Abstract
A xylose reductase (XR) gene was identified from the Neurospora crassa whole-genome sequence, expressed heterologously in Escherichia coli, and purified as a His6-tagged fusion in high yield. This enzyme is one of the most active XRs thus far characterized and may be used for the in vitro production of xylitol.
Xylose reductase (XR) is commonly found in yeasts and filamentous fungi, often with several isozymes in one species (21, 24, 33). XR catalyzes the first step of d-xylose metabolism, reducing xylose to xylitol with concomitant NAD(P)H oxidation. In general, XR is specific for NADPH, but in some cases, it utilizes both NADPH and NADH (7, 21, 23, 30) and in at least one case prefers NADH over NADPH (20). The difference in cofactor specificities is proposed to maintain the redox balance between nicotinamide cofactors under a variety of growth conditions (8, 24). Based on sequence and structure similarities, XR belongs to the aldose reductase family (EC 1.1.1.21). The majority of its family members are monomeric, but some have quaternary structural organizations (6, 17), including XR, which has a unique dimeric interface (15).
XRs have gained interest because of their importance both in the fermentation of plant biomass to ethanol and in the production of xylitol, a low-calorie anticariogenic natural sweetener. An economical means of producing xylitol from xylose in vitro consists of coupling XR with a cofactor regeneration system (25, 26). Similar processes have also been proposed for converting glucose into sorbitol (10). Highly active and easily obtainable XRs are desirable for use in such processes.
The genetic sequences of many XRs have been determined and several have been cloned and expressed in a variety of hosts (1, 7, 9, 13, 18, 20, 21, 23, 24, 33), but many XR genes are still uncharacterized or unidentified. Such is the case with Neurospora crassa, which is known to convert plant biomass to ethanol (3, 22, 27), suggesting the presence of d-xylose-metabolizing enzymes (28) though no XR gene has been identified. Only one XR protein has been isolated, but no genetic or protein sequence was determined (29), thus limiting the study and eliminating the possibilities of recombinant expression and large-scale purification.
N. crassa XR gene identification.
Utilizing the recently completed 40-Mb whole-genome sequence of N. crassa (5), we discovered one XR-encoding gene from the 10,082 predicted genes. XRs from Candida tenuis (GenBank accession number AAC25601.1) and Candida tropicalis I-II (BAA19476.1) were used as templates for a protein BLAST search (www.ncbi.nlm.nih.gov). Of four protein sequences with >35% sequence identity, hypothetical protein NCU 08384.1 (EAA34695.1) had the greatest sequence identity (>52%). This protein (referred to as N. crassa XR hereafter) had significant homology (51 to 66% identity) with other XRs (supplemental data). Among the conserved residues was the catalytic triad of lysine, tyrosine, and aspartate, along with a conserved histidine that positions xylose (11, 12, 15). Based on this information, we postulated that this hypothetical protein was an XR.
N. crassa RNA purification, reverse transcription-PCR, cloning, and N. crassa XR purification.
For experimental details, see the supplemental material. Reverse transcription-PCR performed on total RNA isolated from xylose-induced N. crassa 10333 showed the expected product and established that this gene is transcribed into mRNA. The reverse transcription-PCR product was subcloned into pET15b and pET26b(+) vectors (Novagen) using NdeI and BamHI restriction sites and were used to transform Escherichia coli BL21(DE3). The first construct (pET15b) encoded N. crassa XR as an N-terminal His6-tagged fusion with a thrombin cleavage site, while the second vector [pET26b(+)] encoded only N. crassa XR.
These two constructs were used to compare the activities of the recombinant enzyme with and without a fusion tag. Cell lysates of IPTG (isopropyl-α-d-thiogalactopyranoside)-induced cultures of these cells were prepared, analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and assayed for XR activities. Both tagged and nontagged constructs produced soluble XR at >25% of the total cellular protein, with slightly better overall expression of the His6-tagged XR. The nontagged XR had about 25%-greater specific activity than the tagged XR, as determined by the ratio of total lysate activity to expressed soluble protein. However, most of this lost activity was regained by cleaving the His6 tag from the protein by use of thrombin. The His6-tagged XR was chosen for purification and characterization due to its higher expression level, as well as because of the ease of purifying His6-tagged proteins by immobilized metal affinity chromatography (IMAC).
