Abstract
Since high hydrostatic pressure is becoming increasingly important in modern food preservation, its potential effects on microorganisms need to be thoroughly investigated. In this context, mild pressures (<200 MPa) have recently been shown to induce an SOS response in Escherichia coli MG1655. Due to this response, we observed a RecA- and LexA-dependent induction of lambda prophage upon treating E. coli lysogens with sublethal pressures. In this report, we extend this observation to lambdoid Shiga toxin (Stx)-converting bacteriophages in MG1655, which constitute an important virulence trait in Stx-producing E. coli strains (STEC). The window of pressures capable of inducing Stx phages correlated well with the window of bacterial survival. When pressure treatments were conducted in whole milk, which is known to promote bacterial survival, Stx phage induction could be observed at up to 250 MPa in E. coli MG1655 and at up to 300 MPa in a pressure-resistant mutant of this strain. In addition, we found that the intrinsic pressure resistance of two types of Stx phages was very different, with one type surviving relatively well treatments of up to 400 MPa for 15 min at 20°C. Interestingly, and in contrast to UV irradiation or mitomycin C treatment, pressure was not able to induce Stx prophage or an SOS response in several natural Stx-producing STEC isolates.
Shiga toxin (Stx)-producing Escherichia coli (STEC) strains have been recognized since the early 1980s as food-borne pathogens causing gastrointestinal illnesses that vary from uncomplicated diarrhea to hemorrhagic colitis and a severe complication called hemolytic-uremic syndrome (39). A major virulence factor of these bacteria are the Stxs, also called verotoxins, which comprise a family of heteromultimeric proteins belonging to the AB family of toxins and are part of the potent class 2 ribosome-inhibiting proteins (18, 35). Most human cases have been associated with serotype O157:H7, but stx genes are widespread in many other serotypes (28). In fact, as their name suggests, Stxs were first identified in Shigella dysenteriae type 1, which causes similar illnesses (29).
In contrast to the stx genes of S. dysenteriae, which are fixed chromosomally in the vicinity of phage remnants, the stx variants in STEC strains typically reside on intact lambdoid prophages (26, 41). Two major types of such phages have been characterized, encoding, respectively, stx1 (e.g., phage H-19B), resembling the original stx regulon of S. dysenteriae type 1, and the slightly distinct stx2 (e.g., phage 933W), and more recently additional variants of the stx genes have also been isolated (31, 36, 46). The expression of both stx1 and stx2 was found to be coregulated with transcription of late phage genes associated with the lytic cycle, and, in the case of stx1, could also be induced by low-iron conditions (9, 20, 42, 43). The location of these stx genes on a lambdoid phage genome integrated in the E. coli chromosome and their expression upon induction of the lytic cycle have some important implications. Because of the capacity of these lambdoid bacteriophages to infect E. coli and perhaps some other related bacteria, the production of Stxs is a virulence trait that can be easily transmitted horizontally in a process termed lysogenic conversion (37). Further, the lytic cycle and the late phage genes of lambdoid phages are known to be induced as a result of the SOS response, a well-characterized genetic regulatory mechanism that is triggered not only by DNA damage (12, 21) but also by several antibiotics (25, 47). The use of this type of antibiotics for treatment of diarrhea caused by STEC strains is therefore to be avoided. It has even been suggested that the introduction of trimethoprim and 4-quinolones in human and veterinary clinical practice may have led to or have stimulated the appearance of STEC as a food-borne human pathogen (19, 20).
Modern food preservation increasingly depends on hurdle technology and aims to achieve microbiological safety by an intelligent, synergistic combination of mild stress factors (15, 22). In this context, high hydrostatic pressure (HHP) treatment is a promising new technique for inactivating food-borne microorganisms while maintaining a fresh food quality (10). However, the fundamental perception of HHP stress by microorganisms remains poorly understood, and the genetic response induced by sublethal pressurization is only recently beginning to be determined and involves several stress response pathways, of which in bacteria the heat shock regulon is best documented (2, 17, 44). Recently, we also demonstrated the induction of the SOS response and concomitant induction of λ lysogens upon mild HHP treatment in E. coli MG1655 (3). Therefore, in the present study, we investigated the effect of HHP treatment on lysogenic lambdoid Stx-converting bacteriophages. The implications of our findings for food safety are discussed.
MATERIALS AND METHODS
Strains and growth conditions.
E. coli strain MG1655 (8) and one of its pressure-resistant mutants, LMM1010 (16), were lysogenized with H-19B and 933W, obtained from C600 H-19B and C600 933W, respectively, which were kindly donated by Lothar Beutin (Robert Koch Institute, Berlin, Germany) (7). Lysogens were selected, and their carriage of stx was confirmed by PCR amplification of the stxA1 or stxA2 promoter with the following primers: 5′-CAGTGGATCCTGGCACGGAAACATGGGT-3′ and 5′-TCAGTCTAGATTACGTCTTTGCAGTCGAGAAGT-3′ for stxA1 and 5′-GTCAGGATCCCTCATAATCGCCAGGTCGCT-3′ and 5′-CAGTTCTAGATTACAGTAACAGGCACAGTACCC-3′ for stxA2. All lysogens could be induced to produce phage particles by UV irradiation and mitomycin C treatment.
