Abstract
Mitochondria are densely packed with proteins, of which most are involved physically or more transiently in protein–protein interactions (PPIs). Mitochondria host among others all enzymes of the Krebs cycle and the oxidative phosphorylation pathway and are foremost associated with cellular bioenergetics. However, mitochondria are also important contributors to apoptotic cell death and contain their own genome indicating that they play additionally an eminent role in processes beyond bioenergetics. Despite intense efforts in identifying and characterizing mitochondrial protein complexes by structural biology and proteomics techniques, many PPIs have remained elusive. Several of these (membrane embedded) PPIs are less stable in vitro hampering their characterization by most contemporary methods in structural biology. Particularly in these cases, cross-linking mass spectrometry (XL-MS) has proven valuable for the in-depth characterization of mitochondrial protein complexes in situ. Here, we highlight experimental strategies for the analysis of proteome-wide PPIs in mitochondria using XL-MS. We showcase the ability of in situ XL-MS as a tool to map suborganelle interactions and topologies and aid in refining structural models of protein complexes. We describe some of the most recent technological advances in XL-MS that may benefit the in situ characterization of PPIs even further, especially when combined with electron microscopy and structural modeling.
Keywords: in situ cross-linking mass spectrometry, mitochondria, cross-linking reagents, membrane complexes, protein localization, protein–protein interactions
Graphical Abstract
Highlights
-
•
Cross-linking mass spectrometry (XL-MS) can be applied in situ in naïve mitochondria.
-
•
In situ XL-MS provides maps for suborganelle interactions and topologies.
-
•
XL-MS can be used to refine structural models of mitochondrial protein complexes.
-
•
XL-MS provides complementary data compared with other MS-based approaches.
-
•
XL-MS provides complementary data compared with cryo-EM and cryo-electron tomography.
In Brief
Mitochondria are vital organelles in eukaryotic cells, densely packed with proteins, of which most are involved physically in protein–protein interactions, and sublocalized over distinct subcompartments. In this perspective, the current state of crosslinking mass spectrometry (MS) as applied to detail the interactome and structures of protein complexes in mitochondria is highlighted, thereby placing this approach also in the context of other MS-based methods (e.g., affinity purification–MS and BioID) and complementary structural biology approaches such as cryogenic electron tomography.
Mitochondria are crucial cellular organelles that besides being instrumental in the generation of most of the cellular ATP participate in various cellular processes such as apoptosis and the immune response (1, 2, 3, 4, 5, 6, 7). This multifaceted role is supported by their complex ultrastructure, hallmarked by an elaborate two-membrane system providing compositionally and functionally distinct subcompartments (Fig. 1A) (8, 9). Mitochondria are densely packed with proteins, and different proteins and protein complexes are specifically located in one or more of these subcompartments or play a role in making connections in between them. Given the intricate nature of mitochondria as versatile organelles crucial in diverse cellular processes, it may not come as a surprise that several diseases have been linked to mitochondrial dysfunction, often stemming from deficiencies in mitochondrial enzymes and/or protein complexes, as reviewed by Javadov et al. and Nunnari et al. (10, 11). By unraveling the intricacies of these complexes, mass spectrometry (MS)–based techniques, such as cross-linking mass spectrometry (XL-MS), can help to pinpoint aberrations that underlie such dysfunction, potentially offering insights into a disease origin (12). To set the stage for this review on in situ XL-MS, we first concisely describe mitochondrial subcompartmentalization and some of the key protein complexes located in or between these subcompartments.
Fig. 1.
Mitochondrial subcompartmentalization and characteristic features of specific mitochondrial proteins and protein complexes.A, schematic of the mitochondrial subcompartmentalization with illustrative protein complexes indicated. Compartments are defined and restricted by the outer mitochondrial membrane (OMM) and the functionally divided inner membrane (inner boundary membrane [IBM], cristae membrane [CM], and cristae junction [CJ]). Aqueous compartments include the intermembrane space (IMS), the intracristal space (ICS), and the mitochondrial matrix (matrix). Characteristic proteins and protein complexes include the pore-forming membrane proteins like the translocases of the outer membrane and inner membrane (TOM and TIM), the voltage-dependent anion-selective channel (VDAC), the mitochondrial contact site and cristae organizing system (MICOS), optic atrophy 1 protein (OPA1), and OXPHOS complexes (CI, CII, CIII, CIV, and CV). B, a cryotomogram of a HeLa cell thinned by cryo-FIB milling, showing the ultrastructure of typical mitochondria (adapted from (35) (catalog no.: EMD-4491). 3D segmentation reveals details about the ultrastructure of two mitochondria and their interaction with the cytoskeleton (blue tube). Reconstructed membranes (orange and green) show an oval shape, with partially resolved cristae structures forming long tubes with parallel membranes (scale bar represents 250 nm). OXPHOS, oxidative phosphorylation.
Mitochondrial Subcompartmentalization
The so-called outer mitochondrial membrane (OMM) encapsulates the whole organelle. The OMM hosts important pore-forming membrane proteins such as the voltage-dependent anion-selective channels, several protein translocases of the outer membrane enabling the traverse of ions, small uncharged molecules and mitochondrial precursor proteins, and proteins involved in cellular signaling (e.g., mitofusins). In contrast, the inner mitochondrial membrane (IMM) forms a tight diffusion barrier for all ions and molecules and can even be further subdivided into the inner boundary membrane (IBM) and the cristae membrane (CM) (13, 14). The IBM is nearest to the OMM, forming functional contact sites, thereby facilitating the import of ions and molecules via specific transporters (e.g., protein translocases of the inner membrane that interact with the OMM porins (15, 16). The CM are flat or tubulovesicular invaginations of the IMM (17) and form the principal site of energy conversion as they accommodate all protein complexes involved in mitochondrial oxidative phosphorylation (OXPHOS) (18).
The two-membrane system and the subcompartmentalized IMM define several functionally distinct compartments. The intermembrane space (IMS) is the region between the OMM and IBM, hosting proteins participating in protein import and folding, signaling events, and transport of molecules (e.g., lipids, ions, and metabolites) (19, 20). Membrane protein complexes such as the mitochondrial contact site and cristae organizing system and the optic atrophy 1 protein (OPA1) connect the CM and IBM by stabilizing narrow openings called cristae junctions (21, 22). Cristae junctions function as an additional diffusion barrier, creating the intracristal space, proposed to be important for signaling events and bioenergetics (23, 24, 25). The innermost compartment, the mitochondrial matrix, is enclosed by the IMM. The matrix includes proteins participating in key biosynthetic pathways (e.g., synthesis of fatty acids, amino acids, proteins) as well as proteins of the tricarboxylic acid (TCA)–Krebs cycle, which provide electron carriers that are subsequently fed into the OXPHOS system (26, 27).
As outlined previously, the function of a mitochondrial protein is directly linked to the subcompartment it resides in. Over the last decades, fluorescence microscopy (28, 29), MS-based techniques, and computational prediction methods (30) have yielded insights into the localization of many well-known mitochondrial proteins and protein complexes (31). A remaining challenge lies within determining the more spatiotemporal organization of proteins and their interactions, as several highly dynamic processes maintain the functionality of mitochondria. While high-resolution fluorescence microscopy enables the detailed tracking of a handful of proteins at the time (32), recent technological innovations in the field of MS have advanced this landscape, facilitating not only the concurrent identification of numerous proteins in spatiotemporal context (33) but also revealing insights into their interaction landscape (34), as described in more detail in the following sections.
Structural Analysis on Mitochondrial Protein Complexes
Advances in structural biology and microscopy, and most recently in cryo-electron tomography (cryo-ET), have played a pivotal role in unraveling mitochondrial ultrastructure (Fig. 1B) (35, 36). Notwithstanding, the intricate morphology and the high protein density of a wide variety of distinct proteins represent a major challenge for cryo-ET (18, 37). Consequently, for multiple protein–protein interactions (PPIs) and protein complexes found within the two-membrane system, we still lack solid structural and functional annotations. To tackle this complexity from an alternative angle, several MS-based approaches have been developed, aiming at identifying and spatially resolving proteins and their interactions in mitochondria.
Affinity purification–mass spectrometry (AP–MS) facilitates the identification of PPIs (38) and has made several seminal contributions to identifying a protein suborganellar localization as well as its interaction partners in mitochondria (39, 40). The method relies most often on the recombinant expression of a protein of interest (bait), coupled to an epitope tag (e.g., Strep-tag), enabling the subsequent purification of the bait protein together with its interactors (preys). Alternatively, baits and respective preys can be enriched using a ligand or an antibody, coupled to a solid support (41). Limitations of AP–MS, such as the potential generation of false-positive protein interactions, occurring during the cell lysis step in sample preparation and the inability to identify weaker and transient interactions (42, 43) can be partially overcome by proximity labeling approaches, such as proximity-dependent biotin identification (BioID) and ascorbic acid peroxidase (APEX) (44, 45). Both BioID and APEX rely on a dedicated mutated enzyme that is fused to the protein bait of interest (biotin ligase; BioID, ascorbic acid peroxidase; APEX) and converts exogenously supplied biotin, resulting in the covalent biotinylation of proteins that are within close proximity of the tagged bait protein. Subsequently, biotinylated proteins can be enriched and identified by MS. When applied to mitochondria, BioID and APEX generated comprehensive mitochondrial interaction networks, with high subcompartment resolution (46, 47). Besides being able to detect transient and weak interactions, the targeted component of BioID and APEX makes purification of mitochondria (or other subcellular regions) for the analysis obsolete, thereby avoiding the detection of artifacts that are sometimes introduced by purification protocols (48). A disadvantage that all proximity labeling–based approaches have in common is that they cannot distinguish between directly interacting proteins and two proteins that are in close proximity, for example, because they reside within the same subcompartment. By integrating proximity labeling with complementary methodologies such as AP–MS (49) or XL-MS (50), these limitations may be partially addressed, making this combination of approaches an effective tool to study PPIs in mitochondria. While both AP–MS and proximity labeling approaches provide powerful platforms to interrogate PPIs with high spatial resolution, gaining insights into the temporal context in which interactions occur remains very challenging. However, the efficiency and inducibility of APEX labeling makes it a great candidate to tackle this challenge. Lobingier et al. (33) combined APEX with quantitative proteomics and a system of spatial references, thereby gaining insights into the spatial and temporal context of the interaction network of G-protein–coupled receptors. Such an approach would technically also be feasible in mitochondria; however, it is limited to proteins for which prior information about temporal redistributions exist.
