Abstract
Histamine production from histidine in fermented food products by lactic acid bacteria results in food spoilage and is harmful to consumers. We have isolated a histamine-producing lactic acid bacterium, Lactobacillus hilgardii strain IOEB 0006, which could retain or lose the ability to produce histamine depending on culture conditions. The hdcA gene, coding for the histidine decarboxylase of L. hilgardii IOEB 0006, was located on an 80-kb plasmid that proved to be unstable. Sequencing of the hdcA locus disclosed a four-gene cluster encoding the histidine decarboxylase, a protein of unknown function, a histidyl-tRNA synthetase, and a protein, which we named HdcP, showing similarities to integral membrane transporters driving substrate/product exchange. The gene coding for HdcP was cloned downstream of a sequence specifying a histidine tag and expressed in Lactococcus lactis. The recombinant HdcP could drive the uptake of histidine into the cell and the exchange of histidine and histamine. The combination of HdcP and the histidine decarboxylase forms a typical bacterial decarboxylation pathway that may generate metabolic energy or be involved in the acid stress response. Analyses of sequences present in databases suggest that the other two proteins have dispensable functions. These results describe for the first time the genes encoding a histamine-producing pathway and provide clues to the parsimonious distribution and the instability of histamine-producing lactic acid bacteria.
Histamine is normally present at low levels in the human body and participates in diverse key functions, including vascular permeability, neurotransmission, and the allergic response (27). The concentration of histamine in blood may increase substantially after ingestion of foods containing high doses of histamine. Substances inhibiting histamine metabolism, such as ethanol present in alcohols, may strengthen this increase. Histamine then triggers harmful effects referred to as false food allergies and characterized by headaches, itching, nausea, abdominal cramps, vomiting, or more severe symptoms (40). Although it is a natural constituent of foods such as chocolate and tomato, histamine is a contaminant that appears in many products during growth of undesirable bacteria. It is well documented that gram-negative bacteria form histamine in raw fish and meat following temperature abuse and that gram-positive bacteria cause histamine spoilage of fermented foods such as cheese, sausage, miso, soy sauce, beer, and wine (references 21 and 36 and references therein). Identification of histamine-producing (histidine decarboxylase-positive [HDC+])bacteria is difficult since they belong to diverse species and only some strains of a given species are histamine producers. HDC+ bacteria differ from non-histamine-producing (HDC−) bacteria by the presence of HDC, the enzyme that converts histidine into histamine and CO2. Two different kinds of HDCs were found in gram-negative and gram-positive bacteria. HDCs of gram-negative bacteria use pyridoxal phosphate as a cofactor for activity, whereas HDCs of gram-positive bacteria use a different catalytic mechanism based on a pyruvoyl group linked at the active site (39, 43).
The best-studied histidine decarboxylases are those from gram-positive bacteria. Six pyruvoyl-dependent HDCs were purified and characterized; these were from Clostridium perfringens strain ATCC 13124, Lactobacillus strain 30a (ATCC 33222), Lactobacillus buchneri strain ST2A, Oenococcus oeni strain IOEB 9204 (formerly Leuconostoc oenos 9204), a Micrococcus sp., and Tetragenococcus muriaticus strain JCM 10006 (7, 15, 29, 30, 31). The proteins are synthesized in an inactive form of about 310 amino acids that undergoes autoserinolysis, yielding an α chain of about 230 residues containing the pyruvoyl group at the N terminus and a β chain of about 80 residues (30). The subunits associate into active trimeric (αβ)3 or hexameric (αβ)6 complexes (14). The X-ray structure of the HDC complex of Lactobacillus strain 30a revealed that it acts as a trimer with three active sites (9, 28). Comparison of structures solved at different pH showed that the enzyme folds into the active form at acidic pH, whereas neutral and alkaline pH induce structural changes at the substrate binding site that greatly reduce activity (34). The gram-positive HDC preproteins are encoded by the hdcA genes that have been identified in Lactobacillus strain 30a (42), C. perfringens 13124 (44), O. oeni 9204 (7) and T. muriaticus (unpublished, acc no AB125629). In Lactobacillus strain 30a, the hdcA gene is cotranscribed in tandem with a downstream gene, hdcB, encoding a protein of unknown function (6).
The source of histidine used to produce histamine is believed to originate from the extracellular medium, and therefore it is anticipated that the histidine decarboxylase works in cooperation with a transporter protein driving histidine uptake and histamine extrusion (precursor/product exchange). Although the transporter protein was never identified, an exchange of histidine and histamine was detected in membrane vesicles prepared from L. buchneri ST2A (26). In whole cells of this bacterium, the coupled reactions of histidine decarboxylation and histidine/histamine exchange generated a transmembrane pH gradient (inside alkaline) and an electrical potential (inside negative), i.e., a proton motive force (PMF) (secondary metabolic energy generation [20]). Those authors suggested that the physiological function of the system might be metabolic energy generation or intracellular pH regulation, thus allowing HDC+ bacteria to better survive in poor or acidic environments.
This study was undertaken to identify the genes involved in the histamine-producing pathway of a gram-positive bacterium of wine. Screening of a collection of wine lactic acid bacteria allowed identification of a new HDC+ strain, Lactobacillus hilgardii IOEB 0006 (L. hilgardii 0006). Unexpectedly, the phenotype was lost depending on culture conditions. Loss of HDC activity corresponded to loss of a large plasmid. The hdcA locus on the plasmid was identified and shown to be part of a four-gene cluster most likely involved in the histamine-producing pathway. One of the gene products is an integral membrane protein that was shown to catalyze the exchange of histidine and histamine.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. hilgardii 0006 was from the collection of the Faculty of Enology of Bordeaux (Talence, France). It was grown at 25°C on standard 1111 medium (33), on 1111 medium supplemented with 100 mM histidine (1111-M), or on Carr-M medium, which is derived from Carr medium (22) and contains no malic acid, 0.5% glucose, 0.5% fructose, and 100 mM histidine and is adjusted to pH 3.8. Lactococcus lactis strains were grown at 30°C in half-strength M17 broth (41) containing 0.5% glucose and 5 μg of chloramphenicol per ml. Escherichia coli DH5α cells harboring the pCR-XL-TOPO vector (Invitrogen) and derivatives were grown at 37°C on Luria-Bertani medium (25) in the presence of 50 μg of kanamycin per ml.
Identification of HDC+ bacteria and stability assay.