Natural STEC isolates 86-24 (O157:H7, stx2+), 493/98 (O157:H−, stx2+), 5905 (O55:H7, stx2+), and H19 (O26:H11, stx1+) were kindly donated by Laurence Van Melderen (Université Libre de Bruxelles, Brussels, Belgium) and are available at the STEC center (The National Food Safety and Toxicology Center, Michigan State University [http://www.shigatox.net]). STEC EH367 (O111:H−, stx1+ was isolated and kindly provided by Denis Piérard (Vrije Universiteit Brussel, Brussels, Belgium), and STEC EDL 933 (ATCC 43895; O157:H7, stx1+, stx2+) was obtained from the Belgian Coordinated Collections of Microorganisms (University of Ghent, Ghent, Belgium).
Stationary-phase cultures were obtained by growth in Luria-Bertani broth (LB) (34) for 21 h at 37°C under well-aerated conditions.
Phage induction by pressure, UV, or mitomycin C.
Bacterial suspensions for phage induction were prepared by diluting overnight cultures 1/100 in fresh prewarmed LB, followed by further incubation until late exponential phase (optical density at 600 nm [OD600] of 0.6), collection by centrifugation (5 min at 6,000 × g), and resuspension in fresh LB or whole milk. Induction by HHP was done as described before (3). Briefly, 1 to 2 ml of a suspension was sealed without air bubbles in a polyethylene bag and then pressurized for 15 min in an 8-ml pressure vessel, maintained at 20°C with an external cooling circuit (Resato, Roden, The Netherlands). The pressure was increased by ca. 100 MPa/min, while decompression occurred in less than 1 s. For UV induction, 1-ml portions were poured in a petri dish and irradiated (0.2 kJ/m2) in a UV oven, equipped with five fluorescent lamps (8 W each) and emitting from 180 to 280 nm with a peak at 254 nm (Bio-Link, Vilber Lourmat, France). UV doses were programmed and are controlled by a radiometer that constantly monitors the UV light emission. The distance between the lamps and the plates was 14 cm. Mitomycin C induction was achieved by the addition of mitomycin C to a final concentration of 0.5 μg/ml.
After induction, cultures were maintained shaking at 37°C. At different time points after treatment, 30 μl of chloroform was added to 500-μl portions of treated cells and untreated control cells and, after vortexing, the suspension was centrifuged (5 min at 24,000 × g). This treatment completely killed the bacterial suspension. Dilutions of the supernatant were then added to 1 ml of stationary phase C600 cells, which were a more efficient plating host than MG1655. Then, 3 ml of LB top agar supplemented with MgSO4 (10 mM) and CaCl2 (5 mM) was added to this culture, and the mixture was poured on an LB plate. After 24 h, the plaques were counted, and the phage titer was calculated as PFU per milliliter of the original culture. When appropriate, phage production was expressed as the log10 of the induction factor, which was obtained by dividing phage titers of treated and untreated samples at the same time point after induction. Viability was also assayed at the same time points by diluting cells in potassium phosphate buffer (10 mM, pH 7), followed by plating on LB agar plates.
Pressure inactivation of phages H-19B and 933W.
Confluently lysed top agar plates of H-19B and 933W grown on C600 were used for phage extraction (27), and the resulting phage stocks were diluted 10-fold in phage buffer (10 mM MgSO4, 5 mM CaCl2). Aliquots of this solution were sealed in polyethylene bags and pressurized as described above. After pressure treatment, the infective phage titer was determined by adding appropriate dilutions of the samples to 1 ml of C600 and adding top agar as described above. The obtained plaque count was recalculated to the percent survival of the respective phage.
Measurement of recA expression.
A gfp transcription fusion to the promoter of MG1655 recA was constructed earlier (3) and transformed to E. coli MG1655 H-19B, MG1655 933W, EDL 933, and H19. After induction with HHP, UV, or mitomycin C as described above, 300-μl samples were transferred to microplate wells and placed in a fluorescence reader (Fluoroscan Ascent FL; Thermolabsystems, Brussels, Belgium). Fluorescence at 520 nm was then measured at 30-min intervals with intermittent shaking (every 5 min) at 37°C, by using an excitation wavelength of 480 nm. At the same time the OD600 was measured, and fluorescence was expressed per unit of OD600.
RESULTS
HHP induction of H-19B and 933W prophages in MG1655.