Besides, AP–MS and the proximity labeling approaches, complexome profiling–MS (CP–MS) provides an alternative powerful method for the characterization of mitochondrial protein complexes. CP combines mild separation techniques, such as native gel electrophoresis, size-exclusion chromatography (SEC), or density gradient centrifugation with quantitative MS, enabling the characterization of coeluting proteins, putatively forming larger protein complexes. CP–MS has facilitated in-depth investigations of key mitochondrial complexes, including the elucidation of the assembly process for the respiratory chain complex I. In addition, it yielded insights into protein complex–related dysfunctions associated with diseases, as extensively reviewed by Wittig et al. (51) and Cabrera-Orefice et al. (52). In contrast to AP–MS and the proximity labeling approaches, CP–MS cannot be performed in situ as mitochondria have to be solubilized prior to separation. To achieve this, mild detergents such as digitonin or n-dodecyl β-d-maltoside are typically used, preserving most protein complexes in their near physiological state (53, 54). Although all separation techniques have advantages and disadvantages, blue native polyacrylamide gel electrophoresis (BN-PAGE) has become a robust separation method that provides great resolution for the analysis of several mitochondrial complexes. CP–MS in conjunction with BN-PAGE is somewhat limited to assemblies that can be separated by electrophoresis. Consequently, in mitochondria, it has been mostly used for the characterization of large protein complexes, notably the important family of respiratory chain complexes.
Cumulatively, although these methods are all conceptually different, they all enhanced the identification and localization of PPIs within mitochondria, revealing also numerous new PPIs (46, 47, 55, 56, 57, 58). However, these approaches do not provide detailed insights into structural features, such as protein complex topology and interaction interfaces. In addition, AP–MS and CP–MS are somewhat insensitive to weaker and transient interactions (41, 42), as these may disassemble during the analysis.
At the center of this review is another MS-based approach, namely XL-MS (59, 60, 61), which uses chemical crosslinkers to covalently link interacting proteins. In contrast to AP–MS, BioID, and APEX, XL-MS is commonly not performed on intact cells. However, for some organelles, XL-MS can be well executed in situ, as demonstrated by several groups for intact naïve mitochondria. In this manner, XL-MS can be used to provide detailed structural and topological data and elucidate mitochondrial interaction networks in situ (34, 62, 63, 64).
Probing Mitochondrial PPIs With XL-MS
XL-MS relies on a chemical reagent (crosslinker), which covalently links two proximal amino acids, enabling the identification of PPIs. Furthermore, by considering the length of the cross-link reagent as well as the linked amino acids, distance restraints are generated that can be used to decipher protein structure and protein complex topology (65, 66).
Available Cross-Link Reagents
While XL-MS was initially mostly applied to purified proteins and protein complexes, advances in instrumentation, bioinformatics, and cross-linker design led to an increase in XL-MS studies for the PPI analysis in complex systems, such as whole organelles and cells (61). To date, a variety of cross-link reagents have been introduced, typically sharing a similar basic structural design: a spacer arm connecting two reactive moieties. The reactive group specifies which amino acids are targeted, with N-hydroxysuccinimide-reactive moieties targeting primary amines (e.g., between lysine side chains) being the most frequently used reactive group (67). The spacer arm affects the spatial resolution and cross-link density, with a longer spacer increasing the amount of proximal residues and therefore crosslinkable residues. This is advantageous when aiming at investigating proximal proteins for a comprehensive interaction network but somewhat disadvantageous for retrieving structural information to decipher protein complex topology (68). Besides differently sized spacer arms, developments regarding the spacer arm composition significantly improved the application of XL-MS in mitochondria. For instance, implementing affinity handles, such as biotin (e.g., Leiker), phosphonic acid (e.g., PhoX), as well as click chemistry–based enrichable azides or alkynes (e.g., disuccinimidyl bis-sulfoxide, cliXlink) into the spacer arm, can be used to tackle the low efficiency of the cross-linking reaction (69) and increase the number of identified cross-linked peptides (70, 71, 72, 73). Likewise, improving the membrane permeability of the reagent (e.g., tBu-PhoX) has been shown to lead to an increase in the cross-link density in mitochondria (74). Further improvements were achieved by integrating a labile moiety (e.g., sulfoxide, disuccinimidyl sulfoxide) into the spacer arm (75). So called “cleavable crosslinkers” can be cleaved in the gas phase during MS analysis, breaking a cross-linked peptide into its two entities and thereby facilitating peptide sequencing and cross-link identification. Besides modified spacer arms, crosslinkers with reactive groups harboring a specificity beyond lysine–lysine interactions such as 4-(4,6-dimethoxy[1,3,5]triazin-2-yl)-4-methyl-morpholinium chloride (76) and sulfosuccinimidyl 4,4′-azipentanoate (77) can improve the cross-link density in comparison to the most frequently used N-hydroxysuccinimide-reactive crosslinkers (e.g., disuccinimidyl sulfoxide, BS3) (78, 79).
Selection of a Suitable Crosslinker for the In Situ Analysis of PPIs in Mitochondria
Given the complexity of mitochondria, the selection of a suitable membrane-permeable crosslinker prior to MS analysis is extremely important when seeking to probe PPIs in situ. Crosslinkers with an enrichment handle or with a gas phase–cleavable spacer arm can simplify the subsequent MS analysis, increasing the number of identified crosslinks. Likewise, cross-link reagents with an alternative amino acid reactivity are of particular interest for applications in mitochondria, as hydrophobic patches of, for example, membrane proteins that often lack lysine residues can be targeted (Fig. 2A). Importantly, before each application, cross-linking conditions need to be carefully optimized (61). It is advisable to initially validate chosen cross-link conditions using SDS-PAGE or BN-PAGE (59, 78) to avoid over crosslinking and thus the generation of false-positive cross-linked proteins. In practice, multiple cross-linking reagents with varying spacer arm compositions and residue specificities are often combined, as each of them comes with certain pros and cons (34, 62, 63, 64). The parallel use of more than one cross-link reagent, with different residue specificities, has been shown to be especially beneficial for the structural characterization of mitochondrial protein complexes, providing more details on interaction interfaces (78, 80).
Fig. 2.
Experimental strategies for the analysis of proteome-wide protein–protein interactions in mitochondria.A, cross-link reagents comprise various chemistries. For cross-linking mitochondria, reagents with a cleavable spacer arm, enrichable handle, and/or an alternative amino-acid reactivity help to increase cross-link identifications. The DSSO reagent is depicted as example of a cleavable crosslinker. PhoX is depicted as an example for a crosslinker with an enrichment handle (phosphonic acid), and DMTMM is depicted as an example for a crosslinker with an alternative amino acid reactivity (targets carboxyl groups [glutamic acid and aspartic acid] and primary amines [lysines]). For each application, conditions such as reagent concentration and reaction time need to be optimized to achieve optimal crosslinking. B, after solubilization of mitochondrial membranes, proteins are enzymatically digested, producing a mixture of cross-linked and background linear peptides. Crosslinks can be formed within protein side chains (intralink) and between side chains of different proteins (inter link). In addition, linear peptides with the cross-linker attached (monolinks) are formed. C, to increase mass spectrometry (MS) identification, cross-linked peptides are separated from linear peptides using chromatographic methods, such as SCX, SEC, or Fe3+ IMAC enrichment. D, depending on the chosen crosslinker (e.g., gas phase–cleavable or noncleavable), different MS acquisition strategies can be applied to increase the likelihood of identifying a cross-linked peptide. Figure adapted with permission from Ref. (152). DMTMM, 4-(4,6-dimethoxy[1,3,5]triazin-2-yl)-4-methyl-morpholinium chloride. DSSO, disuccinimidyl sulfoxide; IMAC, immobilized metal affinity chromatography; SCX, strong-cation-exchange chromatography; SEC, size exclusion chromatography.
Protein Digestion
After the cross-linking reaction, mitochondria are commonly disrupted either mechanically (e.g., sonication) or by using detergents (e.g., Triton X-100, digitonin), and respective soluble proteins are enzymatically digested into peptides, generating intra cross-linked peptides (linked peptides from the same protein) and interprotein crosslinks (linked peptides from two distinct proteins). The majority of produced peptides, however, consist of linear (not crosslinked) and “monolinked” (linear peptide captured by only one “war-head” of the cross-linking reagent, with the other end being hydrolyzed) (69, 81) (Fig. 2B). Trypsin is typically used for the enzymatic digestion. Ryl et al. (62) introduced a sequential digestion workflow for in situ analysis of PPIs in mitochondria, in which tryptic peptides are additionally incubated with a second protease. To date, the use of a variety of different proteases as secondary digestion enzyme (e.g., elastase, AspN, GluC) has helped the identification of additional cross-linking sites, which were not accessible when using trypsin alone (82, 83, 84).