Detection of HDC+ bacteria was performed by a colorimetric method (5). A total of 345 strains from the collection of the Faculty of Enology of Bordeaux were screened. Bacteria were inoculated in the liquid colorimetric medium and incubated at 25°C. After several days of growth, HDC+ strains induced a color change of the medium from yellow to purple. Alternatively, bacteria were plated on indicator plates based on the colorimetric medium. After 5 to 15 days of incubation at 25°C, purple or colorless bacterial colonies appeared on the plate, depending on whether they produced histamine or not, respectively.
The stability of HDC+ cells was examined in 1111-M and Carr-M broths. HDC+ bacteria were obtained from a purple colony isolated as described above. The colony was resuspended in 500 μl of sterile water, and aliquots were used to inoculate 5-ml cultures in 1111-M and Carr-M broths. When the cultures reached an optical density at 600 nm (OD600) of 0.5 to 2.0, they were diluted 1000-fold in fresh broth. The growth and dilution cycle was repeated eight times, with each cycle corresponding to approximately 10 bacterial generations. At the inoculation time and after each cycle, aliquots of the cultures were analyzed on the indicator plates to determine the proportions of HDC+ and HDC− cells remaining in the cultures.
Determination of histamine levels.
Bacteria were grown in 1111 broth to the stationary phase of growth. Cells were removed by centrifugation, and the level of histamine present in the supernatant was measured with the enzyme-linked immunosorbent assay of the Ridascreen-Histamin kit (r-Biopharm) according to the instructions of the manufacturer.
Preparation of genomic and plasmid DNAs.
High-molecular-weight genomic DNA was prepared with the Wizard genomic DNA purification kit (Promega). Plasmids of L. hilgardii 0006 were isolated by the method of Anderson and McKay (3) modified as follows. Cells of an exponentially growing culture were harvested, washed once with 500 μl of 50 mM Tris-HCl (pH 8)-50 mM NaCl-5 mM EDTA-25% sucrose, resuspended in 200 μl of the same buffer containing 40 mg of lysozyme per ml, and incubated for 1 h at 37°C. Cell lysis was initiated by adding 400 μl of a freshly prepared solution of 0.2 N NaOH-1% sodium dodecyl sulfate (SDS). After 1 h of incubation at 37°C, lysis was stopped with 300 μl of ice-cold 3 M potassium acetate (pH 4.8). Cell debris was removed by centrifugation, and plasmids in the supernatant were phenol extracted, ethanol precipitated, air dried, and dissolved in 20 μl of water. RNA contaminants were eliminated by treatment with DNase-free RNase (Promega).
Pulse-field gel electrophoresis and Southern blotting.
Bacteria grown in 1111 broth were harvested during the exponential phase of growth, washed twice with TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]), resuspended in T100E buffer (10 mM Tris-HCl, 100 mM EDTA [pH 7.5]), and embedded in 1% agarose slices. DNA was isolated by incubating the gel slices for 8 h at 37°C in T100E containing 10 mg of lysozyme per ml, followed by 16 h at 37°C in TE supplemented with 1.5% N-lauryl sarcosine and 2 mg of pronase per ml. The gel slices were subsequently transferred in T100E and stored at 4°C until used. To obtain NotI digests, the gel slices were washed four times with TE, rinsed with water, and incubated for 16 h at 25°C in 120-μl reaction mixtures containing 150 U of NotI (New England Biolabs) according to the manufacturer's instructions. Pulse-field gel electrophoresis was performed in a 1% agarose gel with the CHEF-DRIII System (Bio-Rad) with pulse times of 1 to 25 s for 20 h at 6 V/cm and 15°C in 0.5× TEB buffer (45 mM Tris-OH [pH 8], 45 mM boric acid, 1 mM EDTA). DNAs were transferred onto a Hybond-N+ membrane (Amersham Biosciences) and hybridized as described by Maniatis et al. (25). The DNA probe corresponded to a 437-bp internal region of the hdcA gene amplified by PCR with total DNA of L. hilgardii 0006 HDC+ cells and the primers hdc3 (5′-GATGGTATTGTTTCKTATGA) and hdc4 (5′-CCAAACACCAGCATCTTC), provided by E. Coton (M. Coton and E. Coton, unpublished data). The probe was labeled with digoxigenin-11-dUTP by using the DIG-DNA labeling kit (Roche), and detection was by chemiluminescence with an antidigoxigenin antibody and CDP-Star (Roche).
Cloning and expression of HdcP.
The gene encoding HdcP was amplified by PCR with total plasmid DNA of L. hilgardii 0006, an upstream primer (5′-GTCTGATCCATGGACACGGCTGAAC) designed to introduce an NcoI site (boldface) at the initiation codon, and a downstream primer (5′-GTTGCCGCGAATCTAGAATC) located 88 bp downstream of the stop codon and creating an XbaI site (boldface).The PCR product was ligated into the vector pCR-XL-TOPO (Invitrogen) and introduced into E. coli DH5α. Subsequently, the insert was recovered by digestion with NcoI and XbaI, gel purified, and cloned into the corresponding restriction sites of the vector pNZ8048 (18). The resulting plasmid, named pNZhdcP, codes for HdcP extended with a 10-histidine tag at the N terminus. The plasmid insert was sequenced (ServiceXS B.V., Leiden, The Netherlands) to ensure that no mutations occurred in the hdcP gene and that it was in frame with the 10-histidine tag. The plasmid was subsequently introduced into L. lactis strain NZ9000, which allows expression of genes under control of the tightly regulated nisA promoter (8). L. lactis cells harboring pNZhdcP or the control vector pNZ8048 were grown to an OD660 of 0.6 prior to induction by adding 0.1% of the supernatant of an overnight culture of the nisin A-producing strain L. lactis NZ9700 (17, 18). Subsequently, cells were allowed to grown for another hour and were harvested by centrifugation.
Preparation of membrane vesicles and immunoblotting.
Cells of L. lactis were resuspended in 50 mM KPi (pH 6) and disrupted in a French pressure cell operated at 20,000 lb/in2. Intact cells and debris were removed by centrifugation for 15 min at 2,910 × g, after which membranes were recovered from the supernatant by ultracentrifugation for 30 min at 288,000 × g. Membranes were resuspended in the same buffer, and the protein concentration was determined with the DC protein assay kit (Bio-Rad). Membrane proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride membrane (Roche) by semidry electroblotting. His-tagged proteins were detected with a primary monoclonal anti-His antibody (Amersham Biosciences, product number 27-4710-01) and a secondary anti-mouse antibody coupled to alkaline phosphatase (Sigma, product number A-1293), followed by chemiluminescent detection with CDP-Star (Roche).