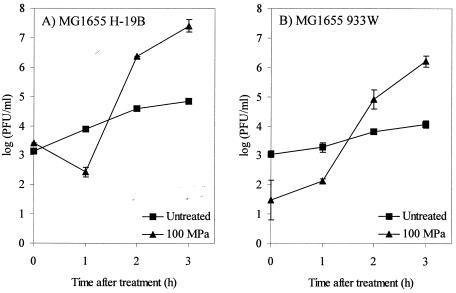
We demonstrated previously that HHP induces lysogenic λ phage in E. coli MG1655 by eliciting an SOS response (3). Since Stx phages are lambdoid phages, we investigated whether they are also induced by HHP. To make the results of this investigation maximally comparable to those obtained with λ, we lysogenized MG1655 with Stx phages H-19B and 933W and pressurized late-exponential-phase cultures of the resulting lysogens for 15 min at 100 MPa, which is the treatment that was found to work well for phage λ. This pressure treatment caused only very little inactivation of MG1655 (<0.6 log10 units), and Fig. 1 shows that between 1 and 2 h after pressure treatment the numbers of phage particles in the culture supernatants of the pressurized cultures started to increase. Also, in the unpressurized control cultures, a significant increase in phage titer occurred during the 3-h incubation period, but the increase in phage titer was at least 100-fold higher in the pressurized than in the unpressurized cultures, indicating that the lytic cycle of both Stx phages can be activated by HHP. The drop in 933W phage titer immediately after HHP treatment (Fig. 1B) can be attributed to the outspoken pressure sensitivity of the phage particles (see below).
FIG. 1.
Induction of H-19B (A) and 933W (B) lysogens of MG1655 by HHP treatment (100 MPa, 15 min, 20°C). The evolution of phage particle count [log(PFU/milliliter)] in untreated (▪) and high-pressure-treated (▴) cell suspensions of MG1655 is shown. The results are expressed as the means ± standard deviations of three experiments.
Induction of Stx prophages at higher pressures.
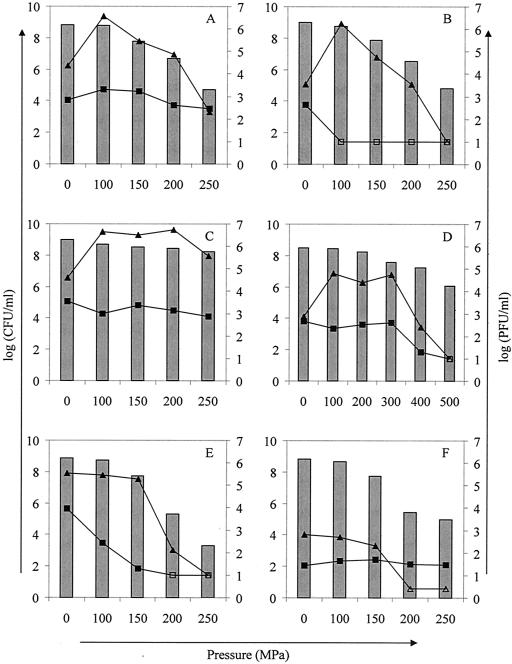
Since most HHP food applications operate at pressures well above 100 MPa, we investigated phage induction in both lysogenic strains by comparing phage titers immediately after and 3 h after treatment at different pressures (0 to 250 MPa) (Fig. 2A and B). Again, we observed an increase of ca. 1 and 2 log10 units in the phage titers of the untreated control cultures of the 933W and H-19B lysogens, respectively, after the 3-h incubation period, and an additional induction of at least 100-fold upon treatment at 100 MPa. Treatment at pressures of >100 MPa resulted in lower phage titers than treatment at 100 MPa. However, since these pressures cause partial inactivation of the bacteria and the preexisting virions in the culture, particularly in the case of phage 933W (see below), it is more difficult to judge whether actual lysogen induction has taken place at these higher pressures. Therefore, we apply here a very conservative judgement of what is to be considered phage induction: only when the phage titer 3 h after HHP treatment is consistently higher than that of the untreated (0 MPa) control culture after 3 h of incubation will the pressure treatment be considered to have caused lysogen induction. By this criterion, phage induction occurred at up to 150 MPa for both phages in MG1655 (Fig. 2A and B). At higher pressures, the phage titer measured 3 h after treatment was not consistently higher than the titer of the untreated (0 MPa) control culture at the same time, and we can therefore not conclude with certainty that phage induction has taken place.
FIG. 2.
Phage counts [log(PFU/milliliter), plotted on the right y axis] immediately (▪) and 3 h (▴) after induction at different pressures (15 min, 20°C) of MG1655 H-19B in LB (A), MG1655 933W in LB (B), MG1655 H-19B in whole milk (C), LMM1010 H-19B in whole milk (D), EDL 933 (O157:H7) in LB (E), and H19 (O26:H11) in LB (F). Open symbols represent counts below the detection limit. Bars represent bacterial survival after pressure treatment [log(CFU/milliliter), plotted on the left y axis]. Representative results of at least three experiments are shown.
Since it is known that bacteria can be strongly protected from HHP inactivation in a food matrix such as milk (14), we investigated whether milk can influence the window of pressure-induced phage production. Exponential-phase cells of MG1655 H-19B were resuspended in whole milk and subjected to the same pressure range (0 to 250 MPa) (Fig. 2C). It can be seen that the use of milk as a matrix not only enhanced the pressure resistance of the MG1655 lysogen but also extended the Stx prophage induction to 250 MPa, which is an increase of 100 MPa compared to the same experiment done in LB broth (compare Fig. 2A and C).