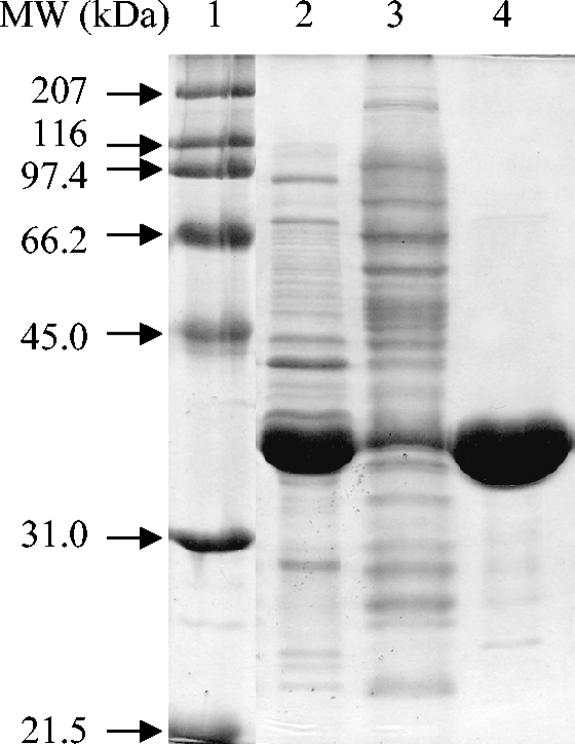
The His6-tagged N. crassa XR was expressed and purified in a single step with a previously described IMAC purification protocol (32) by using a BioLogic LP fast-performance liquid chromatography system (Bio-Rad) and a column packed with 10 ml of IMAC resin (Talon). The purified protein was desalted by ultrafiltration with several washes of 50 mM MOPS (morpholinepropanesulfonic acid) buffer (pH 7.25) and frozen in 10% glycerol. Protein concentrations were determined by the Bradford method (2) and by using an estimated extinction coefficient (San Diego Supercomputer Center Biology Workbench [http://workbench.sdsc.edu]) of 56 mM−1 at 280 nm with similar results. The purity of the protein was analyzed by SDS-polyacrylamide gel electrophoresis (19), and the gel was stained with Coomassie brilliant blue (Fig. 1). The induced cells showed soluble XR expression that accounted for nearly 50% of the total cellular protein. The final yield of protein was 68 mg (45 mg/liter of culture or 13 mg/g of E. coli) of >95%-pure XR with a molecular mass of ∼37 kDa, consistent with the predicted value of 38.5 kDa. The yield was high for a heterologously expressed protein, and it may be even further enhanced by fermentation techniques and expression optimization. However, this purification method is convenient and already high yielding.
FIG. 1.
Purification of heterologously expressed XR-His6-tagged fusion protein. Lane 1, the molecular weight marker; lane 2, the soluble fraction of the induced cells; lane 3, the flowthrough during purification; lane 4, final purified XR. The soluble expression of the XR reaches very high levels of around 50% of the total cellular protein. MW, molecular weight.
Steady-state kinetics.
Initial rates were determined as described previously (23) using a Cary 100 Bio UV-visible spectrophotometer (Varian) at 25°C in 50 mM MOPS (pH 6.3). Purified N. crassa XR had activity both with NADH and with NADPH as the cofactor (Table 1). NADPH is clearly favored over NADH, with 100-fold-better catalytic efficiency (kcat/Km). This difference is a function of N. crassa XR having both a higher kcat (3,600 min−1, compared to 312 min−1) and a lower Km (1.8 μM, compared to 16 μM) with NADPH than with NADH. Table 2 displays the kinetic constants of several other sugar substrates accepted by N. crassa XR. d-Ribose, l-arabinose, d-arabinose, d-galactose, sucrose, d-glucose, and d-fructose were all examined as alternative substrates for N. crassa XR with NADPH as the cofactor. Of those, d-ribose, l-arabinose, d-galactose, and d-glucose acted as substrates. Five carbon sugars acted as the best substrates, and d-glucose had the lowest turnover rate (1,320 min−1) and the highest Km (360 mM). This pattern of substrate promiscuity is typical for XRs isolated from other sources (18, 20, 23, 24).