Uptake and exchange in whole cells.
L. lactic cells were washed once with ice-cold 100 mM KPi (pH 6), and resuspended to an OD660 of 2.0. After the addition of 0.2% glucose, 100-μl samples were incubated for 5 min at 30°C with constant stirring. At time zero, l-[U-14C]histidine (308 mCi/mmol; Amersham LifeScience) was added to a final concentration of 1.55 μM. In the exchange experiments, 10 μl of a histamine solution (or buffer) was added after 30 s of uptake, yielding the indicated concentrations. Uptake was stopped by the addition of 2 ml of ice-cold 0.1 M LiCl solution, which was immediately followed by filtration through a 0.45-μm-pore-size nitrocellulose filter (BA 85; Schleicher & Schuell GmbH). The filter was washed once with 2 ml of ice-cold 0.1 M LiCl and submerged in Emulsifier Scintillator Plus scintillation fluid (Packard BioScience), and the retained radioactivity was counted in a Tri-Carb 2000CA liquid scintillation analyzer (Packard Instrumentation). The background was estimated by adding the radiolabeled substrate to the cell suspension after the addition of 2 ml of ice-cold LiCl, which was immediately followed by filtering.
Sequencing of the hdcA locus.
The sequence of the hdcA locus was determined by the linker-mediated PCR strategy with the Topo-Walker kit (Invitrogen). Plasmids purified from HDC+ cells of L. hilgardii 0006 by the procedure of Anderson and McKay (3) modified as described above were used to draw a restriction map of the hdcA locus in order to identify restriction enzyme cleavage sites located between 1 and 5 kb from the 437-bp internal fragment of the hdcA gene amplified with primers hdc3 and hdc4. Two primers located inside this fragment and directed to the upstream (uptopo, 5′-GAACAGTTCCACCAACACCAGAG) or downstream (downtopo, 5′-GTACTCAAGTATGTACGTTG) region were used to elongate 2.9- and 3.4-kb DNA fragments containing an ApaI site and a BamHI site at their extremities, respectively. The Topo linker was added at the extremities of the elongated DNAs by topoisomerase-mediated ligation, providing two DNA templates that were PCR amplified with the primer uptopo or downtopo and a primer located inside the Topo linker. The two PCR products were ligated into the vector pCR-XL-TOPO (Invitrogen), and the constructs were transformed into E. coli DH5α. Sequencing of the plasmid inserts was performed by Millegen (France).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this study have been deposited in the DDJB/EMBL/GenBank databases under accession number AY651779.
RESULTS
Instability of HDC+ phenotype of L. hilgardii 0006.
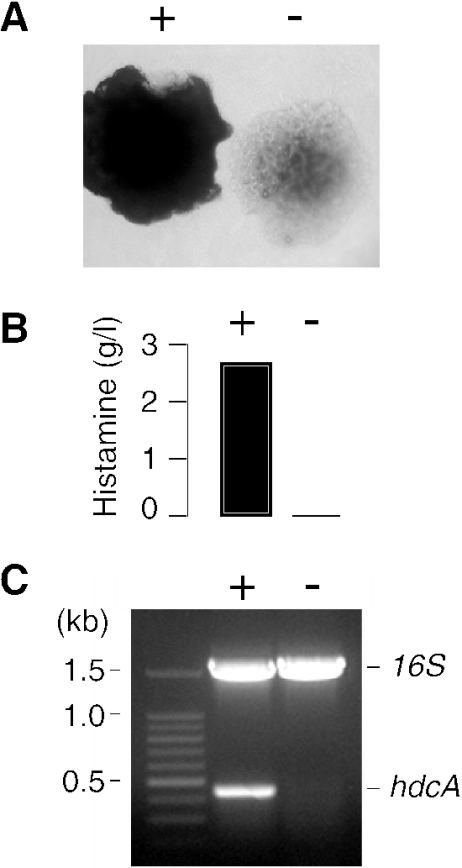
A total of 345 strains of lactic acid bacteria isolated from wines from the collection of the Faculty of Enology of Bordeaux were tested for their ability to produce histamine by the colorimetric method described by Bover-Cid and Holzapfel (5). The strains were inoculated in a medium containing histidine and a pH indicator that changes color from yellow to purple following the increase of pH associated with histamine production. Very few strains scored positive, among which was L. hilgardii 0006 (data not shown). Surprisingly, when this strain was analyzed on indicator plates based on the colorimetric medium, a mixture of positive and negative colonies appeared (Fig. 1A). Replating a positive colony yielded the same mixture of the two types of colonies, excluding the possibility of a contaminating strain and indicating that HDC+ cells generated HDC− mutants spontaneously (not shown). The levels of histamine produced in cultures inoculated with a purple colony and a colorless colony confirmed that they derived from HDC+ and HDC− cells, respectively (Fig. 1B). Moreover, a PCR performed with the primers hdc3 and hdc4, which are specific for an internal region of the hdcA gene coding for the histidine decarboxylase, showed that hdcA was present in HDC+ cells but not in HDC− cells (Fig. 1C). The results indicate that the hdcA gene is lost spontaneously during the growth of L. hilgardii 0006 on the indicator plates.
FIG. 1.
(A) Two kinds of colonies formed by L. hilgardii 0006 on histamine indicator plates. Cells producing histamine raise the pH of the surrounding medium, which turns from yellow to purple (dark colony) (+), whereas HDC− cells form a colorless colony (−). (B) The histamine produced in bacterial cultures inoculated with a purple colony (+) and a colorless colony (−) of L. hilgardii 0006 were determined after 3 days of growth. (C) PCR analysis of total DNA isolated from HDC+ and HDC− cells (lanes + and −, respectively). A multiplex PCR was performed with two pairs of primers, hdc3-hdc4 and 16Srrna1-16Srrna2, which amplify 437- and 1,534-bp internal regions of the hdcA and 16S rRNA genes, respectively.
