Abstract
Frailty is an aging-related clinical phenotype defined as a state in which there is an increase in a person’s vulnerability for dependency and/or mortality when exposed to a stressor. While underlying mechanisms leading to the occurrence of frailty are complex, the importance of genetic factors has not been fully investigated. We conducted a large-scale genome-wide association study (GWAS) of frailty, as defined by the five criteria (weight loss, exhaustion, physical activity, walking speed, and grip strength) captured in the Fried Frailty Score (FFS), in 386,565 European descent participants enrolled in the UK Biobank (mean age 57 [SD 8] years, 208,481 [54%] females). We identified 37 independent, novel loci associated with the FFS (p < 5 × 10–8), including seven loci without prior described associations with other traits. The variants associated with FFS were significantly enriched in brain tissues as well as aging-related pathways. Our post-GWAS bioinformatic analyses revealed significant genetic correlations between FFS and cardiovascular-, neurological-, and inflammation-related diseases/traits, and subsequent Mendelian Randomization analyses identified causal associations with chronic pain, obesity, diabetes, education-related traits, joint disorders, and depressive/neurological, metabolic, and respiratory diseases. The GWAS signals were replicated in the Health and Retirement Study (HRS, n = 9,720, mean age 73 [SD 7], 5,582 [57%] females), where the polygenic risk score built from UKB GWAS was significantly associated with the FFS in HRS individuals (OR per SD of the score 1.27, 95% CI 1.22–1.31, p = 1.3 × 10–11). These results provide new insight into the biology of frailty by comprehensively evaluating its genetic architecture.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-023-00771-z.
Keywords: Frailty, GWAS, Genetic correlation, Polygenic risk score, Aging
Introduction
As the proportion of older adults rise around the globe, frailty has increasingly become the focus of attention among researchers and clinicians. Frailty can be defined as a medical condition characterized by a progressive decline in physiological systems that increases an individual’s vulnerability to adverse health outcomes when exposed to a stressor [1]. Most prevalent in the elderly, frailty is not an inevitable consequence of the aging process; rather, frailty can be viewed as an accelerated or pathological state of aging. Frailty is associated with higher risks of disability, dementia, hospitalization, mortality, and poor outcomes after surgery, a constellation of findings that reflect both the importance and phenotypic complexity of this phenotype [1]. Moreover, frailty is related to increased healthcare costs and poor prognosis in surgical patients as well as with chronic renal failure, liver disease or cardiological conditions [2]. Novel preventive and therapeutic strategies are urgently needed to reduce the socioeconomic burden associated with the development and progression of frailty.
The main systems identified to date in the pathophysiology of frailty include the endocrine axis [3], inflammation/immune system [4], stress response [5] and metabolic pathways [6]. Multiple age-related hormone changes have been associated with frailty [3]. It has also been suggested that the immune system is involved in the development of frailty. Particularly, serum levels of the proinflammatory cytokine interleukin IL-6 [7], procalcitonin [7] and C-reactive protein (CRP) [7, 8], as well as white blood cell and monocyte counts [8], are elevated in frail adults. Other stress response and metabolic systems include altered glucose metabolism [9], dysregulation of the autonomic nervous system [10] and age-related changes in the renin-angiotensin system [11]. Given the observational design of most of the studies outlined above, it is difficult to judge whether the associations with these pathophysiological mechanisms represent true causal links or just the co-occurrence of frailty with the numerous disease that are highly prevalent in older adults.
In addition to the aforementioned observations, genetic studies using a candidate-gene design identified several genes associated with frailty, mostly related to inflammation, protein regulation, and cognitive function (with the epsilon variants within APOE playing a prominent role) [12, 13]. Importantly, previous genetic studies of frailty identified loci associated with traits such as body mass index (BMI), cardiovascular disease, smoking, HLA proteins, neuropathy, endocrine system, depression and neuroticism [13, 14]. However, these studies were limited by small sample size and their restriction to incident frailty, leading to diminished statistical power.
Further exploration of the genetic architecture of frailty will provide novel insights on the biological pathways involved in the development and progression of frailty and identify new targets for prevention and treatment. Here, we conducted a large-scale genome-wide association study (GWAS) of frailty in the UK Biobank (UKB) [15] comprising over 380,000 individuals of European ancestry and replicated the associations in 9,720 individuals from the Health and Retirement Study (HRS) [16]. In contrast to a recent genome-wide studies of frailty [17], we used the Fried Frailty Score (FFS) [18] instead of the Frailty Index (FI) to model frailty. While the FI requires a thorough clinical assessment, including laboratory work-up, the FFS uses a small number of signs and symptoms (weight loss, low physical activity, exhaustion, slowness, and weakness), thus facilitating applicability and reproducibility [19]. In addition, compared to this recent frailty GWAS, we maximized the discovery power by avoiding an age restriction and including more samples. The applicability of our findings to older adults was validated in a landmark study of aging—HRS.
Subjects and methods
Study design
We conducted a two-stage (discovery and replication) GWAS using publicly available data from two established observational studies: the UKB and HRS. Research activities were approved by the corresponding local IRBs and written informed consent was obtained from all study participants or their legally appointed representatives. With the GWAS, we then performed a series of post-GWAS analyses to gain deeper understanding of the genetic basis of frailty, as illustrated in the flowchart in the Supplemental Data (Supp. Figure 1).
The UK biobank
The UKB is a large prospective study established to be a public resource for research into the causes of diseases in the UK population. The study protocol, details of the study design and information on data access are available online [15]. In brief, a total of 502,618 community-dwelling persons aged 40–69 years were recruited from across the United Kingdom between 2006 and 2010. We restricted the analysis to unrelated individuals of genetically-confirmed European ancestry [20].
Fried frailty score
The FFS developed by Fried et al. [18] is a well-established and validated standardized definition of frailty phenotype. It has been widely used in several large-scale studies, including both the UKB and HRS [21, 22]. Other frailty definitions exist, however, these are either lacking in validity evaluations or are too complex in their criteria [23]. The FFS offers a concise and generalizable method for defining frailty which enhances the applicability and reproducibility of GWAS results. Thus, in this study, we adopted the FFS definition to classify frailty status in our participants. Specifically, based on self-reported answers and physical measurements collected at enrollment, participants were assigned an FFS score of 0–5 according to the number of criteria met (weight loss, exhaustion, physical activity, walking speed, and grip strength) [18]. Although frailty is usually modeled as a binary variable, with FFS > = 3 classified as frailty. Here we analyzed this phenotype as an ordinal variable in order to reduce information loss and enhance the statistical power of the analysis. There were 35,588 (7.1%) participants excluded from analysis for missing one or more of the five frailty criteria at baseline.