Finally, we repeated the same experiment with E. coli LMM1010, a previously isolated spontaneous mutant of MG1655 displaying an extreme resistance to HHP, lysogenized with H-19B, and extending the applied pressure range to 500 MPa (Fig. 2D). The results show that a 100-fold increase in phage titer compared to untreated cells (3 h after HHP treatment) is possible at up to 300 MPa in this strain in whole milk.
Intrinsic HHP resistance of phages H-19B and 933W.
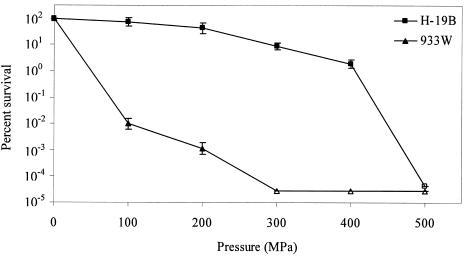
As can be deduced from Fig. 2A and B, looking at the PFU per milliliter in the culture supernatant directly after pressure treatment, a difference in pressure sensitivity exists between H-19B and 933W. This disparity in pressure resistance was investigated in more detail in cell-free phage suspensions at a range of pressures (0 to 500 MPa) (Fig. 3). At 300 MPa (15 min, 20°C), phage 933W was inactivated more than (3 × 106)-fold compared to only ∼10-fold for phage H-19B. Complete inactivation of phage H-19B required treatment at 500 MPa (15 min, 20°C).
FIG. 3.
Inactivation of isolated H-19B (▪) and 933W (▴) phage particles by pressure treatment (0 to 500 MPa, 15 min, 20°C), as determined by infectivity upon plating on C600. Untreated (0-MPa) phage titers correspond to ca. 2.3 × 107 and 3.8 × 107 PFU/ml for H-19B and 933W, respectively. Open symbols represent counts below the detection limit. The results are expressed as means ± the standard deviations of three experiments.
HHP induction of Stx prophages in natural STEC isolates.
To demonstrate whether pressure mediated Stx prophage induction occurs also in naturally occurring STEC strains, we first examined E. coli EDL 933 (Fig. 2E) and H19 (Fig. 2F) at pressures of up to 250 MPa. Although the basal phage titers for both strains differed considerably [4 log10(PFU/ml) and 1.5 log10(PFU/ml) for EDL 933 and H19, respectively], both unpressurized control cultures showed an increase in phage titer of ∼1.5 log10(PFU/ml) during the 3-h incubation period in fresh LB medium. Similar observations of spontaneous Stx phage titer increases in untreated cultures have been described earlier (24, 45). However, as opposed to what was observed for the MG1655 lysogens (Fig. 2A and B), a 100-MPa treatment did not cause any induction in either of the STEC strains (Fig. 2E and F). At 150 MPa, there was a decrease in the bacterial plate count of about 1 log10 unit, and since the phage titers after 3 h remained more or less stable, this may be an indication of weak phage induction. However, this cannot be concluded with certainty because there may be a fraction of injured cells that can repair during further incubation of the suspension for 3 h and yet cannot form colonies when plated immediately. Additional, molecular, evidence that lysogen induction did not take place in both STEC strains is given below. Although EDL 933 contains both Stx2 (933W) and Stx1 (933V) prophages (30), the latter is cryptic, and only 933W is capable of lytic growth and production of infectious particles (32). The presence of 933W is reflected in the typically pressure sensitive inactivation pattern of the preexisting phage particles (Fig. 2E). Moreover, PCR amplification of randomly picked plaques from EDL 933 with stx1- and stx2-specific primers confirmed the presence of 933W only (data not shown).
Because of this failure to induce Stx phages from these strains with HHP, four additional STEC isolates were tested for HHP induction of Stx prophage and compared to MG1655 H-19B and MG1655 933W. At the same time, HHP treatment was compared to well-established phage-inducing treatments such as UV irradiation (0.2 kJ/m2) and mitomycin C (0.5 μg/ml) treatment (Table 1). It should be noted that induction was scored 5 h after treatment in this case, since phage induction by mitomycin C treatment was delayed to some extent compared to HHP or UV (Fig. 4). Surprisingly, none of the tested STEC strains proved to be responsive toward HHP treatment, whereas all strains showed phage induction upon UV or mitomycin C treatment.
TABLE 1.