TABLE 1.
Parameters for N. crassa XRa
| N. crassa XR with indicated coenzyme | Km for NAD(P)H (mean ± SD) (μM) | kcat (mean ± SD) (min−1) | kcat/Km for NAD(P)H (μM−1 min−1) | Km for xylose (mean ± SD) (mM) |
|---|---|---|---|---|
| NADPH | 1.8 ± 0.5 | 3,600 ± 100 | 2,000 | 34 ± 4 |
| NADH | 16 ± 4 | 310 ± 10 | 19 | 37 ± 7 |
All assays were performed at 25°C in 50 mM MOPS, pH 6.3.
TABLE 2.
Parameters for N. crassa XR with other substratesa
| N. crassa XR with indicated substrate | kcat (mean ± SD) (min−1) | Km (mean ± SD) (mM) | kcat/Km (mM−1 min−1) | % Efficiency |
|---|---|---|---|---|
| d-Xylose | 3,600 ± 200 | 34 ± 4 | 110 | 100 |
| d-Ribose | 3,120 ± 100 | 70 ± 10 | 45 | 41 |
| l-Arabinose | 1,800 ± 100 | 40 ± 10 | 45 | 41 |
| d-Galactose | 1,800 ± 100 | 180 ± 30 | 10 | 9.1 |
| d-Glucose | 1,320 ± 100 | 360 ± 60 | 3 | 3.3 |
All assays were performed at 25°C in 50 mM MOPS, pH 6.3.
Table 3 displays the kinetic characteristics of purified and characterized XRs from Candida intermedia, Candida parapsilosis, C. tropicalis, C. tenuis, Pachysolen tannophilus, Pichia stipitus, and Saccharomyces cerevisiae (4, 7, 11, 20, 21, 23, 24, 30, 33). Compared to these enzymes, N. crassa XR has a higher kcat and higher catalytic efficiencies with respect to xylose and NADPH. Its kcat is >100% higher than the closest NADPH-dependent enzyme (P. stipitus XR) (30) and 16% higher than the NADH-dependent C. parapsilosis XR (20). Its catalytic efficiency with respect to NADPH was 7-fold higher than that of the next closest enzyme (C. tenuis XR) (23) and 11-fold higher than that of any of the other XRs. The XR gene heterologously expressed and characterized here does not appear to encode an XR previously isolated and characterized from N. crassa NCIM 870 (29). Due to the lack of genetic and sequence information from that protein, it is not certain that they are different, but there are several differences between the two proteins. The subunit weights and apparent native weights of the two enzymes are significantly dissimilar (Table 3) (29) , and the steady-state kinetic constants are different (the previously isolated XR has a fourfold-lower catalytic efficiency with respect to NADPH). Additionally, the previously isolated XR showed no activity with NADH, and the two enzymes differ in their pH optima and Km values for xylose (29). Although these differences could be due to alternative mRNA splicing, posttranslational modifications in N. crassa, or the fusion of the His6 tag, the multiplicity of XRs is common in yeasts and filamentous fungi, and there are several other open reading frames in the N. crassa genome with significant sequence identity to XRs. The most logical conclusion is that these are different proteins from different genes.
TABLE 3.
Properties of XR from various organisms
| Organism (reference[s]) | Subunit mol mass (kDa) | Native mol mass (kDa) | kcat (min−1)a | Km for xylose (mM)a | kcat/Km for xylose (mM−1 min−1)a | Km for NADPH (μM) | kcat/Km for NADPH (μM−1 min−1) | Km for NADH (μM) | Optimal pH |
|---|---|---|---|---|---|---|---|---|---|
| N. crassa (this work) | 38.4 | 53 | 3,600 | 34 | 106 | 1.8 | 2,000 | 16 | 5.5 |
| C. intermedia (21, 24) | 36 | 58 | 900 | 50 | 18 | 56 | 16 | 28 | 6.0 |
| C. parapsilosis (20) | 36.6 | 69 | 3,100b | 32b | 98b | 37 | 939b | 3.3 | 6.0 |
| C. tropicalis (33) | 36.5 | 58 | NDc | 30-37 | ND | 9-18 | ND | ND | 6.0 |
| C. tenuis (7, 23) | 36 | 60 | 1,300 | 72 | 18 | 4.8 | 271 | 25 | 6.0 |
| P. tannophilus (4) | 38 | 38 | 600 | 162 | 4 | 59 | 10 | ND | 7.0 |
| P. stipitus (30) | 34 | 65 | 1,500 | 42 | 36 | 9 | 167 | 21 | 6.0 |
| S. cerevisiae (11) | 33 | 33 | 860 | 13.6 | 63 | 7.6 | 113 | ND | ND |
With NADPH as a cofactor, except for C. parapsilosis.