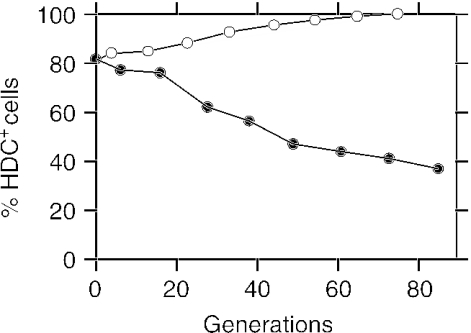
The instability of the HDC+ phenotype in L. hilgardii 0006 was surprising, as histamine-producing bacteria are frequently encountered in natural environments such as in wines. Thus, we examined whether culture conditions could cause the loss of the gene. A positive colony from the indicator plates was used to inoculate a standard rich medium (1111-M; 20% glucose, pH 5.4) and a minimal, acidic medium (Carr-M; 1% glucose, pH 3.8) that mimics the conditions in wine (22). Both media were supplemented with a large excess of histidine in order to allow the histidine decarboxylation system to function. The percentage of HDC+ cells in the two cultures was monitored for more than 80 generations (Fig. 2). At the inoculation time, 82% of the bacteria were HDC+, indicating that HDC− mutants already appeared in the colonies on the colorimetric medium. Subsequently, in the rich 1111-M medium the percentage of HDC+ bacteria decreased continuously, reaching 36% after 80 generations. In contrast, in the poor Carr-M medium the proportion of HDC+ bacteria increased constantly. After 70 generations, almost all of the bacteria present in this medium were histamine producers. These results show that the propagation or the loss of hdcA in L. hilgardii 0006 is triggered by culture conditions.
FIG. 2.
The percentage of L. hilgardii 0006 cells that retain the ability to produce histamine was examined during growth in 1111-M medium (filled circles) and in Carr-M medium (empty circles). Growth rates were approximately 6 and 14 h per generation in 1111-M and Carr-M broths, respectively.
Genetic location of hdcA.
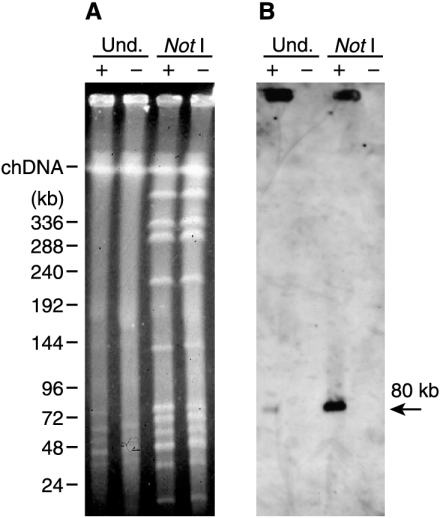
The instability of the hdcA gene suggested that it was located on a plasmid or in a mobile region of the chromosome. Analysis of L. hilgardii 0006 total DNA by standard gel electrophoresis revealed the presence of several small plasmids of 2 to 20 kb in size, but, as revealed by Southern hybridization, none of these contained hdcA (not shown). Subsequently, native and NotI-digested total DNAs of HDC+ and HDC− cells were analyzed by pulse-field gel electrophoresis (Fig. 3A). In addition to chromosomal DNA, undigested DNA revealed several large plasmids that migrate at the position of linear DNAs of 30 to 80 kb. Hybridization with a probe targeted to an internal fragment of hdcA revealed a weak signal at the level of an 80-kb molecule in the HDC+ cells but not in the HDC− cells (Fig. 3B). No hybridization was observed with the chromosomal DNA. It is concluded that hdcA is located on a plasmid, which we named pHDC. It is well documented that only the linear form of large plasmids migrates in pulse-field gels, with the circular forms being trapped in the sample wells (4). Accordingly, the strong hybridization signal detected in the wells could come from the circular forms of pHDC. During the sequencing of the hdcA locus (see below), we found that the rare-cutter enzyme NotI has a cleavage site near the hdcA gene. As seen in Fig. 3B, digestion by NotI strongly enhanced the signal seen at 80 kb. Since one form of the undigested plasmid (presumably the linear form) and the NotI-digested plasmid migrate at the same position, we concluded that pHDC is a circular plasmid of 80 kb.
FIG. 3.
(A) Pulse-field gel electrophoresis of undigested (Und.) and NotI-digested total DNAs of HDC+ and HDC− cells of L. hilgardii 0006 (+ and −, respectively). Sizes of linear DNA standards are shown on the left along with the position of native chromosomal DNA (chDNA). (B) Southern hybridization analysis of total DNA prepared for panel A, using an internal fragment of the hdcA gene as a probe.
Characterization of the hdcA locus.
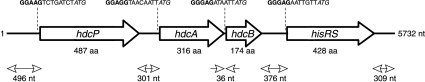
To identify the proteins involved in histamine production in L. hilgardii 0006, a sequence of 5,732 bp surrounding the hdcA gene on plasmid pHDC was determined by a linker-mediated PCR strategy, starting from an initial 437-bp internal region of hdcA obtained as described in Fig. 1C. The genetic organization of the sequence is depicted in Fig. 4. Four complete open reading frames corresponding to proteins larger than 50 amino acids were identified. There is no other potential open reading frame of significant size in the 5′ and 3′ extremities of the sequence (496 and 309 bp, respectively). The four genes are oriented in the same direction and are preceded by putative ribosome binding sites located 6 to 8 bp upstream of the start codons. The genes were named hdcP, hdcA, hdcB, and hisRS.
FIG. 4.
Genetic organization of the DNA region surrounding the hdcA gene of L. hilgardii 0006. Large arrows represent putative open reading frames. The sequences of ribosome binding sites (boldface) situated upstream of the start codons (italic) are indicated. aa, amino acid; nt, nucleotide.
The hdcA gene of L. hilgardii 0006 codes for a protein with 316 amino acids and a calculated molecular mass of 34.3 kDa. This is the fifth gene encoding an HDC of a gram-positive bacterium reported to date. A multiple-amino-acid-sequence alignment shows that the proenzymes of L. hilgardii 0006, O. oeni 9204, and T. muriaticus are remarkably similar, differing only by two or three residues. The three sequences are also close to the pro-HDC of Lactobacillus strain 30a, sharing 79% sequence identity, and are somewhat more distantly related to the C. perfringens proenzyme, with 40% sequence identity.
