Abstract
The effect of short-term ammonia starvation on Nitrosospira briensis was investigated. The ammonia-oxidizing activity was determined in a concentrated cell suspension with a NOx biosensor. The apparent half-saturation constant [Km(app)] value of the NH3 oxidation of N. briensis was 3 μM NH3 for cultures grown both in continuous and batch cultures as determined by a NOx biosensor. Cells grown on the wall of the vessel had a lower Km(app) value of 1.8 μM NH3. Nonstarving cultures of N. briensis showed potential ammonia-oxidizing activities of between 200 to 250 μM N h−1, and this activity decreased only slowly during starvation up to 10 days. Within 10 min after the addition of fresh NH4+, 100% activity was regained. Parallel with activity measurements, amoA mRNA and 16S rRNA were investigated. No changes were observed in the 16S rRNA, but a relative decrease of amoA mRNA was observed during the starvation period. During resuscitation, an increase in amoA mRNA expression was detected simultaneously. The patterns of the soluble protein fraction of a 2-week-starved culture of N. briensis showed only small differences in comparison to a nonstarved control. From these results we conclude that N. briensis cells remain in a state allowing fast recovery of ammonia-oxidizing activity after addition of NH4+ to a starved culture. Maintaining cells in this kind of active state could be the survival strategy of ammonia-oxidizing bacteria in nature under fluctuating NH4+ availability.
Chemolithoautotrophic ammonia-oxidizing bacteria (AOB) generate their energy by oxidizing ammonia (NH3) to nitrite (NO2−) and fix carbon via the Calvin cycle (3, 53). The oxidation of NH3 to NO2− is a two-step process, where NH3 is first oxidized to hydroxylamine (NH2OH) catalyzed by ammonia monooxygenase. The NH2OH is further oxidized to NO2− catalyzed by the hydroxylamine oxidoreductase, which is the energy-generating step of the ammonia oxidation (3, 53). AOB often live in close proximity to nitrite-oxidizing bacteria and together they convert the most reduced form of nitrogen (NH4+) to the most oxidized (NO3−) (40).
In nature, AOB often face longer periods of NH4+ starvation and limitation due to low nitrogen input, low mineralization rates, or competition with other AOB (8), heterotrophic bacteria (48, 49), or plants (5, 6, 50). In order to respond rapidly when NH4+ becomes available, AOB must maintain their ability to oxidize NH4+ during these periods.
With the exception of a few marine strains within the genus Nitrosococcus (of the γ-subclass of the Proteobacteria), all known AOB belong to a distinct clade within the β-subclass of the Proteobacteria (13), which comprises 11 clusters (37). By using 16S rRNA gene and more recently amoA gene sequencing, directly from environmental samples, the distribution of the members of the different clusters of AOB has been correlated to the characteristics of the environments (29, 37). The starvation behavior of several AOB belonging to different phylogenetic groups has previously been investigated. Nitrosomonas europaea affiliated with Nitrosomonas cluster 7—a group of AOB detected in environments with high NH4+ availability like wastewater (36, 40, 51)—rapidly became active again after periods of starvation in batch and retentostat experiments (8, 31, 46, 52), and the marine AOB, Nitrosomonas cryotolerans, showed a similarly rapid response to the presence of ammonia (22, 23, 24). On the other hand, members of Nitrosomonas cluster 6a (Nitrosomonas oligotropha group), often found in freshwater environments (7, 12, 43), and Nitrosospira briensis, often found in terrestrial habitats, regain their activity slower than Nitrosomonas europaea after long-term starvation of 10 weeks or 4 months (8, 32).
Up to now, members of the Nitrosospira clusters have not been investigated in detail with respect to short-term ammonia starvation. Therefore, we present a detailed investigation of the starvation response of N. briensis on the cellular and subcellular level. The activity of the N. briensis was followed online using a NOx biosensor. Additionally, we investigated the influence of starvation on both the amoA mRNA and protein expression patterns.
MATERIALS AND METHODS
Microorganisms.
The experiments were performed with N. briensis ATCC 25971 and Nitrobacter winogradskyi ATCC 25391.
Medium.
Mineral salt medium (MS medium) containing 3 mM (NH4)2SO4, 10 mM NaCl, 1 mM KCl, 0.2 mM MgSO4 · 7H2O, 1 mM CaCl2 · 2H2O, 0.4 mM KH2PO4, and 1-ml/liter trace element solution (49) in distilled water was used for all experiments. For the batch incubations, 30 mM HEPES [4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid] was added to keep the pH constant. The pH was adjusted to 7.8 with NaOH before autoclaving. The phosphate solution was autoclaved separately and added at room temperature.
Continuous culture cultivation.
