Abstract
Microorganisms, such as bacteria, which might be present as contamination inside an industrial food or pharmaceutical clean room process need to be identified on short time scales in order to minimize possible health hazards as well as production downtimes causing financial deficits. Here we describe the first results of single-particle micro-Raman measurements in combination with a classification method, the so-called support vector machine technique, allowing for a fast, reliable, and nondestructive online identification method for single bacteria.
Microbial contamination not only is a medical problem but also plays a large role in pharmaceutical clean-room production and food-processing technology. For all these fields, a fast and nonambiguous identification of pathogenic and nonpathogenic microorganisms is required. Conventional bacterial identification methods are based on morphological evaluation of the microorganisms and their ability to grow in various media under different conditions (23). Depending on the type of bacteria, the identification process may take at least 1 day but generally takes much longer (2). Bacterial detection methods have to be rapid and very sensitive because even a single pathogenic organism may be an infectious dose (23). To address the requirements, new analysis methods such as mass spectroscopy, PCR, flow cytometry and fluorescence spectroscopy were developed, allowing a fast and reliable identification (2, 23).
An alternative approach to the analysis of microorganisms is the application of vibrational spectroscopic techniques (infrared [IR] and Raman spectroscopy), which have a long tradition since the vibrational spectrum displays a fingerprint of the chemical composition of each bacterium (35). While an IR spectroscopic investigation of microorganisms requires a few hundred cells from controlled cultivation conditions for an analysis and a drying step (37), this is not necessary when applying Raman spectroscopy (36). In particular, when only a small sample amount is available, a special Raman technique called SERS spectroscopy (surface-enhanced Raman scattering spectroscopy) is especially suited. For an investigation of bacteria, various different SERS substrates or SERS microchips in combination with antibodies were tested (14, 15, 19, 24, 25, 53, 54, 63). By applying UV-resonance Raman spectroscopy, direct investigation of macromolecules such as proteins or DNA becomes possible. However, this Raman technique involves extensive experimental costs and extremely careful sample handling (4, 7, 8, 13, 18, 26, 28-30, 38, 39, 58, 59). In 1990, Puppels et al. developed a confocal Raman microscope, capable of recording Raman spectra of single human cells and polytene chromosomes (42). Since then, many biological phenomena in single human cells have been studied by Puppel's group; e.g., Raman spectra of the cell nucleus and the cell cytoplasm in human white blood cells were obtained (43-46). Various groups have reported the classification of bacteria by means of Raman spectroscopy (3, 11, 12, 17, 27, 31-34, 47). Very recently, Maquelin et al. (31) performed for the first time a clinical Fourier transform IR and near-IR-Raman study of bacterial contamination in blood cultures by using microcolonies obtained after 6 to 8 h of cultivation. Other papers have reported Raman and SERS investigations of single yeast cells, bacteria, or spores (1, 10, 21, 60-62). Various investigations of cell components of single bacteria or spores by means of Raman spectroscopy have also been reported (16, 20, 22, 40, 50, 51). In this paper we describe, to the best of our knowledge for the first time, a fast, nondestructive, and very reliable approach to the identification of bacteria on a single-microparticle level by means of a combination of a micro-Raman analysis together with a data classification approach, the so-called support vector machine (SVM) technique.
MATERIALS AND METHODS
Spectroscopic instrumentation.
The Raman spectra were obtained with a micro-Raman setup (HR LabRam invers, Jobin-Yvon-Horiba). The spectrometer has an entrance slit of 100 μm and a focal length of 800 mm and is equipped with a 300-lines/mm grating. As excitation wavelengths, the 532-nm line of a frequency-doubled Nd:YAG laser (Coherent Compass) with a laser power of 10 mW incident on the sample were used. The Raman-scattered light was detected by a charge-coupled-device camera operating at 220 K. A Leica PLFluoar 100× objective focused the laser light onto the samples (ca. 0.7-μm focus diameter). An integration time of 60 s was used both for the bulk and single-bacterium spectra. For the spatially resolved measurements, an x/y motorized stage (Merzhäuser) with a minimal possible step size of 0.1 μm was used. The z displacement was controlled by a piezo-transducer on the objective.
Bacteria and growth conditions.
The microorganisms were chosen according to the conditions present in clean rooms. The microorganisms Micrococcus luteus (DSM 348 and DSM 20030), Micrococcus lylae (DSM 20315 and DSM 20318), Bacillus subtilis (DSM 10 and DSM 347), Bacillus pumilus (DSM 27 and DSM 361), Bacillus sphaericus (DSM 28 and DSM 396), Escherichia coli (DSM 423, DSM 498, and DSM 499), Staphylococcus cohnii (DSM 6669, DSM 20260, DSM 6718, and DSM 6719), Staphylococcus warneri (DSM 20036 and DSM 20316), and Staphylococcus epidermidis (RP 62A) were purchased from the Deutsche Sammelstelle für Mikroorganismen und Zellkulturen and from the Institut für Infektionsbiologie, Universität Würzburg. They were cultivated on a standard or nutrition agar (Micrococcus and Bacillus) or on caseine-peptone soymeal-peptone agar (Staphylococcus) for different growing conditions, such as growing time and temperature, respectively. To simulate samples from clean rooms, the Raman measurements were directly performed on single cells from smears on fused silica plates.
SVMs.
The analysis of Raman spectra was performed in two steps: (i) preprocessing of the spectra and (ii) classification by using SVMs. The preprocessing was tested by different methods such as baseline correction, normalization, first derivative, and median filtering. Normalization for bulk data and median filtering for both bulk and single-bacterium analysis have obtained the best results. The classification was based on the regions from 850 to 1,750 and 2,650 to 3,150 cm−1. The limits of those regions were chosen by using the optimization procedure of linear SVMs described below.
The classification step was achieved by using SVM. These large-margin classifiers are widely used in pattern analysis and are already well understood (5, 57). It has been shown that standard SVMs perform as well as or better than neural networks (NN) even in domains such as hand-written character recognition, where several years of research were spent to optimize the NN for a certain problem (48).
Since a classification task can always be broken down into several two-class problems (using a one-versus-one approach), a SVM solves only a two-class problem. The basic idea is as follows. The traditional approach to classification usually tries to build a model during the training step for each class independently from the other classes. The classifier then tests how well an unknown spectrum matches the different models and assigns the spectrum to the best-fitting one. This could also be interpreted as first using the two models for calculating a border between the two classes and second testing on which side of this border the spectrum lies. The idea of an SVM is therefore to combine these two steps and directly model the border between the two classes. This omits modeling of irrelevant parts and therefore needs much less training data. Since there are many possible hyperplanes, which separate the two classes in the feature space (Fig. 1A), the distance to the training samples is introduced as a quality criterion. With this criterion, there is only one global optimum—the hyperplane with the largest margin—which could reliably be found in the training process. This is a big advantage of SVMs compared to NN, where several suboptimal solutions are found during the training process. The samples that are touched by the margin of the hyperplane are called “support vectors.” Therefore, for training and classification, only these support vectors are necessary, while all other vectors could be removed from the training set without changing the result. If an SVM is trained, for example, to classify yeasts and bacteria, it will select the most yeast-like bacteria and the most bacterium-like yeasts as support vectors and will use only those to classify an unknown microorganism. The optimal separation plane with the largest margin and the support vectors (adjacent training samples marked by circles) are shown in Fig. 1B. In cases where the training set includes outliers, i.e., samples that are beyond the separating plane, a cost value is introduced to give those data points a disadvantage. In that way, SVM can model a real-world training data set very efficiently.