The protein encoded by hdcB (HdcB) consists of 174 amino acid residues with a calculated molecular mass of 19.5 kDa. The hdcB gene is homologous to hdcB genes of Lactobacillus strain 30a, O. oeni 9204, and T. muriaticus. Surprisingly, an alignment of the four HdcB protein sequences (partial sequence of 92 residues in O. oeni 9204) revealed that the L. hilgardii 0006, O. oeni 9204, and T. muriaticus proteins are 100% identical. They are more distantly related to the Lactobacillus strain 30a protein (48% identical and 83% similar).
Homology searches with BLAST (2) indicated that the protein of 428 amino acids encoded by the hisRS gene is most likely a histidyl-tRNA synthetase. The closest homologue found was HisRS of Lactobacillus plantarum, which shared 64% sequence identity. Many other HisRS proteins of gram-positive and gram-negative bacteria showed significant levels of similarity with the protein. L. hilgardii HisRS contains all of the essential amino acid residues identified in functional histidyl-tRNA synthetases (37).
The gene hdcP codes for a hydrophobic protein of 487 amino acid residues that contains the typical features of an integral membrane protein. Secondary structure prediction by the TMHMM program (16) revealed the presence of 13 transmembrane segments long enough to span the membrane in an α-helical conformation. BLAST searches showed that the protein is a member of the basic amino acid/polyamine antiporter (APA) family in the amino acid/polyamine/organocation superfamily (http://tcdb.ucsd.edu/), which contains many amino acid transporters and amino acid/amine exchangers. The highest sequence identity was observed with a protein from C. perfringens (42% sequence identity) that was annotated as an arginine/ornithine antiporter. Given that the hdcP gene is located in the vicinity of the hdcA gene, it is likely that it in fact encodes a histidine/histamine exchanger.
Functional expression of hdcP.
The hdcP gene was amplified by PCR with total DNA from L. hilgardii 0006 and ligated into the pNZ8048 vector downstream of the nisin-inducible promoter. The resulting plasmid, named pNZhdcP, codes for a recombinant HdcP containing a 10-histidine tag at the N terminus and with a calculated molecular mass of 55 kDa. The plasmid was introduced in the expression strain L. lactis NZ9000 (8, 18).
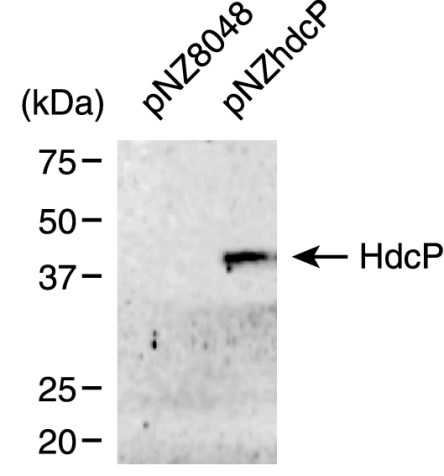
L. lactis NZ9000 cells harboring pNZhdcP or the control vector pNZ8048 were grown in the presence of nisin after which cytoplasmic membranes were isolated. Membrane proteins were separated by SDS-PAGE and detected by immunoblotting with an antibody directed against the His tag (Fig. 5). A single band corresponding to a protein with an apparent molecular mass of 40 kDa was expressed in the cells containing pNZhdcP and not in the control cells. Integral membrane proteins are known to have a higher mobility on SDS-PAGE, which explains the apparent molecular mass of 40 kDa while the calculated molecular weight is 55 kDa.
FIG. 5.
Immunoblot of membrane proteins prepared from L. lactis NZ9000 cells harboring the vector pNZ8048 (15 μg of protein) or pNZhdcP (7.5 μg of protein). The sizes of protein standards are indicated on the left.
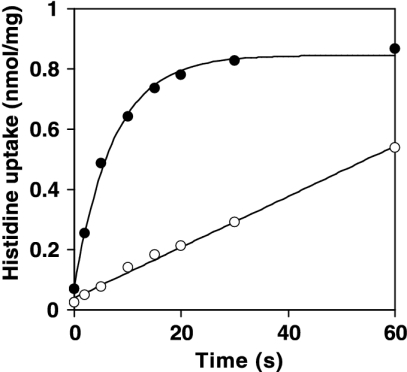
The ability of HdcP to act as a histidine transporter was examined by measuring the uptake of [U-14C]histidine in resting cells of L. lactis NZ9000 containing either pNZ8048 or pNZhdcP. At a concentration of 1.5 μM [U-14C]histidine, the control cells took up histidine at an initial rate of 0.0084 nmol/s/mg of cell protein (Fig. 6). Apparently, under the growth conditions used, L. lactis expresses an endogenous histidine transporter. However, in cells containing HdcP, the initial rate of histidine uptake increased by a factor of 12 to 0.0991 nmol/s/mg of cell protein (Fig. 6), showing that HdcP is a histidine transporter.
FIG. 6.
The uptake of histidine by resting cells of L. lactis NZ9000 harboring either pNZ8048 (open circles) or pNZhdcP (filled circles) was monitored after addition of 1.55 μM l-[U-14C]histidine to the cell suspensions.
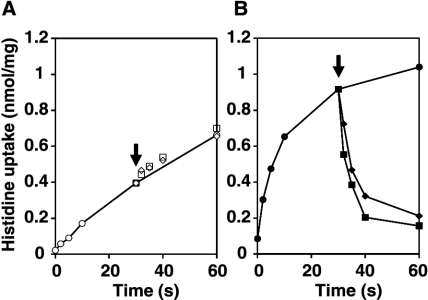
The ability of HdcP to catalyze histidine/histamine exchange was analyzed in a chase experiment (Fig. 7). Cells were allowed to take up [U-14C]histidine, after which an excess of unlabeled histamine was added to the medium. In the control cells, concentrations of 5 and 50 μM histamine (and up to 500 μM histamine [not shown]) did not affect the uptake of [U-14C]histidine, showing that the endogenous histidine transporter that is responsible for the uptake has no affinity for histamine (Fig. 7A). In contrast, addition of histamine at concentrations of as low as 5 μM to the cells expressing HdcP resulted in a rapid release of previously accumulated histidine from the cells (Fig. 7B). A concentration of 1 μM histamine already had a clear effect on the histidine uptake (not shown). HdcP catalyzes exchange of histidine and histamine.
FIG. 7.