Enrichment of Cross-Linked Peptides
Because of the substoichiometric nature of the cross-linking reaction, linear and monolinked peptides (“background peptides”) are far more frequent and typically more abundant than cross-linked peptides (70), which significantly obstructs their MS-based identification of the latter. To alleviate the detection, cross-linked peptides can be enriched from the background peptides (Fig. 2C). For complex mitochondrial samples, this is commonly achieved by chromatographic methods such as strong-cation-exchange chromatography (SCX) (34, 64) or SEC (62, 63). SCX separates the typically doubly charged tryptic background peptides from the higher charged cross-linked peptides, whereas SEC separates smaller, linear, and monolinked peptides, from larger cross-linked peptides. A recently described two-dimensional fractionation workflow, which couples SEC and tip-based high pH reverse-phase fractionation, seems to further improve the cross-link identification in complex samples (85). These separation-based enrichment methods require quite some input material and also put quite a demand on MS instrument time, requiring the analysis of multiple fractions. Therefore, several groups have invested in designing cross-linking reagents with an affinity handle, such as a biotin group, a phosphonate group (PhoX, tBu-PhoX), or a click chemistry–based enrichable alkyne (cliXlink). Making use of these reagents, the cross-linked peptides can be enriched using a suitable material (e.g., streptavidin, Fe3+-immobilized metal affinity chromatography), thereby depleting the majority of background peptides (70, 73, 74, 86), and reducing the LC–MS analysis to just a single run. Unfortunately, AP does not separate monolinked peptides from cross-linked peptide pairs, whereas chromatographic separation methods like SCX and SEC do enable such a separation. Consequently, an improvement can be made by combining AP in conjunction with ion-exchange chromatography or SEC (64).
MS Acquisition and Software Analysis
Following enrichment, cross-linked peptides are subsequently subjected to LC–MS analysis for which different data acquisition strategies (MS2 or MS3 based) have been described to enable efficient identification (Fig. 2D). In contrast to classical bottom–up proteomics, where linear peptides are identified based on their accurate mass (measured on MS1 level) and sequence-specific fragment ions (measured on MS2 level), in XL-MS, two (covalently bound) linear peptides are cofragmented in the MS2, which significantly complicates the identification. The cofragmentation and the covalent linkage often impair an efficient fragmentation resulting in a compromised sequence coverage, hampering cross-link identification (87, 88). To enhance identification, MS2-based XL-MS acquisition methods commonly utilize multiple collision energies (collision-induced dissociation/higher-energy collisional dissociation) or combine complementary fragmentation methods such as collision-induced dissociation/higher-energy collisional dissociation with electron transfer dissociation. The optimal (combination of) fragmentation methods and activation energies are different for different cross-linker reagents (89, 90, 91). Besides MS2-based acquisition, MS3-based methods were introduced with the aim to achieve a more comprehensive sequence coverage (87) enhancing unambiguous identification of the cross-linked peptides. The introduced MS3 methods rely on the crosslinker harboring a gas phase–cleavable spacer arm (92), which is targeted in an initial survey scan (MS2) with a dissociation energy that specifically breaks the crosslinker and thus enables the individual sequencing of the initially linked peptides in a subsequent MS3 scan. Although MS3 acquisition methods hold a great potential for mitochondrial XL-MS studies by simplifying the data analysis (34, 64, 78), they currently do not yet outperform MS2 methods. As a result of decreased acquisition speed, and low sensitivity (88, 93), only a small number of cross-linked peptides (∼40%) are selected for subsequent MS3 fragmentation. However, recently, a novel MS3 triggering algorithm was introduced, which significantly enhances the selection of cross-linked peptides for MS3 analysis and subsequently identified crosslinks (94). In the final step, recorded XL-MS spectra can be analyzed by using various software suits, such as pLink2 (95), MeroX/StavroX (96, 97), Xi (98), Kojak (99), and XlinkX (59). Importantly, not all software solutions can analyze all MS-acquisition strategies or available cross-link reagents. Therefore, software capabilities should be cautiously taken into consideration when designing an XL-MS experiment.
Applications of XL-MS for the In Situ Characterization of Proteins and Protein Complexes in Mitochondria
To better understand biological processes within mitochondria, it is essential to untangle the plethora of known and unknown PPIs present in mitochondria and annotate their subcompartmental localization. As described previously, in many structural biology approaches, the in situ characterization of mitochondrial PPIs remains challenging. Therefore, the ability of XL-MS to covalently lock proximal proteins and to provide information regarding the spatial organization of amino acid residues in situ presents putative advantages. Although, most cross-linking reagents are not (fully) cell permeable, having issues in crossing the cell membrane, it has been shown that several reagents used do enter the mitochondrial (sub) compartments, when these are carefully isolated intact and naïve from cells or tissue (62, 64). When probing the in situ interaction landscape, it is important to ensure the purity and integrity of isolated mitochondria. Preparations with unwanted protein contaminations, for example, stemming from the copurification of endoplasmic reticula or ribosomes can severely reduce the cross-linking efficiency (100). Disruption of the mitochondrial structural integrity may cause proteins to (partially) lose their designated subcompartmental locations, thereby leading to the loss of PPIs and potentially also to an increase in non-natural “new” interactions. For this reason, additional quality control algorithms, like scoring pipelines developed for AP–MS experiments such as SAINT (101) or COMPASS (102), are beneficial to provide a metrics for confidence. Routinely, the quality of a proteome-wide XL-MS experiment (besides conventional false discovery rate calculations) is controlled by validating observed crosslinks against experimentally known protein structures. While this works well for purified protein complexes or less complex samples, this structure-based validation approach can lead to a significant underestimation of false positives in proteome-wide XL-MS studies (103). By implementing a set of four metrices, Yugandhar et al. (103) recently reported a comprehensive framework that facilitates the estimation of error rates in proteome-wide XL-MS experiments. Metrices include the validation of observed PPIs against databases containing known interactions along with the confirmation of newly observed interactions with ortholog experimental assays, such as yeast two-hybrid screening (104) or AP–MS, proximity labeling, or CP–MS. A vast amount of data (originating from AP–MS, BioID, APEX, CP–MS, and XL-MS experiments on mitochondria) has therefore been accumulated and deposited in databases such as CEDAR (https://www3.cmbi.umcn.nl/cedar/browse), PRIDE (https://www.ebi.ac.uk/pride/), MitCoM (https://viewer.complexomics.org/mitcom/), and several publications (39, 40, 46), forming a great resource for the validation of PPIs. Likewise, databases such as MitoCarta3 (31) and MitoMiner (105) provide valuable information and data to validate and enhance the characterization of identified PPIs.
The purity of mitochondria can be assessed by quantitation of cytosolic, nuclear, and mitochondrial marker proteins. In addition, the purity and structural integrity of mitochondria is often assessed by EM (34, 106) or by using activity assays like substrate-driven respiration (64) or luciferase-based measurements of ATP production rates (107). Furthermore, other functional assays such as measuring the ADP stimulation or the mitochondrial Ca2+-buffering capacities have been performed to test the integrity and activity of the purified mitochondria (108). Although functional assays provide direct evidence about functional integrity, these assays may still be blind, for observing that part of the isolated mitochondrial population has lost its structural integrity.
Deciphering the Suborganelle Interaction Landscape in Mitochondria
So far, several in situ PPI studies for a variety of different mitochondria have been performed, unraveling comprehensive networks with novel insights into mitochondrial complex topology. In human and mouse mitochondria for instance, XL-MS identified new interactions within complexes of the respiratory chain (34, 62), the TCA cycle as well as the mitochondrial contact site and mitochondrial contact site and cristae organizing system complex (64). Furthermore, by focusing on PPIs including proteins with known subcellular localization, XL-MS can yield detailed information about the relative subcellular localization for many proteins in the PPI network (109) (Fig. 3A).
Fig. 3.