GWAS analysis
We used phase three genotype data released by UKB [20] where the participants underwent genotyping with one of two closely related Affymetrix microarrays (UK BiLEVE Axiom Array or UK Biobank Axiom Array) for ~ 820,000 variants. Additional genotypes were imputed centrally using the 1000 Genomes [24] and Haplotype Reference Consortium (HRC) [25] reference panels, yielding ~ 93 million variants for each individual. We restricted the analysis to 8,883,488 autosomal variants with imputation quality score > 0.9, Hardy–Weinberg p-value > 1 10–6, and minor allele frequency (MAF) > 0.01 and genotyping missing rate < 0.1. European samples were identified by a combination of self-reported and genetically confirmed ancestry based on principal component (PC) analysis of individuals’ genotypes. Further sample exclusion criteria included poor heterozygosity or missingness, sex chromosome aneuploidy, and withdrawal of informed consent. We used BOLT-LMM (v2.3.2) to perform single-variant genome-wide association testing [26], including age, sex and the first 20 genetic principal components as covariates. We used Linkage Disequilibrium Score Regression (LDSC, v1.0.0) [27] to perform SNP-based heritability estimation. The level of bias in GWAS was estimated using both LDSC intercept and lambda GC.
Genomic risk loci definition
To identify independent associations, we conducted a conditional and joint (COJO) analysis of the GWAS results as implemented by GCTA, accounting for the correlation structure between single nucleotide polymorphisms (SNPs) within a 10-Mb window and using a random subset of 10,000 unrelated Europeans from the UKB as linkage disequilibrium (LD) reference. For comparison, we used PLINK [28] to identify regional lead SNPs for genome-wide significant loci, and subsequently clumped variants with this lead SNP if they were located less than 10 Mb away and had r2 > 0.01 with the index variant. Since GCTA-COJO accounts for long-range LD and interaction effects between variants, it tends to be more conservative, reporting fewer but more valid independent loci, we use it as primary method to identify the independent genomic risk loci.
Pathway analysis
We used MAGENTA (Meta-Analysis Gene-set Enrichment of Variant Associations) to conduct a pathway-wide association analysis [29]. MAGENTA implements gene set enrichment analysis (GSEA) [30] from across five databases to test a total of 3,224 pathways and provides GSEA p-values for both 95th percentile and 75th percentile cutoffs. We chose the latter option, as it provides more statistical power when analyzing highly polygenic diseases [29]. Biological pathways were considered suggestively significant if their GSEA marginal P value was less than 0.05.
Tissue and cell-type enrichment analysis
We applied LDSC to perform tissue and cell-type enrichment analysis for FFS. We first used dichotomized annotations and 1000 Genomes European reference panels to estimate annotation-stratified LD scores. Enrichment was then defined as the ratio of the percentage of heritability explained by variants in each annotated category versus the percentage of variants covered by that category. In addition to the 53 baseline annotations [27] for diverse genomic features as suggested in the LDSC user manual and GenoCanyon general functionality scores [31], two tiers of annotations of different resolutions were further considered in enrichment analyses: 1) GenoSkyline-Plus functionality scores [32] of seven broad tissue clusters (immune, brain, cardiovascular, muscle, gastrointestinal tract, epithelial, other) and 2) GenoSkyline-Plus functionality scores of 66 tissue and cell types. For comparison, we also used MAGMA [33] to perform the tissue enrichment analysis, where we used annotation data from 30 general tissues types and 54 specific tissue types (GTEx project. v.8) [34] as provided by the FUMA (Functional Mapping and Annotation) platform [35]. Bonferroni multiple-testing correction was applied for each tier of enrichment analysis, separately.
Genetic correlation
To investigate potential shared underlying molecular mechanisms, we applied GNOVA [36], an annotation-stratified genetic covariance analyzer, to test the genetic correlation between FFS and a wide range of traits. We extracted GWAS summary statistics without large sample overlap with UKB from the LD Hub (http://ldsc.broadinstitute.org/gwashare/), the FinnGen cohort (https://finngen.gitbook.io/documentation/data-download), the Philadelphia Neurodevelopmental cohort (https://www.med.upenn.edu/bbl/philadelphianeurodevelopmentalcohort.html), and other large GWAS consortiums as organized previously [37], retaining available summary data for 2,086 traits in total. A Bonferroni-corrected p < 2.4 × 10–5 (corrected for 2,086 tests) was considered significant. Specifically, we also tested the genetic correlation between hearing loss [38] and frailty, as a strong epidemiological association between these two has long been observed [39].
Mendelian randomization
Mendelian randomization (MR) analysis is widely used to explore the potential causal relations between phenotypic traits and health-related outcomes. For the traits that had significant genetic correlation with FFS, we further investigated their causal associations with FFS. Specifically, we used inverse variance weighted MR (IVW-MR) [40] as the initial approach since it usually provides greater power to detect potential causal relationships. For traits with significant causal associations with FFS based on the IVW-MR, we also used other MR approaches in sensitivity analyses, including MR PRESSO [41], MR‐Egger regression [42], weighted median MR [40] and mode‐based methods [43] (e.g., simple mode MR and weighted mode MR). The 37 independent leading SNPs were used as instrumental variables.
Replication GWAS
To evaluate the consistency of our GWAS discovery results in an independent sample, we used data from the HRS (http://www.nia.nih.gov/research/resource/health-and-retirement-study-hrs) to conduct replication analyses. HRS is a nationally representative longitudinal survey of individuals over age 50 in 23,000 households in the USA. We included individuals from Waves 8–13 (2006–2016) of the HRS, as these waves contained physical measurement data for calculating the frailty score. A subgroup of 15,567 study participants underwent genome-wide genotyping using Illumina's Human Omni2.5-Quad (Omni2.5) BeadChip. The current analysis was restricted to study participants of genetically confirmed European ancestry. The quality control, imputation, post-imputation filtering, and association testing were similar to the one used in the discovery cohort. Specifically, for variants that were genome-wide significant in UKB GWAS but not available or with low quality in HRS, we selected the variant having the highest LD, with the target variant within a 10-Mb window as the proxy-variant.