With NADH as a cofactor.
ND, not determined.
Temperature dependence.
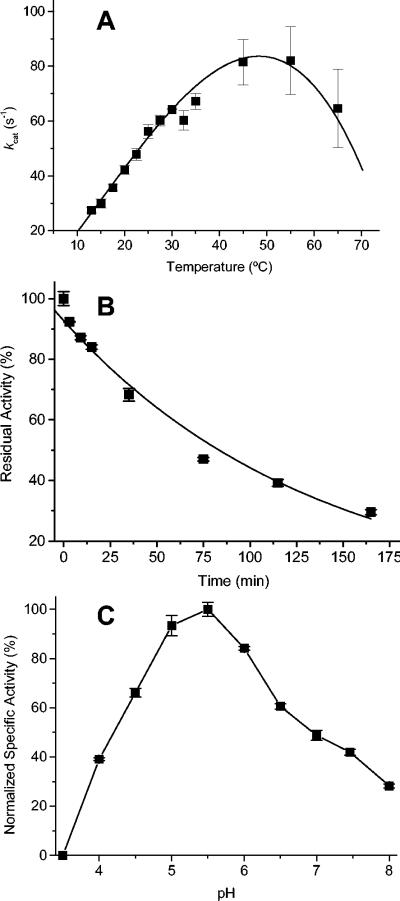
The optimal temperature of turnover was determined by assaying XR activities at temperatures ranging from 13 to 65°C by using a recirculating water bath with a jacketed cuvette holder. The data show the optimum turnover temperature to be between 45 and 55°C (Fig. 2A). At higher temperatures, the enzyme rapidly inactivates, and at lower temperatures, the rate decreases with temperature according to the Arrhenius equation. The energy of activation for xylose reduction by N. crassa XR was determined to be 37.3 kJ/mol by fitting the data from 13 to 30°C to the Arrhenius equation. Like many other mesophilic (α/β)8 proteins, the natural stability for N. crassa XR was high, as it retained activity at room temperature for longer than 1 month and at 4°C for several months. Thermal inactivation was studied by incubating N. crassa XR (27 ng/μl) in a heating block with a heated lid at 40°C in 50 mM MOPS (pH 6.3), and samples were removed at various times and assayed. The data were plotted as percentages of residual activity versus incubation times (Fig. 2B) and followed a first-order exponential decay with a half-life of 94 min.
FIG. 2.
(A) kcat dependence on temperature. XR was assayed at different temperatures from 13 to 65°C. (B) Thermal inactivation of XR at 40°C. The heat inactivation at 40°C was irreversible and followed first-order kinetics with a half-life of 94 min. (C) pH rate profile. Saturating concentrations of 1 M xylose and 200 μM NADPH were used to measure the activity in a universal buffer at various pH values from 3.5 to 8.0.
pH rate profile.
In order to determine the optimal pH and range for activity, a pH rate profile was obtained (Fig. 2C) with a universal buffer consisting of 25 mM MES (morpholinoethanesulfonic acid), 50 mM Tris, and 25 mM acetate. Activity was measured at pH values between 3.5 and 8.0 under saturating concentrations of NADPH (200 μM) and xylose (1 M). The pH range for N. crassa XR activity was large, with >25% of the activity occurring with pH values of 4.0 to 8.0. The pH optimum was around pH 5.5, and >60% of the activity remained in the 2 pH-unit span from 4.5 to 6.5. The pH optimum of N. crassa XR is slightly lower than the optima of other XRs (Table 3), but its profile is similar to those of many other XRs (20, 23).