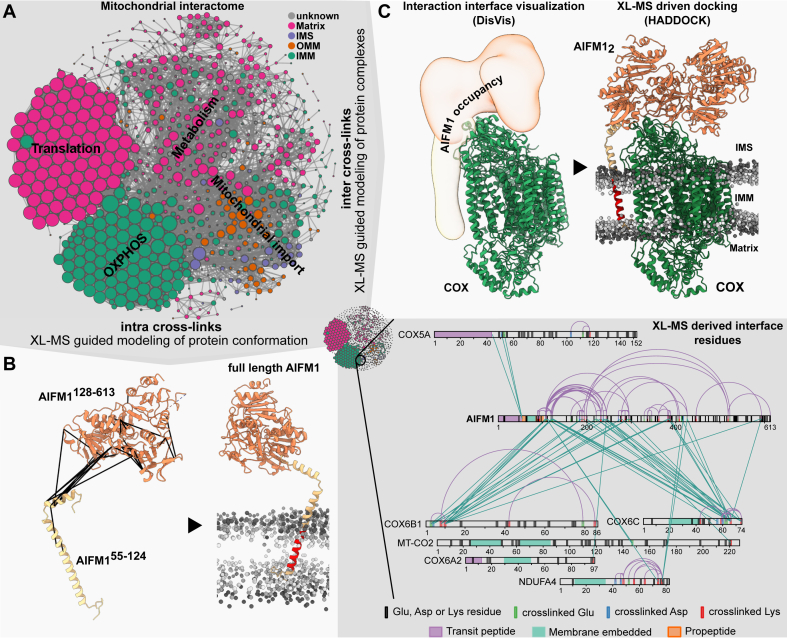
Applications of XL-MS for the characterization of proteins and protein complexes in mitochondria.A, overview of a mitochondrial protein–protein interaction (PPI) network. Node color represents the subcompartmental localization of proteins in mitochondria, whereas node size reflects its connectivity (the bigger the more interactions are reported). In the network, functionally related proteins tend to interact more frequently (see annotated clusters). The network was build using STRING interaction data (153) (minimum score = 0.8) for proteins that are listed in the MitoCarta 3.0 repository (31). XL-MS data also provide insights into the interaction interface of detected PPIs, as illustrated for the recently reported COX–AIFM1 interaction (bottom right). Purple colored links indicate intralinks. Green colored links indicate interlinks. Respective sequence and cross-link features are indicated accordingly. B, detected intra crosslinks mapped onto the structural models of the AIFM1 monomer as well as the de novo modeled N-terminal domain of AIFM1. Monomeric AIFM1 (dark orange) was modeled based on the previously resolved human homolog (Protein Data Bank: 4BUR, residues 128–516, 551–613). The N-terminal domain of AIFM1 (bright orange) (residues 55–124) was generated using trRosetta. The predicted transmembrane (TM) domain is highlighted in red. Intra crosslinks are used to generate a full-length structural model of AIFM1 (right model), highlighting how AIFM1 is integrated into the IMM. C, detected inter crosslinks can be used to structurally model protein complexes as shown for reported COX–AIFM12 complex. Interface residues are identified using DisVis and subsequently used in conjunction with the inter crosslinks to drive protein docking using Haddock. Adapted with permission from Ref. (78). AIFM1, apoptosis-inducing factor 1; COX, cytochrome c oxidase; IMM, inner mitochondrial membrane; XL-MS, cross-linking mass spectrometry.
Information about interface residues (Fig. 3A), together with higher spatial resolution of <3 nm compared with BioID (∼10–20 nm) and APEX (∼10–15 nm) (44, 110, 111), render XL-MS especially useful to decipher topologies of protein complexes across two compartments (e.g., IMM matrix, IMS matrix). Illustratively, XL-MS enabled to map the molecular environment of the IMS interface of the TOM–translocase of the inner membrane protein import machinery in vitro (112). Observed crosslinks provided an interaction map of the active translocase, with novel mechanistic insights regarding the protein precursor transport across the OMM and IMM. Likewise, XL-MS provided new insights into the arrangement of the OXPHOS complexes in mitochondria of Saccharomyces cerevisiae. In contrast to mammalian mitochondria, yeast lack a membrane-bound proton pumping complex I and instead possess an IMS (Nde1-2p) and matrix (Nde1p) residing NADH:ubiquinone oxidoreductase (113, 114). In situ cross-linking experiments suggested that, although located in the matrix, Nde1p is tethered to the IMM where it associates with complex III and IV into a supercomplex (Nde1p–CIII2–CIV2), comparable to the mammalian respirasome (supercomplex of complex I, III, and IV [CI–CIII2–CIV]) (63). Beyond deciphering the structural organization of mitochondrial complexes, XL-MS was recently applied to understand the mechanistic details of SS-31, a therapeutic peptide currently undergoing clinical trials for the use against multiple mitochondrial disorders (115). Observed inter crosslinks identified components relevant for the ATP production and the 2-oxoglutarate metabolism as frequent SS-31 interactors, thereby providing details regarding its mechanism of action and laying the foundation for follow-up functional studies (116).
Structural Characterization of Mitochondrial Protein Complexes
Apart from capturing the protein interaction landscape in mitochondria, XL-MS data provide structural information in terms of distance restraints than can be used to guide the structural characterization of proteins and protein complexes using computational modeling (68, 117). Homology modeling software such as I-Tasser (118), Robetta (119), and Modeller (120) utilize homologous protein structures as templates but in addition allow the utilization of crosslinks as spatial restraints for model building. In contrast, de novo structural prediction tools, such as AlphaFold2 (121) and RoseTTA-fold (122), enable the generation of structural models based solely on the sequence of a protein. However, for both, de novo or homology modeling approaches, finding the biological relevant conformation as well as validation of the retrieved models can be time consuming and challenging with only having the provided output scores at hand. Accordingly, intra crosslinks are a valuable resource to aid the modeling and validation process as well as to pinpoint toward biologically relevant structural conformations (123, 124). Unfortunately, XL-MS does not (yet) provide atomic-scale resolution, and therefore, modeling does mostly provide data on for instance the relative orientation of globular domains within a protein rather than providing atomic details on secondary and tertiary structural elements (125). The benefit of sourcing XL-MS data for the structural characterization of mitochondrial proteins was recently shown for several heat shock proteins (62) and the apoptosis-inducing factor 1 (AIFM1) (78). For the latter, XL restraints were used to guide the modeling of the full-length protein, including the hitherto structurally unresolved N-terminal region (residues 55–124), revealing how AIFM1 might be N-terminally tethered to the IMM (Fig. 3B).
Inter crosslinks provide information about the arrangement of proteins within a complex, which can be used to guide or validate structural models (126). These crosslinks are especially useful for ab initio protein–protein docking (127, 128), as it simplifies the sampling and scoring of the theoretically possible conformational spaces (127, 129). Accordingly, inter crosslinks between cytochrome c oxidase (COX) and AIFM1 were utilized to elucidate active interface residues and subsequently to model the respective COX–AIFM1 complex (Fig. 3C) (78). The final structural model contradicted direct or indirect (via Cytc) electron transport between AIFM1 and COX, excluding a functional role within the active ETC but rather hinting towards an involvement in OXPHOS biogenesis or apoptosis. Likewise, inter crosslinks in combination with protein–protein docking revealed novel insights into the OXPHOS biogenesis in S. cerevisiae, where Min8 seems to promote COX subunit 12 assembly into COX (63).
Besides guiding protein–protein docking, cross-linking data were shown to benefit artificial intelligence–driven de novo complex predictions (130). Alphafold2-Multimer (131) in combination with cross-linking data was most recently used to explore the architecture of the 2-oxoglutarate dehydrogenase complex in bovine heart mitochondria (80). Next to its three catalytically active subunits (E1, E2, and E3), XL-MS confirmed that the miss-named mitochondrial ribosomal protein S36 (MRPS36) is actually an ubiquitous fourth member, exclusive to the eukaryotic 2-oxoglutarate dehydrogenase complex, as also suggested by a previous study (132). The generated model highlights how E1, E3, and MRPS36 are organized with respect to the octahedral E2 core and how all components assemble into a functional metabolon of over approximately 3.45 MDa to act as a crucial component of the TCA cycle. Notwithstanding, experimental cross-linking restraints in combination with de novo modeling have only been used to guide candidate selection for complex prediction and output validation (80, 130, 133). Most recently, AlphaLink, a framework directly incorporating crosslinks as additional restraints into the AlphaFold2 protein prediction algorithm, was introduced (134). AlphaLink provides improved model predictions in respect to the artificial intelligence–only assisted workflow, potentially enabling the prediction of specific structural states of protein and protein complexes.
Apart from being a valuable input for the introduced computational modeling approaches, cross-linking restraints were used to support the structural analysis of mitochondrial protein complexes via cryo-EM/tomography (135). XL-MS in combination with cryo-EM unveiled stoichiometries and proximity principles of the mammalian pyruvate dehydrogenase complex (136). Moreover, XL-MS aided the assignment of ambiguous densities obtained from in situ cryo-ET, deciphering structural details of the cytoskeleton (137) and the mitochondria–cytoskeleton interface in mammalian sperm (138). In the latter study, subtomogram averaging identified ordered protein arrays on the surface of sperm mitochondria, which guided by XL-MS could be identified to consist of glycerol kinase-like proteins anchored on lattices of voltage-dependent anion-selective channel proteins in the OMM.
Remaining Challenges and Future Perspectives for the XL-MS-Based In Situ Characterization of Protein Complexes in Mitochondria
In this review, we described and highlighted several in situ XL-MS studies that have contributed toward a deeper understanding of how the mitochondrial proteome is wired. Notwithstanding these successes, several challenges remain, some of them more generic for XL-MS, and others more specific for the in situ crosslinking of mitochondria.
As mentioned earlier, the substoichiometric reaction efficiencies as well as the impaired MS fragmentation and thus identification of covalently linked peptides remain major challenges for XL-MS in general and thus also for in situ cross-linking studies. Consequently, identification of crosslinks is, as in many proteomics experiments, highly biased toward the more abundant proteins, such as those belonging to the OXPHOS or TCA cycle (Fig. 4) (34, 62, 78). In line, conducting XL-MS analysis on mitochondria is presently constrained to using purified mitochondria. This limitation arises because of the increased complexity when working with intact cells, which includes the less permeable cell membrane and the presence of numerous highly abundant complexes in the cytoplasm (e.g., ribosomes), both of which are likely to impede the access of cross-linking reagents to mitochondria. To address these challenges, further optimizations and innovations are needed. Developments aiming at improving the selectivity and fragmentation of cross-linked peptides within the mass spectrometer (93, 94), as well as innovative data analysis strategies (139), may hold potential to increase our ability to detect crosslinks in situ, especially across a broader protein abundance range. In addition, the recent development of a targeted cross-linker delivery methodology enabled the in situ crosslinking of mitochondria in living cells. Here, functionalized nanoparticles are used to successfully deliver cross-link reagents into mitochondria, resulting in the identification of several novel PPIs, which very likely only exist in the context of an intact cellular environment (140).
Fig. 4.