Polygenic risk score analysis
Polygenic risk scores (PRS) quantify the inherited risk for an individual by aggregating information from GWAS summary statistics. Here we applied PRS-CS [44] to generate weights for each genetic variant based on our GWAS summary statistics and 1000 Genomes LD reference panel. PRS for each individual in the replicated dataset was then calculated as the weighted sum of the genetic variants. The genetic variants we used were the overlaps between the reference panel, UKB and HRS genetic data. We calculated both odds ratio (OR) per standard deviation (SD) and area under ROC curve (AUC) to evaluate the prediction performance of PRS after adjustments for age, sex and first 10 PCs in a logistic regression model, where we used the binary frailty phenotype (FFS > = 3 is classified as frailty) as the outcome during the PRS evaluation.
Statistical analysis
In our replication analysis, we not only evaluated the associations of individual SNPs in the HRS cohort, but also conducted a sign test to compare the leading SNPs in both the UKBB and HRS cohorts. Specifically, we employed a binomial sign test with the null hypothesis that the probability of having the same sign in both cohorts is 0.5. The significance threshold was set at 0.05. To better visualize the results of the PRS analysis, we divided the HRS population into 20 groups based on PRS quantiles and calculated the mean frailty scores within each group. We then plotted the mean frailty scores against PRS to easily visualize the correlation between them. All statistical analyses in this study, aside from the genetic analysis tools mentioned previously, were performed using R software version 3.5.
Results
Characteristics of participants
The discovery phase included 386,565 participants of European ancestry with complete data for frailty criteria from the UKB (mean age 57 years [SD 8], 208,481 (54%) were female). The distribution of the FFS (from 0–5) in the UKB participants was: 231,629 (60%), 110,651 (29%), 31,561 (8%), 9,564 (2%), 2,801 (1%), and 359 (~ 0%), respectively. A comparison of the basic characteristics among the non-frail, pre-frail, and frail people is summarized in Table 1. The replication phase included 9,720 HRS participants with complete data for the FFS. This population was older than the UKB cohort (mean age 73 years, [SD 7]), with over half (n = 5,582, 57%) being female. The distribution of the FFS (from 0–5) in the HRS was: 3,734 (38%), 2,948 (30%), 1,769 (18%), 891 (9%), 357 (4%), and 21 (~ 0%), respectively.
Table 1.
Basic characteristics in the UK Biobank
| Characteristics | All (n = 386,565) |
Non-frail (n = 231,629) |
Pre-frail (n = 142,212) |
Frail (n = 12,724) |
|---|---|---|---|---|
| Age (years), mean (SD) | 56.9 (8.0) | 56.6 (8.0) | 57.2 (8.0) | 58.5 (7.4) |
| Sex, (Female, %) | 208,481 (54%) | 119,427 (52%) | 81,067 (57%) | 7,942 (62%) |
| BMI, mean (SD) | 27.4 (4.7) | 26.6 (4.1) | 28.3 (5.1) | 31.3 (6.7) |
| Hypertension, n (%) | 212,532 (55%) | 121,716 (53%) | 82,001 (58%) | 8,815 (69%) |
| T2D, n (%) | 29,670 (8%) | 11,323 (5%) | 15,017 (11%) | 3,330 (26%) |
| Lipid medication, n (%) | 66,937 (17%) | 32,509 (14%) | 29,741 (21%) | 4,687 (37%) |
| Current smoking, n (%) | 38,397 (10%) | 19,472 (8%) | 16,359 (12%) | 2,566 (20%) |
| Previous smoking, n (%) | 136,282 (35%) | 80,199 (35%) | 51,400 (36%) | 4,683 (37%) |
| CAD, n (%) | 34,829 (9%) | 18,145 (8%) | 14,505 (10%) | 2,179 (17%) |
| AF, n (%) | 20,278 (5%) | 10,299 (4%) | 8,577 (6%) | 1,402 (11%) |
T2D Type 2 diabetes; CAD Coronary artery disease; AF Atrial fibrillation
GWAS
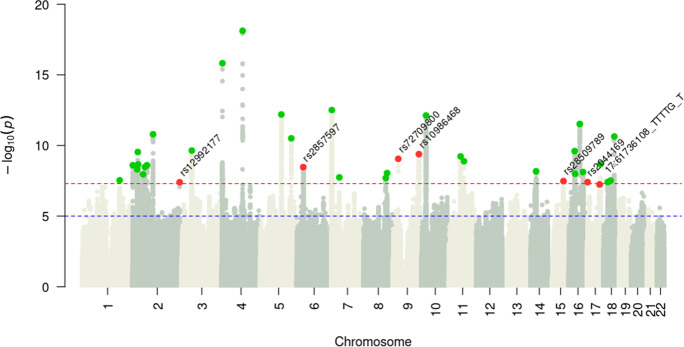
In the GWAS analysis (Fig. 1), a total of 8,883,488 SNPs with a MAF greater than 0.01 were examined. The estimated SNP heritability (h2) for FFS based on these SNPs in our data was 0.062 ± 0.003. LDSC produced a genomic inflation factor (λGC) estimate of 1.37 with an intercept of 1.03 0.01 prior to inflation correction (Supp. Figure 2), suggesting that the inflation was probably due to polygenic signals and unlikely to be confounded by population structure [27]. Using COJO as our primary method, we identified 37 independent novel risk loci for FFS (Supp. Table 1). We obtained similar results when utilizing a clumping approach in PLINK (Supp. Table 2). Of these 37 novel loci for FFS, 30 have been previously reported to be associated with other traits (Supp. Table 3) and 7 have not been associated with any other traits (Supp. Figure 3). Among these 7 loci without previously described associations, the top associated variants within each locus included two intergenic variants (rs12992177 [AC017104.2, AC017104.4] and rs72709800 [RN7SKP120, TUSC1], four in protein-coding genes, including one splicing variant (rs10986468 in ARPC5L), one exonic variant (rs28509789 in C15orf39), and two intronic variants (rs2044169 in METTL16 and 17:61,736,108:TTTTG:T in MAP3K3); and the remaining variant rs2857597 located 500b downstream from AIF1.
Fig. 1.