Chemical stability.
The specific activity of XR in the presence of several chemicals was measured. Incubating N. crassa XR with 1 mM EDTA showed a slight activation of 5%, suggesting that it is not metal dependent. The reduction of cysteine residues by sulfhydryl compounds has previously been shown to enhance the activity of XR from C. tenuis (23) and C. parapsilosis (20); however, neither 1 mM dithiothreitol nor 1 mM tri(2-carboxyethyl)phosphine hydrochloride had any significant effect on the activity of N. crassa XR. Cu2+ was reported to inactivate C. parapsilosis XR (20); however, incubation with 1 mM CuSO4 had no effect on N. crassa XR activity. The addition of 0.1% (wt/vol) sodium azide also had no effect on N. crassa XR, allowing its antimicrobial use for future bioreactor studies. Bovine serum albumin activated N. crassa XR by approximately 30% when added at a concentration of 1 mg/ml, while 50 mM NaCl reduced activity by about 25%. Finally, activity was reduced only 9 or 37% in the presence of sodium phosphite at 50 and 200 mM, respectively. This result suggests that N. crassa XR is compatible with phosphite dehydrogenase, a potentially efficient in situ NADPH regeneration system (31, 32).
Determination of mass and quaternary structure.
To determine the quaternary structure, size exclusion high-performance liquid chromatography was performed using an Agilent 1100 series high-performance liquid chromatography system with a Bio-Sil SEC-250 column (300 by 7.8 mm) and a mobile phase of 0.1 M Na2PO4, 0.15 M NaCl, and 0.01 M NaN3, pH 6.8. A Bio-Rad standard was used to standardize the column's retention time with respect to molecular mass (supplemental data). All experiments were repeated twice with <0.05 min of deviation in the retention times. The molecular mass of XR was calculated from its retention time to be ∼53 kDa. Monomerization could be induced in the presence of 15% SDS, causing XR to elute as a single peak, with a retention time corresponding to a molecular mass of ∼34 kDa. The data suggest that the native XR is a noncovalently linked dimer. The significant difference between the calculated molecular weight of the dimer and its apparent molecular weight is not uncommon for XRs from other species (7, 21, 33), which also typically exist as dimers. However, to be certain of the protein size, N. crassa XR was subjected to mass analysis by electrospray ionization-quadrupole time of flight mass spectrometry. The highest abundance peak had a value of 38,381 m/z, corresponding exactly with the predicted molecular mass for the His6-tagged XR with the N-terminal fMet removed (supplemental data). Additionally, a 2M+ peak of about 20% abundance at 76,759 m/z corresponds well with the predicted mass of the dimeric form of the enzyme (76,762 Da).
Homology modeling.
Using the coordinates (Protein Data Bank [www.pdb.org]) for XR from C. tenuis (PDB accession code 1MI3) (14) and human aldose reductase (PDB accession code 2ALR), a homology model was created with Insight II software. The model was docked with NADPH and subjected to energy minimization by using the Molecular Operating Environment (MOE) program. The model was verified for consistency with known protein folds and allowed φ and ψ angles (supplemental data).
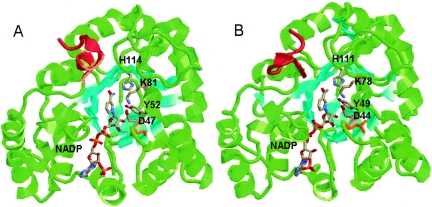
The resulting model was very similar to the C. tenuis XR crystal structure in overall fold and binding of coenzyme, as depicted in Fig. 3. The only major deviation between the α-carbons of these two structures is the C-terminal region of amino acids 311 through 322 (Fig. 3). This fact is interesting, as the C-terminal region of C. tenuis XR contains a conserved dimerization domain (14, 16). Additionally, dimerization domains in helix 5 and helix 6 (14, 16) are not found in the N. crassa XR sequence, yet N. crassa XR is a dimeric protein. Therefore, it is likely that N. crassa XR forms a dimer with the same type of interface but without the conserved sequence.
FIG. 3.
