Abstract
We report a pronounced diel rhythm in ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) gene expression in a natural population of the coccolithophorid Coccolithus pelagicus sampled during a Lagrangian experiment in the Northeast Atlantic. Our observations show that there is greater heterogeneity in the temporal regulation of RubisCO expression among planktonic chromophytes than has been reported hitherto.
Despite the importance of the oceans in the global carbon cycle, comparatively little is known of the regulation of photosynthetic carbon fixation in marine phytoplankton. As in higher plants, the bulk of CO2 is fixed via the Calvin cycle enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) (10) in these organisms. Therefore, it is of considerable interest to understand how the environment structures the expression of this key enzyme. In the present study, we investigated the diel variability in rbcL mRNA abundance in a natural population of the coccolithophorid Coccolithus pelagicus.
Observations were made in subpolar waters to the south of Iceland within an anticyclonic eddy with a cold surface temperature anomaly (6, 12). A patch of the tracer SF6 was deployed at the eddy center (5), and the research vessel operated in Lagrangian mode by analyzing surface SF6 concentrations continuously (4). Seawater samples were obtained during conductivity-temperature-depth hydrocasts, and the abundance of C. pelagicus cells was determined by flow cytometry (13). For RNA, near-surface (∼2.5-m depth) seawater samples (10 to 20 liters) were rapidly filtered (<10 min) onto 90-mm-diameter Whatman GF/C filters, and the filters were stored at −70°C in extraction buffer (15) following snap-freezing in a propan-2-ol cooling bath. RNA was isolated and prepared for Northern analysis (16) under the hybridization conditions described below.
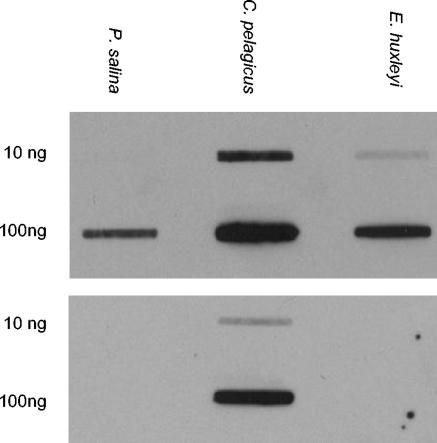
An internal region of rbcL was amplified from DNA isolated from C. pelagicus cells collected at the study site (16) and from laboratory cultures of the haptophytes Emiliania huxleyi and Pavlova salina. The identity of the products was confirmed by comparison to previously published sequences (2, 3), and sense strand in vitro transcription products were synthesized as previously described (16). Northern slot blots were hybridized at 55°C in Easy-Hyb solution (Roche) amended with 25 ng of C. pelagicus rbcL probe DNA (labeled with digoxigenin-dUTP) ml−1. Stringency washes were performed on the following day with 0.05× SSPE (1× SSPE is 150 μM NaCl, 10 μM Na2HPO4, 1 μM EDTA)-0.1% sodium dodecyl sulfate at 68°C, and hybrids were detected with alkaline phosphatase-conjugated antidigoxigenin in conjunction with the chemiluminescent substrate CDP-Star (Roche). This hybridization protocol enables the specific detection of C. pelagicus rbcL mRNA and was optimized empirically by testing in vitro transcription products under conditions of increasing stringency (Fig. 1). Like the rbcL genes of the other coccolithophorids that have been characterized to date, the nucleotide sequence of the fragment from E. huxleyi is 88 to 89% identical to that of C. pelagicus whereas rbcL from noncalcifying P. salina is less closely related (86% identical).
FIG. 1.
Northern slot blot assays of sense strand in vitro rbcL transcription products probed with a digoxigenin-labeled fragment of rbcL from C. pelagicus. The upper luminograph shows the results obtained under suboptimal posthybridization wash conditions (0.2× rather than 0.05× SSPE in the stringency washes), while the lower luminograph shows the high specificity of the probe once the optimal hybridization protocol (see text) had been established.
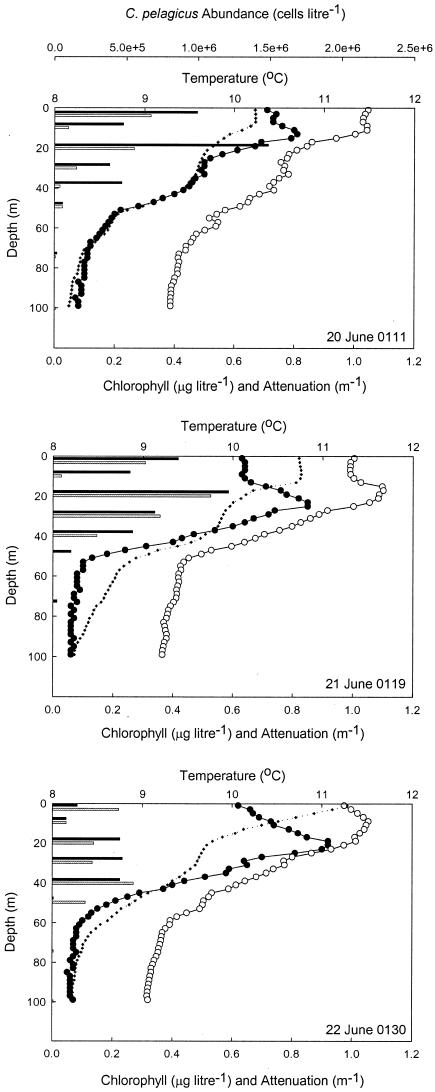
Field observations and the sampling program for this study were confined to the first 5 days after the tracer release. During this time, the SF6 patch remained coherent and the surface waters within the eddy retained their identity (6). C. pelagicus accounted for approximately 30% of the phytoplankton biomass within the eddy but was absent from surrounding waters (13), highlighting not only its distinct biological signature but also its physical resolution. Two distinct C. pelagicus population maxima were evident: a near-surface maximum and a somewhat deeper population at ∼20 m that extended to a depth of 40 to 50 m (Fig. 2).