Manhattan plot of the frailty GWAS in the UK Biobank. Manhattan plot for the discovery GWAS of frailty (n = 386,565). Green dots indicate independent significant leading SNPs that have been reported to be associated with traits other than frailty; Red dots indicate independent significant leading SNPs that have never been reported before
Replication
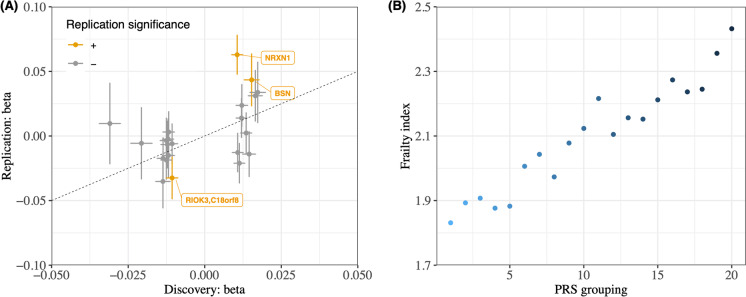
In the replication phase, we analyzed 9,720 individuals from the HRS cohort. Of the 37 lead SNPs, 22 were eligible or well proxied by an eligible SNP in the HRS cohort after genetic QC. Of the 22 eligible SNPs, 18 were found to have consistent direction of effect in both UKB and HRS datasets, resulting in a highly significant P value of 0.00042 in the binomial sign test. Notably, three of these 18 SNPs retained significance in their individual associations with frailty when corrected for a 5% false discovery rate in the HRS (Fig. 2a). Taking the overlap between UKB genetic data and quality controlled HRS genetic data, we built a PRS based on 890,487 SNPs for 9,720 HRS individuals. After adjusting for age, sex and the first 10 PCs, the PRS was significantly associated with the frailty phenotype (OR per SD 1.27, 95% CI 1.22–1.31, p = 1.3 × 10–11). The prediction performance of raw PRS was moderate with AUC being 0.57 (95% CI 0.56–0.59). When combined with age, sex and first 10 PCs, the prediction of frailty PRS achieved an AUC of 0.72 (95% CI 0.70–0.73) (Fig. 2b). The improvement in AUC when including the 10 PCs suggested that there may be sub-populations within the European British population that are captured by the PCs.
Fig. 2.
Replication in the Health and Retirement Study. (a) We plotted the effect sizes of the SNPs identified in the discovery GWAS (UKBB) against their effect sizes in the replication GWAS (HRS). The points represent significant leading SNPs identified in the UKBB. The x-axis and y-axis depict the effect sizes of the SNPs in the UKBB and HRS, respectively. The colored points indicate SNPs that retained significance in the HRS. Specifically, out of the 22 SNPs in the plot, 18 had consistent directions of effects in both studies and three remained significant in HRS. (b) We visualized the association between the frailty phenotype and the frailty PRS in HRS by plotting the mean FFS in 20 groups, which were binned according to the percentile of the frailty PRS in HRS. The PRS was generated using the UKBB data. The plot shows a significant association between the frailty PRS and the frailty phenotype in the HRS cohort, further supporting that the genetic findings from our discovery GWAS are replicable
Gene set enrichment/pathway analysis
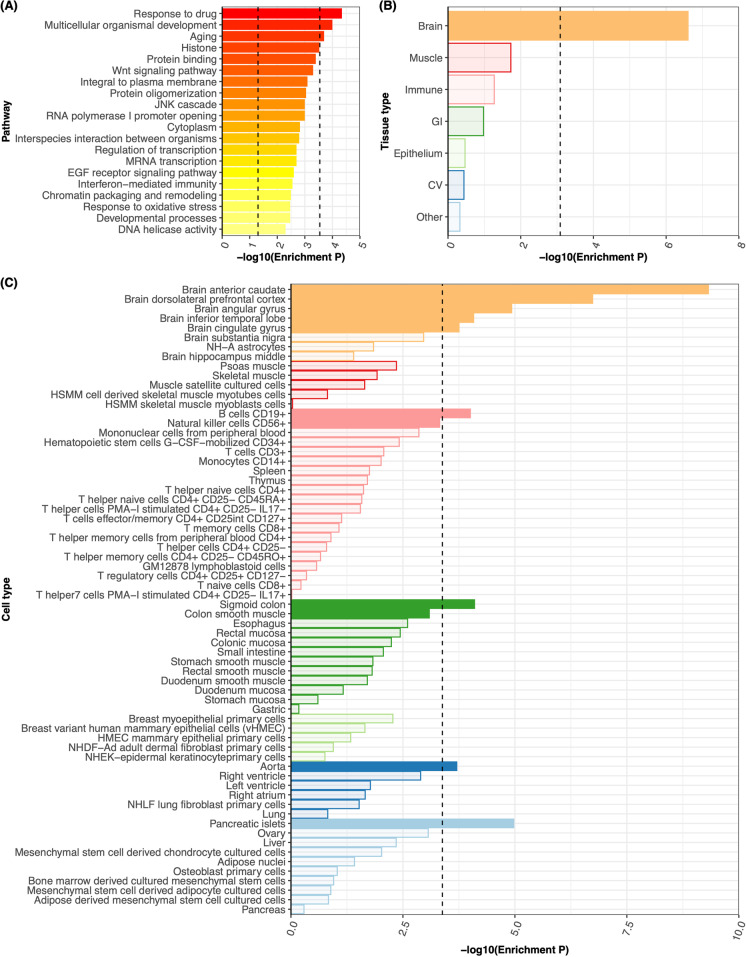
176 gene sets were marginally significantly (P < 0.05) associated with the FFS using MAGENTA (Supp. Table 4), three of them remained significant after multiple testing correction, including response to drugs, multicellular organismal development, and aging. (Fig. 3a).
Fig. 3.