(A) Crystal structure of C. tenuis XR with bound NADPH (1K8C). (B) Homology model of N. crassa XR with bound NADPH, built using the Insight II and MOE programs. The β-sheets are colored cyan to aid in the visualization of the (α/β)8 barrel. The catalytic tyrosine, lysine, and aspartate, as well as the bound cofactors and conserved histidine, are colored by atom type. The C-terminal regions involved in dimerization are colored red. Cys23 of C. tenuis XR (A) and Leu20 of N. crassa XR (B) are colored orange.
The conserved catalytic residues His114, Lys81, Tyr52, and Asp47 (11, 12, 15) from C. tenuis XR (Fig. 3A) have similar orientations and locations in the N. crassa XR model (Fig. 3B). However, there are small variances in residues involved in substrate binding. The xylose binding pocket for N. crassa XR is slightly more hydrophilic due to the presence of Asn167 and the lack of two C. tenuis XR hydrophobic residues (Phe132 and Trp315). This increased hydrophilicity is most likely the cause of N. crassa XR having lower Km values for xylose than C. tenuis XR. One major difference between the C. tenuis XR and N. crassa XR NADPH binding pockets (14) is the replacement of Cys23 by Leu20 in N. crassa XR (Fig. 3). Since C. tenuis XR is sensitive to cysteine-reducing conditions and N. crassa XR is not, Cys23 may be the responsible residue.
In conclusion, we have identified a gene encoding XR from N. crassa. This gene can be heterologously expressed in E. coli as a His6-tagged fusion in high yield. The enzyme is not susceptible to cysteine oxidation, is rather stable, acts on several sugar substrates, and operates over a wide pH range. The ease of isolating this enzyme, coupled with its high activity and catalytic efficiency, may prove useful in the in vitro production of xylitol (or other sugar alcohols like sorbitol). In the future, this enzyme will be tested for its ability to produce sugar alcohols with in situ cofactor regeneration.
Supplementary Material
Acknowledgments
Support for this research was provided by the Biotechnology Research and Development Consortium (project 2-4-121). A Q-Tof Ultima mass spectrometer was purchased in part with a grant from the National Science Foundation, Division of Biological Infrastructure (DBI-0100085).
Electrospray ionization-quadrupole time of flight mass spectrometry measurements were made by the Mass Spectrometry Laboratory at the University of Illinois. Access to Insight II and MOE programs was provided by the University of Illinois School of Chemical Sciences' Computer Application and Network Services. We thank Bill Metcalf for initially providing E. coli BW 25141 utilized in genetic manipulations. Finally, we thank Tyler W. Johannes for his help with N. crassa growth and genomic DNA manipulation.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Amore, R., P. Kotter, C. Kuster, M. Ciriacy, and C. P. Hollenberg. 1991. Cloning and expression in Saccharomyces cerevisiae of the NAD(P)H-dependent xylose reductase-encoding gene (Xyl1) from the xylose-assimilating yeast Pichia stipitis. Gene 109:89-97. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Deshpande, V., S. Keskar, C. Mishra, and M. Rao. 1986. Direct conversion of cellulose hemicellulose to ethanol by Neurospora crassa. Enzyme Microb. Technol. 8:149-152. [Google Scholar]
- 4.Ditzelmuller, G., C. P. Kubicek, W. Wohrer, and M. Rohr. 1984. Xylose metabolism in Pachysolen tannophilus purification and properties of xylose reductase. Can. J. Microbiol. 30:1330-1336. [Google Scholar]