The continuous culture experiments were carried out in a chemostat composed of a 3-liter glass vessel, a stirrer, a pH control unit, an aeration unit, and a peristaltic pump. The cell suspension (approximately 2 liters) was kept at a temperature of 25°C. The stirrer speed was adjusted to 300 rpm and the pH was adjusted continuously to 7.5 ± 0.2 by adding 5% Na2CO3. The culture was aerated with 1 liter of air min−1. The chemostats were inoculated with actively growing batch cultures of a coculture of N. briensis and N. winogradskyi. For 5 days the chemostats ran as batch cultures. When the cells had consumed 80 to 90% of the NH4+, fresh MS medium containing 5 mM NH4+ was pumped into the chemostats and the growth rate was adjusted to 0.014 h−1. The chemostats were sampled regularly to determine the NH4+, NO2−, and NO3− concentrations.
Starvation and resuscitation experiments.
The experiments were carried out in a 3-liter glass batch reactor, equipped with a stirrer and an aeration unit. Approximately 2 liters of culture were kept at 25°C with a stirrer speed of 200 rpm and were aerated with 200 ml of air min−1. The reactors were inoculated with actively growing batch cultures of a coculture of N. briensis and N. winogradskyi and sampled daily to determine the NH4+, NO2−, and NO3− concentrations. Onset of starvation was defined as the time where NH4+ was consumed completely. To resuscitate the cultures NH4+ was added to a final concentration of 5 mM. During the experiments 100-ml samples were taken, centrifuged (20 min, 22,000 × g, 4°C), washed once in 2 ml of MS medium without NH4+, and resuspended in 2 ml of MS medium without NH4+. A 1.5-ml sample was used for activity measurement and two times 0.2 ml was frozen in liquid nitrogen and stored at −80°C for molecular analysis.
Acetylene treatment.
A starved coculture of N. briensis and N. winogradskyi was incubated overnight in the presence of acetylene to inhibit ammonia monooxygenase (18). After aerating the culture for 1 h with air to remove the acetylene, fresh NH4+ was added. As a control, a cell suspension without acetylene treatment was used. Samples were taken and treated the same way as in the other starvation-resuscitation experiments.
Determination of the potential ammonia-oxidizing activity and the Km(app) value.
For the determination of the apparent half-saturation constant [Km(app)] value, a 1-liter sample was taken from the chemostat or batch cultures, centrifuged (20 min, 22,000 × g, 4°C), washed with NH4+-free MS medium, concentrated, and used within 2 h for the measurements. The Km(app) of the attached cells was determined from the biomass that was scrubbed off the wall and homogenized at the end of one run. A concentrated culture sample (1.5 ml) was added to 13.5 ml of MS medium without NH4+ (pH 7.5) to determine the Km(app) and the potential ammonia-oxidizing activity. The mixture was aerated for 10 min. Then, 0.15 ml of concentrated NH4+ solution was added. The NO2−/NO3− production was followed by using the NOx biosensor and recorded with a strip chart recorder.
The Km(app) value of the NH3 oxidation of N. briensis was determined by adding different concentrations of NH4+, corresponding to NH3 concentrations between 0.5 and 10 μM, to the cell suspension. The Km(app) was calculated by nonlinear regression based on the Michaelis Menten kinetics (10).
The measurements of the potential ammonia-oxidizing activity during the starvation and resuscitation experiments were done by adding NH4+ to obtain a concentration of 10 μM NH3. The high NH4+ concentration was chosen to ensure saturation of the ammonia oxidation.
NO2−/NO3− production measurement.
The NO2−/NO3− production was determined with a NOx biosensor (Unisense, Aarhus, Denmark). This sensor contains a denitrifying bacterial culture that reduces NO3− and NO2− to N2O, which is then detected by an electrochemical N2O sensor (33). Calibrations of the sensors were done at the beginning and at the end of the experiments. All experiments were done at 25°C in glass vials, which were kept dark.
O2 consumption measurement.
The O2 consumption was measured with a Clark-type O2 sensor in a self-constructed setup (26). The samples were prepared and treated the same way as for the measurements of the NO2−/NO3− production.
Chemical analysis.
Samples for measuring NH4+ were analyzed immediately or stored at −20°C. The NH4+ concentration was determined colorimetrically (25).
RNA analysis.
RNA was extracted with a RNeasy Protect Bacteria kit (QIAGEN) using mechanical disruption of the cells by beat beating. The obtained RNA was treated with DNase (Ambion, Austin, Tex.). The absence of DNA contamination was tested by PCR directly using 1 μl of RNA extract as template. A two-step reverse transcription (RT)-PCR was performed: first the RNA was reverse transcribed to produce cDNA, which was then amplified by PCR in a second step. Two different primer pairs were used: an AOB-specific 16S rRNA primer pair (28) and an amoA primer pair, targeting the gene coding for the subunit A of the ammonia monooxygenase. N. briensis-specific amoA primers were designed based on the amoA sequence of N. briensis obtained from a public database (amoA-109F, 5′-GTT GGA ACC TAC CAC ATG CA-3′, and amoA-608R, 5′-TCT GAG TGA GCC TTG TTC GA-3′). No quantification of the RNA prior to amplification was done. The RT reactions were done in 5-μl reactions with a RT kit from Ambion or Amersham Bioscience according to the instructions of the manufacturer, by using the amoA-608R or the 16S rRNA reverse primers.