Pathway and tissue enrichment. (a) The top 20 pathways associated with frailty were identified using pathway analysis based on our discovery GWAS. The left vertical dotted line represents a P-value of 0.05 for suggestive enrichment and the right vertical dotted line represents the Bonferroni-adjusted P-value for significant enrichment. We found that three pathways were significantly associated with frailty after Bonferroni correction. (b) and (c) show the results from tissue enrichment analysis based on LDSC. Specifically, (b) illustrates the enrichment results across 7 tissues, including brain, muscle, immune, gastrointestinal (GI), epithelium, cardiovascular (CV), and other tissues as annotated using GenoSkyline-Plus annotations. (c) provides a closer look at the stratified enrichment by sub-tissues or cell-types. In both panels (b) and (c), the vertical dotted line represents the Bonferroni-adjusted P-value for significant enrichment. The analysis revealed that brain tissues were significantly associated with frailty
Tissue and cell-type enrichment
We used both LDSC and MAGMA to test whether genetic associations with FFS were enriched in human tissues and cell-types. Both LDSC and MAGMA indicated that brain tissues were significantly associated with frailty (Fig. 3b, Supp. Figure 4A, Supp. Table 5-8). Specifically, in the LDSC analysis, anterior caudate, dorsolateral prefrontal cortex, angular gyrus, inferior temporal lobe and cingulate gyrus in brain tissue were shown to be significantly enriched with frailty GWAS signals (Fig. 3c, Supp. Table 6). In the MAGMA analysis, frailty genetic associations were significantly enriched in cerebellar hemisphere, frontal cortex BA9, cerebellum, anterior cingulate cortex BA24 and the nucleus accumbens within the basal ganglia (Supp. Figure 4B, Supp. Table 8).
Genetic correlation
Of the 2,086 traits tested for genetic correlation with FFS, 591 had significant genetic correlations after Bonferroni correction (Supp. Table 9). The most significant of these were multisite chronic pain (genetic correlation (rg) = 1.079, P = 3.95 × 10–264), BMI (rg = 0.714, P = 3.01 × 10–196) and depressive symptoms (rg = 1.116, P = 3.23 × 10–189). Among the other most interesting statistically significant genetic correlations were education attainment (rg = -0.601, P = 5.36 × 10–168), age of first birth (rg = -0.692, P = 3.60 × 10–95), insomnia (rg = 0.824, P = 7.82 × 10–95), Attention-Deficit/Hyperactivity Disorder (ADHD, rg = 0.535, P = 6.05 × 10–94), type 2 diabetes (rg = 0.416, P = 4.16 × 10–72), smoking behavior (rg = 0.418, P = 1.14 × 10–71), neuroticism (rg = 0.525, P = 4.52 × 10–70), cognitive performance (rg = -0.347, P = 5.40 × 10–64), neurological diseases (0.484, P = 7.56 × 10–57), and hypertensive diseases (0.376, P = 1.46 × 10–56). Of the four types of hearing loss tested, both low frequency and low-to-medium frequency hearing loss were marginally significantly genetically correlated with FFS, while there was no evidence suggesting genetic correlation between high frequency hearing loss and FFS (Table 2). And possibly due to the small sample size of the available hearing loss GWAS, these genetic correlations do not pass the multiple testing correlation.
Table 2.
Genetic correlation between frailty and hearing loss
| Hearing loss type | Correlation coefficient | Standard error | P |
|---|---|---|---|
| HML: HIGH – LOW | 0.0031 | 0.005 | 0.53 |
| HIGH: 4&8 kHz | 0.0095 | 0.005 | 0.052 |
| LOW: 0.5, 1, 2 kHz | 0.0111 | 0.005 | 0.025 |
| WHO: 0.5, 1, 2, 4 kHz | 0.012 | 0.005 | 0.010 |
We highlight the P values passing significant threshold using bold formats
HML Patients with high frequency hearing loss without low frequency hearing loss; HIGH Patients with high frequency hearing loss; LOW Patients with low frequency hearing loss; WHO Patients with low-to-median hearing loss
Mendelian randomization
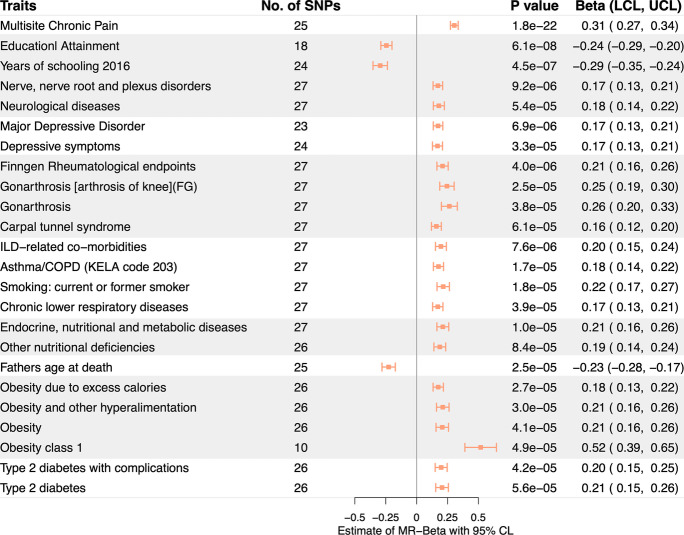
Of the 591 traits with significant genetic correlation with FFS, 8 could not be evaluated using MR analyses due to the lack of at least 10 valid instruments in the corresponding GWAS. Of the 583 traits evaluated via IVW-MR, 24 were causally associated with FFS after Bonferroni correction, including multisite chronic pain, education-related traits, joint disorders (e.g. rheumatism and carpal tunnel syndrome), depression (e.g. major depressive disorders, depressive symptom), interstitial lung disease related comorbidities, neurological diseases (e.g., nerve/nerve root/plexus disorders), endocrine/nutritional/metabolic diseases, respiratory diseases (e.g. asthma/chronic obstructive pulmonary disease, smoking: current or former smoker), father’s age at death, obesity, and diabetes. (Fig. 4, Supp. Table 10). 17 of these causal associations remained significant when evaluated using the MR PRESSO. (Supp. Table 11).
Fig. 4.
Mendelian randomization analysis estimates. Effect sizes (beta) and 95% confidence intervals of the associations between FFS and significantly causally associated traits are provided (based on MR-IVW method)
Discussion
We report the largest-to-date GWAS of frailty, an important phenotype in aging research. Our study included over 380,000 individuals from two cohort studies and identified 37 novel susceptibility risk loci for frailty. Thirty of these newly found loci have been previously reported in GWASs of other traits, including obesity/BMI, lipids, coronary artery disease, hypertension, diabetes, and cancer. These results provide important confirmatory evidence for the associations of these traits with frailty found in observational and clinical studies [45–47]. Importantly, genetic correlation and MR analyses also revealed significant associations for pain, neuropsychiatric, cardiovascular, and inflammation-related traits, adding important evidence to support a causal link between these pathways and frailty.
