to sequence determination of the N-terminal 10 amino acid residues.
TABLE 3.
N-terminal sequences of φSMA5 virion proteins
| Virion protein | Mol wt | Sequence |
|---|---|---|
| P1 | 60 | 1 GQYMAPGVYT 10 |
| P2 | 43 | 1 ATFSQNLGAL 10 |
| P3 | 20 | 1 GLNIDALLVG 10 |
| P4 | 18.5 | 1 AEDLSYNYAE 10 |
Posttranslational processing by proteolysis is necessary for the morphogenesis of many bacteriophages (7). For examples, the N termini of several of the φKZ major virion proteins are removed, including, among others, the major capsid protein (gp120, the most abundant protein, with the first 163 residues cleaved off) and gp145 (the second most predominant protein, which is possibly the contractile tail sheath, with only the fMet cleaved off).
φSMA5 does not infect the bacteria other than the S. maltophilia tested.
To test for host range, lawns of all of our S. maltophilia strains isolated from hospital samples were subjected to spot tests with φSMA5. After 24 h, 61 (70.1%) and 21 (24.1%) strains showed clear and turbid clearing zones, respectively. Five strains (5.8%) were resistant to φSMA5 and gave no clearing zones at all. In contrast to the high phenotypic homogeneity, it is known that the clinical and environmental S. maltophilia isolates are in fact very heterogeneous genotypically, as revealed by studies using phenotypic profiling, sequencing of the 16S rRNA genes, and various molecular methods (6, 20). The high degrees of intraspecies heterogeneity may account for our observation that φSMA5 could infect only a portion of the clinical isolates of S. maltophilia. Therefore, for effective phage therapy, it was obvious that we need to isolate different phages and use them as cocktails to kill the heterogeneous S. maltophilia strains existing (9, 16, 36).
Several bacteria other than S. maltophilia were subjected to spot tests, which included E. coli, E. cloacae, K. pneumoniae, P. aeruginosa, P. mirabilis, S. marcescens, Xanthomonas campestris pv. campestris, Xanthomonas axonopodis pv. vesicatoria, and Xanthomonas oryzae pv. oryzae (Table 1). None of these bacteria were susceptible to φSMA5, indicating that this phage has a narrow host range.
Acknowledgments
This work was supported by grants from Taichung Veterans General Hospital and National Chung Hsing University (TCVGH-NCHU-927616) and the National Science Council, Republic of China (no. NSC-92-2311-B005-029).
REFERENCES
- 1.Ackermann, H. W. 2001. Frequency of morphological phage descriptions in the year 2000. Arch. Virol. 146:843-857. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann, H. W. 2003. Bacteriophage observations and evolution. Res. Microbiol. 154:245-251. [DOI] [PubMed] [Google Scholar]
- 3.Aznar, R., E. Alcaide, and E. Garay. 1992. Numerical taxonomy of pseudomonads isolated from water, sediment and eels. Syst. Appl. Microbiol. 14:235-246. [Google Scholar]
- 4.Ballestero, S., I. Virseda, H. Escobar, L. Suarez, and F. Baquero. 1995. Stenotrophomonas maltophilia in cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 14:728-729. [DOI] [PubMed] [Google Scholar]
- 5.Barrangou, R., S.-S. Yoon, F. Breidt, Jr., H. P. Fleming, and T. R. Klaenhammer. 2002. Characterization of six Leuconostoc fallax bacteriophages isolated from an industrial sauerkraut fermentation. Appl. Environ. Microbiol. 68:5452-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, G., N. Roskot, and K. Smalla. 1999. Genotypic and phenotypic relationships between clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 37:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black, L. W., M. K. Showe, and A. C. Steven. 1994. Morphogenesis of the T4 head, p. 218-258. In J. Karam, J. W. Drake, K. N. Kreuzer, G. Mosig, D. H. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 8.Boonchan, S., M. L. Britz, and G. A. Stanley. 1998. Surfactant-enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia. Biotechnol. Bioeng. 59:482-494. [DOI] [PubMed] [Google Scholar]