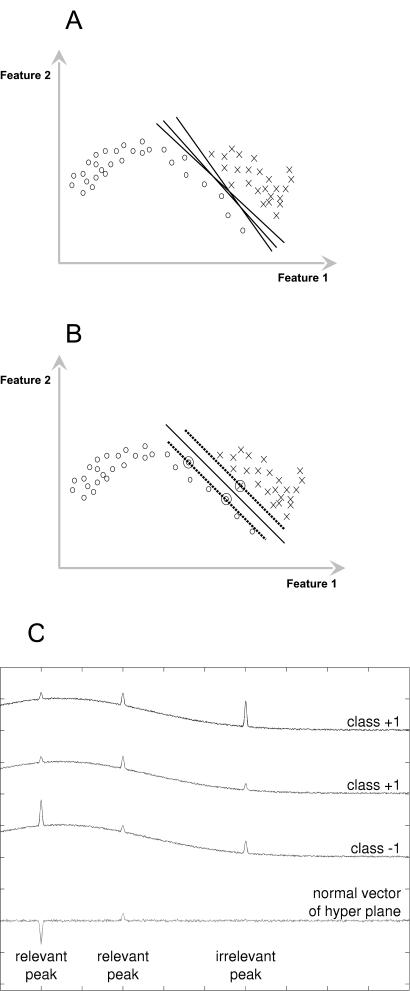
FIG. 1.
(A) Possible planes for separating the two classes. (B) The optimal separation plane has the largest margin and is defined only by the adjacent training samples. Support vectors are marked by circles. (C) Classification of simulated spectra. The SVM automatically detects relevant and irrelevant peaks. The third peaks of class +1 differ in size, and so compared to the third peak in class −1, those peaks contain no discriminative information and are irrelevant for the SVM classification.
For classification, we used an SVM template library that was developed at our institute and is based on the libsvm library (9). Due to the high dimensionality of the data, the border could be defined by a linear function yielding a linear SVM. Applying other (nonlinear) SVMs to the classification problem did not improve the recognition result significantly. We tried one-versus-one and one-versus-rest test setups separately; we found that for our data, the one-versus-one test performed slightly better. Therefore, all the reported results were acquired with a linear SVM and a one-versus-one method for training and classification, while the cost value was set to 90,000.
The output of linear SVMs can be interpreted geometrically, so that one can find out which parts of the spectrum were used for the classification by looking at the direction of the normal vector of the separating hyperplane. In that way, one can identify relevant and irrelevant peaks; this is one of the main advantages of using SVM for classification of spectral data.
This is shown in a simple simulation in Fig. 1C, where three spectra (two of class +1 and one of class −1) and a plot of the normal vector are given. For the spectra, only the first two peaks contain relevant information, while the third does not. Training an SVM with those three spectra, the SVM will automatically find the relevant parts of the spectrum and ignore the irrelevant parts. The height of the peaks in the hyperplane plot shows how important this peak is for the classification, whereas the sign of the peak tells whether it belongs to class +1 or class −1.
Leave-one-out test.
For the estimation of the classification error probability of the final system (which will use all recorded spectra as the training set), the leave-one-out error was chosen (which uses N − 1 samples as the training set) instead of the widely used “holdout method” (which uses only a certain fraction, e.g., 50%, of the samples as the training set). While it is mathematically proven that the leave-one-out error is an “almost unbiased” estimate for the real classification error probability (49), the holdout method is proven to always return a biased (too high) estimate of the classification error probability (55). (The term “almost” refers to the fact that the leave-one-out error provides an estimate for training on sets of size N − 1 rather than N [49].)
Since the a priori probability for the occurrence of each cell species may vary from clean room to clean room, the reported “average recognition rate” is always the arithmetic mean of the recognition rates for each species and therefore equalizes the varying number of samples per species in our database.
RESULTS AND DISCUSSION
In clean-room applications not only the composition of dust but also the number of particles differ from those in the regular environment (56). Usually, production in the pharmaceutical industry takes place in class A or B clean rooms (3,500 0.5-μm particles per m3 and 0 5-μm particles per m3). The main components of dust are metal particles, rubber abrasion, skin, hair, and a few microorganisms. Conventional investigation methods count the total number of particles or the colony-building units (56). A complete investigation of the origin of particle contamination would be extremely beneficial, since this would lead to a rapid identification of the source of the contamination. Since clean-room conditions differ from a natural environment, bacteria that can be found in a clean room depend on the purpose for which the room is being used. Furthermore, the total number of bacteria is fairly small, and only a limited number of species are detected compared to a natural environment (H.-W. Motzkus, personal communication).
Bulk spectra.
Since the origin of the microorganisms present in a clean room is unknown, a single-bacterium analysis requires careful testing of various parameters such as different culture media or growth times. In the experiments described below, typical clean-room samples are modeled as smears of various microorganisms on fused silica plates. In a first approach, 20 different strains of nine bacterial species which are typical of clean-room contaminants (Motzkus, personal communication) were chosen. Among the chosen bacteria, both colored and noncolored species can be found. Colored bacteria can be easily identified by the presence of carotenoids, which is the pigment in most colored microorganisms. However, this does not allow a distinct identification of the species or strain that is present. Identification of the noncolored bacteria is expected to be even more difficult. Different regions can be found on such a smear: (i) multilayer regions, which can be used to record bulk spectra, and (ii) regions with isolated single cells, where single-bacterium investigations can be performed.
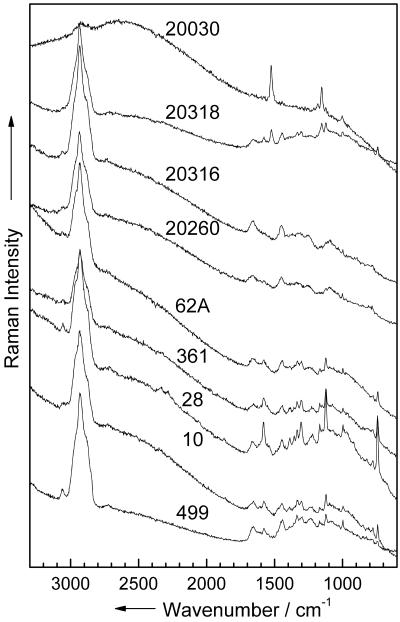
In a first attempt, Raman measurements within a multilayer region on the smear were recorded in order to obtain bulk Raman spectra. Figure 2 shows the Raman spectra of nine different strains, typical of each species. The spectra were recorded with an integration time of 60 s on different multilayer regions on the plate (10 to 20 repetitions [Table 1 ]). The spectrum of the colored strain (M. luteus DSM 20030) is dominated by the carotene bands at 1,525, 1,154, and 1,002 cm−1. Almost no vibrations due to the cell matrix can be seen. For the noncolored strains, the Raman spectra of the four genera are very similar. The Raman spectrum of E. coli DSM 499 reveals higher intensities of the amid-I band than do the spectra of the Bacillus strains, as well as a very intense C-H band (around 2,900 cm−1). The three Bacillus strains (B. subtilis DSM 10, B. pumilus DSM 361, and B. sphaericus DSM 28) exhibit very similar spectra. The signal-to-noise ratio of the three Staphylococcus spectra is very low, which is due to the high fluorescence background of these strains (S. warneri DSM 20316, S. cohnii DSM 20260, and S. epidermidis RP 62 A). For a distinct identification of the strains, a reliable data analysis method is required. Therefore, a chemometric data analysis was performed.
FIG. 2.