- 5.Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read, D. Jaffe, W. FitzHugh, L. J. Ma, S. Smirnov, S. Purcell, B. Rehman, T. Elkins, R. Engels, S. G. Wang, C. B. Nielsen, J. Butler, M. Endrizzi, D. Y. Qui, P. Ianakiev, D. B. Pedersen, M. A. Nelson, M. Werner-Washburne, C. P. Selitrennikoff, J. A. Kinsey, E. L. Braun, A. Zelter, U. Schulte, G. O. Kothe, G. Jedd, W. Mewes, C. Staben, E. Marcotte, D. Greenberg, A. Roy, K. Foley, J. Naylor, N. Stabge-Thomann, R. Barrett, S. Gnerre, M. Kamal, M. Kamvysselis, E. Mauceli, C. Bielke, S. Rudd, D. Frishman, S. Krystofova, C. Rasmussen, R. L. Metzenberg, D. D. Perkins, S. Kroken, C. Cogoni, G. Macino, D. Catcheside, W. X. Li, R. J. Pratt, S. A. Osmani, C. P. C. DeSouza, L. Glass, M. J. Orbach, J. A. Berglund, R. Voelker, O. Yarden, M. Plamann, S. Seller, J. Dunlap, A. Radford, R. Aramayo, D. O. Natvig, L. A. Alex, G. Mannhaupt, D. J. Ebbole, M. Freitag, I. Paulsen, M. S. Sachs, E. S. Lander, C. Nusbaum, and B. Birren. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859-868. [DOI] [PubMed] [Google Scholar]
- 6.Gulbis, J. M., S. Mann, and R. MacKinnon. 1999. Structure of a voltage-dependent K+ channel beta subunit. Cell 97:943-952. [DOI] [PubMed] [Google Scholar]
- 7.Hacker, B., A. Habenicht, M. Kiess, and R. Mattes. 1999. Xylose utilisation: cloning and characterisation of the xylose reductase from Candida tenuis. Biol. Chem. 380:1395-1403. [DOI] [PubMed] [Google Scholar]
- 8.Hahn-Hagerdal, B., C. F. Wahlbom, M. Gardonyi, W. H. van Zyl, R. R. Cordero Otero, and L. J. Jonsson. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73:53-84. [DOI] [PubMed] [Google Scholar]
- 9.Ho, N. W., F. P. Lin, S. Huang, P. C. Andrews, and G. T. Tsao. 1990. Purification, characterization, and amino terminal sequence of xylose reductase from Candida shehatae. Enzyme Microb. Technol. 12:33-39. [DOI] [PubMed] [Google Scholar]
- 10.Ikemi, M., N. Koizumi, and Y. Ishimatsu. 1990. Sorbitol production in charged membrane bioreactor with coenzyme regeneration system. 1. Selective retainment of NADP(H) in a continuous reaction. Biotechnol. Bioeng. 36:149-154. [DOI] [PubMed] [Google Scholar]
- 11.Jeong, E. Y., I. S. Kim, and H. Lee. 2002. Identification of lysine-78 as an essential residue in the Saccharomyces cerevisiae xylose reductase. FEMS Microbiol. Lett. 209:223-228. [DOI] [PubMed] [Google Scholar]
- 12.Jeong, E. Y., C. Sopher, I. S. Kim, and H. Lee. 2001. Mutational study of the role of tyrosine-49 in the Saccharomyces cerevisiae xylose reductase. Yeast 18:1081-1089. [PubMed] [Google Scholar]
- 13.Kang, M. H., H. Ni, and T. W. Jeffries. 2003. Molecular characterization of a gene for aldose reductase (CbXYL1) from Candida boidinii and its expression in Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 106:265-276. [DOI] [PubMed] [Google Scholar]
- 14.Kavanagh, K. L., M. Klimacek, B. Nidetzky, and D. K. Wilson. 2002. The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis. Biochemistry 41:8785-8795. [DOI] [PubMed] [Google Scholar]
- 15.Klimacek, M., M. Szekely, R. Griessler, and B. Nidetzky. 2001. Exploring the active site of yeast xylose reductase by site-directed mutagenesis of sequence motifs characteristic of two dehydrogenase/reductase family types. FEBS Lett. 500:149-152. [DOI] [PubMed] [Google Scholar]
- 16.Klimacek, M., F. Wuhrer, K. L. Kavanagh, D. K. Wilson, and B. Nidetzky. 2003. Altering dimer contacts in xylose reductase from Candida tenuis by site-directed mutagenesis: structural and functional properties of R180A mutant. Chem. Biol. Interact. 143-144:523-532. [DOI] [PubMed] [Google Scholar]