Effect of addition of histamine during histidine uptake experiments. Uptake of histidine was monitored after addition of 1.55 μM l-[U-14C]histidine to L. lactis NZ9000 cells harboring the control vector pNZ8048 (A) or the HdcP expression vector pNZhdcP (B). After 30 s (arrow), histamine was added to a final concentration of 0 (circles), 5 (diamonds), or 50 (squares) μM.
DISCUSSION
Genetic organization of histidine decarboxylation systems.
In this study we identified the genes involved in the histidine decarboxylation system of the wine lactic acid bacterium L. hilgardii 0006, which is responsible for the production of the biogenic amine histamine. The genes hdcP, hdcA, and hisRS code for a histidine/histamine exchanger, a histidine decarboxylase, and a histidyl-tRNA synthetase, respectively, while the function of the hdcB product is unknown. Nevertheless, the involvement of the hdcB product in the histidine decarboxylation system is strongly suggested by (i) the position of hdcB close to the hdcA gene, (ii) the conservation of the gene in HDC+ strains (see below), and (iii) a previous study that showed that hdcA and hdcB are cotranscribed in Lactobacillus strain 30a (6). While the hdcA and hdcB genes are separated by only 33 nucleotides, the hdcP gene upstream of hdcA and the hisRS gene downstream of hdcB are separated by larger intergenic regions of 301 and 376 bp, respectively (Fig. 8). Given that the hdcB-hisRS intergenic region can fold into the terminator-antiterminator structure characteristic of the RNA leader regions of aminoacyl-tRNA synthetase genes (not shown), the hisRS gene is most likely transcribed from its own promoter.
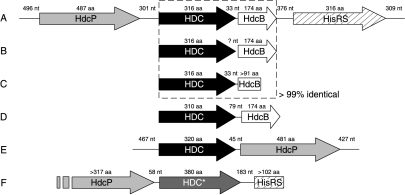
FIG. 8.
Genetic organizations of bacterial histidine decarboxylase loci. Shading scheme: black, HDC (pyruvoyl dependent); dark gray, HDC* (pyridoxal phosphate dependent); white, HdcB (protein of unknown function); light gray, HdcP (histidine/histamine exchanger); hatched, HisRS (histidyl-tRNA synthetase). The dashed box indicates a set of genes that are more than 99% identical. nt, nucleotides; aa, amino acids. (A) L. hilgardii 0006 (accession no. AY651779); (B) T. muriaticus (accession no. AB125629 and AB125630); (C) O. oeni 9204 (accession no. U58865); (D) Lactobacillus strain 30a (accession no. J02613); (E) C. perfringens strain 13 (accession no. NC_003366); (F) P. phosphoreum strain RHE01 (accession no. AY223843).
The hdcA-hdcB gene pair is also found in Lactobacillus strain 30a, T. muriaticus, and O. oeni 9204, suggesting that the hdcP-hdcA-hdcB-hisRS gene cluster may also be present in these organisms. Remarkably, the sequences of the hdcA and hdcB genes are almost identical in L. hilgardii 0006, T. muriaticus, and O. oeni 9204, suggesting that the distribution of the genes occurred only recently (Fig. 8). A different genetic organization is observed on the genome of the histamine producer C. perfringens strain 13 (35). The hdcA gene and a homologue of hdcP are arranged in the reverse order, and no homologues of hdcB and hisRS are present. The lack of the latter two indicates that they are not essential to the histidine decarboxylation pathway and are likely to code for accessory functions. It may also be interesting that the genes are encoded on different genetic elements: a plasmid in L. hilgardii 0006 and the chromosome in C. perfringens.
The hdcA gene products belong to the pyruvoyl-dependent HDCs of gram-positive bacteria. Gram-negative bacteria use a different type of HDC that is pyridoxal phosphate dependent (39, 43). In Photobacterium phosphoreum, a known gram-negative histamine producer found in spoiled fish (13), the HDC-encoding gene is preceded by a sequence coding for a 187-amino-acid gene product annotated as a putative amino acid permease. Closer inspection of the upstream region revealed an open reading frame coding for a 317-residue protein when the stop codon was ignored. The translated protein was homologous to several precursor/product exchangers (not shown). Furthermore, 183 nucleotides downstream of the stop codon of the HDC-encoding gene, a sequence encoding the first 102 amino acids of a protein clearly identified as a histidyl-tRNA synthetase was found (Fig. 8). It follows that in this gram-negative organism that uses a different HDC, the genetic organization of the histidine decarboxylation pathway is similar to the one observed in gram-positive organisms.
The histidine decarboxylation pathway.
The combination of a histidine/histamine exchanger and a histidine decarboxylase forms a typical decarboxylation pathway in bacteria. The transporter transports the substrate (precursor) from the medium into the cell, where it is decarboxylated by the decarboxylase, followed by excretion of the product out of the cell by the same transporter. Importantly, uptake of the substrate and excretion of the product are coupled events (precursor/product exchange). Similar pathways have been described for a number of other amino acids and for di- and tricarboxylates (20). The pathways generate a PMF by an indirect mechanism that was termed secondary PMF generation. The two components of the PMF are generated in separate steps. The membrane potential is generated by the transporter because of a charge difference between precursor and product, i.e., monovalent histidine and divalent histamine. The pH gradient is generated through scalar proton consumption in the decarboxylation reaction catalyzed by the decarboxylase. PMF generation by histidine decarboxylation and electrogenic histidine/histamine exchange was demonstrated in L. buchneri (26). Generation of metabolic energy may be the primary function of the pathways, but alternatively, the pathways may be involved in the acid stress response (24).
The hdcP gene of L. hilgardii 0006 is the first gene that was experimentally demonstrated to code for a histidine/histamine exchanger (Fig. 6 and 7). HdcP belongs to the basic APA family in the amino acid/polyamine/organocation superfamily (http://tcdb.ucsd.edu/tcdb/). The translated protein contains most of the proposed signature sequence for the APA family (12). The APA family contains a number of other experimentally verified precursor/product exchangers, such as the arginine/ornithine exchanger ArcD of Pseudomonas aeruginosa (45) and the putrescine/ornithine antiporter PotE (14), the arginine/agmatine antiporter AdiC (11), and the cadaverine/lysine exchanger CadB (38), all from E. coli. The highest sequence identity was observed with a protein of C. perfringens strain 13 whose genome was sequenced recently (42% identity) (35). The protein was annotated as an arginine/ornithine antiporter. However the gene encoding this protein is located only 46 bp upstream of a gene encoding a histidine decarboxylase (discussed above), strongly suggesting that, in fact, it is a histidine/histamine antiporter. At present, we are addressing this issue experimentally.