FIG. 2.
Depth profiles of temperature (⧫), chlorophyll concentrations (•), and beam attenuation (○) through the upper 100 m of the water column at the eddy center on 20, 21, and 22 June 1996. The histograms show the vertical distribution of C. pelagicus biomass observed at discrete depths sampled during the midday (gray bars) and midnight (black bars) hydrocasts. Greenwich mean times are shown.
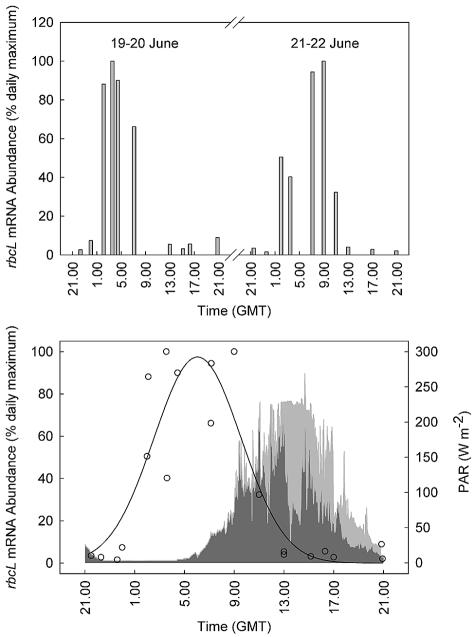
Considerable variability was observed in the abundance of C. pelagicus rbcL mRNA over the course of both diel cycles (Fig. 3, top panel) that was unrelated to temporal changes in the size of the C. pelagicus population, which generally increased at night (Fig. 2). RubisCO expression was greatest at around daybreak but declined by at least an order of magnitude during the latter half of the day and into the early nighttime period. Because of gaps in the sampling regimen when the research vessel was surveying waters outside of the SF6 patch (particularly on the morning of 20 June), the two data sets were combined (Fig. 3, bottom panel) and analyzed by Rayleigh's test (1). The mean vector determined for those samples (n = 8), when the mRNA abundance was ≥40% of the daily maximum, was found to be 4:48 a.m. (r = 0.812, 0.002 < P < 0.005), indicating that there was significant clustering around the daily peak in gene expression during both diel cycles. The excellent reproducibility of the rhythm between days was confirmed also by nonlinear regression analysis (r2 = 0.805, P < 0.0001).
FIG. 3.
Diel variability in the abundance of C. pelagicus rbcL mRNA normalized to the maximum hybridization signal recorded during each diel cycle (top panel). The bottom panel shows a three-parameter Gaussian regression (SigmaPlot for Windows, version 8.02) of the combined data set from both diel cycles and the temporal variability in photosynthetically active radiation (PAR) in the 24-h period between 2100 on 19 June and 2100 on 20 June (light gray) and the 24-h period between 2100 on 21 June and 2100 on 22 June (dark gray). The temporal resolution of the irradiance data is 1 min and was logged with a 4π collector mounted on the unshaded upper deck of the research vessel. GMT, Greenwich mean time.
Previous studies have examined temporal variability in rbcL mRNA abundance in mixed communities of marine phytoplankton (8, 9, 15, 16), but this is the first report of species- rather than group-specific determinations of RubisCO gene expression in a natural population. Intriguingly, the diel rhythm in C. pelagicus rbcL mRNA abundance found was quite distinct from that observed in the haptophyte Pavlova gyrans or in natural phytoplankton populations at lower latitudes with a chromophyte-specific rbcL probe (7, 8). In both cases, rbcL mRNA abundance maxima were observed toward the end of the diurnal period whereas in Heterosigma carterae and the diatom Phaeodactylum triconutum, a broad mid-morning maximum in rbcL mRNA abundance has been reported (11, 14). Like the natural population of C. pelagicus studied here, rbcL mRNA began to accumulate in the dark prior to the onset of illumination. No evidence of a nighttime increase in transcriptional activity was found in P. gyrans (7) or the natural communities of diatoms and picoeukaryotes investigated previously (8).