PCR using the 16S rRNA primers was done in 10-μl reactions with 25 cycles as described by Kowalchuk et al. (28). The PCR with the amoA-specific primers was done in 10-μl reactions containing 1.25 nmol of each dNTP, 1.5 mM Mg2+, 3 μg of bovine serum albumin, and 5 pmol of each primer. Thermocycling was done with an initial step at 92°C for 1 min, 40 cycles of 92°C for 30 s of denaturation, 57°C for 30 s of annealing, and 72°C for 45 s plus 1 s/cycle extension; the last cycle had a 5-min final extension step.
All RT-PCR products were separated on 2% agarose gels containing 0.5 μg of ethidium bromide ml−1 and visualized by UV translumination (Gel Doc 2000; Bio-Rad, Hercules, Calif.). Digital images were acquired with a charge-coupled-device camera controlled by the software Quantity One (Bio-Rad). Quantification of band intensities was done manually by eye, and in order to evaluate the relative differences in band intensities correctly, RT-PCRs were done on undiluted, 10× and 100× diluted RNA extracts.
2D gel electrophoresis.
For the analysis of the soluble protein fraction consisting of the cytoplasmatic and the periplasmatic proteins, two reactors with N. briensis were grown up (without N. winogradskyi). One reactor was harvested at the end of the logarithmic phase, the other after 2 weeks of starvation. Cells were harvested by centrifugation (22,000 × g, 20 min, 4°C), washed two times with sterile filtered tap water, resuspended in 1 ml of tap water, frozen in liquid nitrogen, and stored at −80°C until analysis. Sample preparation and electrophoresis were done as described by Schmidt et al. (39).
Proteins in the gel were then fixed and stained in the gels by silver staining. The digitalization of the two-dimensional (2D) protein patterns was done using a Sharp JX scanner interfaced with the Image Master 2D Elite software (Amersham Bioscience).
RESULTS
Km(app) and Vmax(app) of NH3 oxidation.
To validate the use of the NO2−/NO3− biosensor approach to estimate Km(app) values, we compared obtained values with values based on the standard method of O2 consumption measurements. The Km(app) and the Vmax(app) were determined based on NO2−/NO3− production and O2 consumption, respectively (Table 1). The Km(app) values of the ammonia oxidation for pelagic cells grown in continuous and batch cultures were higher than the Km(app) for cells growing in biofilms on the vessel wall (Table 2).
TABLE 1.
Km(app) and Vmax(app) of the NH3 oxidation of N. briensis grown in batch culturea
| Determination | Km(app) (μM NH3) | Vmax(app) (μM N h−1) |
|---|---|---|
| NO2−/NO3− biosensor | 2.9 (±0.4) | 248 (±12) |
| O2 sensor | 2.4 (±0.8) | 303 (±42) |
Determined by NO2− production using a NO2−/NO3− biosensor and O2 consumption using an O2 sensor. Values in parentheses are standard errors of the nonlinear regression of the NO2−/NO3− production or O2 consumption versus the NH3 concentrations (n = 7, r2 > 0.93).
TABLE 2.
Km(app) for the NH3 oxidation of N. briensis cultured under different conditionsa
| Culture conditions | Km(app) (μM NH3) |
|---|---|
| Batch culture (N. briensis) | 2.9 |
| Continuous culture (N. briensis and N. winogradskyi) | 3.2 ± 0.4b |
| Wall growth in one of the continuous cultures | |
| (N. briensis and N. winogradskyi) | 1.8 |
Determined with the NOx biosensor.
Value represents mean ± standard deviation of three independent measurements of the Km(app).
Starvation experiment.
The start of the starvation period was defined as the time when NH4+ was completely consumed in the coculture of N. briensis and N. winogradskyi and was found to be 7 days after inoculation (Fig. 1). The potential ammonia-oxidizing activity was around 200 μM N h−1 at the end of the growth phase and at the beginning of the starvation period but decreased to 60 μM N h−1 during a 2-week period of starvation.
FIG. 1.
NH4+ dynamics (•) and potential ammonia-oxidizing activity (○) of a coculture of N. briensis and N. winogradskyi during growth and starvation (reactor 1).