Micro-Raman spectra of nine different strains (bulk). The numbers in the figure are the strain numbers.
TABLE 1.
Recognition rate for bulk Raman spectra of various bacterial strains
| Strain | Total no. of spectra | No. of wrongly classified strain spectra | Recognition rate for strains (%) | No. of wrongly classified species spectra | Recognition rate for species (%) |
|---|---|---|---|---|---|
| B. pumilus DSM 27 | 12 | 0 | 100.0 | 0 | 100.0 |
| B. pumilus DSM 361 | 12 | 0 | 100.0 | 0 | 100.0 |
| B. sphaericus DSM 28 | 14 | 0 | 100.0 | 0 | 100.0 |
| B. sphaericus DSM 396 | 16 | 1 | 93.8 | 0 | 100.0 |
| B. subtilis subsp. subtilis DSM 10 | 16 | 0 | 100.0 | 0 | 100.0 |
| B. subtilis subsp. spizizenii DSM 347 | 10 | 0 | 100.0 | 0 | 100.0 |
| E. coli DSM 423 | 12 | 0 | 100.0 | 0 | 100.0 |
| E. coli DSM 498 | 20 | 1 | 95.0 | 0 | 100.0 |
| E. coli DSM 499 | 20 | 0 | 100.0 | 0 | 100.0 |
| M. luteus DSM 348 | 12 | 0 | 100.0 | 0 | 100.0 |
| M. luteus DSM 20030 | 20 | 0 | 100.0 | 0 | 100.0 |
| M. lylae DSM 20315 | 21 | 0 | 100.0 | 0 | 100.0 |
| M. lylae DSM 20318 | 22 | 0 | 100.0 | 0 | 100.0 |
| S. cohnii subsp. cohnii DSM 6669 | 20 | 2 | 90.0 | 2 | 90.0 |
| S. cohnii subsp. cohnii DSM 20260 | 13 | 1 | 92.3 | 1 | 92.3 |
| S. cohnii subsp. urealyticum DSM 6718 | 16 | 0 | 100.0 | 0 | 100.0 |
| S. cohnii subsp. urealyticum DSM 6719 | 18 | 1 | 94.4 | 0 | 100.0 |
| S. epidermidis RP 62A | 25 | 0 | 100.0 | 0 | 100.0 |
| S. warneri DSM 20036 | 20 | 1 | 95.0 | 1 | 95.0 |
| S. warneri DSM 20316 | 20 | 0 | 100.0 | 0 | 100.0 |
| Average recognition rate | 98.0 | 98.9 |
Data analysis of bulk spectra.
Since the Raman intensities differ between two successive measurements due to slightly different experimental conditions, a normalization procedure is required. The Raman spectra were normalized on the intensity of the CH peak because this signal represents the total amount of organic compounds in the cells. Not only is the C-H vibration characteristic of one special component, as, for example, fatty acids, but also it corresponds to the sum of all saturated organic material. (Note that it has been found that using C-H vibrations for normalization yields the best results of classification and testing.)
The recognition rate (median filtering, normalization, linear SVM, leave-one-out test) shows very good results (98.0%; Table 1) for all bacterial strains. Of the 339 Raman spectra, 7 were misclassified; e.g., within E. coli, one DSM 498 strain spectrum was not classified correctly but was assigned as a spectrum of E. coli DSM 499 (i.e., the species was identified correctly whereas the assigned strain was wrong). Therefore, the overall identification at the species level reveals 98.9%. These results nicely prove that a reliable identification can be obtained for microcultures. However, it requires up to 6 h to obtain those microcultures by cultivation. It would be greatly preferable to identify single bacteria by means of micro-Raman spectroscopy. Therefore, experiments with isolated single cells were performed to test if a reliable identification of the bacteria is possible on the single-cell level.
Single-cell spectra.
Bulk Raman spectra are the result of an averaging over several bacteria. However, for isolated single bacteria, individual variations within the various cells need to be considered. Before performing a single-bacterium analysis, various experiments are needed to investigate if and how different parameters influence the identification of microorganisms on the single-cell level. As already mentioned, no information about the origin of the bacteria is available. Therefore, to create a reliable data set, the variation of different parameters, e.g., nutrition, temperature, and growth time, needs to be considered. Furthermore, it is also necessary to investigate the spatial heterogeneity within a single microorganism, i.e., whether there are any variations within the Raman spectra if the laser focus slightly shifts on the investigated single bacterium.
Raman mapping (heterogeneity effect).
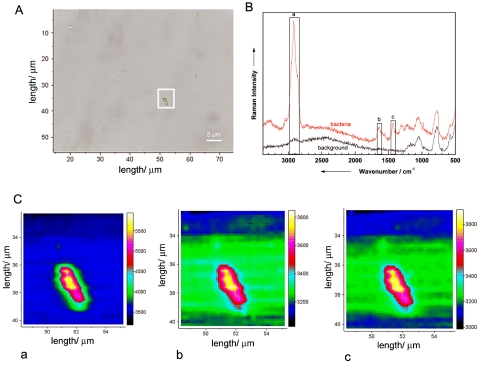
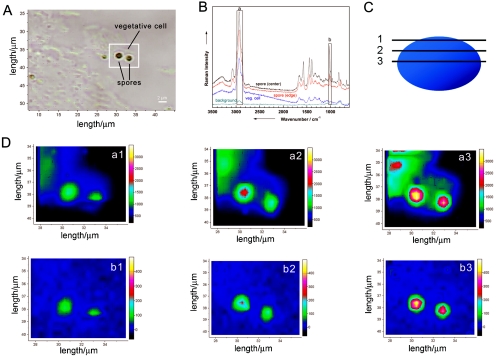
Figure 3A shows a micrograph of an isolated single B. sphaericus DSM 28 cell on a fused silica plate. Figure 3B displays a Raman spectrum of this bacterium in comparison with a background spectrum, which was recorded beside the microorganism. To test if the Raman spectrum depends on the spatial position of the focus within the bacterium, Raman mapping experiments over the area displayed by the white square in Fig. 3A were performed. For the mapping experiments, a step size of 0.3 by 0.3 μm2 (total, 20 by 28 points) was chosen. These parameters are smaller than the spatial resolution of the Raman microscope (0.7 μm) but were chosen to increase the spatial overlap of the Raman mapping experiments. Each spectrum was measured with an integration time of 120 s, which leads to a total measuring time of 20 h. To minimize the background of fused silica, a pinhole of 500 μm was used.
FIG. 3.
Raman mapping experiment of a single bacterium (B. sphaericus DSM 28). (A) Micrograph of a single bacterium. The white frame indicates the mapping area (0.3 by 0.3 μm2) for taking the Raman images shown in panel C. (B) Micro-Raman spectra from selected positions within the marked scan area. The marked bands are used to calculate the Raman images plotted in panel C. (C) Raman maps for three different wavenumber regions labeled in panel B: a, 2,851 to 2,964 cm−1; b, 1,604 to 1,671 cm−1; and c, 1,410 to 1,455 cm−1. For details, see the text.