Challenges for the in situ characterization of protein complexes in mitochondria using XL-MS.A, detected cross-linked proteins are generally more abundant. Boxplot with protein abundances for all identified proteins, all cross-linked proteins, and proteins that have either only inter-, intra-, or both (inter and intra) links. The number of proteins and the median iBAQ is indicated on top of each box. B, when performing XL-MS in situ in mitochondria, inter crosslinks are observed for over more than 1000 binary pairs. However, pairs identified with at least five unique inter crosslinks involve exclusively proteins belonging to the abundant OXPHOS or the TCA cycle assemblies, highlighted with red dots. Adapted with permission from Ref. (78). iBAQ, intensity-Based Absolute Quantification; OXPHOS, oxidative phosphorylation; TCA, tricarboxylic acid; XL-MS, cross-linking mass spectrometry.
Another limiting factor for in situ XL-MS is the diverse protein complex landscape within mitochondria, whereby certain proteins can be found across several assemblies, as observed for the well-characterized respiratory supercomplexes (54). While cross-link restraints provide information about the physical interaction for an observed PPI, the identification of respective assembly state(s) and interaction modules is often hampered (141). As understanding the exact stoichiometry is important to guarantee the accurate structural characterization and thus functional characterization of protein complexes, methods aiming at supporting the confident assignment of complex stoichiometries are needed. A promising, time-effective, and cost-effective approach involves the combination of XL-MS with BN-PAGE–based CP–MS. This consolidation not only provided detailed insights into the macromolecular organization of mitochondrial complexes but also enabled the generation of assembly-specific cross-link restraints as recently reviewed (51, 52). Other native-like separation techniques, such as SEC or density gradient ultracentrifugation, have been used to provide detailed information about the composition and abundance of co-occurring assembly states of complexes (142, 143). BN-PAGE has the advantage of providing high resolution and great separation and can be performed with very small amounts of sample (∼10 μg of protein), which is especially beneficial when analyzing dysfunctional mitochondria from patient samples for which usually only very limited amounts are available. SEC and density gradient ultracentrifugation might also be beneficial for low abundant protein complexes in which scenario the limited loading capacity of a gel could be a disadvantage. In addition, the separated protein complexes stay in solution and are therefore readily available for additional analysis, for example, by cryo-EM (144), while in the case of BN-PAGE, the complexes would need to be extracted from the gel pieces.
Besides isolating specific assembly states, combining APEX with XL-MS (as mentioned earlier in this review) provides a great tool for retrieving subcompartment specific crosslinks for PPIs and protein complexes (50, 145). This is especially useful when interested in the structural analysis of low abundant proteins and protein complexes residing in subcellular compartments such as the mitochondrial IMS that are hard to purify. With no purification possible, low abundant proteins and protein complexes would likely not be detected in a proteome-wide XL-MS experiment, but these could be specifically targeted by using a combined APEX-XL-MS (APEX-CXMS) approach (50).
Another promising direction of development regards the (relative) quantification of cross-linked peptides, enabling the monitoring of changes observed for PPIs between different states of a system (e.g., drug treatment or a disease state). First data have already been reported by Chavez et al. (146), who introduced a stable isotope–labeled crosslink reagent, used to investigate the dynamic nature of mitochondrial PPIs within differently aged skeletal muscle tissue (147). Although less accurate, also label-free quantification strategies can be applied, that may be performed with all available crosslinkers, as reviewed (98, 148). For label-free quantification, MS1 signals of individual cross-linked peptides are integrated over their chromatographic elution profile. While stable isotope–labeled crosslinkers are mostly limited to pairwise comparisons, the low abundant nature of cross-linked peptides significantly complicates label-free MS1-based quantification. Likewise, the lack of automated quantification tools is restricting the frequent usage of quantitative XL-MS (98). Introducing novel workflows, for example, utilizing tandem mass tags for the MS2-based quantitation as demonstrated recently (149) or performing more targeted MS approaches, for example, data-independent acquisition (150) or parallel reaction monitoring (151), could provide alternative promising approaches toward reliable quantitation of crosslinks.
Conclusion
As highlighted in this review, technical advances and new workflows have made in situ XL-MS a powerful tool to explore and characterize the complex mitochondrial PPI landscape. Compared with the other MS-based methods for the detection of PPIs in mitochondria, like AP–MS or BioID, XL-MS is conceptually quite different as it provides narrowly restricted distance restraints, which are advantageous for the annotation of a protein’s subcellular localization. Likewise, distance restraints can be leveraged for the structural characterization of proteins and protein complexes, especially when used in combination with structural modeling. Current limitations include the low reaction efficiency, resulting in only a limited number of detectable crosslinks, mainly observed for the most abundant assemblies. Nevertheless, the design of innovative cross-link reagents and workflows already provided valuable insights into the mitochondrial PPI network with several novel discoveries. As XL-MS is rapidly gaining attention across different scientific communities, it is likely that more innovations will follow soon, further strengthening the capacities of XL-MS to study the mitochondrial interactome.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We acknowledge support from the Netherlands Organization for Scientific Research (NWO) funding the Netherlands Proteomics Centre through the X-omics Road Map program (project no.: 184.034.019).
Funding and additional information
A. J. R. H. acknowledges further support by NWO through the Spinoza Award SPI.2017.028.
Author contributions
J. F. H. and A. J. R. H. writing–review & editing.
References
- 1.Giacomello M., Pyakurel A., Glytsou C., Scorrano L. The cell biology of mitochondrial membrane dynamics. Nat. Rev. Mol. Cell Biol. 2020;21:204–224. doi: 10.1038/s41580-020-0210-7. [DOI] [PubMed] [Google Scholar]
- 2.Guo R.Y., Gu J.K., Wu M., Yang M.J. Amazing structure of respirasome: unveiling the secrets of cell respiration. Protein Cell. 2016;7:854–865. doi: 10.1007/s13238-016-0329-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X.S., Kim C.N., Yang J., Jemmerson R., Wang X.D. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 4.Nass M.M.K., Nass S. Intramitochondrial fibers with DNA characteristics.1. Fixation and electron staining reactions. J. Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belenguer P., Duarte J.M.N., Schuck P.F., Ferreira G.C. Mitochondria and the brain: bioenergetics and beyond. Neurotox. Res. 2019;36:219–238. doi: 10.1007/s12640-019-00061-7. [DOI] [PubMed] [Google Scholar]
- 6.Mohanty A., Tiwari-Pandey R., Pandey N.R. Mitochondria: the indispensable players in innate immunity and guardians of the inflammatory response. J. Cell Commun. Signal. 2019;13:303–318. doi: 10.1007/s12079-019-00507-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C.X., Youle R.J. The role of mitochondria in apoptosis. Annu. Rev. Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iovine J.C., Claypool S.M., Alder N.N. Mitochondrial compartmentalization: emerging themes in structure and function. Trends Biochem. Sci. 2021;46:902–917. doi: 10.1016/j.tibs.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J.S., Kim J., Park S., Jeon J., Shin Y.E., Kim S. Spatial and functional organization of mitochondrial protein network. Sci. Rep. 2013;3:1403. doi: 10.1038/srep01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Javadov S., Kozlov A.V., Camara A.K.S. Mitochondria in health and diseases. Cells-Basel. 2020;9:1177. doi: 10.3390/cells9051177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milenkovic D., Misic J., Hevler J.F., Molinié T., Chung I., Atanassov I., et al. Preserved respiratory chain capacity and physiology in mice with profoundly reduced levels of mitochondrial respirasomes. Cell Metab. 2023;35:1799–1813.e7. doi: 10.1016/j.cmet.2023.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Vogel F., Bornhovd C., Neupert W., Reichert A.S. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wurm C.A., Jakobs S. Differential protein distributions define two sub-compartments of the mitochondrial inner membrane in yeast. FEBS Lett. 2006;580:5628–5634. doi: 10.1016/j.febslet.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Dolder M., Wendt S., Wallimann T. Mitochondrial creatine kinase in contact sites: interaction with porin and adenine nucleotide translocase, role in permeability transition and sensitivity to oxidative damage. Biol. Signals Recept. 2001;10:93–111. doi: 10.1159/000046878. [DOI] [PubMed] [Google Scholar]
- 16.Schwaiger M., Herzog V., Neupert W. Characterization of translocation contact sites involved in the import of mitochondrial proteins. J. Cell Biol. 1987;105:235–246. doi: 10.1083/jcb.105.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panek T., Elias M., Vancova M., Lukes J., Hashimi H. Returning to the fold for lessons in mitochondrial crista diversity and evolution. Curr. Biol. 2020;30:R575–R588. doi: 10.1016/j.cub.2020.02.053. [DOI] [PubMed] [Google Scholar]