Parallel to the potential ammonia-oxidizing activity measurements, samples for RNA extraction were taken and analyzed for the presence of amoA mRNA and 16S rRNA (Fig. 2). The amplification products obtained with the 16S rRNA-specific primers were used as an internal standard, as AOB are known to keep their ribosomes intact during starvation (23, 51). The band intensities of the 16S rRNA amplicons were stable, but the intensities of the amoA mRNA products were decreasing over time, indicating a relative decrease of the amoA mRNA. During the growth phase of N. briensis, the amoA mRNA level appeared constant, whereas it constantly decreased over a period of 12 days of starvation.
FIG. 2.
amoA mRNA and 16S rRNA RT-PCR products of N. briensis from the reactor 1 during starvation for NH4+.
Resuscitation experiment.
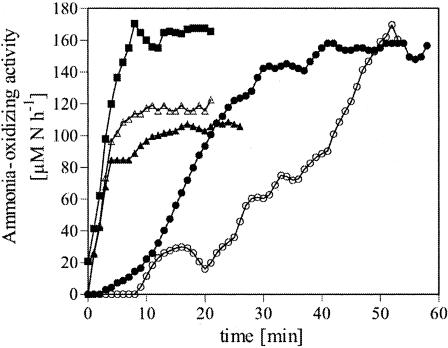
Starvation of N. briensis for 1 week resulted in a decrease of the potential ammonia-oxidizing activity from 200 to 150 μM N h−1 (Fig. 3). After the addition of fresh substrate, NH4+ consumption started immediately, whereas the activity remained constant on the first day and increased later. The potential ammonia-oxidizing activity data shown in Fig. 1 and 3 represent the maximum activities of the cultures after addition of fresh NH4+ to the samples during the potential ammonia-oxidizing activity measurement. In addition to these maximal values, we determined the ammonia-oxidizing activity minute by minute by calculating the NO2−/NO3− production rate at intervals of 1 min by taking into account the slope of the NO2−/NO3− concentration for the 2 min before and after each time point. Using this approach it was possible to track the development of the ammonia-oxidizing activity over time after the addition of fresh NH4+ to the concentrated cell suspensions (Fig. 4). Growing cells reached their maximum ammonia-oxidizing activity 5 min after the addition of NH4+. When the cells were starved for 3 and 7 days, they needed 30 and 50 min, respectively, to regain their maximum activity. However, if the culture was resuscitated just before, it took only 5 to 10 min until the maximum ammonia-oxidizing activity was regained (Fig. 4). The amoA mRNA concentration decreased during the starvation period (Fig. 5). After the addition of NH4+ to the culture, an increase in amoA mRNA was detectable after 10 min, but the relative amount was very low and the level of amoA mRNA increased slowly, regaining the level for unstarved cells by the final sample (245 min).
FIG. 3.
NH4+ dynamics (•) and potential ammonia-oxidizing activity (○) of a coculture of N. briensis and N. winogradskyi during growth, starvation, and resuscitation (reactor 2). A, overall picture; B, detailed graph for the first hours of resuscitation.
FIG. 4.
Ammonia-oxidizing activity over time in the concentrated samples used to measure the potential ammonia-oxidizing activity at the different time points during growth, starvation, and resuscitation (reactor 2). The ammonia-oxidizing activity was calculated for every minute as the slope of the NO2−/NO3− production within the 2 min before and after each time point. ▪, during growth; •, 3 days starved; ○, 7 days starved; ▴, after 10 min fresh NH4+; ▵, after 4 h fresh NH4+.
FIG. 5.
amoA mRNA and 16S rRNA RT-PCR products of N. briensis from the reactor 2 during starvation and resuscitation.
Protein pattern.
We compared the soluble protein fraction of a growing and a 2-week-starved N. briensis culture by using 2D gel electrophoresis (Fig. 6). The protein pattern of the starved and growing N. briensis cultures showed only small changes, with five spots disappearing and two spots appearing after starvation.
FIG. 6.
Comparison of the protein pattern of the soluble protein fraction of a growing and a 2-week-starved culture of N. briensis. Circles with solid lines indicate protein spots disappearing and circles with dashed lines indicate spots where protein spots are appearing during starvation. MW, molecular weight.
Treatment with acetylene.
A starved coculture of N. briensis and N. winogradskyi was incubated with acetylene to inhibit the ammonia monooxygenase and subsequently resuscitated by the addition of fresh NH4+ parallel with an untreated control culture (Fig. 7). NH4+ was consumed in the noninhibited culture immediately after the addition of fresh NH4+ and the potential ammonia-oxidizing activity remained at the same high level. In the acetylene-treated culture, the potential ammonia-oxidizing activity decreased to almost zero. After the addition of NH4+ the potential ammonia-oxidizing activity increased slowly but constantly.
FIG. 7.