Induction of phage particles 5 h after treatment with pressure, UV, or mitomycin C
| E. coli strain | Mean fold induction (log10) ± SEM after treatment with:
|
||
|---|---|---|---|
| Pressure (100 MPa) | UV (0.2 kJ/m2) | Mitomycin C (0.5 μg/ml) | |
| MG1655 H-19B | 2.4 ± 0.1 | 2.0 ± 0.3 | 2.7 ± 0.4 |
| MG1655 933W | 3.5 ± 0.1 | 3.9 ± 0.1 | 3.6 ± 0.1 |
| EDL 933 | −0.2 ± 0.0 | 1.3 ± 0.1 | 1.8 ± 0.1 |
| H19 | −0.2 ± 0.1 | 2.7 ± 0.2 | 2.3 ± 0.2 |
| 5905 | −0.6 ± 0.6 | 1.2 ± 0.3 | 1.4 ± 0.6 |
| EH 367 | 0.0 ± 0.0 | 1.3 ± 0.2 | 2.1 ± 0.0 |
| 493/89 | −0.1 ± 0.2 | 2.3 ± 0.3 | 2.9 ± 0.2 |
| 86-24 | −0.3 ± 0.0 | 2.8 ± 0.4 | 2.4 ± 0.1 |
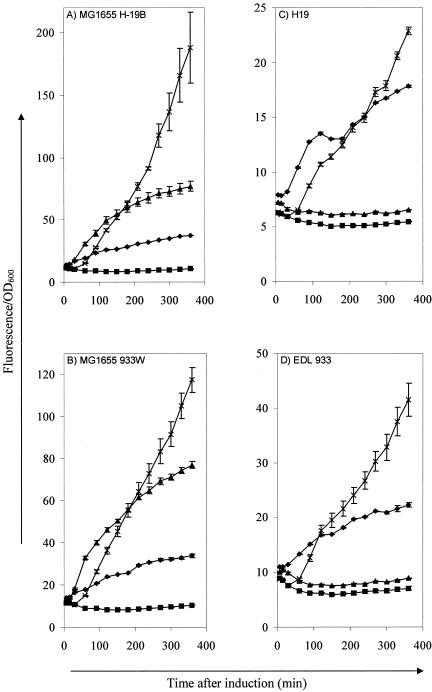
FIG. 4.
Determination of SOS promoter activity in MG1655 H-19B (A), MG1655 933W (B), H19 (O26:H11) (C), and EDL 933 (O157:H7) (D) after treatment with pressure (▴; 100 MPa, 15 min, 20°C), UV (♦; 0.2 kJ/m2), or mitomycin C (×; 0.5 μg/ml) compared to untreated cells (▪). SOS promoter activity was measured with a recA promoter transcription fusion to gfp and is expressed as fluorescence per unit of OD600. The results are expressed as means ± the standard deviations.
Pressure-induced SOS response in natural STEC isolates.
To investigate whether the inability of HHP to induce the lytic cycle of Stx prophage in natural STEC isolates stems from the lack of an SOS response after HHP treatment, a plasmid carrying a transcriptional fusion of the MG1655 recA promoter (PrecA) to gfp was transformed to MG1655 H-19B, MG1655 933W, H19, and EDL 933 as a reporter of the SOS response. After the corresponding induction treatments with HHP (100 MPa, 15 min), UV (0.2 kJ/m2), or mitomycin C (0.5 μg/ml), the time course of Gfp expression was followed (Fig. 4). Although PrecA was induced by all three treatments in both lysogens of MG1655 (Fig. 4A and B), it was only induced by UV and mitomycin C, and not by HHP, in H19 and EDL 933 (Fig. 4C and D, respectively). Treatments at 150 and 200 MPa also did not induce PrecA (data not shown). Thus, the response of the recA promoter corresponded well with Stx phage induction, indicating that the high-pressure-induced SOS response, described earlier (3), and the concomitant induction of Stx prophage, as described here, are not present or at least are not at a detectable level in the six natural STEC isolates tested in the present study.
DISCUSSION
We have recently described induction of the SOS response in E. coli MG1655 upon sublethal (100- to 150-MPa) high-pressure treatment. This SOS response proved to be RecA and LexA dependent and triggered the induction of the lytic cycle of a λ prophage in MG1655 (3). Since HHP treatment has been used in food preservation, we wanted to investigate whether lambdoid Stx-converting bacteriophages could also be induced in E. coli strains by high pressure.
First, we conducted a number of experiments in MG1655 strains lysogenized, respectively, with phages H-19B (stx1) and 933W (stx2) and found that, similar to λ prophage, the lytic cycle of both H-19B and 933W Stx prophage could be triggered by treatment at 100 MPa (Fig. 1). Despite the significant ∼100-fold increase in Stx phage particle count 3 h after pressure treatment, no eminent cell lysis of the culture or decrease in the viable plate count was observed then or at later time points, indicating that only a relatively small subpopulation of the lysogens is induced by high pressure. If we assume a burst size comparable to that of phage λ, ca. 100 to 200, a final phage titer of ca. 106 (933W in Fig. 1B) or 107 (H-19B in Fig. 1A) would correspond to an induction rate of ca. 0.01 to 0.1% of the population. Since hydrostatic pressure is transmitted directly and evenly through a bacterial suspension and thus delivers an equal amount of stress to every cell, it seems that some cellular factors that are heterogeneous throughout the population influence the induction of lytic development by HHP.