- 18.Kuhlbrandt W. Structure and function of mitochondrial membrane protein complexes. BMC Biol. 2015;13:89. doi: 10.1186/s12915-015-0201-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards R., Eaglesfield R., Tokatlidis K. The mitochondrial intermembrane space: the most constricted mitochondrial sub-compartment with the largest variety of protein import pathways. Open Biol. 2021;11 doi: 10.1098/rsob.210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrmann J.M., Riemer J. The intermembrane space of mitochondria. Antioxid. Redox Signal. 2010;13:1341–1358. doi: 10.1089/ars.2009.3063. [DOI] [PubMed] [Google Scholar]
- 21.Stephan T., Brüser C., Deckers M., Steyer A.M., Balzarotti F., Barbot M., et al. MICOS assembly controls mitochondrial inner membrane remodeling and crista junction redistribution to mediate cristae formation. EMBO J. 2020;39 doi: 10.15252/embj.2019104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 23.Frey T.G., Mannella C.A. The internal structure of mitochondria. Trends Biochem. Sci. 2000;25:319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 24.Gottschalk B., Madreiter-Sokolowski C.T., Graier W.F. Cristae junction as a fundamental switchboard for mitochondrial ion signaling and bioenergetics. Cell Calcium. 2022;101 doi: 10.1016/j.ceca.2021.102517. [DOI] [PubMed] [Google Scholar]
- 25.Rampelt H., Zerbes R.M., van der Laan M., Pfanner N. Role of the mitochondrial contact site and cristae organizing system in membrane architecture and dynamics. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:737–746. doi: 10.1016/j.bbamcr.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Reyes I., Chandel N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spinelli J.B., Haigis M.C. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018;20:745–754. doi: 10.1038/s41556-018-0124-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anna S., Ludovic R., Alexandre S., Yves M., Ivan T. Suborganellar localization of mitochondrial proteins and transcripts in human cells. Methods Mol. Biol. 2021;2277:157–173. doi: 10.1007/978-1-0716-1270-5_11. [DOI] [PubMed] [Google Scholar]
- 29.Appelhans T., Busch K.B. Dynamic imaging of mitochondrial membrane proteins in specific sub-organelle membrane locations. Biophys. Rev. 2017;9:345–352. doi: 10.1007/s12551-017-0287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emanuelsson O., Nielsen H., Brunak S., von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 31.Rath S., Sharma R., Gupta R., Ast T., Chan C., Durham T.J., et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541–D1547. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Appelhans T., Richter C.P., Wilkens V., Hess S.T., Piehler J., Busch K.B. Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy. Nano Lett. 2012;12:610–616. doi: 10.1021/nl203343a. [DOI] [PubMed] [Google Scholar]
- 33.Lobingier B.T., Hüttenhain R., Eichel K., Miller K.B., Ting A.Y., von Zastrow M., Krogan N.J. An approach to spatiotemporally resolve protein interaction networks in living cells. Cell. 2017;169:350–360.e12. doi: 10.1016/j.cell.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F., Lossl P., Rabbitts B.M., Balaban R.S., Heck A.J.R. The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol. Cell. Proteomics. 2018;17:216–232. doi: 10.1074/mcp.RA117.000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ader N.R., Hoffmann P.C., Ganeva I., Borgeaud A.C., Wang C., Youle R.J., Kukulski W. Molecular and topological reorganizations in mitochondrial architecture interplay during Bax-mediated steps of apoptosis. Elife. 2019;8 doi: 10.7554/eLife.40712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y.F., Lin S.S., Chen J.M., Tsai H.Z., Hsieh T.S., Fu C.Y. Electron tomographic analysis reveals ultrastructural features of mitochondrial cristae architecture which reflect energetic state and aging. Sci. Rep. 2017;7 doi: 10.1038/srep45474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hylton R.K., Swulius M.T. Challenges and triumphs in cryo-electron tomography. iScience. 2021;24 doi: 10.1016/j.isci.2021.102959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 39.Floyd B.J., Wilkerson E.M., Veling M.T., Minogue C.E., Xia C., Beebe E.T., et al. Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol. Cell. 2016;63:621–632. doi: 10.1016/j.molcel.2016.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgenstern M., Peikert C.D., Lübbert P., Suppanz I., Klemm C., Alka O., et al. Quantitative high-confidence human mitochondrial proteome and its dynamics in cellular context. Cell Metab. 2021;33:2464–2483.e18. doi: 10.1016/j.cmet.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunham W.H., Mullin M., Gingras A.C. Affinity-purification coupled to mass spectrometry: basic principles and strategies. Proteomics. 2012;12:1576–1590. doi: 10.1002/pmic.201100523. [DOI] [PubMed] [Google Scholar]
- 42.Herrmann J.M., Westermann B. Analysis of protein-protein interactions in mitochondria. Methods Cell Biol. 2007;80:743–759. doi: 10.1016/S0091-679X(06)80034-8. [DOI] [PubMed] [Google Scholar]
- 43.Yugandhar K., Gupta S., Yu H. Inferring protein-protein interaction networks from mass spectrometry-based proteomic approaches: a mini-review. Comput. Struct. Biotechnol. J. 2019;17:805–811. doi: 10.1016/j.csbj.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martell J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonicka H., Lin Z.Y., Janer A., Aaltonen M.J., Weraarpachai W., Gingras A.C., Shoubridge E.A. A high-density human mitochondrial proximity interaction network. Cell Metab. 2020;32:479–497. doi: 10.1016/j.cmet.2020.07.017. [DOI] [PubMed] [Google Scholar]
- 47.Rhee H.W., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C.L., Perrimon N. Proximity-dependent labeling methods for proteomic profiling in living cells. Wiley Interdiscip. Rev. Dev. Biol. 2017 doi: 10.1002/wdev.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X., Salokas K., Weldatsadik R.G., Gawriyski L., Varjosalo M. Combined proximity labeling and affinity purification-mass spectrometry workflow for mapping and visualizing protein interaction networks. Nat. Protoc. 2020;15:3182–3211. doi: 10.1038/s41596-020-0365-x. [DOI] [PubMed] [Google Scholar]
- 50.Sun M., Yuan F., Tang Y., Zou P., Lei X. Subcellular interactomes revealed by merging APEX with cross-linking mass spectrometry. Anal. Chem. 2022;94:14878–14888. doi: 10.1021/acs.analchem.2c02116. [DOI] [PubMed] [Google Scholar]
- 51.Wittig I., Malacarne P.F. Complexome profiling: assembly and remodeling of protein complexes. Int. J. Mol. Sci. 2021;22:7809. doi: 10.3390/ijms22157809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabrera-Orefice A., Potter A., Evers F., Hevler J.F., Guerrero-Castillo S. Complexome profiling-exploring mitochondrial protein complexes in health and disease. Front. Cell Dev. Biol. 2022;9 doi: 10.3389/fcell.2021.796128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulte U., den Brave F., Haupt A., Gupta A., Song J., Müller C.S., et al. Mitochondrial complexome reveals quality-control pathways of protein import. Nature. 2023;614:153–159. doi: 10.1038/s41586-022-05641-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schagger H., Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshinaka T., Kosako H., Yoshizumi T., Furukawa R., Hirano Y., Kuge O., et al. Structural basis of mitochondrial scaffolds by prohibitin complexes: insight into a role of the coiled-coil region. iScience. 2019;19:1065–1078. doi: 10.1016/j.isci.2019.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin C.S., Meng S., Garbis S.D., Moradian A., Taylor R.W., Sweredoski M.J., et al. LONP1 and mtHSP70 cooperate to promote mitochondrial protein folding. Nat. Commun. 2021;12:265. doi: 10.1038/s41467-020-20597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X.N., Salokas K., Tamene F., Jiu Y., Weldatsadik R.G., Öhman T., Varjosalo M. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 2018;9:1188. doi: 10.1038/s41467-018-03523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heide H., Bleier L., Steger M., Ackermann J., Dröse S., Schwamb B., et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell Metab. 2012;16:538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Klykov O., Steigenberger B., Pektaş S., Fasci D., Heck A.J.R., Scheltema R.A. Efficient and robust proteome-wide approaches for cross-linking mass spectrometry. Nat. Protoc. 2018;13:2964–2990. doi: 10.1038/s41596-018-0074-x. [DOI] [PubMed] [Google Scholar]
- 60.Piersimoni L., Kastritis P.L., Arlt C., Sinz A. Cross-linking mass spectrometry for investigating protein conformations and protein-protein interactions-A method for all seasons. Chem. Rev. 2022;122:7500–7531. doi: 10.1021/acs.chemrev.1c00786. [DOI] [PubMed] [Google Scholar]
- 61.O'Reilly F.J., Rappsilber J. Cross-linking mass spectrometry: methods and applications in structural, molecular and systems biology. Nat. Struct. Mol. Biol. 2018;25:1000–1008. doi: 10.1038/s41594-018-0147-0. [DOI] [PubMed] [Google Scholar]
- 62.Ryl P.S.J., Bohlke-Schneider M., Lenz S., Fischer L., Budzinski L., Stuiver M., et al. In situ structural restraints from cross-linking mass spectrometry in human mitochondria. J. Proteome Res. 2020;19:327–336. doi: 10.1021/acs.jproteome.9b00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linden A., Deckers M., Parfentev I., Pflanz R., Homberg B., Neumann P., et al. A cross-linking mass spectrometry approach defines protein interactions in yeast mitochondria. Mol. Cell. Proteomics. 2020;19:1161–1178. doi: 10.1074/mcp.RA120.002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schweppe D.K., Chavez J.D., Lee C.F., Caudal A., Kruse S.E., Stuppard R., et al. Mitochondrial protein interactome elucidated by chemical cross-linking mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2017;114:1732–1737. doi: 10.1073/pnas.1617220114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rappsilber J., Siniossoglou S., Hurt E.C., Mann M. A generic strategy to analyze the spatial organization of multi-protein complexes by cross-linking and mass spectrometry. Anal. Chem. 2000;72:267–275. doi: 10.1021/ac991081o. [DOI] [PubMed] [Google Scholar]