We provide important new evidence related to the genetic underpinnings of frailty, an important phenotype in aging research that has been minimally investigated from a genomics perspective. Existing candidate-gene studies represented important first steps to identify genetic risk factors for frailty but are limited in scope, tend to yield inconsistent results, and many reported signals failed to reach statistical significance [12, 48]. The first GWAS study on frailty, conducted by Mekli et al. [14], was hampered by its relatively limited sample sizes (n = 8532) and the leading SNPs identified in their study were not replicable in the current study. Nevertheless, it is noteworthy that one of the two SNPs identified by Mekli et al. is related to brain development, synaptic plasticity, and cognition, which is consistent with our findings. A more recent GWAS conducted by Atkins et al. is more comparable to the current study [17], as they also used the UKBB as the discovery dataset. However, their study was restricted to older individuals, which resulted in a smaller sample size (n = 164,610) compared to this study. Additionally, the study by Atkins et al. used the FI instead of the FFS to define the frailty phenotype. The FI is a count of health-related problems, which is distinct from the physical frailty definition used in the current study by FFS. As a result of these differences, most of the loci identified by Atkins et al. were not present in our study. However, there was one notable exception: both studies identified a genetic locus located in the gene HTT, which has previously been linked to factors such as waist-hip ratio, LDL levels, and longevity [17].
Many novel loci with no reported associations are located at or close to protein-coding genes, pointing to novel pathophysiological pathways for frailty. rs10986469 is a splice site variant within ARPC5L, the gene that codes for Actin Related Protein 2/3 Complex Subunit 5 Like, a protein that regulates actin polymerization and branched actin networks. Similarly, rs2857597 is 500b downstream of AIF1, which encodes an actin-binding protein. MAP3K3, where the intronic variant 17:61,736,108:TTTTG:T resides, is a component of protein kinase signal transduction cascades, mediating activation of NF-kappa-B, AP1, and DDIT3 transcription regulators. METTL16, containing the intronic rs2044169, encodes a methyltransferase involved in RNA modification and gene expression. These genes have been linked to cancer and inflammation pathways [49], two groups of diseases with strong associations reported in observational studies [50].
Important to note is the relationship of frailty with the central nervous system, observed when using different, albeit complementary, analytical tools. Our tissue and cell-type enrichment analyses revealed that the genomic regions where frailty-associated SNPs are mapped, as determined by LD, are more likely to exhibit higher functional expression in brain tissues compared to other tissues. In addition, Genetic correlation analyses indicated a significant correlation between frailty variants and both neurological (especially cognitive decline) and psychiatric (depression, insomnia, ADHD, and neuroticism) diseases, with Mendelian randomization further implying the potential causality. These genetic findings are aligned with the results of existing cognitive and neuroanatomical studies showing that frailty is an early predictor of dementia and Alzheimer disease and is associated with reduced total brain volume, reduced gray matter volume, and increased cortical brain infarcts. [51] Our results provide important new evidence supporting these connections between frailty and disease involving the central nervous system. The specific underlying mechanisms are not yet fully understood, but previous research has identified shared biological pathways between frailty and cognitive impairment [52]. A more recent study found that the association between frailty and cognitive function persisted even after adjusting for potential confounders, suggesting that other mechanisms may be involved [53]. Further research is needed to fully understand the underlying mechanisms.
Aging research is increasingly interested in identifying biological pathways and therapeutic targets involved in resilience rather than risk of disease [54]. This notion is particularly important for traits like frailty, that capture a person’s general functional status and likely lie downstream in the causal pathway of several different specific diseases. Our study provides important novel results on this front, including the identification of genetic loci for frailty known to associate with intelligence as well as protective causal associations with years of schooling and educational attainment using Mendelian Randomization analyses. These causal associations are particularly important, as prior observational studies could not differentiate whether educational phenotypes constitute a true causal protective factor or a proxy for better health and socioeconomic status. Akin to cognitive reserve as a protective factor for dementia, these findings support the concept of “functional reserve” as a protective factor against frailty: it is possible that prolonged exposures to certain beneficial exposures, in this case educational activities, will improve a person’s overall physical status resulting in a delay or avoidance of frailty.
Our study features many strengths, which include a large sample size of middle- and older-aged participants in our discovery and replication cohorts. We utilize the FFS, a physical model of frailty that can be easily assessed in the clinic, has been validated across multiple studies and populations [55], but has not yet been utilized to define frailty in published GWAS. A limitation of our study is the inclusion of only European-ancestry individuals. Participants of the UKB are also healthier and less socioeconomically deprived than the average population and thus do not reflect frail individuals who tend to be of low socioeconomic status and/or ethnic minorities [56]. In addition, many SNPs significantly associated with frailty in the UKB were not able to be replicated in HRS. This discrepancy may be due to certain leading SNPs being removed during quality control, or the proxy SNPs used in HRS not having sufficient power to reach statistical significance. Also, it is important to note that much of the variations in frailty are non-genetic, and various population-specific factors—both genetic and non-genetic—can affect the outcome. The small sample size of HRS also resulted in a heritability estimate with large uncertainty in the replication GWAS. Despite these limitations, three significant loci were replicated, and a PRS of all associated loci demonstrated good prediction of frailty status in HRS. While we have identified many possible causal relations between frailty and common diseases/traits using multiple MR methods, it is important to note that MR is an epidemiological tool and further experimental validation is necessary to confirm these findings. Contributions to frailty from epigenetics or rare variants (MAF < 0.01) should also be considered in future studies.
In conclusion, the genetic architecture of frailty is complex. Identification of individuals with a genetic predisposition to frailty provides an opportunity for the prevention and screening of frailty earlier in life and can inform clinical strategies in a myriad of specialties, from cardiology to oncology, to identify at-risk populations. Increasing efforts to address diabetes, obesity, and blood pressure, preventable and modifiable diseases, in midlife and later life may reduce the burden of frailty in our aging society.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We sincerely thank many GWAS consortia for making their GWAS summary data publicly accessible. We conducted the research using the UK Biobank resource under an approved data request (ref: 34763) and the HRS resource approved via dbGap.
Funding
GJF is supported by the National Institutes of Health (K76AG059992, R03NS112859 and P30AG021342), the American Heart Association (18IDDG34280056 and 817874) and the Neurocritical Care Society Research Fellowship. YY and HZ were supported in part by the National Institutes of Health (R01 GM134005) and National Science Foundation (DMS 1902903).