At pressures of 150 MPa or higher, the MG1655 lysogens were inactivated by about 1 log10 unit or more (Fig. 2A and B). This represents a direct inactivation by the HHP treatment and not by phage burst, since there was no increase in phage titers and because nonlysogenic MG1655 was equally pressure sensitive (data not shown). However, it was shown that both the protective effect of the food matrix and the intrinsic pressure resistance of E. coli could extend the window of Stx prophage induction by 100 to 200 MPa. In LB broth, the pressure range leading to activation of H-19B and 933W prophage was ca. 100 to 150 MPa (Fig. 2A and B), whereas in whole milk, in which the pressure resistance of MG1655 is enhanced, activation of H-19B occurred up to 250 MPa (Fig. 2C). Moreover, when a H-19B lysogen of LMM1010, a pressure-resistant derivative of MG1655 (16), was pressure treated in whole milk, induction accompanied by a 100-fold increase of phage particles was observed at up to 300 MPa (Fig. 2D). Although food pasteurization by HHP will generally use higher pressures, pressures of this magnitude are relevant in a number of current or proposed industrial applications of HHP treatment, such as shucking of oysters and pressure-assisted freezing or thawing. In addition, some E. coli strains, including STEC, have a very high natural resistance to HHP (4, 6). Based on our findings under laboratory conditions, it would be interesting to investigate whether mild HHP treatments might promote the spread of Stx phages and their associated stx virulence genes in real foods under more realistic conditions. Moreover, in addition to Stx phages, other virulence- or resistance-conferring genetic elements exist, the horizontal dissemination of which is enhanced by the SOS response. Most recently, transfer of SXT, an integrating conjugative element derived from Vibrio cholerae and encoding genes that confer resistance to chloramphenicol, sulfamethoxazole, trimethoprim, and streptomycin, has also been shown to be induced by the SOS response evoked by a variety of environmental factors and antibiotics (5). Whether HHP treatment of foods under industrial conditions can induce a measurable increase in the natural rate of transfer of genes or phages between bacteria remains to be demonstrated. It can be anticipated, however, that the level of Stx phage induction in industrial processes will be much lower than the one reported here because these processes are generally conducted at higher pressures, resulting at least in a partial inactivation of E. coli bacteria, and because foods contain much lower densities of E. coli than the suspensions used here.
The pressure stability of the Stx virions themselves is of course also relevant in the context of food safety. Throughout our experiments, it became clear that H-19B and 933W virions displayed a significantly dissimilar pressure stability (Fig. 2 and 3). Whereas H-19B showed little loss of activity at pressures up to 400 MPa (in phage buffer), 933W was readily inactivated several log10 cycles at 100 MPa (Fig. 3). Despite the fact that both Stx phages belong to the same family of siphoviridae and share extensive overall genetic homology, no detailed comparison has yet been made between their capsid proteins. However, electron micrographs show that their morphology differs: 933W consists of regular hexagonal heads with short contractile tails, whereas H-19B has elongated hexagonal heads with long noncontractile tails (33, 37). Although H-19B shares most homology with phage λ, its pressure stability appears to be higher than that reported by Chen et al. (11) for bacteriophage λ. Where phage λ was completely eliminated (>7 log10 cycles) at 400 MPa for 5 min (22°C), H-19B was only inactivated by <2 log10 cycles at 400 MPa for 15 min (20°C). Phage 933W, on the other hand, is considerably more pressure sensitive than phage λ, showing a >6-log10 cycle inactivation at 300 MPa (15 min, 20°C). However, there seems to be no general correlation between morphology and pressure stability of viruses, and it is likely that very subtle differences in amino acid sequence of coat proteins may strongly affect virion stability under high pressure (23).
Surprisingly, HHP treatment failed to induce Stx phage production in E. coli EDL 933 and H19 (Fig. 2E and F) and in all of the other natural STEC tested (Table 1), whereas in these strains Stx prophage was inducible with UV irradiation and mitomycin C treatment. Looking further into this apparent anomaly, we also observed the lack of recA promoter activation by HHP in EDL 933 and H19, as opposed to MG1655 H-19B and MG1655 933W, whereas in all four strains the recA promoter was quite responsive to UV and mitomycin C (Fig. 4). Although the lack of an HHP-induced SOS response in these two strains confirms and explains the absence of Stx phage induction, the reason for this failed SOS response is nevertheless puzzling. Moreover, as discussed previously (3), the mechanism of SOS induction by high pressure itself is elusive to date and seems unlike that underlying the classical induction by UV irradiation or mitomycin C treatment. Notably, we observed no evidence for DNA damage to occur at high pressures, since no enhanced mutation rate could be observed after pressure treatment, and recA or lexA1 strains, which are known to be hypersensitive to DNA damage inflicted by UV irradiation, proved as resistant to HHP as did the wild type (3). The fact that high-pressure treatment fails to induce cleavage of both CI, which is responsible for the lysogenic state of Stx prophage, and LexA, which is responsible for repression of SOS promoters such as PrecA, in natural STEC strains suggests that the RecA protein is not being activated by pressure in these strains. It is therefore tempting to speculate that the high-pressure activation of RecA observed in MG1655 deviates from the known activation pathway involving exposure of single-stranded damaged DNA and might be due to a more direct effect of high pressure on the structure of the MG1655 RecA protein. According to this hypothesis, although the RecA proteins from MG1655 and EDL 933 only differ in one or perhaps two amino acids (based on their DNA sequences in GenBank), it could be that EDL 933 RecA is not converted to an active conformation by HHP. This possible mechanism is currently the topic of ongoing research in our laboratory.