The histidyl-tRNA synthetase (HisRS) coded by the hisRS gene does not seem to have a direct role in the histidine decarboxylation pathway. Although the presence of this enzyme was unexpected, it is consistent with a recent result showing that a tyrosyl-tRNA synthetase is part of the tyrosine decarboxylation system of gram-positive bacteria, which results in the production of the biogenic amine tyramine (23). Expression of HisRS may be essential to provide additional capacity to synthesize hystidyl-tRNA, which is necessary for protein synthesis under conditions when the histidine decarboxylase depletes the internal histidine pool.
The unstable plasmid pHDC.
Plasmids ranging in size from 3 to 200 kb are common in lactic acid bacteria. L. hilgardii 0006 contains several small plasmids and at least four that exceed 30 kb (Fig. 2A). Southern hybridizations showed that the histidine decarboxylation system is encoded on one of these plasmids, which was named pHDC and estimated to have a size of approximately 80 kb. Known bacterial amino acid decarboxylation systems are found on the chromosome, with the exception of an aspartate decarboxylase operon detected on a 25-kb plasmid in the lactic acid bacterium Tetragenococcus halophila (1). A plasmidic location may explain why HDC+ bacteria are parsimoniously distributed. Until now, very few strains of HDC+ lactic acid bacteria were identified in the genera Lactobacillus, Oenococcus, Tetragenococcus, Pediococcus, and Leuconostoc (19). Given that lactobacilli could transfer a conjugative plasmid to bacteria of the same or different genera (10), a plasmid-encoded histidine decarboxylation system could be transferred horizontally, which would be in agreement with the 99 to 100% identical hdcA- and hdcB-encoded proteins of L. hilgardii 0006, T. muriaticus, and O. oeni 9204 (Fig. 8). Such a degree of identity strongly suggests that the genes were recently transferred in the three bacteria. It is very likely that T. muriaticus and O. oeni 9204 in fact harbor the same 80-kb plasmid as found in L. hilgardii 0006.
The instability of HDC+ cells of L. hilgardii 0006 is easily explained by the loss of pHDC, which depends greatly on bacterial culture conditions (Fig. 2). The increase of the population of HDC+ cells in a poor and acidic medium may be attributed to a growth advantage of HDC+ cells, given that histidine decarboxylation and the exchange of histidine and histamine could provide metabolic energy and help the organism to better survive in acidic environments (see above). The advantage would be lost in rich medium with a mild pH, leading to HDC− cells in which pHDC presumably is not transmitted during cell division. Interestingly, HDC− mutants of Lactobacillus strain 30a were previously obtained by chemical mutagenesis (32). Those authors compared the properties of wild-type and mutant cells and showed that a low extracellular pH limited the growth of HDC− mutants, while HDC+ cells grow well. Moreover, it was reported that HDC+ cells of O. oeni isolated from wine rapidly lost the capacity to form histamine when they were grown in a synthetic medium (22). This finding strengthens not only the importance of growth conditions but also the hypothesis that O. oeni carries the same 80-kb plasmid as L. hilgardii 0006.
Acknowledgments
This work was supported by the European Commission, contract number QLK1-CT-2002-02388.
We thank E. Coton (Adria Normandie, Villers-Bocage, France) for providing the sequences of universal PCR primers specific for the hdcA gene.
REFERENCES
- 1.Abe, K., F. Ohnishi, K. Yagi, T. Nakajima, T. Higuchi, M. Sano, M. Machida, R. I. Sarker, and P. C. Maloney. 2002. Plasmid-encoded asp operon confers a proton motive metabolic cycle catalyzed by an aspartate-alanine exchange reaction. J. Bacteriol. 184:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton, B. M., Harding, G. P., and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226:235-240. [DOI] [PubMed] [Google Scholar]
- 5.Bover-Cid, S., and W. H. Holzapfel. 1999. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 53:33-41. [DOI] [PubMed] [Google Scholar]
- 6.Copeland, W. C., J. D. Domena, and J. D. Robertus. 1989. The molecular cloning, sequence and expression of the hdcB gene from Lactobacillus 30a. Gene 85:259-265. [DOI] [PubMed] [Google Scholar]
- 7.Coton, E., G. C. Rollan, and A. Lonvaud-Funel. 1998. Histidine carboxylase of Leuconostoc oenos 9204: purification, kinetic properties, cloning and nucleotide sequence of the hdc gene. J. Appl. Microbiol. 84:143-151. [DOI] [PubMed] [Google Scholar]
- 8.de Ruyter, P. G., O. P. Kuipers, and W. M. De Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher, T., D. Rozwarski, S. R. Ernst, and M. L. Hackert. 1993. Refined structure of pyruvoyl-dependent histidine decarboxylase from Lactobacillus 30a. J. Mol. Biol. 230:516-528. [DOI] [PubMed] [Google Scholar]
- 10.Gevers, D., G. Huys, and J. Swings. 2003. In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other Gram-positive bacteria. FEMS Microbiol. Lett. 225:125-130. [DOI] [PubMed] [Google Scholar]
- 11.Gong, S., H. Richard, and J. W. Foster. 2003. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jack, D. L., I. T. Paulsen, and M. H. Saier. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology 146:1797-1814. [DOI] [PubMed] [Google Scholar]
- 13.Kanki, M., T. Yoda, M. Ishibashi, and T. Tsukamoto. 2004. Photobacterium phosphoreum caused a histamine fish poisoning incident. Int. J. Food Microbiol. 92:79-87. [DOI] [PubMed] [Google Scholar]
- 14.Kashiwagi, K., S. Miyamoto, F. Suzuki, H. Kobayashi, and K. Igarashi. 1992. Excretion of putrescine by the putrescine-ornithine antiporter encoded by the potE gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 89:4529-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konagaya, Y., B. Kimura, M. Ishida, and T. Fujii. 2002. Purification and properties of a histidine decarboxylase from Tetragenococcus muriaticus, a halophilic lactic acid bacterium. J. Appl. Microbiol. 92:1136-1142. [DOI] [PubMed] [Google Scholar]
- 16.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 17.Kuipers, O. P., M. M. Beerthuyzen, R. J. Siezen, and W. M. De Vos. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281-291. [DOI] [PubMed] [Google Scholar]
- 18.Kunji, E. R. S., D. J. Slotboom, and B. Poolman. 2003. Lactococcus lactis as host for overproduction of functional membrane proteins. Biochem. Biophys. Acta 1610:97-108. [DOI] [PubMed] [Google Scholar]
- 19.Le Jeune, C., A. Lonvaud-Funel, B. Ten Brink, H. Hofstra, and J. M. B. M. van der Vossen. 1995. Development of a detection system for histidine decarboxylating lactic acid bacteria based on DNA probes, PCR and activity test. J. Appl. Bacteriol. 78:316-326. [DOI] [PubMed] [Google Scholar]
- 20.Lolkema, J. S., B. Poolman, and W. N. Konings. 1996. Secondary transporters and metabolic energy generation in bacteria, p. 229-260. In W. N. Konings, H. R. Kaback, and J. S. Lolkema (ed.), Handbook of biophysics. Elsevier Science, Amsterdam, The Netherlands.