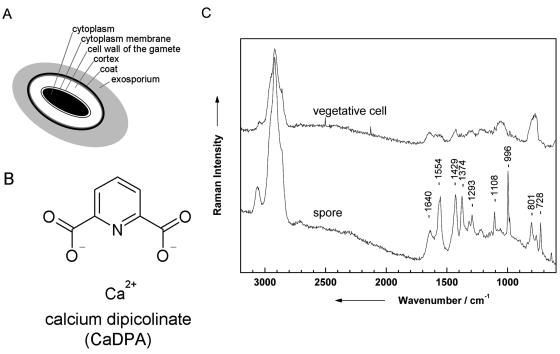
Figure 3C shows three Raman images of the three representative bands labeled in Fig. 3B, namely, the C-H stretch vibrations at ∼2,900 cm−1 (a), the amide I vibration at ∼1,650 cm−1 (b), and the CH2 deformation vibration at ∼1,420 cm−1 (c). As can be clearly seen in the Raman images, no dependency on the spatial position of the measurement could be observed; i.e., the bacterium shows spatial homogeneity. This can be explained by the fact that bacteria normally have no compartments. Some bacteria might contain vesicles where, for example, sulfur or poly-β-hydroxybutyric acid is stored. As was shown by Schuster et al. (50, 51), Raman spectra of single Clostridium cells differ with different amounts of starchlike granulose. When line scans over the cell axis were used, no variations with the measuring position could be observed. Another example of structured bacteria involves resistant dominant bodies (spores), which are known to be more complex than vegetative cells since they exhibit several layers, which are schematically displayed in Fig. 4A. The marker substance of bacterial spores is calcium dipicolinate (CaDPA; the structure is shown in Fig. 4B). In Fig. 4C, two spectra of isolated B. sphaericus DSM 28 cells (vegetative cell and spore) are displayed. Distinct differences due to CaDPA can be observed in the spectra. The Raman spectrum of the spore shows a band at 1,651 cm−1, which is due to the amide I band. The very intense signals at 1,565, 1,440, 1,383, and 1,007 cm−1 can be assigned to CaDPA (6, 16, 18, 30, 41, 53). According to Carmona (6), the vibration at 1,007 cm−1 can be assigned to the ring-breathing vibration of the pyridine ring. The C-O-C stretching vibration can be observed at 1,385 cm−1, whereas both signals at 1,440 and 1,565 cm−1 can be assigned to ring vibrations.
FIG. 4.
(A) Schematic diagram of a spore. (B) Chemical structure of CaDPA, which is a marker substance and can be found in all spores. (C) Raman spectra of a vegetative cell and a spore of B. sphaericus DSM 28.
Since bacterial spores are highly heterogeneous (Fig. 4A), it is expected that the Raman spectra of such spores will depend on the spatial position where the Raman spectrum was recorded. Figure 5A shows a microphotograph of several vegetative cells and two spores of B. sphaericus DSM 28. Spores, which are highly refractive bodies, can be distinguished from vegetative cells under a light microscope (Fig. 5A). In Fig. 5B, Raman spectra taken at four different spatial positions within the white square are shown. The spectrum at the bottom corresponds to a background spectrum, while the other spectra are from a vegetative cell or at two different positions within the spore, respectively. As already shown in Fig. 4C, the spectra of vegetative cells and spores differ due to the different concentrations of CaDPA. Additionally, the Raman spectra of the spore recorded at two different positions show subtle variations. This becomes even more obvious in Fig. 5D, where various confocal Raman images are shown. The images were recorded over the area displayed by the white square shown in the microphotograph in Fig. 5A. Furthermore, a depth profiling has been performed by measuring the images at three different depths shown in the schematic sketch in Fig. 5C. For the three-dimensional Raman mapping, a lateral and axial spatial resolution of 0.5 μm was used, which leads to a total volume of 22 by 16 by 3 points. Each spectrum was measured with an integration time of 300 s, resulting in a total time of approximately 90 h. For the confocal measurements, a hole of 200 μm and a slit of 50 μm was used. The laser power at the sample was ca. 2.5 mW. The images a1, a2, and a3 map the intensity of the C-H stretch vibrations (2,871 to 2,991 cm−1 in Fig. 5B, region a) recorded for the three different depth positions 1, 2, and 3, as shown in Fig. 5C inside the mapping area. As can be seen from the Raman spectra (Fig. 5B), C-H stretch vibrations can be found in both vegetative cells and spores. However, when looking at the confocal Raman images displayed in Fig. 5D (a1, a2, and a3), it is evident that spores exhibit a higher degree of ellipticity because at depth position 1 the CH intensity can be found only at positions where the spores are located (image a1 in Fig. 5D). For positions 2 and 3, CH intensity also occurs where the vegetative cells can be found (images a2 and a3 in Fig. 5D). This becomes even more obvious when mapping a Raman band which can be only found in the Raman spectrum of the spores. Images b1, b2, and b3 in Fig. 5D map the intensity of the ring-breathing mode of the pyridine ring of CaDPA (993 to 1,034 cm−1 in Fig. 5B, region b), which is a characteristic component of spores and which does not appear in vegetative cells. Images b1, b2, and b3 in Fig. 5D map the spores exclusively. The spore images recorded by integrating over the C-H stretch vibrations (a1, a2, and a3 in Fig. 5D) show subtle differences from the images obtained by integrating over the pyridine ring breathing mode (b1, b2, and b3 in Fig. 5D). CaDPA can be found only within the cortex layer of the spore (Fig. 4A). The C-H stretch vibrations result from all biological components (proteins, lipids, DNA, etc.) within the vegetative cells or spores, respectively. Their distribution within the spores differs from that of CaDPA.
FIG. 5.
Raman mapping experiment on single spores and vegetative cells (B. sphaericus DSM 28). (A) Micrograph of two spores surrounded by vegetative cells. The white frame indicates the mapping area. (B) Micro-Raman spectra from selected positions within the marked scan area. The spectrum at the bottom corresponds to a background spectrum, while the other spectra are taken from a vegetative cell or at two different positions within the spore, respectively. The marked bands are used to calculate the Raman images plotted in panel D. (D) Raman maps for the two different wavenumber regions labeled in panel B (a, 2,871 to 2,991 cm−1; b, 993 to 1,034 cm−1) for three different depths positions indicated by the three horizontal lines within the schematic sketch of a spore shown in panel C. Position 3, −1.0 μm; position 2, −0.5 μm; position 1, 0 μm. For details, see the text.
To establish an automatic analytical procedure for the identification of single bacteria by means of Raman spectroscopy, these observations have to be taken into account; i.e., to identify heterogeneous bacteria such as single spores, measurements at three to five different positions should be performed (S. Hofer et al., 23 February 2004, German Patent Office).
Cultivation conditions.
Another issue which might be of relevance for an analysis at the single-cell level is that of different nutritional conditions. This has also been tested by measuring Raman spectra of various single bacteria of different strains (not shown here) in different culture media and included in the identification data set.
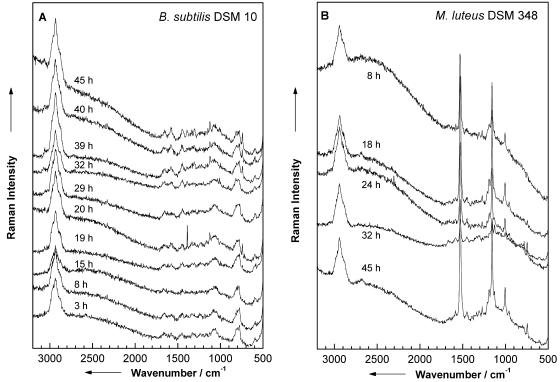
Furthermore, the influence of the growth time on the Raman spectrum needs to be evaluated. Figure 6 shows representative Raman spectra of single B. subtilis DSM 10 (Fig. 6A) and M. luteus DSM 348 (Fig. 6B) cells recorded for different growth times as indicated. The spectra of single cells from very young cultures exhibit a low signal-to-noise ratio with broad bands, while Raman spectra of cells of older cultures show sharp distinct signals. These signals belong to vegetative cells and not to spores (compare Fig. 4C). For an unambiguous analysis on the single-cell level, these variations have to be taken into account, being included in the applied database for the chemometric identification (see below).