Evidently, there is considerable heterogeneity in the diel regulation of RubisCO gene expression between and within different taxonomic classes of marine chromophytic algae. To what extent this reflects differences in genetic controls, the life cycle of individual species, or environmental variability is worthy of further investigation to establish its ecological significance. It has been proposed, for instance, that differences in the diel pattern of RubisCO expression between chromophytes and the picocyanobacteria indicate that they occupy different temporal niches (8). This study has shown that this may not always be the case since the diel rhythm in C. pelagicus RubisCO expression reported here is very similar to that of natural populations of Synechococcus spp. from the same oceanic region (15).
Acknowledgments
This research was supported by a PRIME special topic grant (GST/02/1082) awarded to M.W. and D.A.P. by the Natural Environment Research Council of the United Kingdom.
We thank the officers, crew, and technicians of RRS Discovery for logistical support and outstanding service during cruise D221. Kirsty Park (University of Stirling) gave valuable advice and assistance with the statistical analyses.
Footnotes
Dedicated to the memory of contributing author John T. Davies, who tragically passed away in 2003 after years of struggle in an imperfect world.
REFERENCES
- 1.Batschelet, E. 1981. Circular statistics in biology. Academic Press, Inc., New York, N.Y.
- 2.Daugbjerg, N., and R. A. Andersen. 1997. Phylogenetic analyses of the rbcL sequences from haptophytes and heterokont algae suggest their chloroplasts are unrelated. Mol. Biol. Evol. 14:1242-1251. [DOI] [PubMed] [Google Scholar]
- 3.Fujiwara, S., M. Sawada, J. Someya, N. Minaka, M. Kawachi, and I. Inouye. 1994. Molecular phylogenetic analysis of rbcL in the Prymnesiophyta. J. Phycol. 30:863-871. [Google Scholar]
- 4.Law, C. S., A. J. Watson, M. I. Liddicoat, and T. Stanton. 1998. Sulphur hexafluoride as a tracer of biogeochemical and physical processes in an open ocean iron fertilization experiment. Deep-Sea Res. II 45:977-994. [Google Scholar]
- 5.Law, C. S., A. P. Martin, M. I. Liddicoat, A. J. Watson, K. J. Richards, and E. M. S. Woodward. 2001. A Lagrangian SF6 tracer study of an anticyclonic eddy in the North Atlantic: patch evolution, vertical mixing and nutrient supply to the mixed layer. Deep-Sea Res. II 48:705-724. [Google Scholar]
- 6.Martin, A. P., I. P. Wade, K. J. Richards, and K. J. Heywood. 1998. The PRIME eddy. J. Mar. Res. 56:439-462. [Google Scholar]
- 7.Paul, J. H., J. B. Kang, and F. R. Tabita. 2000. Diel patterns of regulation of rbcL transcription in a cyanobacterium and a prymnesiophyte. Mar. Biotechnol. 2:429-436. [DOI] [PubMed] [Google Scholar]
- 8.Paul, J. H., S. L. Pichard, J. B. Kang, G. M. F. Watson, and F. R. Tabita. 1999. Evidence for a clade-specific temporal and spatial separation in ribulose bisphosphate carboxylase gene expression in phytoplankton populations off Cape Hatteras and Bermuda. Limnol. Oceanogr. 44:12-23. [Google Scholar]
- 9.Pichard, S. L., L. Campbell, J. B. Kang, F. R. Tabita, and J. H. Paul. 1996. Regulation of ribulose bisphosphate carboxylase expression in natural phytoplankton communities. I. Diel rhythms. Mar. Ecol. Prog. Ser. 139:257-265. [Google Scholar]
- 10.Raven, J. A. 1996. Inorganic carbon assimilation by marine biota. J. Exp. Mar. Biol. Ecol. 203:39-47. [Google Scholar]
- 11.Reynolds, A. E., B. L. McConaughy, and R. A. Cattolico. 1993. Chloroplast genes of the marine alga Heterosigma carterae are transcriptionally regulated during a light/dark cycle. Mol. Mar. Biol. Biotechnol. 2:121-128. [Google Scholar]
- 12.Savidge, G., and P. J. L. Williams. 2001. The PRIME 1996 cruise: an overview. Deep-Sea Res. II 48:687-704. [Google Scholar]
- 13.Tarran, G. A., M. V. Zubkov, M. A. Sleigh, P. H. Burkill, and M. Yallop. 2001. Microbial community structure and standing stocks in the N. E. Atlantic in June and July 1996. Deep-Sea Res. II 48:963-985. [Google Scholar]
- 14.Wawrik, B., J. H. Paul, and F. R. Tabita. 2002. Real-time PCR quantification of rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes. Appl. Environ. Microbiol. 68:3771-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyman, M. 1999. Diel rhythms in ribulose-1,5-bisphosphate carboxylase/oxygenase and glutamine synthetase gene expression in a natural population of marine picoplanktonic cyanobacteria (Synechococcus spp.). Appl. Environ. Microbiol. 65:3651-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyman, M., J. T. Davies, D. W. Crawford, and D. A. Purdie. 2000. Molecular and physiological responses of two classes of marine chromophytic phytoplankton (diatoms and prymnesiophytes) during the development of nutrient-stimulated blooms. Appl. Environ. Microbiol. 66:2349-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]