With respect to the food safety, it can of course not be excluded that other natural STEC strains may exist that do react to HHP treatment with increased production of Stx phages, as was seen with MG1655 H-19B or 933W. Even if this is not the case, naturally generated Stx lysogens of environmental E. coli strains may be present in foods and, if they are HHP inducible, contribute to the spread of the Stx phages. It has indeed become extensively clear that Stx phages can convert Stx-negative E. coli strains in vivo and in the natural environment (1, 13, 38, 40). Based on these findings, it is at least theoretically possible that treatment of foods with sublethal pressures may, in the long run, select for pressure-resistant E. coli strains carrying lysogenic Stx phages. To prevent this selection, only pressure treatments with sufficient lethality should be used. However, it should be noted that, other than stx genes, additional virulence factors are required to confer pathogenicity and that the majority of stx carrying strains have never been associated with disease.
In summary, we have demonstrated that HHP can initiate the lytic cycle of Stx prophages in some E. coli hosts and that this induction persists at pressures relevant in the food industry when the pressure resistance of lysogens was increased either by the surrounding food matrix or intrinsically by mutation. Moreover, when the intrinsic pressure resistance of the two best-characterized Stx phage types, H-19B and 933W, was assessed, we found remarkable differences in pressure resistance. Both the HHP-induced increased production of Stx phages and the remarkable HHP resistance of H-19B raise the possibility of increased horizontal spread of the virulence trait carried by these phages. However, further studies are required to adequately address this possibility and to evaluate the eventual risk for food safety in a food processing environment. Finally, it became clear that the HHP-induced SOS response that is underlying phage induction is not a universal feature shared by all E. coli strains. A further study of the reasons for the lack of HHP-mediated SOS and prophage induction in the natural STEC strains tested here will contribute to the understanding of the molecular effects of high pressure on microorganisms.
Acknowledgments
We acknowledge financial support by research grants OT/01/35 from the K. U. Leuven Research Fund and G.0195.02 from F. W. O. Vlaanderen.
We thank Lothar Beutin, Laurence Van Melderen, and Denis Piérard for kindly providing the natural STEC strains.
REFERENCES
- 1.Acheson, D. W., J. Reidl, X. Zhang, G. T. Keusch, J. J. Mekalanos, and M. K. Waldor. 1998. In vivo transduction with Shiga toxin 1-encoding phage. Infect. Immun. 66:4496-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aertsen, A., K. Vanoirbeek, P. De Spiegeleer, J. Sermon, K. Hauben, A. Farewell, T. Nyström, and C. W. Michiels. 2004. Heat shock protein mediated resistance toward high hydrostatic pressure in Escherichia coli. Appl. Environ. Microbiol. 70:2660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aertsen, A., R. Van Houdt, K. Vanoirbeek, and C. W. Michiels. 2004. An SOS response induced by high pressure in Escherichia coli. J. Bacteriol. 186:6133-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alpas, H., N. Kalchayanand, F. Bozoglu, A. Sikes, C. P. Dunne, and B. Ray. 1999. Variation in resistance to hydrostatic pressure among strains of food-borne pathogens. Appl. Environ. Microbiol. 65:4248-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72-74. [DOI] [PubMed] [Google Scholar]
- 6.Benito, A., G. Ventoura, M. Casadei, T. Robinson, and B. Mackey. 1999. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl. Environ. Microbiol. 65:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutin, L., D. Geier, S. Zimmermann, S. Aleksic, H. A. Gillespie, and T. S. Whittam. 1997. Epidemiological relatedness and clonal types of natural populations of Escherichia coli strains producing Shiga toxins in separate populations of cattle and sheep. Appl. Environ. Microbiol. 63:2175-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 9.Calderwood, S. B., and J. J. Mekalanos. 1987. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169:4759-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheftel, J. C. 1995. Review: high-pressure, microbial inactivation and food preservation. Food Sci. Technol. Int. 1:75-90. [Google Scholar]
- 11.Chen, H., R. D. Joerger, D. H. Kingsley, and D. G. Hoover. 2004. Pressure inactivation kinetics of phage lambda cI857. J. Food. Prot. 67:505-511. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 13.Gamage, S. D., J. E. Strasser, C. L. Chalk, and A. A. Weiss. 2003. Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect. Immun. 71:3107-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Graells, C., B. Masschalck, and C. W. Michiels. 1999. Inactivation of Escherichia coli in milk by high-hydrostatic-pressure treatment in combination with antimicrobial peptides. J. Food Prot. 62:1248-1254. [DOI] [PubMed] [Google Scholar]