FIG. 6.
(A) Raman spectra of single B. subtilis cells recorded for different growth times as indicated. (B) Raman spectra of single colored M. luteus bacteria for various growth times as indicated.
M. luteus is a colored bacterium, in which the pigment comes from the presence of the carotenoid sarcinaxanthin exhibiting an absorption maximum around 500 nm. Therefore, the Raman spectra of M. luteus recorded for an excitation wavelength of 532 nm are resonantly enhanced. The strong bands at 1,532, 1,157, and 1,005 cm−1 (Fig. 6B) are due to C=C stretch, C—C stretch, and C—CH3 deformation modes of sarcinaxanthin. As can be seen in Fig. 6B, the concentration of this chromophore varies with the cultivation age. For 32 h, the lowest concentration of sarcinaxanthin can be observed, while for 45 h, a maximum is reached. Overall, the concentration distribution of sarcinaxanthin does not show a linear relationship to cultivation age.
Photobleaching.
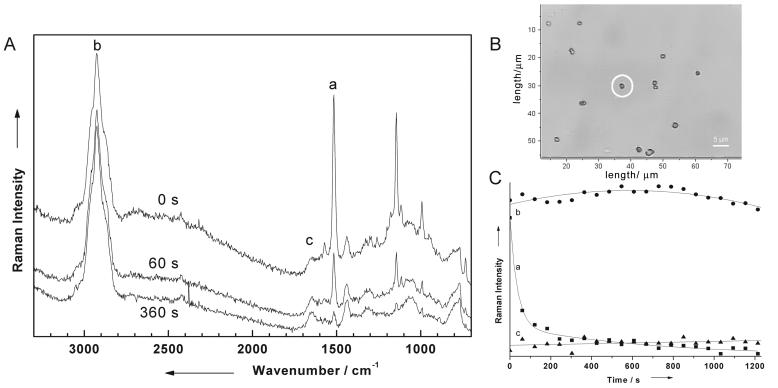
The concentration variation of colored bacteria is accompanied by bleaching effects, which occur exclusively when working with a single colored bacterium and when the Raman excitation laser lies within the absorption band of the bacterium's chromophore. This is illustrated in Fig. 7A, where Raman spectra of a single M. luteus cell (cell marked by a circle in Fig. 7B, showing a microphotograph of several M. luteus DSM 348 cells) are plotted for three different irradiation times, as indicated. Each Raman spectrum was recorded with an integration time of 60 s. The top spectrum shows the initial Raman spectrum, while the two other spectra were recorded directly after irradiating the same single M. luteus bacterium after 60 and 360 s, respectively, with the 532-nm laser. The intensity of the carotenoid bands at 1,532, 1,157, and 1,005 cm−1 decreases with increasing irradiation time. Figure 7C shows the dependency of the intensity of three different bands labeled by a, b, and c in Fig. 7A as a function of the irradiation time. As can be clearly seen from the marked signals, only the mode corresponding to the C=C stretch vibration of sarcinaxanthin at 1,532 cm−1 (band a), which is resonantly enhanced, shows a time dependency. The intensities of the other two bands, b and c, which are not chromophore vibrations, are almost unaffected by the irradiation process. These measurements have been repeated several times, and the same time behavior could always be observed; i.e., this irradiation time behavior is absolutely reproducible. However, this bleaching effect of the chromophore modes is advantageous for a single-cell analysis, since after the bleaching has taken place, only the characteristic Raman bands due to the cell matrix are left. This is especially important since many bacteria produce pigment structures of the carotenoid type and since the production of these structures depends strongly on the cultivation and growth state. Thus, pigmentation of bacteria alone is generally useless for microbiological identification, but when it is used in combination with the information about the cell matrix obtained after bleaching, an exact identification can be made.
FIG. 7.
(A) Micro-Raman spectra of M. luteus DSM 348 recorded after irradiating the single M. luteus cell at 0, 60, and 360 s with the 532-nm laser, which is resonant with an electronic absorption of the chromophore sacinaxanthin of this microorganism. (B) Micrograph of various single M. luteus bacteria. The cell with which the Raman spectra in panel A was obtained is marked by a circle. (C) Dependency of the three bands labeled a, b, and c in panel A on the irradiation time. Only the band which corresponds to the C=C stretch vibration of the protection pigment sacinaxanthin of M. luteus shows a bleaching effect, while the other two vibrations, namely, the C-H vibration (b) and the amide-I band (c), are almost not affected by irradiation with 532 nm. For details, see the text.
All these results demonstrate that the above-mentioned parameters, such as heterogeneity, growth time, bleaching effects, and nutrition conditions, might lead to more or fewer variations within the Raman spectra of single cells. Most of those variations have already been included in our database as well as in our classification procedure for an unambiguous assignment of bacteria on a single-cell level.
Single-cell identification.
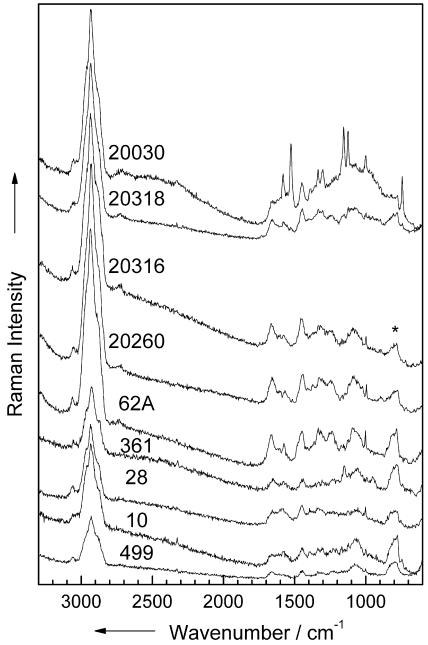
In Fig. 8, examples of micro-Raman spectra of single cells of nine representative different species are shown, with an integration time of 60 s per spectrum. The spectra show characteristic differences from the corresponding bulk spectra plotted in Fig. 2: a lower background, a lower signal-to-noise ratio, or additional signals due to the fused silica plate (asterisk in Fig. 8), which all occur from the very low sample volume of 0.5 μm3 (Micrococcus and Staphylococcus) to 2.5 μm3 (Bacillus and E. coli) of a single bacterium. The poor signal-to-noise ratio for each Raman spectrum of the various single cells is a result of the short integration time. However, the quality of the single-cell spectra shown in Fig. 8 is sufficient for an identification of the bacteria by means of an SVM (see below) (Table 2). Since time is a critical issue for the analysis of clean-room samples, the overall investigation time should be kept as short as possible.
FIG. 8.
Micro-Raman spectra of single living bacteria of nine different strains. The numbers in the figure are the strain numbers. *, fused silica.
TABLE 2.