NH4+ dynamics and potential ammonia-oxidizing activity of a coculture of N. briensis and N. winogradskyi during resuscitation after starvation treated with and without acetylene before addition of fresh NH4+ (reactor 3). □, NH4+ (without acetylene treatment); ▪, NH4+ (after acetylene treatment); ▵, potential ammonia-oxidizing activity (without acetylene treatment); ▴, potential ammonia-oxidizing activity (after acetylene treatment).
DISCUSSION
In summary, we demonstrated that N. briensis is able to recover rapidly after periods of starvation up to 2 weeks. The culture responded within minutes to the addition of fresh NH4+ and the maximum potential ammonia-oxidizing activity was reached after 30 to 60 min (Fig. 4). A fast recovery after starvation within the first weeks has been observed for other AOB, e.g., Nitrosomonas europaea, Nitrosomonas cryotolerans, and the culture G5-7, closely related to Nitrosomonas oligotropha (8, 17, 23, 31, 46). Recovery after longer starvation periods revealed differences between the AOB strains; Nitrosomonas europaea and Nitrosomonas cryotolerans were recovering very fast after >10 weeks of starvation, whereas the culture G5-7 and N. briensis showed a longer lag time before they regained their activity (8, 23, 32). Despite this recovery after longer starvation periods of N. briensis that was slower than with other ammonia-oxidizing bacteria, N. briensis showed recovery patterns similar to other AOB with respect to short-term starvation. The delay in reaching the maximum potential ammonia-oxidizing activity after starvation (Fig. 4) could be explained by the need for key molecules necessary for the metabolism. The addition of the intermediate NH2OH to a starved culture of Nitrosomonas europaea reduced the time delay before reaching the maximum activity (17). As the oxidation of NH2OH delivers electrons back to the ammonia monooxygenase in addition to electrons for ATP generation (53), the observed delay in the ammonia oxidation could be due to a lack of reducing equivalents for the ammonia monooxygenase. A similar observation was made with methanotrophic bacteria, where the addition of the intermediate methanol was found to improve CH4 uptake (20) due to provision of energy derived from the oxidation of methanol.
In contrast to many other bacteria, the rRNA content of AOB is kept at a high level during starvation (23, 51). Furthermore, our results indicate that not only ribosomes are retained but that there is a general retention of functionality by the organisms even during starvation periods. We found that amoA mRNA was still present after a starvation period of 12 days, although the concentration was much lower than in a growing cultures (Fig. 2 and 5). In contrast, Sayavedra-Soto et al. (38) did not detect amoA mRNA in Nitrosomonas europaea when using Northern blot hybridization after 8 to 12 h of starvation. This might, however, be explained by differences in the sensitivity of the detection of mRNA, as Northern blot hybridization has no PCR amplification step prior to detection. Although also possible, differences in the stability of the mRNA molecules for Nitrosomonas europaea and N. briensis are unlikely as both AOB show the same pattern in the response to short-term starvation.
The detection of amoA mRNA has been used to measure the activity of AOB (11), but the possibility that amoA mRNA would still be detectable after 12 days of starvation makes this approach questionable and care should be taken in order to make a direct correlation between mRNA detection and in situ activity of the cells.
The half-life of mRNA in most bacteria has an average of 3 min (0.5 to 50 min) (45). In the heterotrophic bacteria Vibrio angustum S14 (1, 2) and Rhizobium leguminosarum (47), starvation leads to an increase in mRNA half-life. Thus, it could be speculated that starvation leads to a stabilization of the amoA mRNA in AOB as well.
In the presence of acetylene, the activity is almost zero but increases slowly after the addition of NH4+, as the inhibition is irreversible and new ammonia monooxygenase has to be synthesized to regain activity (Fig. 7). In the starved and resuscitated cultures without acetylene treatment, the ammonia-oxidizing activity reached a maximum almost immediately (Fig. 3 and 7). The pattern of the soluble protein fraction of actively growing cells and 2-week-starved cells showed only small differences (Fig. 6), indicating that the overall change to function of the cells is not great and that they maintain much of their metabolic machinery. The observed lack in major changes in the protein pattern of N. briensis is in contrast to the stress and/or starvation response of other bacteria investigated. In Bacillus subtilis, the induction of many different proteins occurs during starvation, including proteins involved in sporulation (14). Several other comparative studies with Vibrio sp. strain S14, Salmonella enterica serovar Typhimurium, and Escherichia coli also demonstrated the induction of proteins necessary for survival during starvation periods (34, 35, 41, 42). In E. coli and other gram-negative bacteria, the stationary-phase response is regulated by the expression of the rpoS gene coding for the σs factor (15, 16). Nitrosomonas europaea lacks rpoS-like genes completely (9), and though the absence of rpoS in N. briensis was not confirmed, the observed induction of new proteins during starvation being less than that for other bacteria indicates that N. briensis might have a different response pattern to starvation stress.