- 15.Gould, G. W. 2000. Preservation: past, present and future. Br. Med. Bull. 56:84-96. [DOI] [PubMed] [Google Scholar]
- 16.Hauben, K. J., D. H. Bartlett, C. C. Soontjens, K. Cornelis, E. Y. Wuytack, and C. W. Michiels. 1997. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Appl. Environ. Microbiol. 63:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karatzas, K. A., J. A. Wouters, C. G. Gahan, C. Hill, T. Abee, and M. H. Bennik. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility, and virulence. Mol. Microbiol. 49:1227-1238. [DOI] [PubMed] [Google Scholar]
- 18.Karmali, M. A. 2004. Infection by Shiga toxin-producing Escherichia coli: an overview. Mol. Biotechnol. 26:117-122. [DOI] [PubMed] [Google Scholar]
- 19.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 1999. Induction of type 2 Shiga toxin synthesis in Escherichia coli O157 by 4-quinolones. Lancet 353:1588-1589. [DOI] [PubMed] [Google Scholar]
- 20.Kimmitt, P. T., C. R. Harwood, and M. R. Barer. 2000. Toxin gene expression by Shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg. Infect. Dis. 6:458-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leistner, L. 2000. Basic aspects of food preservation by hurdle technology. Int. J. Food Microbiol. 55:181-186. [DOI] [PubMed] [Google Scholar]
- 23.Lima, S. M., D. S. Peabody, J. L. Silva, and A. C. de Oliveira. 2004. Mutations in the hydrophobic core and in the protein-RNA interface affect the packing and stability of icosahedral viruses. Eur. J. Biochem. 271:135-145. [PubMed] [Google Scholar]
- 24.Livny, J., and D. I. Friedman. 2004. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 51:1691-1704. [DOI] [PubMed] [Google Scholar]
- 25.Matsushiro, A., K. Sato, H. Miyamoto, T. Yamamura, and T. Honda. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181:2257-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonough, M. A., and J. R. Butterton. 1999. Spontaneous tandem amplification and deletion of the Shiga toxin operon in Shigella dysenteriae 1. Mol. Microbiol. 34:1058-1069. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Loughlin, E. V., and R. M. Robins-Browne. 2001. Effect of Shiga toxin and Shiga-like toxins on eukaryotic cells. Microbes. Infect. 3:493-507. [DOI] [PubMed] [Google Scholar]
- 30.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 31.Piérard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rietra, P. J., G. A. Willshaw, H. R. Smith, A. M. Field, S. M. Scotland, and B. Rowe. 1989. Comparison of Vero-cytotoxin-encoding phages from Escherichia coli of human and bovine origin. J. Gen. Microbiol. 135:2307-2318. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sandvig, K. 2001. Shiga toxins. Toxicon 39:1629-1635. [DOI] [PubMed] [Google Scholar]
- 36.Scheutz, F., L. Beutin, D. Piérard, and H. R. Smith. 2001. Appendix: nomenclature of verocytotoxins, p. 447-452. In G. Duffy, P. Garvey, and D. A. McDowell (ed.), Verocytotoxigenic Escherichia coli. Food & Nutrition Press, Inc., Trumbull, Conn.
- 37.Schmidt, H. 2001. Shiga-toxin-converting bacteriophages. Res. Microbiol. 152:687-695. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt, H., M. Bielaszewska, and H. Karch. 1999. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorpe, C. M. 2004. Shiga toxin-producing Escherichia coli infection. Clin. Infect. Dis. 38:1298-1303. [DOI] [PubMed] [Google Scholar]
- 40.Toth, I., H. Schmidt, M. Dow, A. Malik, E. Oswald, and B. Nagy. 2003. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a Shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69:7242-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner, P. L., M. N. Neely, X. Zhang, D. W. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner, P. L., J. Livny, M. N. Neely, D. W. Acheson, D. I. Friedman, and M. K. Waldor. 2002. Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol. Microbiol. 44:957-970. [DOI] [PubMed] [Google Scholar]
- 44.Welch, T. J., A. Farewell, F. C. Neidhardt, and D. H. Bartlett. 1993. Stress response of Escherichia coli to elevated hydrostatic pressure. J. Bacteriol. 175:7170-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto, T., S. Kojio, I. Taneike, S. Nakagawa, N. Iwakura, and N. Wakisaka-Saito. 60Co irradiation of Shiga toxin (Stx)-producing Escherichia coli induces Stx phage. FEMS Microbiol. Lett. 222:115-121. [DOI] [PubMed]
- 46.Zhang, W., M. Bielaszewska, T. Kuczius, and H. Karch. 2002. Identification, characterization, and distribution of a Shiga toxin 1 gene variant (stx1c) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. Acheson. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]