Recognition rate for Raman spectra of single bacteria
| Strain | Total no. of spectra | No. of wrongly classified strain spectra | Recognition rate for strains (%) | No. of wrongly classified species spectra | Recognition rate for species (%) |
|---|---|---|---|---|---|
| B. pumilus DSM 27 | 57 | 11 | 80.7 | 10 | 82.5 |
| B. pumilus DSM 361 | 43 | 10 | 76.7 | 5 | 88.4 |
| B. sphaericus DSM 28 | 53 | 8 | 84.9 | 5 | 90.6 |
| B. sphaericus DSM 396 | 42 | 7 | 83.3 | 7 | 83.3 |
| B. subtilis subsp. subtilis DSM 10 | 306 | 10 | 96.7 | 8 | 97.4 |
| B. subtilis subsp. spizizenii DSM 347 | 42 | 8 | 81.0 | 3 | 92.9 |
| E. coli DSM 423 | 51 | 4 | 92.2 | 3 | 94.1 |
| E. coli DSM 498 | 21 | 1 | 95.2 | 0 | 100.0 |
| E. coli DSM 499 | 20 | 3 | 85.0 | 1 | 95.0 |
| M. luteus DSM 348 | 619 | 1 | 99.8 | 1 | 99.8 |
| M. luteus DSM 20030 | 48 | 7 | 85.4 | 4 | 91.6 |
| M. lylae DSM 20315 | 20 | 0 | 100.0 | 0 | 100.0 |
| M. lylae DSM 20318 | 20 | 2 | 90.0 | 2 | 90.0 |
| S. cohnii subsp. cohnii DSM 6669 | 67 | 3 | 95.5 | 2 | 97.0 |
| S. cohnii subsp. cohnii DSM 20260 | 65 | 2 | 96.9 | 0 | 100.0 |
| S. cohnii subsp. urealyticum DSM 6718 | 65 | 14 | 78.5 | 7 | 89.2 |
| S. cohnii subsp. urealyticum DSM 6719 | 63 | 8 | 87.3 | 2 | 96.8 |
| S. epidermidis RP 62A | 517 | 6 | 98.8 | 6 | 98.8 |
| S. warneri DSM 20036 | 67 | 6 | 91.0 | 4 | 94.0 |
| S. warneri DSM 20316 | 71 | 10 | 85.2 | 7 | 90.1 |
| Average recognition rate | 89.2 | 93.6 |
Additionally, some other features appear in the spectra, e.g., additional protein signals at 1,655 and 1,452 cm−1 in the spectra of M. luteus DSM 20030. The differences in the three Raman spectra of Bacillus strains as well as the E. coli spectrum are less pronounced than for to the corresponding bulk spectra plotted in Fig. 2. The three Staphylococcus spectra as well as the M. lylae spectrum are of much better quality than the bulk spectra in Fig. 2.
For an identification, spectra from single vegetative cells of different agar types and growth times are used and each single bacterium is represented by one spectrum; however, for M. luteus, three spectra recorded in a row were always used for the identification. Performing a classification with 2,257 spectra of the 20 different strains from nine species (median filtering, no normalization, linear SVM, leave-one-out test), we obtain 2,136 correctly identified spectra at the strain level (average recognition rate, 89.2%) and 2,180 correctly identified spectra at the species level (average recognition rate, 93.6%). The lowest recognition rate for strains was obtained for B. sphaericus DSM 362, with 76.7%, and that for species was for B. sphaericus DSM 27, with 82.5%. All the results at the single-cell level are summarized in Table 2. The decrease in recognition rate for single cells compared to the bulk samples was mostly because of less characteristic spectra and low signal-to-noise ratios.
With this first approach, it can be shown that micro-Raman spectroscopy in connection with SVMs is capable of rapid identification of bacteria at the single-cell level. When going from a bulk environment to a single-cell analysis, several points need to be considered. For this reason, different culture methods, which include different growth times and different agar types, are used to maximize the variation in the single strains. Furthermore, possible heterogeneity effects within single cells were evaluated, and it could be shown that single bacteria exhibit a spatial homogeneity. This is not the case for spores. If spores or bacteria with, for example, poly-β-hydroxybutyric acid inclusions are included in the data set, more than one sample spot is necessary. It is possible, for example, to identify the principal axis of spores and to measure three times along this axis. For an investigation of single colored bacteria, where the Raman laser is resonant with the electronic absorption of the pigment, possible bleaching effects must be taken into account. However, it could be shown that such bleaching effects are advantageous since, after the bleaching process, only the Raman bands corresponding to the cell matrix necessary for an unambiguous identification of single cells are left. Overall, a total of 2,257 Raman spectra of single cells were used to differentiate among 20 strains belonging to nine different species, and a recognition rate of 89.2% for strains and 93.6% for species using an SVM technique could be achieved. These results make us hopeful that by increasing the number of species, a reliable database allowing for a rapid identification of bacteria in clean rooms can be established.
Within the scope of the main research “Biophotonic” supported by the German Ministry of Education and Research, we are currently developing a technique for the rapid detection of airborne biological contaminations within clean rooms (S. Hofer et al., 23 February 2004, German Patent Office). In this technique, the airborne microparticles are deposited on special filters and, in a successive monitoring step, the particles are differentiated into biological and nonbiological particles by means of fluorescence detection. Once the biological particles have been identified on the filter, the actual identification step by means of Raman spectroscopy and SVM can take place. The investigated basic principles of this method are supported by the companies Kayser-Threde (Munich) and RapId (Berlin). With these companies, a first functional model has already been realized.
The presented method can be readily used for all fields where a limited number of bacteria need to be identified. The ultimate goal of our work, however, is a generalization of the technique to all applications, e.g., food-processing technologies and medical applications, where microorganism contaminations are troublesome. To reach this goal, the diversity of microorganism needs to be extended.
Acknowledgments
Funding of research project FKZ 13N8369, 13N8365, and 13N8379 within the framework “Biophotonik” from the Federal Ministry of Education and Research, Germany (BMBF), is gratefully acknowledged.
We thank W. Kiefer for helpful scientific discussions. We are most grateful to D. Naumann and G. Puppels for many helpful and fruitful discussions and for the thorough review of the manuscript.
REFERENCES
- 1.Alexander, T. A., P. M. Pellegrino, and J. B. Gillespie. 2003. Near-infrared surface-enhanced-Raman-scattering (SERS) mediated identification of single optically trapped bacterial spores. Proc. SPIE 5085:91-100. [DOI] [PubMed] [Google Scholar]
- 2.Al-Khaldi, S. F., and M. M. Mossoba. 2004. Gene and bacterial identification using high-throughput technologies: genomics, proteomics, and phenomics. Nutrition 20:32-38. [DOI] [PubMed] [Google Scholar]
- 3.Berger, A. J., and Q. Zhu. 2003. Identification of oral bacteria by Raman microspectroscopy. J. Mod. Optics 50:2375-2380. [Google Scholar]
- 4.Britton, K. A., R. A. Dalterio, W. H. Nelson, D. Britt, and J. F. Sperry. 1988. Ultraviolet resonance Raman spectra of Escherichia coli with 222.5-251.0 nm pulsed laser excitation. Appl. Spectrosc. 42:782-788. [Google Scholar]
- 5.Burges, C. J. C. 1998. A tutorial on support vector machines for pattern recognition. Data Mining Knowledge Disc. 2:121-167. [Google Scholar]
- 6.Carmona, P. 1980. Vibrational spectra and structure of crystalline dipicolinic acid and calcium dipicolinate trihydrate. Spectrochim. Acta Ser. A 36:705-712. [Google Scholar]
- 7.Chadha, S., R. Manoharan, P. Moenne-Loccoz, W. H. Nelson, W. L. Peticolas, and J. F. Sperry. 1993. Comparison of the UV resonance Raman spectra of bacteria, bacterial cell walls, and ribosomes excited in the deep UV. Appl. Spectrosc. 47:38-43. [Google Scholar]
- 8.Chadha, S., W. H. Nelson, and J. F. Sperry. 1993. Ultraviolet micro-Raman spectrograph for the detection of small numbers of bacterial cells. Rev. Sci. Instrum. 64:3088-3093. [Google Scholar]
- 9.Chang, C.-C., and C.-J. Lin. 2001. LIBSVM: a library for support vector machines. [Online.] http://www.csie.ntu.edu.tw/∼cjlin/libsvm.