Data availability
The UKBB data are available through the UK Biobank Access Management System. The HRS data are accessible on dbGap with accession number phs000428.v2.p2. A reporting summary for this article is available in the supplementary tables. The full GWAS summary statistics produced by this study are freely available on figshare (https://figshare.com/s/6683396c68807fe4e729).
Declarations
Web resources
Finngen, https://www.finngen.fi/en/access_results
FUMA, https://fuma.ctglab.nl
GNOVA, https://github.com/xtonyjiang/GNOVA
HRS, http://www.nia.nih.gov/research/resource/health-and-retirement-study-hrs
LDhub, http://ldsc.broadinstitute.org/ldhub/
LDSC, https://github.com/bulik/ldsc
MAGENTA, https://software.broadinstitute.org/mpg/magenta/
PLINK, https://www.cog-genomics.org/plink/
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Hongyu Zhao and Guido J. Falcone jointly supervised this work
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yixuan Ye and Rommell B. Noche contributed equally to this work.
Contributor Information
Hongyu Zhao, Email: hongyu.zhao@yale.edu.
Guido J. Falcone, Email: guido.falcone@yale.edu
References
- 1.Cesari M, Calvani R, Marzetti E. Frailty in older persons. Clin Geriatr Med. 2017;33:293–303. doi: 10.1016/j.cger.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Mondor L, Maxwell CJ, Hogan DB, Bronskill SE, Campitelli MA, Seitz DP, Wodchis WP. The incremental health care costs of frailty among home care recipients with and without dementia in Ontario, Canada: a cohort study. Med Care. 2019;57:512–520. doi: 10.1097/MLR.0000000000001139. [DOI] [PubMed] [Google Scholar]
- 3.Clegg A, Hassan-Smith Z. Frailty and the endocrine system. Lancet Diabetes Endocrinol. 2018;6(9):743–752. doi: 10.1016/S2213-8587(18)30110-4. [DOI] [PubMed] [Google Scholar]
- 4.Soysal P, Stubbs B, Lucato P, Luchini C, Solmi M, Peluso R, Sergi G, Isik AT, Manzato E, Maggi S, et al. Inflammation and frailty in the elderly: a systematic review and meta-analysis. Ageing Res Rev. 2016;31:1–8. doi: 10.1016/j.arr.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Soysal P, Isik AT, Carvalho AF, Fernandes BS, Solmi M, Schofield P, Veronese N, Stubbs B. Oxidative stress and frailty: a systematic review and synthesis of the best evidence. Maturitas. 2017;99:66–72. doi: 10.1016/j.maturitas.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Kameda M, Teruya T, Yanagida M, Kondoh H. Frailty markers comprise blood metabolites involved in antioxidation, cognition, and mobility. Proc Natl Acad Sci U S A. 2020;117:9483–9489. doi: 10.1073/pnas.1920795117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Hao Q, Flaherty JH, Cao L, Zhou J, Su L, Shen Y, Dong B. Comparison of procalcitonin, a potentially new inflammatory biomarker of frailty, to interleukin-6 and C-reactive protein among older Chinese hospitalized patients. Aging Clin Exp Res. 2018;30:1459–1464. doi: 10.1007/s40520-018-0964-3. [DOI] [PubMed] [Google Scholar]
- 8.Samson LD, Boots AMH, Verschuren WMM, Picavet HSJ, Engelfriet P, Buisman AM. Frailty is associated with elevated CRP trajectories and higher numbers of neutrophils and monocytes. Exp Gerontol. 2019;125:110674. [DOI] [PubMed]
- 9.Zaslavsky O, Walker RL, Crane PK, Gray SL, Larson EB. Glucose levels and risk of frailty. J Gerontol A Biol Sci Med Sci. 2016;71(9):1223–1229. doi: 10.1093/gerona/glw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parvaneh S, Howe CL, Toosizadeh N, Honarvar B, Slepian MJ, Fain M, Mohler J, Najafi B. Regulation of cardiac autonomic nervous system control across frailty status: a systematic review. Gerontology. 2015;62:3. doi: 10.1159/000431285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abadir PM. The frail renin-angiotensin system. Clin Geriatr Med. 2011;27:53. doi: 10.1016/j.cger.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathyan S, Verghese J. Genetics of frailty: a longevity perspective. Transl Res. 2020;221:83–96. doi: 10.1016/j.trsl.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sathyan S, Barzilai N, Atzmon G, Milman S, Ayers E, Verghese J. Genetic insights into frailty: association of 9p21-23 locus with frailty. Front Med. 2018;5:105. doi: 10.3389/fmed.2018.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mekli K, Stevens A, Marshall AD, Arpawong TE, Phillips DF, Tampubolon G, Lee J, Prescott CA, Nazroo JY, Pendleton N. Frailty Index associates with GRIN2B in two representative samples from the United States and the United Kingdom. PLoS One. 2018;13(11):e0207824. [DOI] [PMC free article] [PubMed]
- 15.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Collins R. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher GG, Ryan LH. Overview of the Health and Retirement Study and introduction to the Special Issue. Work Aging Retire. 2018;4:1–9. doi: 10.1093/workar/wax032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atkins JL, Jylhävä J, Pedersen NL, Magnusson PK, Lu Y, Wang Y, Hägg S, Melzer D, Williams DM, Pilling LC. A genome-wide association study of the frailty index highlights brain pathways in ageing. Aging Cell. 2021;20:e13459. doi: 10.1111/acel.13459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fried LP, Tangen CM, Walston J, Newman AB, Hirsch C, Gottdiener J, Seeman T, Tracy R, Kop WJ, Burke G, et al. Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56(3):M146–56. [DOI] [PubMed]
- 19.Livshits G, Ni Lochlainn M, Malkin I, Bowyer R, Verdi S, Steves CJ, Williams FMK. Shared genetic influence on frailty and chronic widespread pain: a study from TwinsUK. Age Ageing. 2018;47:119–125. doi: 10.1093/ageing/afx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanlon P, Nicholl BI, Jani BD, Lee D, McQueenie R, Mair FS. Frailty and pre-frailty in middle-aged and older adults and its association with multimorbidity and mortality: a prospective analysis of 493 737 UK Biobank participants. Lancet Public Health. 2018;3:e323–e332. doi: 10.1016/S2468-2667(18)30091-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cigolle CT, Ofstedal MB, Tian Z, Blaum CS. Comparing models of frailty: the Health and Retirement Study. J Am Geriatr Soc. 2009;57:830–839. doi: 10.1111/j.1532-5415.2009.02225.x. [DOI] [PubMed] [Google Scholar]