- 21.Lonvaud-Funel, A. 2001. Biogenic amines in wines: role of lactic acid bacteria. FEMS Microbiol. Rev. 199:9-13. [DOI] [PubMed] [Google Scholar]
- 22.Lonvaud-Funel, A., and A. Joyeux. 1994. Histamine production by wine lactic acid bacteria. Isolation of a histamine-producing strain of Leuconostoc oenos. J. Appl. Bacteriol. 77:401-407. [DOI] [PubMed] [Google Scholar]
- 23.Lucas, P., J. Landete, M. Coton, E. Coton, and A. Lonvaud-Funel. 2003. The tyrosine decarboxylase operon of Lactobacillus brevis IOEB 9809: characterization and conservation in tyramine-producing bacteria. FEMS Microbiol. Lett. 229:65-71. [DOI] [PubMed] [Google Scholar]
- 24.Magni, C., D. Mendoze, W. N. Konings, and J. S. Lolkema. 1999. Mechanism of citrate metabolism in Lactococcus lactis. Resistance against lactate toxicity at low pH. J. Bacteriol. 181:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Molenaar, D., J. S. Bosscher, B. TenBrink, A. J. M. Driessen, and W. N. Konings. 1993. Generation of a proton motive force by histidine decarboxylation and electrogenic histidine/histamine antiport in Lactobacillus buchneri. J. Bacteriol. 175:2864-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohtsu, H., and T. Watanabe. 2003. New functions of histamine found in histidine decarboxylase gene knockout mice. Biochem. Biophys. Res. Commun. 305:443-447. [DOI] [PubMed] [Google Scholar]
- 28.Parks, E. H., S. R. Ernst, R. Hamlin, H. Xuong Ng, and M. L. Hackert. 1985. Structure determination of histidine decarboxylase from Lactobacillus 30a at 3 Å resolution. J. Mol. Biol. 182:455-465. [DOI] [PubMed] [Google Scholar]
- 29.Prozorovski, V., and H. J.örnvall. 1975. Structural studies of histidine decarboxylase from Micrococcus sp. n. Eur. J. Biochem. 53:169-174. [DOI] [PubMed] [Google Scholar]
- 30.Recsei, P. A., W. M. Moore, and E. E. Snell. 1983. Pyruvoyl-dependent histidine decarboxylase from Clostridium perfringens and Lactobacillus buchneri. Comparative structure and properties. J. Biol. Chem. 258:439-444. [PubMed] [Google Scholar]
- 31.Recsei, P. A., and E. E. Snell. 1970. Histidine decarboxylase of Lactobacillus 30a. VI. Mechanism of action and kinetic properties. Biochemistry 9:1492-1497. [DOI] [PubMed] [Google Scholar]
- 32.Recsei, P. A., and E. E. Snell. 1972. Histidine decarboxylaseless mutants of Lactobacillus 30a: isolation and growth properties. J. Bacteriol. 112:624-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rollan, G. C., E. Coton, and A. Lonvaud-Funel. 1995. Histidine decarboxylase activity of Leuconostoc oenos 9204. Food Microbiol. 12:455-461. [Google Scholar]
- 34.Schelp, E., S. Worley, A. F. Monzigo, S. Ernst, and J. D. Robertus. 2001. pH-induced structural changes regulate histidine decarboxylase activity in Lactobacillus 30a. J. Mol. Biol. 306:727-732. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silla Santos, M. H. 1996. Biogenic amines: their importance in foods. Int. J. Food Microbiol. 29:213-231. [DOI] [PubMed] [Google Scholar]
- 37.Sissler, M., C. Delorme, J. Bond, S. D. Ehrlich, P. Renault, and C. Francklyn. 1999. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. USA 96:8985-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soksawatmaekhin, W., A. Kuraishi, K. Sakata, K. Kashiwagi, and K. Igarashi. 2004. Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol. Microbiol. 51:1401-1412. [DOI] [PubMed] [Google Scholar]
- 39.Tanase, S., B. M. Guirard, and E. E. Snell. 1985. Purification and properties of a pyridoxal 5′-phosphate-dependent histidine decarboxylase from Morganella morganii AM-15. J. Biol. Chem. 260:6738-6746. [PubMed] [Google Scholar]
- 40.Taylor, S. L. 1986. Histamine food poisoning: toxicology and clinical aspects. Crit. Rev. Toxicol. 17:91-128. [DOI] [PubMed] [Google Scholar]
- 41.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanderslice, P., W. C. Copeland, and J. D. Robertus. 1986. Cloning and nucleotide sequence of wild type and a mutant histidine decarboxylase from Lactobacillus 30a. J. Biol. Chem. 261:15186-15191. [PubMed] [Google Scholar]
- 43.van Poelje, P. D., and E. E. Snell. 1990. Pyruvoyl-dependent enzymes. Annu. Rev. Biochem. 59:29-59. [DOI] [PubMed] [Google Scholar]
- 44.van Poelje, P. D., and E. E. Snell. 1990. Cloning, sequencing, expression, and site-directed mutagenesis of the gene from Clostridium perfringens encoding pyruvoyl-dependent histidine decarboxylase. Biochemistry 29:132-139. [DOI] [PubMed] [Google Scholar]
- 45.Verhoogt, H. J., H. Smit, T. Abee, M. Gamper, A. J. Driessen, D. Haas, and W. N. Konings. 1992. arcD, the first gene of the arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa, encodes an arginine-ornithine exchanger. J. Bacteriol. 174:1568-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]