- 10.Chan, J. W., A. P. Esposito, C. E. Talley, C. W. Hollars, S. M. Lane, and T. Huser. 2004. Reagentless identification of single bacterial spores in aqueous solution by confocal laser tweezers Raman spectroscopy. Anal. Chem. 76:599-603. [DOI] [PubMed] [Google Scholar]
- 11.Choo-Smith, L. P., K. Maquelin, H. P. Endtz, H. A. Bruining, and G. J. Puppels. 1999. A novel method for rapid identification of micro-organisms using confocal Raman microspectroscopy. Spectrosc. Biol. Mol. New Dir. 8:537-540. [Google Scholar]
- 12.Choo-Smith, L. P., K. Maquelin, T. Van Vreeswijk, H. A. Bruining, G. J. Puppels, N. A. N. Thi, C. Kirschner, D. Naumann, D. Ami, A. M. Villa, F. Orsini, S. M. Doglia, H. Lamfarraj, G. D. Sockalingum, M. Manfait, P. Allouch, and H. P. Endtz. 2001. Investigating microbial (micro)colony heterogeneity by vibrational spectroscopy. Appl. Environ. Microbiol. 67:1461-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalterio, R. A., W. H. Nelson, D. Britt, and J. F. Sperry. 1987. An ultraviolet (242 nm excitation) resonance Raman study of live bacteria and bacterial components. Appl. Spectrosc. 41:417-422. [Google Scholar]
- 14.Efrima, S., and B. V. Bronk. 1998. Silver colloids impregnating or coating bacteria. J. Phys. Chem. Ser. B 102:5947-5950. [Google Scholar]
- 15.Efrima, S., B. V. Bronk, and J. Czege. 1999. Surface-enhanced Raman spectroscopy of bacteria coated by silver. Proc. SPIE 3602:164-171. [Google Scholar]
- 16.Esposito, A. P., C. E. Talley, T. Huser, C. W. Hollars, C. M. Schaldach, and S. M. Lane. 2003. Analysis of single bacterial spores by micro-Raman spectroscopy. Appl. Spectrosc. 57:868-871. [DOI] [PubMed] [Google Scholar]
- 17.Fehrmann, A., M. Franz, A. Hoffmann, L. Rudzik, and E. Wust. 1995. Dairy product analysis: identification of microorganisms by mid-infrared spectroscopy and determination of constituents by Raman spectroscopy. J. AOAC Int. 78:1537-1542. [PubMed] [Google Scholar]
- 18.Ghiamati, E., R. Manoharan, W. H. Nelson, and J. F. Sperry. 1992. UV resonance Raman spectra of Bacillus spores. Appl. Spectrosc. 46:357-364. [Google Scholar]
- 19.Grow, A. E., L. L. Wood, J. L. Claycomb, and P. A. Thompson. 2003. New biochip technology for label-free detection of pathogens and their toxins. J. Microbiol. Methods 53:221-233. [DOI] [PubMed] [Google Scholar]
- 20.Huang, W. E., R. I. Griffiths, I. P. Thompson, M. J. Bailey, and A. S. Whiteley. 2004. Raman microscopic analysis of single microbial cells. Anal. Chem. 76:4452-4458. [DOI] [PubMed] [Google Scholar]
- 21.Huang, Y.-S., T. Karashima, M. Yamamoto, and H.-O. Hamaguchi. 2003. Molecular-level pursuit of yeast mitosis by time- and space-resolved Raman spectroscopy. J. Raman Spectrosc. 34:1-3. [Google Scholar]
- 22.Huang, Y.-S., T. Karashima, M. Yamamoto, T. Ogura, and H.-O. Hamaguchi. 2004. Raman spectroscopic signature of life in a living yeast cell. J. Raman Spectrosc. 35:525-526. [Google Scholar]
- 23.Ivnitski, D., I. Abdel-Hamid, P. Atanasov, and E. Wilkins. 1999. Biosensors for detection of pathogenic bacteria. Biosens. Bioelectron. 14:599-624. [DOI] [PubMed] [Google Scholar]
- 24.Jarvis, R. M., A. Brooker, and R. Goodacre. 2004. Surface-enhanced Raman spectroscopy for bacterial discrimination utilizing a scanning electron microscope with a Raman spectroscopy interface. Anal. Chem. 76:5198-5202. [DOI] [PubMed] [Google Scholar]
- 25.Jarvis, R. M., and R. Goodacre. 2004. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal. Chem. 76:40-47. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis, R. M., and R. Goodacre. 2004. Ultra-violet resonance Raman spectroscopy for the rapid discrimination of urinary tract infection bacteria. FEMS Microbiol. Lett. 232:127-132. [DOI] [PubMed] [Google Scholar]
- 27.Kirschner, C., K. Maquelin, P. Pina, N. A. N. Thi, L. P. Choo-Smith, G. D. Sockalingum, C. Sandt, D. Ami, F. Orsini, S. M. Doglia, P. Allouch, M. Mainfait, G. J. Puppels, and D. Naumann. 2001. Classification and identification of enterococci: a comparative phenotypic, genotypic, and vibrational spectroscopic study. J. Clin. Microbiol. 39:1763-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Diez, E. C., and R. Goodacre. 2004. Characterization of microorganisms using UV resonance Raman spectroscopy and chemometrics. Anal. Chem. 76:585-591. [DOI] [PubMed] [Google Scholar]
- 29.Manoharan, R., E. Ghiamati, R. A. Dalterio, K. A. Britton, W. H. Nelson, and J. F. Sperry. 1990. UV resonance Raman spectra of bacteria, bacterial spores, protoplasts, and calcium dipicolinate. J. Microbiol. Methods 11:1-15. [Google Scholar]
- 30.Manoharan, R., E. Ghiamati, S. Chadha, W. H. Nelson, and J. F. Sperry. 1993. Effect of cultural conditions of deep UV resonance Raman spectra of bacteria. Appl. Spectrosc. 47:2145-2150. [Google Scholar]
- 31.Maquelin, K., C. Kirschner, L. P. Choo-Smith, N. A. Ngo-Thi, T. van Vreeswijk, M. Stammler, H. P. Endtz, H. A. Bruining, D. Naumann, and G. J. Puppels. 2003. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 41:324-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maquelin, K., C. Kirschner, L. P. Choo-Smith, N. van den Braak, H. P. Endtz, D. Naumann, and G. J. Puppels. 2002. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 51:255-271. [DOI] [PubMed] [Google Scholar]
- 33.Maquelin, K., L. P. Choo-Smith, H. P. Endtz, H. A. Bruining, and G. J. Puppels. 2002. Rapid identification of Candida species by confocal Raman microspectroscopy J. Clin. Microbiol. 40:594-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maquelin, K., L. P. Choo-Smith, T. V. Vreeswijk, H. P. Endtz, B. Smith, R. Bennett, H. A. Bruining, and G. J. Pupples. 2000. Raman spectroscopic method for identification of clinically relevant microorganisms growing on solid culture medium. Anal. Chem. 72:12-19. [DOI] [PubMed] [Google Scholar]
- 35.Naumann, D. 2000. Infrared spectroscopy in microbiology, p. 102-131. In R. A. Meyers (ed.), Encyclopedia of analytical chemistry. John Wiley & Sons, Ltd., Chichester, United Kingdom.