- 67.Steigenberger B., Albanese P., Heck A.J.R., Scheltema R.A. To cleave or not to cleave in XL-MS? J. Am. Soc. Mass Spectrom. 2020;31:196–206. doi: 10.1021/jasms.9b00085. [DOI] [PubMed] [Google Scholar]
- 68.Orban-Nemeth Z., Beveridge R., Hollenstein D.M., Rampler E., Stranzl T., Hudecz O., et al. Structural prediction of protein models using distance restraints derived from cross-linking mass spectrometry data. Nat. Protoc. 2018;13:478–494. doi: 10.1038/nprot.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leitner A., Walzthoeni T., Kahraman A., Herzog F., Rinner O., Beck M., Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol. Cell. Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steigenberger B., Pieters R.J., Heck A.J.R., Scheltema R.A. PhoX: an IMAC-enrichable cross-linking reagent. ACS Cent. Sci. 2019;5:1514–1522. doi: 10.1021/acscentsci.9b00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matzinger M., Kandioller W., Doppler P., Heiss E.H., Mechtler K. Fast and highly efficient affinity enrichment of azide-A-DSBSO cross-linked peptides. J. Proteome Res. 2020;19:2071–2079. doi: 10.1021/acs.jproteome.0c00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan D., Li Q., Zhang M.J., Liu C., Ma C., Zhang P., et al. Trifunctional cross-linker for mapping protein-protein interaction networks and comparing protein conformational states. Elife. 2016;5 doi: 10.7554/eLife.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stadlmeier M., Runtsch L.S., Streshnev F., Wuhr M., Carell T. A click-chemistry-based enrichable crosslinker for structural and protein interaction analysis by mass spectrometry. Chembiochem. 2020;21:103–107. doi: 10.1002/cbic.201900611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang P.L., Wang C., Diehl A., Viner R., Etienne C., Nandhikonda P., et al. A membrane-permeable and immobilized metal affinity chromatography (IMAC) enrichable cross-linking reagent to advance in vivo cross-linking mass spectrometry. Angew. Chem. Int. Ed. Engl. 2022;61 doi: 10.1002/anie.202113937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kao A., Chiu C.L., Vellucci D., Yang Y., Patel V.R., Guan S., et al. Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leitner A., Joachimiak L.A., Unverdorben P., Walzthoeni T., Frydman J., Förster F., Aebersold R. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomes A.F., Gozzo F.C. Chemical cross-linking with a diazirine photoactivatable cross-linker investigated by MALDI- and ESI-MS/MS. J. Mass Spectrom. 2010;45:892–899. doi: 10.1002/jms.1776. [DOI] [PubMed] [Google Scholar]
- 78.Hevler J.F., Zenezeni Chiozzi R., Cabrera-Orefice A., Brandt U., Arnold S., Heck A.J.R. Molecular characterization of a complex of apoptosis-inducing factor 1 with cytochrome c oxidase of the mitochondrial respiratory chain. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2106950118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Belsom A., Schneider M., Fischer L., Brock O., Rappsilber J. Serum albumin domain structures in human blood serum by mass spectrometry and computational biology. Mol. Cell. Proteomics. 2016;15:1105–1116. doi: 10.1074/mcp.M115.048504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hevler J.F., Albanese P., Cabrera-Orefice A., Potter A., Jankevics A., Misic J., et al. MRPS36 provides a structural link in the eukaryotic 2-oxoglutarate dehydrogenase complex. Open Biol. 2023;13 doi: 10.1098/rsob.220363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sinnott M., Malhotra S., Madhusudhan M.S., Thalassinos K., Topf M. Combining information from crosslinks and monolinks in the modeling of protein structures. Structure. 2020;28:1061–1070.e3. doi: 10.1016/j.str.2020.05.012. [DOI] [PubMed] [Google Scholar]
- 82.Mendes M.L., Fischer L., Chen Z.A., Barbon M., O'Reilly F.J., Giese S.H., et al. An integrated workflow for crosslinking mass spectrometry. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20198994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dau T., Gupta K., Berger I., Rappsilber J. Sequential digestion with trypsin and elastase in cross-linking mass spectrometry. Anal. Chem. 2019;91:4472–4478. doi: 10.1021/acs.analchem.8b05222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leitner A., Reischl R., Walzthoeni T., Herzog F., Bohn S., Förster F., Aebersold R. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiao F.L., Yu C., Wheat A., Wang X., Rychnovsky S.D., Huang L. Two-dimensional fractionation method for proteome-wide cross-linking mass spectrometry analysis. Anal. Chem. 2022;94:4236–4242. doi: 10.1021/acs.analchem.1c04485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kang S., Mou L., Lanman J., Velu S., Brouillette W.J., Prevelige P.E. Synthesis of biotin-tagged chemical cross-linkers and their applications for mass spectrometry. Rapid Commun. Mass Spectrom. 2009;23:1719–1726. doi: 10.1002/rcm.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu F., Lossl P., Scheltema R., Viner R., Heck A.J.R. Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat. Commun. 2017;8 doi: 10.1038/ncomms15473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stieger C.E., Doppler P., Mechtler K. Optimized fragmentation improves the identification of peptides cross-linked by MS-cleavable reagents. J. Proteome Res. 2019;18:1363–1370. doi: 10.1021/acs.jproteome.8b00947. [DOI] [PubMed] [Google Scholar]
- 89.Liu F., Rijkers D.T.S., Post H., Heck A.J.R. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry (vol 12, pg 1179, 2015) Nat. Methods. 2017;12:1179–1184. doi: 10.1038/nmeth.3603. [DOI] [PubMed] [Google Scholar]
- 90.Frese C.K., Altelaar A.F.M., van den Toorn H., Nolting D., Griep-Raming J., Heck A.J.R., Mohammed S. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 2012;84:9668–9673. doi: 10.1021/ac3025366. [DOI] [PubMed] [Google Scholar]
- 91.Campbell J.L., Hager J.W., Le Blanc J.C.Y. On performing simultaneous electron transfer dissociation and collision-induced dissociation on multiply protonated peptides in a linear ion trap. J. Am. Soc. Mass Spectrom. 2009;20:1672–1683. doi: 10.1016/j.jasms.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 92.Sinz A. Divide and conquer: cleavable cross-linkers to study protein conformation and protein-protein interactions. Anal. Bioanal. Chem. 2017;409:33–44. doi: 10.1007/s00216-016-9941-x. [DOI] [PubMed] [Google Scholar]
- 93.Kolbowski L., Lenz S., Fischer L., Sinn L.R., O'Reilly F.J., Rappsilber J. Improved peptide backbone fragmentation is the primary advantage of MS-cleavable crosslinkers. Anal. Chem. 2022;94:7779–7786. doi: 10.1021/acs.analchem.1c05266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kolbowski L., Fischer L., Rappsilber J. Cleavable crosslinkers redefined by novel MS3-trigger algorithm. bioRxiv. 2023 doi: 10.1101/2023.01.26.525676. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen Z.L., Meng J.M., Cao Y., Yin J.L., Fang R.Q., Fan S.B., et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 2019;10:3404. doi: 10.1038/s41467-019-11337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gotze M., Pettelkau J., Schaks S., Bosse K., Ihling C.H., Krauth F., et al. StavroX-A software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 2012;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 97.Gotze M., Pettelkau J., Fritzsche R., Ihling C.H., Schäfer M., Sinz A. Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J. Am. Soc. Mass Spectrom. 2015;26:83–97. doi: 10.1007/s13361-014-1001-1. [DOI] [PubMed] [Google Scholar]
- 98.Chen Z.A., Rappsilber J. Quantitative cross-linking/mass spectrometry to elucidate structural changes in proteins and their complexes. Nat. Protoc. 2019;14:171–201. doi: 10.1038/s41596-018-0089-3. [DOI] [PubMed] [Google Scholar]
- 99.Hoopmann M.R., Zelter A., Johnson R.S., Riffle M., MacCoss M.J., Davis T.N., Moritz R.L. Kojak: efficient analysis of chemically cross-linked protein complexes. J. Proteome Res. 2015;14:2190–2198. doi: 10.1021/pr501321h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fursch J., Kammer K.M., Kreft S.G., Beck M., Stengel F. Proteome-wide structural probing of low-abundant protein interactions by cross-linking mass spectrometry. Anal. Chem. 2020;92:4016–4022. doi: 10.1021/acs.analchem.9b05559. [DOI] [PubMed] [Google Scholar]
- 101.Choi H., Larsen B., Lin Z.Y., Breitkreutz A., Mellacheruvu D., Fermin D., et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yugandhar K., Wang T.Y., Wierbowski S.D., Shayhidin E.E., Yu H. Structure-based validation can drastically underestimate error rate in proteome-wide cross-linking mass spectrometry studies. Nat. Methods. 2020;17:985–988. doi: 10.1038/s41592-020-0959-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rodriguez-Negrete E., Bejarano E.R., Castillo A.G. Using the yeast two-hybrid system to identify protein-protein interactions. Methods Mol. Biol. 2014;1072:241–258. doi: 10.1007/978-1-62703-631-3_18. [DOI] [PubMed] [Google Scholar]