- 9.Carlton, R. M. 1999. Phage therapy: past history and future prospects. Arch. Immunol. Ther. Exp. (Warsaw) 47:267-274. [PubMed] [Google Scholar]
- 10.Chow, M. S., and M. A. Rouf. 1983. Isolation and partial characterization of two Aeromonas hydrophila bacteriophages. Appl. Environ. Microbiol. 45:1670-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denton, M., and K. G. Kerr. 1998. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11:57-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denton, M., N. J. Todd, K. G. Kerr, P. M. Hawkey, and J. M. Littlewood. 1998. Molecular epidemiology of Stenotrophomonas maltophilia isolated from clinical specimens from patients with cystic fibrosis and associated environmental samples. J. Clin. Microbiol. 36:1953-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsner, H. A., U. Duhrsen, B. Hollwitz, P. M. Kaulfers, and D. K. Hossfeld. 1997. Fatal pulmonary hemorrhage in patients with acute leukemia and fulminant pneumonia caused by Stenotrophomonas maltophilia. Ann. Hematol. 74:155-161. [DOI] [PubMed] [Google Scholar]
- 14.Fages, J., and J. F. Arsac. 1991. Sunflower inoculation with Azospirillum and other plant growth promoting rhizobacteria. Plant Soil 137:87-90. [Google Scholar]
- 15.Fisher, M. C., S. S. Long, E. M. Roberts, J. M. Dunn, and R. K. Balsara. 1981. Pseudomonas maltophilia bacteremia in children undergoing open heart surgery. JAMA 246:1571-1574. [PubMed] [Google Scholar]
- 16.Flaherty, J. E., B. K. Harbaugh, J. B. Jones, and G. C. Somodi. 2001. H-mutant bacteriophages as a potential biocontrol of bacterial blight of geranium. HortScience 36:98-100. [Google Scholar]
- 17.Foschino, R., F. Perrone, and A. Galli. 1995. Characterization of two virulent Lactobacillus fermentum bacteriophages isolated from sour dough. J. Appl. Bacteriol. 79:677-683. [Google Scholar]
- 18.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 19.Hatanaka, C., and J. Ozawa. 1971. Pectolytic enzymes of exo-types. Part I. Oligogalacturonide transeliminase of a Pseudomonas. Agric. Biol. Chem. 35:1617-1624. [Google Scholar]
- 20.Hauben, L., L. Vauterin, E. R. Moore, B. Hoste, and J. Swings. 1999. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Bacteriol. 49:1749-1760. [DOI] [PubMed] [Google Scholar]
- 21.Hugh, R., and E. Leifson. 1963. A description of the type strain of Pseudomonas maltophilia. Int. Bull. Bacteriol. Nomencl. Taxon. 13:133-138. [Google Scholar]
- 22.Hugh, R. 1981. Pseudomonas maltophilia sp. nov. nom. rev. Int. J. Syst. Bacteriol. 31:195. [Google Scholar]
- 23.Juhnke, M. E., D. E. Mathre, and D. C. Sands. 1987. Identification and characterization of rhizosphere-competent bacteria of wheat. Appl. Environ. Microbiol. 53:2793-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, T. T., and J. Tu. 1976. Enzymatic synthesis of deoxy-5-methyl-cytidylic acid replacing deoxycytidylic acid in Xanthomonas oryzae phage Xp12 DNA. Nature 263:615. [DOI] [PubMed] [Google Scholar]
- 25.Lambert, T., M.-C. Ploy, F. Denis, and P. Courvalin. 1999. Characterization of the chromosomal aac(6′)-Iz gene of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 43:2366-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livermore, D. M., and N. Woodford. 2000. Carbepenemases: a problem in waiting? Curr. Opin. Microbiol. 3:489-495. [DOI] [PubMed] [Google Scholar]