- 36.Naumann, D., D. Helm, H. Labischinski, and P. Giesbrecht. 1991. The characterization of microorganisms by Fourier-transform infrared spectroscopy (FT-IR), p. 43-96. In. W. H. Nelson (ed.), Modern techniques for rapid microbiological analysis. VCH Publishers, New York, N.Y.
- 37.Naumann, D., S. Keller, D. Helm, C. Schultz, and B. Schrader. 1995. FT-IR spectroscopy and FT-Raman spectroscopy are powerful analytical tools for the noninvasive characterization of intact microbial cells. J. Mol. Struct. 347:399-405. [Google Scholar]
- 38.Nelson, W. H., and J. F. Sperry. 1991. UV resonance Raman spectroscopic detection and identification of bacteria and other microorganisms, p. 97-143. In W. H. Nelson (ed.), Modern techniques for rapid microbiological analysis. VCH Publishers, New York, N.Y.
- 39.Nelson, W. H., R. Manoharan, and J. F. Sperry. 1992. UV resonance Raman studies of bacteria. Appl. Spectrosc. Rev. 27:67-124. [Google Scholar]
- 40.Pasteris, J. D., J. J. Freeman, S. K. Goffredi, and K. R. Buck. 2001. Raman spectroscopic and laser scanning confocal microscopic analysis of sulfur in living sulfur-precipitating marine bacteria. Chem. Geol. 180:3-18. [Google Scholar]
- 41.Phillips, T. E., J. L. Sample, P. F. Scholl, and J. Miragliotta. 2003. The use of surface enhanced Raman scattering for the detection of dipicolinic acid on silver nanoparticles. Mater. Res. Soc. Symp. Proc. 738:227-232. [Google Scholar]
- 42.Puppels, G. J., F. F. M. de Mul, C. Otto, J. Greve, M. Robert-Nicoud, D. J. Arndt-Jovin, and T. M. Jovin. 1990. Studying single living cells and chromosomes by confocal Raman microspectroscopy. Nature 347:301-303. [DOI] [PubMed] [Google Scholar]
- 43.Puppels, G. J., H. S. P. Garritsen, G. M. J. Segers-Nolten, F. F. M. de Mul, and J. Greve. 1991. Raman microspectroscopic approach to the study of human granulocytes. Biophys. J. 60:1046-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puppels, G. J., H. S. P. Garritsen, J. A. Kummer, and J. Greve. 1993. Carotenoids located in human lymphocyte subpopulations and natural killer cells by Raman microscopy. Cytometry 14:251. [DOI] [PubMed] [Google Scholar]
- 45.Puppels, G. J., M. van Rooijen, C. Otto, and J. Greve. 1993. Confocal Raman microscopy, p. 238-258. In W. T. Mason (ed.), Fluorescent and luminescent probes. Confocal Raman microspectroscopy. Academic Press, Ltd., London, United Kingdom.
- 46.Puppels, G. J., T. C. Bakker Schut, N. M. Sijtsema, M. Grond, F. Maraboeuf, C. G. de Grauw, C. G. Figdor, and J. Greve. 1995. Development and application of Raman microspectroscopic and Raman imaging techniques for cell biological studies. J. Mol. Struct. 347:477-484. [Google Scholar]
- 47.Rowe, N. J., J. Tunstall, L. Galbraith, and S. G. Wilkinson. 2000. Lipid composition and taxonomy of [Pseudomonas] echinoides: transfer to the genus Sphingomonas. Microbiology 146:3007-3012. [DOI] [PubMed] [Google Scholar]
- 48.Schölkopf, B., and A. J. Smola. 2002. Learning with kernels, p. 215-222. MIT Press, Cambridge, Mass.
- 49.Schölkopf, B., and A. J. Smola. 2002. Learning with kernels, p. 366-380. MIT Press, Cambridge, Mass.
- 50.Schuster, K. C., E. Urlaub, and J. R. Gapes. 2000. Single-cell analysis of bacteria by Raman microscopy: spectral information on the chemical composition of cells and on the heterogeneity in a culture. J. Microbiol. Methods 42:29-38. [DOI] [PubMed] [Google Scholar]
- 51.Schuster, K. C., I. Reese, E. Urlaub, J. R. Gapes, and B. Lendl. 2000. Multidimensional information on the chemical composition of single bacterial cells by confocal Raman microspectroscopy. Anal. Chem. 72:5529-5534. [DOI] [PubMed] [Google Scholar]
- 52.Shibata, H., S. Yamashita, M. Ohe, and I. Tani. 1986. Laser Raman spectroscopy of lyophilized bacterial spores. Microbiol. Immunol. 30:307-313. [DOI] [PubMed] [Google Scholar]
- 53.Sockalingum, G. D., H. Lamfarraj, A. Beljebbar, P. Pina, M. Delavenne, F. Witthuhn, P. Allouch, and M. Manfait. 1999. Vibrational spectroscopy as a probe to rapidly detect, identify, and characterize micro-organisms. Proc. SPIE 3608:185-194. [Google Scholar]
- 54.Sockalingum, G. D., H. Lamfarraj, A. Beljebbar, P. Pina, P. Allouch, and M. Manfait. 1999. Direct on-plate analysis of microbial cells: a pilot study by surface-enhanced Raman spectroscopy. Spectrosc. Biol. Mol. New Dir. 8:599-600. [Google Scholar]
- 55.Theodoridis, S., and K. Koutroumbas. 1999. Pattern recognition, p. 339-349. Academic Press, Inc., San Diego, Calif.
- 56.Valet, O. Automatische Partikelerkennung in Reinraumtechnik. In D. Buerkle (ed.)., Reinraumtechnik, in press. Carl Hauser Verlag, Munich, Germany.
- 57.Vapnik, V. N. 1995. The nature of statistical learning theory. Springer Verlag, New York, N.Y.
- 58.Wu, Q., T. Hamilton, W. H. Nelson, S. Elliott, J. F. Sperry, and M. Wu. 2001. UV Raman spectral intensities of E. coli and other bacteria excited at 228.9, 244.0, and 248.2 nm. Anal. Chem. 73:3432-3440. [DOI] [PubMed] [Google Scholar]
- 59.Wu, Q., W. H. Nelson, S. Elliot, J. F. Sperry, M. Feld, R. Dasari, and R. Manoharan. 2000. Intensities of E. coli nucleic acid Raman spectra excited selectively from whole cells with 251-nm light. Anal. Chem. 72:2981-2986. [DOI] [PubMed] [Google Scholar]
- 60.Xie, C., and Y.-Q. Li. 2003. Confocal micro-Raman spectroscopy of single biological cells using optical trapping and shifted excitation difference techniques. J. Appl. Physiol. 93:2982-2986. [Google Scholar]
- 61.Xie, C., M. A. Dinno, and Y.-S. Li. 2002. Near-infrared Raman spectroscopy of single optically trapped biological cells. Optics Lett. 27:249-251. [DOI] [PubMed] [Google Scholar]
- 62.Xie, C., Y.-Q. Li, W. Tang, and R. J. Newton. 2003. Study of dynamical process of heat denaturation in optically trapped single microorganisms by near-infrared Raman spectroscopy. J. Appl. Physiol. 94:6138-6142. [Google Scholar]
- 63.Zeiri, L., B. V. Bronk, Y. Shabtai, J. Czege, and S. Efrima. 2002. Silver metal induced surface enhanced Raman of bacteria. Colloids Surf. Ser. 208:357-362. [Google Scholar]