Km(app) values of the NH3 oxidation.
The ammonia-oxidizing activity and the Km(app) values of the NH3 oxidation have often been determined by measuring the O2 uptake in the presence of NH4+ (19, 30, 44). This method has some disadvantages, particularly as O2 is also the substrate of all other oxic processes and the Km(app) can only be measured using this method in pure cultures. In mixed cultures the O2 consumption by non-AOB has to be otherwise inhibited or excluded. A method to measure the ammonia-oxidizing activity and the kinetic parameters of the ammonia oxidation with a NOx biosensor was therefore developed. The comparison of the Km(app) and the Vmax determined with both methods showed differences of approximately 20% (Table 1). The reproducibility of the newly developed method was determined by the threefold determination of the Km(app) value of the NH3 oxidation (Table 2). The standard deviation of the threefold determination indicates that a mistake of 10 to 15% can be expected, and we thus find the difference between the two methods within an expectable range.
The Km(app) value for NH3 oxidation in N. briensis was 3 μM NH3 for cultures grown in continuous and batch cultures, respectively. So the growth mode of N. briensis had no influence on the affinity for NH3. However, the Km(app) values were quite low compared to values of other Nitrosospira species (21). We measured the NO2−/NO3− production rate within 15 min after the addition of NH4+ to the concentrated culture. Jiang and Bakken (21) calculated the starting activity from a long-term experiment and used different initial pH values to determine the different initial NH3 concentrations. Hence, the conditions for the determination of the Km(app) values were very different and could have caused the differences in the results. The Km(app) value was also lower than values determined for members of the Nitrosomonas cluster 7 (relatives of Nitrosomonas europaea), Nitrosomonas cluster 6b (relatives of Nitrosomonas marina), and Nitrosomonas cluster 8 (relatives of Nitrosomonas communis), but they were in the same range as the Km(app) values of members of Nitrosomonas cluster 6a (relatives of Nitrosomonas oligotropha) (27).
The cells from wall growth were found to have a Km(app) value of only 1.8 μM NH3 (Table 2), showing that AOB growing in biofilms have a higher affinity for NH3 than the bacteria in the culture liquid. Biofilm cells of Nitrosomonas europaea have been reported to recover faster from starvation than cells in liquid culture (4, 46), and a lower Km(app) value and better recovery after starvation indicate better competitive abilities of biofilm cells compared to cells in liquid culture.
Conclusion.
The comparison of our observations with published data from studies of several heterotrophic bacteria indicates that AOB may have a unique mechanism to cope with nutrient starvation. They are able to keep their cells in a state where it is possible to start oxidizing NH4+ almost immediately and with the maximum rate after shorter starvation periods, and they are thereby able to respond rapidly to changing environmental conditions. This ability could represent a survival strategy for the chemolithoautotrophic AOB to enable them to be better competitors in the environment. The questions remain whether this is a more widely distributed mechanism among other groups of slow-growing bacteria and how the mechanism behind the ability to maintain this rapidly responsive state might be regulated.
Acknowledgments
We thank Paul Bodelier, Hinnerk Boriss, Riks Laanbroek, and Niels Peter Revsbech for helpful discussions, critical reading of the manuscript, and technical as well as financial support.
The work was supported by a grant from the 5th Framework program of the European Commission (ICON EKV1-CT-2000-00054).
REFERENCES
- 1.Albertson, N. H., T. Nyström, and S. Kjelleberg. 1990. Functional messenger-RNA half-lives in the marine Vibrio sp. S14 during starvation and recovery. J. Gen. Microbiol. 136:2195-2199. [Google Scholar]
- 2.Albertson, N. H., and T. Nyström. 1994. Effects of starvation for exogenous carbon on functional messenger-RNA stability and rate of peptide-chain elongation in Escherichia coli. FEMS Microbiol. Lett. 117:181-188. [DOI] [PubMed] [Google Scholar]
- 3.Arp, D. J., L. A. Sayavedra-Soto, and N. G. Hommes. 2002. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol. 178:250-255. [DOI] [PubMed] [Google Scholar]
- 4.Batchelor, S. E., M. Cooper, S. R. Chhabra, L. A. Glover, G. S. A. B. Stewart, P. Williams, and J. I. Prosser. 1997. Cell density-regulated recovery of starved biofilm populations of ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 63:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodelier, P. L. E., J. A. Libochant, C. W. P. M. Blom, and H. J. Laanbroek. 1996. Dynamics of nitrification and denitrification in root-oxygenated sediments and adaptation of ammonia-oxidizing bacteria to low-oxygen or anoxic habitats. Appl. Environ. Microbiol. 62:4100-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodelier, P. L. E., H. Duyts, C. W. P. M. Blom, and H. J. Laanbroek. 1998. Interactions between nitrifying and denitrifying bacteria in gnotobiotic microcosms planted with the emergent macrophyte glyceria maxima. FEMS Microbiol. Ecol. 25:63-78. [Google Scholar]
- 7.Bollmann, A., and H. J. Laanbroek. 2001. Continuous culture enrichments of ammonia-oxidizing bacteria at low ammonium concentrations. FEMS Microbiol. Ecol. 37:211-221. [Google Scholar]
- 8.Bollmann, A., M.-J. Bär-Gilissen, and H. J. Laanbroek. 2002. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornish-Bowden, A. 1979. Fundamentals of enzyme kinetics. Butterworths, London, United Kingdom.