- 105.Smith A.C., Robinson A.J. MitoMiner v4.0: an updated database of mitochondrial localization evidence, phenotypes and diseases. Nucleic Acids Res. 2019;47:D1225–D1228. doi: 10.1093/nar/gky1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Englmeier R., Pfeffer S., Forster F. Structure of the human mitochondrial ribosome studied in situ by cryoelectron tomography. Structure. 2017;25:1574–1581.e2.. doi: 10.1016/j.str.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 107.Lanza I.R., Nair K.S. Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 2009;457:349–372. doi: 10.1016/S0076-6879(09)05020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liao P.C., Bergamini C., Fato R., Pon L.A., Pallotti F. Isolation of mitochondria from cells and tissues. Mitochondria. 2020;155:3–31. doi: 10.1016/bs.mcb.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhu Y., Akkaya K.C., Borges Lima D., Wang C., Lehmann M., Liu F. Cross-link assisted spatial proteomics to map sub-organelle proteomes and membrane protein topology. bioRxiv. 2022 doi: 10.1101/2022.05.05.490733. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xu Y.F., Fan X.Q., Hu Y. In vivo interactome profiling by enzyme-catalyzed proximity labeling. Cell Biosci. 2021;11:27. doi: 10.1186/s13578-021-00542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Merkley E.D., Rysavy S., Kahraman A., Hafen R.P., Daggett V., Adkins J.N. Distance restraints from crosslinking mass spectrometry: mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 2014;23:747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gomkale R., Linden A., Neumann P., Schendzielorz A.B., Stoldt S., Dybkov O., et al. Mapping protein interactions in the active TOM-TIM23 supercomplex. Nat. Commun. 2021;12:5715. doi: 10.1038/s41467-021-26016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lasserre J.P., Dautant A., Aiyar R.S., Kucharczyk R., Glatigny A., Tribouillard-Tanvier D., et al. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis. Model. Mech. 2015;8:509–526. doi: 10.1242/dmm.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Velazquez I., Pardo J.P. Kinetic characterization of the rotenone-insensitive internal NADH: ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 2001;389:7–14. doi: 10.1006/abbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- 115.Chavez J.D., Tang X., Campbell M.D., Reyes G., Kramer P.A., Stuppard R., et al. Mitochondrial protein interaction landscape of SS-31. Proc. Natl. Acad. Sci. U. S. A. 2020;117:15363–15373. doi: 10.1073/pnas.2002250117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang H.L., Alder N.N., Wang W., Szeto H., Marcinek D.J., Rabinovitch P.S. Reduction of elevated proton leak rejuvenates mitochondria in the aged cardiomyocyte. Elife. 2020;9 doi: 10.7554/eLife.60827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bullock J.M.A., Sen N., Thalassinos K., Topf M. Modeling protein complexes using restraints from crosslinking mass spectrometry. Structure. 2018;26:1015–1024.e2.. doi: 10.1016/j.str.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yang J.Y., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016 doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Baek M., DiMaio F., Anishchenko I., Dauparas J., Ovchinnikov S., Lee G.R., et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lukassen M.V., Franc V., Hevler J.F., Heck A.J.R. Similarities and differences in the structures and proteoform profiles of the complement proteins C6 and C7. Proteomics. 2021;21 doi: 10.1002/pmic.202000310. [DOI] [PubMed] [Google Scholar]
- 124.McCafferty C.L., Pennington E.L., Papoulas O., Taylor D.W., Marcotte E.M. Does AlphaFold2 model proteins' intracellular conformations? an experimental test using cross-linking mass spectrometry of endogenous ciliary proteins. Commun. Biol. 2023;6:421. doi: 10.1038/s42003-023-04773-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ogorzalek T.L., Hura G.L., Belsom A., Burnett K.H., Kryshtafovych A., Tainer J.A., et al. Small angle X-ray scattering and cross-linking for data assisted protein structure prediction in CASP 12 with prospects for improved accuracy. Proteins. 2018;86:202–214. doi: 10.1002/prot.25452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mintseris J., Gygi S.P. High-density chemical cross-linking for modeling protein interactions. Proc. Natl. Acad. Sci. U. S. A. 2020;117:93–102. doi: 10.1073/pnas.1902931116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dominguez C., Boelens R., Bonvin A.M.J.J. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 128.Vakser I.A. Protein-protein docking: from interaction to interactome. Biophys. J. 2014;107:1785–1793. doi: 10.1016/j.bpj.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 130.Burke D.F., Bryant P., Barrio-Hernandez I., Memon D., Pozzati G., Shenoy A., et al. Towards a structurally resolved human protein interaction network. Nat. Struct. Mol. Biol. 2023;30:216–225. doi: 10.1038/s41594-022-00910-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Evans R., O'Neill M., Pritzel A., Antropova N., Andrew, Green T., et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv. 2022 doi: 10.1101/2021.10.04.463034. [preprint] [DOI] [Google Scholar]
- 132.Heublein M., Burguillos M.A., Vögtle F.N., Teixeira P.F., Imhof A., Meisinger C., Ott M. The novel component Kgd4 recruits the E3 subunit to the mitochondrial alpha-ketoglutarate dehydrogenase. Mol. Biol. Cell. 2014;25:3342–3349. doi: 10.1091/mbc.E14-07-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.O'Reilly F.J., Graziadei A., Forbrig C., Bremenkamp R., Charles K., Lenz S., et al. Protein complexes in cells by AI-assisted structural proteomics. Mol. Syst. Biol. 2023;10 doi: 10.15252/msb.202311544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stahl K., Graziadei A., Dau T., Brock O., Rappsilber J. Protein structure prediction with in-cell photo-crosslinking mass spectrometry and deep learning. Nat. Biotechnol. 2023 doi: 10.1038/s41587-023-01704-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.O'Reilly F.J., Xue L., Graziadei A., Sinn L., Lenz S., Tegunov D., et al. In-cell architecture of an actively transcribing-translating expressome. Science. 2020;369:554–557. doi: 10.1126/science.abb3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kyrilis F.L., Semchonok D.A., Skalidis I., Tüting C., Hamdi F., O'Reilly F.J., et al. Integrative structure of a 10-megadalton eukaryotic pyruvate dehydrogenase complex from native cell extracts. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108727. [DOI] [PubMed] [Google Scholar]
- 137.Leung M.R., Zeng J., Wang X., Roelofs M.C., Huang W., Zenezini Chiozzi R., et al. Structural specializations of the sperm tail. Cell. 2023;186:2880–2896.e17. doi: 10.1016/j.cell.2023.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Leung M.R., Zenezini Chiozzi R., Roelofs M.C., Hevler J.F., Ravi R.T., Maitan P., et al. In-cell structures of conserved supramolecular protein arrays at the mitochondria-cytoskeleton interface in mammalian sperm. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2110996118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhou C., Dai S., Lin Y., Lian S., Fan X., Li N., Yu W. Exhaustive cross-linking search with protein feedback. J. Proteome Res. 2022;22:101–113. doi: 10.1021/acs.jproteome.2c00500. [DOI] [PubMed] [Google Scholar]
- 140.Chen Y., Zhou W., Xia Y., Zhang W., Zhao Q., Li X., et al. Targeted cross-linker delivery for the in situ mapping of protein conformations and interactions in mitochondria. Nat. Commun. 2023;14:3882. doi: 10.1038/s41467-023-39485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hevler J.F., Lukassen M.V., Cabrera-Orefice A., Arnold S., Pronker M.F., Franc V., Heck A.J.R. Selective cross-linking of coinciding protein assemblies by in-gel cross-linking mass spectrometry. EMBO J. 2021;40 doi: 10.15252/embj.2020106174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bludau I., Heusel M., Frank M., Rosenberger G., Hafen R., Banaei-Esfahani A., et al. Complex-centric proteome profiling by SEC-SWATH-MS for the parallel detection of hundreds of protein complexes. Nat. Protoc. 2020;15:2341–2386. doi: 10.1038/s41596-020-0332-6. [DOI] [PubMed] [Google Scholar]
- 143.Choi A., Barrientos A. Sucrose gradient sedimentation analysis of mitochondrial ribosomes. Methods Mol. Biol. 2021;2192:211–226. doi: 10.1007/978-1-0716-0834-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Albanese P., Song W., van Keulen S., Koendjbiharie J., Koh F., Steigenberger B., et al. Thriving in the heat – lysine acetylation stabilizes the quaternary structure of a Mega-Dalton hyperthermoactive PEP-synthase. bioRxiv. 2022 doi: 10.1101/2022.08.11.503304. [preprint] [DOI] [Google Scholar]
- 145.An Y., Zhao Q., Gong Z., Zhao L., Li Y., Liang Z., et al. Suborganelle-specific protein complex analysis enabled by in vivo cross-linking coupled with proximal labeling. Anal. Chem. 2022;94:12051–12059. doi: 10.1021/acs.analchem.2c01637. [DOI] [PubMed] [Google Scholar]
- 146.Chavez J.D., Keller A., Mohr J.P., Bruce J.E. Isobaric quantitative protein interaction reporter technology for comparative interactome studies. Anal. Chem. 2020;92:14094–14102. doi: 10.1021/acs.analchem.0c03128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bakhtina A.A., Pharaoh G.A., Campbell M.D., Keller A., Stuppard R.S., Marcinek D.J., et al. Skeletal muscle mitochondrial interactome remodeling is linked to functional decline in aged female mice. Nat. Aging. 2023;3:313–326. doi: 10.1038/s43587-023-00366-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen Z.A., Rappsilber J. Protein dynamics in solution by quantitative crosslinking/mass spectrometry. Trends Biochem. Sci. 2018;43:908–920. doi: 10.1016/j.tibs.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yu C., Huszagh A., Viner R., Novitsky E.J., Rychnovsky S.D., Huang L. Developing a multiplexed quantitative cross-linking mass spectrometry platform for comparative structural analysis of protein complexes. Anal. Chem. 2016;88:10301–10308. doi: 10.1021/acs.analchem.6b03148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Muller F., Kolbowski L., Bernhardt O.M., Reiter L., Rappsilber J. Data-independent acquisition improves quantitative cross-linking mass spectrometry. Mol. Cell. Proteomics. 2019;18:786–795. doi: 10.1074/mcp.TIR118.001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Peterson A.C., Russell J.D., Bailey D.J., Westphall M.S., Coon J.J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hevler J.F. Faculteit Betawetenschappen. Utrecht University; Utrecht, The Netherlands: 2023. Structural characteristics of mitochondrial protein assemblies probed by mass spectrometry. [Google Scholar]
- 153.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]