- 23.Op het Veld LPM, van Rossum E, Kempen GIJM, de Vet HCW, Hajema K, Beurskens AJHM. Fried phenotype of frailty: cross-sectional comparison of three frailty stages on various health domains. BMC Geriatr. 2015;15:77. doi: 10.1186/s12877-015-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genomes Project Consortium. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh P-R, Anttila V, Xu H, Zang C, Farh K, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Segrè AV, DIAGRAM Consortium, MAGIC investigators. Groop L, Mootha VK, Daly MJ, Altshuler D. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Q, Hu Y, Sun J, Cheng Y, Cheung K-H, Zhao H. A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci Rep. 2015;5:10576. doi: 10.1038/srep10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Q, Powles RL, Abdallah S, Ou D, Wang Q, Hu Y, Lu Y, Liu W, Li B, Mukherjee S, et al. Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genet. 2017;13:e1006933. doi: 10.1371/journal.pgen.1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consortium, T.G., and The GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe K, Taskesen E, Bochoven Av, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1–11. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu Q, Li B, Ou D, Erlendsdottir M, Powles RL, Jiang T, Hu Y, Chang D, Jin C, Dai W, et al. A powerful approach to estimating annotation-stratified genetic covariance via GWAS summary statistics. Am J Hum Genet. 2017;101:939. doi: 10.1016/j.ajhg.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye Y, Yang H, Wang Y, Zhao H. A comprehensive genetic and epidemiological association analysis of vitamin D with common diseases/traits in the UK Biobank. Genet Epidemiol. 2021;45:24–35. doi: 10.1002/gepi.22357. [DOI] [PubMed] [Google Scholar]
- 38.Nagtegaal AP, Broer L, Zilhao NR, Jakobsdottir J, Bishop CE, Brumat M, Christiansen MW, Cocca M, Gao Y, Heard-Costa NL, et al. Genome-wide association meta-analysis identifies five novel loci for age-related hearing impairment. Sci Rep. 2019;9:15192. doi: 10.1038/s41598-019-51630-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamil RJ, Li L, Lin FR. Association between hearing impairment and frailty in older adults. J Am Geriatr Soc. 2014;62:1186–1188. doi: 10.1111/jgs.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowden J, Smith GD, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–698. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–1998. doi: 10.1093/ije/dyx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramsay SE, Arianayagam DS, Whincup PH, Lennon LT, Cryer J, Papacosta AO, Iliffe S, Wannamethee SG. Cardiovascular risk profile and frailty in a population-based study of older British men. Heart. 2015;101:616–622. doi: 10.1136/heartjnl-2014-306472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Handforth C, Clegg A, Young C, Simpkins S, Seymour MT, Selby PJ, Young J. The prevalence and outcomes of frailty in older cancer patients: a systematic review. Ann Oncol. 2015;26:1091–1101. doi: 10.1093/annonc/mdu540. [DOI] [PubMed] [Google Scholar]
- 47.Chen C, Winterstein AG, Fillingim RB, Wei Y-J. Body weight, frailty, and chronic pain in older adults: a cross-sectional study. BMC Geriatr. 2019;19:143. doi: 10.1186/s12877-019-1149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melzer D, Ferrucci L. Genetics and mechanisms of human aging and frailty. Innov Aging. 2019;3:S221–S221. [Google Scholar]
- 49.Zhang Y, Wang S-S, Tao L, Pang L-J, Zou H, Liang W-H, Liu Z, Guo S-L, Jiang J-F, Zhang W-J, et al. Overexpression of MAP3K3 promotes tumour growth through activation of the NF-κB signalling pathway in ovarian carcinoma. Sci Rep. 2019;9:8401. doi: 10.1038/s41598-019-44835-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Zhang X, Meng X, Chen Y, Leng SX, Zhang H. The biology of aging and cancer: frailty, inflammation, and immunity. Cancer J. 2017;23:201–205. doi: 10.1097/PPO.0000000000000270. [DOI] [PubMed] [Google Scholar]
- 51.Kant IMJ, de Bresser J, van Montfort SJT, Aarts E, Verlaan J-J, Zacharias N, Winterer G, Spies C, Slooter AJC, Hendrikse J, et al. The association between brain volume, cortical brain infarcts, and physical frailty. Neurobiol Aging. 2018;70:247–253. doi: 10.1016/j.neurobiolaging.2018.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sargent L, Nalls M, Starkweather A, Hobgood S, Thompson H, Amella EJ, Singleton A. Shared biological pathways for frailty and cognitive impairment: a systematic review. Ageing Res Rev. 2018;47:149–158. doi: 10.1016/j.arr.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petermann-Rocha F, Lyall DM, Gray SR, Esteban-Cornejo I, Quinn TJ, Ho FK, Pell JP, Celis-Morales C. Associations between physical frailty and dementia incidence: a prospective study from UK Biobank. Lancet Healthy Longev. 2020;1:e58–e68. doi: 10.1016/S2666-7568(20)30007-6. [DOI] [PubMed] [Google Scholar]
- 54.Fontes AP, Neri AL. Resilience in aging: literature review. Cien Saude Colet. 2015;20:1475–1495. doi: 10.1590/1413-81232015205.00502014. [DOI] [PubMed] [Google Scholar]
- 55.Afilalo J, Lauck S, Kim DH, Lefèvre T, Piazza N, Lachapelle K, Martucci G, Lamy A, Labinaz M, Peterson MD, et al. Frailty in older adults undergoing aortic valve replacement. J Am Coll Cardiol. 2017;70:689–700. doi: 10.1016/j.jacc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 56.Bandeen-Roche K, Seplaki CL, Huang J, Buta B, Kalyani RR, Varadhan R, Xue QL, Walston JD, Kasper JD. Frailty in older adults: a nationally representative profile in the United States. J Gerontol A Biol Sci Med Sci. 2015;70(11):1427–1434. doi: 10.1093/gerona/glv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The UKBB data are available through the UK Biobank Access Management System. The HRS data are accessible on dbGap with accession number phs000428.v2.p2. A reporting summary for this article is available in the supplementary tables. The full GWAS summary statistics produced by this study are freely available on figshare (https://figshare.com/s/6683396c68807fe4e729).