- 11.Ebie, Y., H. Miura, N. Noda, M. Matsumura, S. Tsuneda, A. Hirata, and Y. Inamori. 2002. Detection and quantification of expression of amoA by competitive reverse transcription-PCR. Wat. Sci. Technol. 46:281-288. [PubMed] [Google Scholar]
- 12.Hastings, R. C., J. R. Saunders, G. H. Hall, R. W. Pickup, and A. J. McCarthy. 1998. Application of molecular biological techniques to a seasonal study of ammonia oxidation in a eutrophic freshwater lake. Appl. Environ. Microbiol. 64:3674-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Head, I. M., W. D. Hiorns, T. M. Embley, A. J. McCarthy, and J. R. Saunders. 1993. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16s ribosomal RNA gene sequences. J. Gen. Microbiol. 139:1147-1153. [DOI] [PubMed] [Google Scholar]
- 14.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 15.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 16.Hengge-Aronis, R. 2002. Stationary phase gene regulation: what makes an Escherichia coli promoter sigma(s)-selective? Curr. Opin. Microbiol. 5:591-595. [DOI] [PubMed] [Google Scholar]
- 17.Hooper, A. B. 1969. Lag phase of ammonia oxidation by resting cells of Nitrosomonas europaea. J. Bacteriol. 97:968-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hyman, M. R., and P. M. Wood. 1985. Suicidal inactivation and labelling of ammonia monooxygenase by acetylene. Biochem. J. 227:719-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyman, M. R., and D. J. Arp. 1992. 14C2H2- and 14CO2-labeling studies of the de novo synthesis of polypeptides by Nitrosomonas europaea during recovery from acetylene and light inactivation of ammonia monooxygenase. J. Biol. Chem. 267:1534-1545. [PubMed] [Google Scholar]
- 20.Jensen, S., A. Prieme, and L. R. Bakken. 1998. Methanol improves methane uptake in starved methanotrophic microorganisms. Appl. Environ. Microbiol. 64:1143-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, Q. Q., and L. R. Bakken. 1999. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30:171-186. [DOI] [PubMed] [Google Scholar]
- 22.Johnstone, B. H., and R. D. Jones. 1988. Recovery of a marine chemolithotrophic ammonium-oxidizing bacterium from long-term energy-source deprivation. Can. J. Microbiol. 34:1347-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnstone, B. H., and R. D. Jones. 1988. Physiological-effects of long-term energy-source deprivation on the survival of a marine chemolithotrophic ammonium-oxidizing bacterium. Mar. Ecol. Prog. Ser. 49:295-303. [Google Scholar]
- 24.Jones, R. D., and R. Y. Morita. 1985. Survival of a marine ammonium oxidizer under energy-source deprivation. Mar. Ecol. Prog. Ser. 26:175-179. [Google Scholar]
- 25.Kandeler, E., and H. Gerber. 1988. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils 6:68-72. [Google Scholar]
- 26.Kjeldsen, K. U., C. Joulian, and K. Ingvorsen. Effects of oxygen exposure on respiratory activities of Desulfovibrio desulfuricans strain DvO1 isolated from activated sludge. FEMS Microbiol. Ecol., in press. [DOI] [PubMed]
- 27.Koops, H. P., and A. Pommerening-Röser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 28.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 30.Laanbroek, H. J., P. L. E. Bodelier, and S. Gerards. 1994. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch. Microbiol. 161:156-162. [Google Scholar]
- 31.Laanbroek, H. J., M.-J. Bär-Gilissen, and H. L. Hoogveld. 2002. Nitrite as a stimulus for ammonia-starved Nitrosomonas europaea. Appl. Environ. Microbiol. 68:1454-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laanbroek, H. J., and M.-J. Bär-Gilissen. 2002. Weakened activity of starved ammonia-oxidizing bacteria by presence of pre-activated Nitrobacter winogradskyi. Microbes Environ. 17:122-127. [Google Scholar]
- 33.Larsen, L. H., T. Kjaer, and N. P. Revsbech. 1997. A microscale NO3− biosensor for environmental applications. Anal. Chem. 69:3527-3531. [DOI] [PubMed] [Google Scholar]
- 34.Nyström, T., R. M. Olsson, and S. Kjelleberg. 1992. Survival, stress resistance, and alterations in protein expression in the marine Vibrio sp. strain S14 during starvation for different individual nutrients. Appl. Environ. Microbiol. 58:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nyström, T. 1993. Global systems approach to the physiology of the starved cells, p. 129-150. In S. Kjelleberg (ed.), Starvation in bacteria. Plenum Press, New York, N.Y.