- 27.Mesyanzhinov, V. V., J. Robben, B. Grymonprez, V. A. Kostyuchenko, M. V. Bourkaltseva, N. N. Sykilinda, V. N. Krylov, and G. Volckaert. 2002. The genome of bacteriophage phiKZ of Pseudomonas aeruginosa. J. Mol. Biol. 317:1-19. [DOI] [PubMed] [Google Scholar]
- 28.Miller, E. S., E. Kutter, G. Mosig, F. Arisaka, T. Kunisawa, and W. Rüger. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, E. S., J. F. Heidelberg, J. A. Eisen, W. C. Nelson, A. S. Durkin, A. Ciecko, T. V. Feldblyum, O. White, I. T. Paulsen, W. C. Nierman, J. Lee, B. Szczypinski, and C. M. Fraser. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220-5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1993. Restriction/modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the dairy industry. Appl. Environ. Microbiol. 59:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pajunen, M., S. Kiljunen, and M. Skurnik. 2000. Bacteriophage φYeO3-12, specific for Yersinia enterocolitica serotype O:3, is related to coliphages T3 and T7. J. Bacteriol. 182:5114-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadakis, K. A., S. E. Vartivarian, M. E. Vassilaki, and E. J. Anaissie. 1997. Stenotrophomonas maltophilia meningitis. Report of two cases and review of the literature. J. Neurosurg. 87:106-108. [DOI] [PubMed] [Google Scholar]
- 33.Reller, L. B., F. D. Schoenknecht, M. A. Kenny, and J. C. Sherris. 1974. Antibiotic susceptibility testing of Pseudomonas aeruginosa: selection of a control strain and criteria for magnesium and calcium content in medium. J. Infect. Dis. 130:454-463. [DOI] [PubMed] [Google Scholar]
- 34.Roilides, E., K. M. Butler, R. N. Husson, B. U. Mueller, L. L. Lewis, and P. A. Pizzo. 1992. Pseudomonas infections in children with human immunodeficiency virus infection. Pediatr. Infect. Dis. J. 11:547-553. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Sulakvelidze, A., Z. Alavidze, and J. G. Morris, Jr. 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swings, J., P. De Vos, M. Van den Mooter, and J. De Ley. 1983. Transfer of Pseudomonas maltophilia Hugh 1981 to the genus Xanthomonas as Xanthomonas maltophilia (Hugh 1981) comb. nov. Int. J. Syst. Bacteriol. 33:409-413. [Google Scholar]
- 38.Tseng, Y.-H., K.-T. Choy, C.-H. Hung, N.-T. Lin, J.-Y. Liu, C.-H. Lou, B.-Y. Yang, F.-S. Wen, S.-F. Weng, and J.-R. Wu. 1999. Chromosome map of Xanthomonas campestris pv. campestris 17 with locations of genes involved in xanthan gum synthesis and yellow pigmentation. J. Bacteriol. 181:117-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van den Mooter, M., and J. Swings. 1990. Numerical analysis of 295 phenotypic features of 266 Xanthomonas strains and related strains and an improved taxonomy of the genus. Int. J. Syst. Bacteriol. 40:348-369. [DOI] [PubMed] [Google Scholar]
- 40.Vartivarian, S. E., K. A. Papadakis, and E. J. Anaissie. 1996. Stenotrophomonas (Xanthomonas) maltophilia urinary tract infection. A disease that is usually severe and complicated. Arch. Intern. Med. 156:433-435. [PubMed] [Google Scholar]
- 41.Yang, B. Y., and Y. H. Tseng. 1988. Production of exopolysaccharide and levels of protease and pectinase activity in pathogenic and non-pathogenic strains of Xanthomonas campestris pv. campestris. Bot. Bull. Acad. Sin. 29:93-99. [Google Scholar]
- 42.Yang, B. Y., H. F. Tsai, and Y. H. Tseng. 1988. Broad host range cosmid pLAFR1 and non-mucoid mutant XCP20 provide a suitable vector-host system for cloning genes in Xanthomonas campestris pv. campestris. Chin. J. Microbiol. Immunol. 21:40-49. [PubMed] [Google Scholar]
- 43.Zuravleff, J. J., and V. L. Yu. 1982. Infections caused by Pseudomonas maltophilia with emphasis on bacteremia: case reports and a review of the literature. Rev. Infect. Dis. 4:1236-1246. [DOI] [PubMed] [Google Scholar]