- 17.Kozma, E., E. Brown, E. M. Ellis, and A. J. Lapthorn. 2002. The crystal structure of rat liver AKR7A1. A dimeric member of the aldo-keto reductase superfamily. J. Biol. Chem. 277:16285-16293. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn, A., C. van Zyl, A. van Tonder, and B. A. Prior. 1995. Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl. Environ. Microbiol. 61:1580-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J. K., B. S. Koo, and S. Y. Kim. 2003. Cloning and characterization of the xyl1 gene, encoding an NADH-preferring xylose reductase from Candida parapsilosis, and its functional expression in Candida tropicalis. Appl. Environ. Microbiol. 69:6179-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayr, P., K. Bruggler, K. D. Kulbe, and B. Nidetzky. 2000. d-Xylose metabolism by Candida intermedia: isolation and characterisation of two forms of aldose reductase with different coenzyme specificities. J. Chromatogr. B 737:195-202. [DOI] [PubMed] [Google Scholar]
- 22.Mishra, C., S. Keskar, and M. Rao. 1984. Production and properties of extracellular endoxylanase from Neurospora crassa. Appl. Environ. Microbiol. 48:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neuhauser, W., D. Haltrich, K. D. Kulbe, and B. Nidetzky. 1997. NAD(P)H-dependent aldose reductase from the xylose-assimilating yeast Candida tenuis. Isolation, characterization and biochemical properties of the enzyme. Biochem. J. 326:683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nidetzky, B., K. Bruggler, R. Kratzer, and P. Mayr. 2003. Multiple forms of xylose reductase in Candida intermedia: comparison of their functional properties using quantitative structure-activity relationships, steady-state kinetic analysis, and pH studies. J. Agric. Food Chem. 51:7930-7935. [DOI] [PubMed] [Google Scholar]
- 25.Nidetzky, B., W. Neuhauser, D. Haltrich, and K. D. Kulbe. 1996. Continuous enzymatic production of xylitol with simultaneous coenzyme regeneration in a charged membrane reactor. Biotechnol. Bioeng. 52:387-396. [DOI] [PubMed] [Google Scholar]
- 26.Nidetzky, B., W. Neuhauser, P. Mayr, D. Haltrich, and K. D. Kulbe. 1998. Strategies to an efficient enzymatic production of xylitol. Ann. N. Y. Acad. Sci. 864:442-445. [DOI] [PubMed] [Google Scholar]
- 27.Rao, M., V. Deshpande, S. Keskar, and M. C. Srinivasan. 1983. Cellulase and ethanol production from cellulose by Neurospora crassa. Enzyme Microb. Technol. 5:133-136. [Google Scholar]
- 28.Rawat, U., A. Bodhe, V. Deshpande, and M. Rao. 1993. d-Xylose catabolizing enzymes in Neurospora crassa and their relationship to d-xylose fermentation. Biotechnol. Lett. 15:1173-1178. [Google Scholar]
- 29.Rawat, U. B., and M. B. Rao. 1996. Purification, kinetic characterization and involvement of tryptophan residue at the NADPH binding site of xylose reductase from Neurospora crassa. Biochim. Biophys. Acta 1293:222-230. [DOI] [PubMed] [Google Scholar]
- 30.Verduyn, C., R. Vankleef, J. Frank, H. Schreuder, J. P. Vandijken, and W. A. Scheffers. 1985. Properties of the NAD(P)H-dependent xylose reductase from the xylose-fermenting yeast Pichia stipitis. Biochem. J. 226:669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vrtis, J. M., A. K. White, W. W. Metcalf, and W. A. van der Donk. 2002. Phosphite dehydrogenase: a versatile cofactor-regeneration enzyme. Angew. Chem. Int. Ed. Engl. 41:3257-3259. [DOI] [PubMed] [Google Scholar]
- 32.Woodyer, R., W. A. van der Donk, and H. Zhao. 2003. Relaxing the nicotinamide cofactor specificity of phosphite dehydrogenase by rational design. Biochemistry 42:11604-11614. [DOI] [PubMed] [Google Scholar]
- 33.Yokoyama, S. I., T. Suzuki, K. Kawai, H. Horitsu, and K. Takamizawa. 1995. Purification, characterization and structure analysis of NADPH-dependent d-xylose reductases from Candida tropicalis. J. Ferment. Bioeng. 79:217-223. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.