- 36.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Röser, and H. P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 38.Sayavedra-Soto, L. A., N. G. Hommes, S. A. Russell, and D. J. Arp. 1996. Induction of ammonia monooxygenase and hydroxylamine oxidoreductase mRNAs by ammonium in Nitrosomonas europaea. Mol. Microbiol. 20:541-548. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt, I., P. J. M. Steenbakkers, H. J. M. op den Camp, K. Schmidt, and M. S. M. Jetten. 2004. Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. J. Bacteriol. 186:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schramm, A., L. H. Larsen, N. P. Revsbech, N. B. Ramsing, R. Amann, and K. H. Schleifer. 1996. Structure and function of a nitrifying biofilm as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 62:4641-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spector, M. P., Z. Aliabadi, T. Gonzalez, and J. W. Foster. 1986. Global control in Salmonella typhimurium: two-dimensional gel electrophoretic analysis of starvation-, anaerobiosis-, and heat shock-inducible proteins. J. Bacteriol. 168:420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spector, M. P. 1990. Gene-expression in response to multiple nutrient-starvation conditions in Salmonella typhimurium. FEMS Microbiol. Ecol. 74:175-183. [Google Scholar]
- 43.Speksnijder, A. G. C. L., G. A. Kowalchuk, K. Roest, and H. J. Laanbroek. 1998. Recovery of a Nitrosomonas-like 16s rDNA sequence group from freshwater habitats. Syst. Appl. Microbiol. 21:321-330. [DOI] [PubMed] [Google Scholar]
- 44.Stehr, G., B. Böttcher, P. Dittberner, G. Rath, and H. P. Koops. 1995. The ammonia-oxidizing nitrifying population of the river Elbe estuary. FEMS Microbiol. Ecol. 17:177-186. [Google Scholar]
- 45.Takayama, K., and S. A. Kjelleberg. 2000. The role of RNA stability during bacterial stress responses and starvation. Environ. Microbiol. 2:355-365. [DOI] [PubMed] [Google Scholar]
- 46.Tappe, W., A. Laverman, M. Bohland, M. Braster, S. Rittershaus, J. Groeneweg, and H. W. Van Verseveld. 1999. Maintenance energy demand and starvation recovery dynamics of Nitrosomonas europaea and Nitrobacter winogradskyi cultivated in a retentostat with complete biomass retention. Appl. Environ. Microbiol. 65:2471-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorne, S. H., and H. D. Williams. 1997. Adaptation to nutrient starvation in Rhizobium leguminosarum bv. phaseoli: analysis of survival, stress resistance, and changes in macromolecular synthesis during entry to and exit from stationary phase. J. Bacteriol. 179:6894-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Niel, E. W. J., P. A. M. Arts, B. J. Wesselink, L. A. Robertson, and J. G. Kuenen. 1993. Competition between heterotrophic and autotrophic nitrifiers for ammonia in chemostat cultures. FEMS Microbiol. Ecol. 102:109-118. [Google Scholar]
- 49.Verhagen, F. J. M., and H. J. Laanbroek. 1991. Competition for ammonium between nitrifying and heterotrophic bacteria in dual energy-limited chemostats. Appl. Environ. Microbiol. 57:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhagen, F. J. M., H. J. Laanbroek, and J. W. Woldendorp. 1995. Competition for ammonium between plant roots and nitrifying and heterotrophic bacteria and the effects of protozoan grazing. Plant Soil 170:241-250. [Google Scholar]
- 51.Wagner, M., G. Rath, H. P. Koops, J. Flood, and R. Amann. 1995. In situ analysis of nitrifying bacteria in sewage treatment plants. Wat. Sci. Technol. 34:237-244. [Google Scholar]
- 52.Wilhelm, R., A. Abeliovich, and A. Nejidat. 1998. Effect of long-term ammonia starvation on the oxidation of ammonia and hydroxylamine by Nitrosomonas europaea. J. Biochem. (Tokyo) 124:811-815. [DOI] [PubMed] [Google Scholar]
- 53.Wood, P. M. 1986. Nitrification as a bacterial energy source, p. 39-62. In J. I. Prosser (ed.), Nitrification, vol. 20. IRL Press, Oxford, England. [Google Scholar]