Abstract
Sucrose isomerase (SI) genes from Pantoea dispersa UQ68J, Klebsiella planticola UQ14S, and Erwinia rhapontici WAC2928 were cloned and expressed in Escherichia coli. The predicted products of the UQ14S and WAC2928 genes were similar to known SIs. The UQ68J SI differed substantially, and it showed the highest isomaltulose-producing efficiency in E. coli cells. The purified recombinant WAC2928 SI was unstable, whereas purified UQ68J and UQ14S SIs were very stable. UQ68J SI activity was optimal at pH 5 and 30 to 35°C, and it produced a high ratio of isomaltulose to trehalulose (>22:1) across its pH and temperature ranges for activity (pH 4 to 7 and 20 to 50°C). In contrast, UQ14S SI showed optimal activity at pH 6 and 35°C and produced a lower ratio of isomaltulose to trehalulose (<8:1) across its pH and temperature ranges for activity. UQ68J SI had much higher catalytic efficiency; the Km was 39.9 mM, the Vmax was 638 U mg−1, and the Kcat/Km was 1.79 × 104 M−1 s−1, compared to a Km of 76.0 mM, a Vmax of 423 U mg−1, and a Kcat/Km of 0.62 × 104 M−1 s−1 for UQ14S SI. UQ68J SI also showed no apparent reverse reaction producing glucose, fructose, or trehalulose from isomaltulose. These properties of the P. dispersa UQ68J enzyme are exceptional among purified SIs, and they indicate likely differences in the mechanism at the enzyme active site. They may favor the production of isomaltulose as an inhibitor of competing microbes in high-sucrose environments, and they are likely to be highly beneficial for industrial production of isomaltulose.
Isomaltulose (palatinose, α-d-glucopyranosyl-1,6-d-fructofuranose) is a naturally occurring isomer of sucrose (α-d-glucopyranosyl-1,2-d-fructofuranose) that is valued as an acariogenic sweetener (22). It is a nutritional sugar that is digested more slowly than sucrose, and it has health advantages for diabetics and nondiabetics (8). Greater acid stability and lower hygroscopic tendency than sucrose are advantages for some food applications. Unlike sucrose, isomaltulose is a reducing sugar, which allows different chemical reactivity when it is used as an industrial precursor. Isomaltulose is currently used to manufacture sugar alcohols consumed as low-calorie sweeteners (21), and it is an attractive renewable starting material for the manufacture of biosurfactants and biocompatible polymers (7). It is produced industrially from sucrose by using immobilized cells of bacteria that produce an enzyme designated sucrose isomerase (SI), sucrose mutase, or isomaltulose synthase (EC 5.4.99.11).
SIs have been purified from Erwinia rhapontici NCPPB 1578 (2), Protaminobacter rubrum CBS574.77 (11), Serratia plymuthica ATCC 15928 (10, 24), Agrobacterium radiobacter (13), Pseudomonas mesoacidophila MX-45 (12), Klebsiella sp. (16), and the whitefly Bemisia argentifolii (18). Several of the corresponding genes have been cloned (1, 9, 30), and the gene designations palI, smuA, and mutB have been used. The crystal structure has been elucidated for the SI cloned from Klebsiella sp. strain LX3, which confirmed that there is a TIM (triose phosphate isomerase-like) barrel (β/α)8 core with active site architecture typical of glycoside hydrolase family 13 (28).
In addition to isomaltulose, SIs that have been described produce various proportions of the isomer trehalulose (α-d-glucopyranosyl-1,1-d-fructose) along with glucose and fructose as by-products. Some purified SIs produce predominantly isomaltulose (75 to 85%), and others produce predominantly trehalulose (90%) or only trehalulose in the case of the whitefly enzyme (18). The ratio of the products obtained from bacterial SIs varies with the reaction conditions, particularly the temperature and pH, and under some conditions small quantities of other products, such as isomaltose and isomelezitose, may be formed (24). The formation of multiple products lowers the yield and complicates the recovery of the desired isomer. Slow conversion of sucrose into isomaltulose and a narrow range of optimal reaction conditions also limit the industrial efficiency of isomaltulose production (2, 20). An ideal SI for industrial use would exhibit high speed, complete conversion, high specificity, and a wide window of reaction conditions for isomaltulose production.
Recently, we isolated multiple isomaltulose-producing bacteria by functional screening and found that an isolate of Pantoea dispersa designated UQ68J is exceptionally efficient in sucrose isomerase activity (27). Here, we describe the cloning and characterization of four SIs from isomaltulose-producing bacteria and a comparison of the activities of the purified recombinant enzymes produced in Escherichia coli. The enzyme from P. dispersa proved to be exceptional among the characterized SIs and to have properties highly desirable for use in the industrial biosynthesis of isomaltulose.
MATERIALS AND METHODS
Unless otherwise specified, routine molecular biology procedures were performed as described by Sambrook and Russell (19). Bacteria were isolated and identified as described previously (27).
Cloning of SI genes. (i) Construction and functional screening of bacterial genomic libraries.
High-molecular-weight (∼150-kb) DNA was extracted from E. rhapontici WAC2928, P. dispersa UQ68J, Klebsiella (Raoultella) planticola UQ14S, and unclassified bacterial isolate UQ349J (27) by the method of Priefer et al. (17). Genomic DNA inserts were prepared by partial digestion with restriction endonuclease Sau3A and then ligated into the BamHI site of vector SuperCos 1, in vitro packaged, and transferred into E. coli NM554 by using the manufacturer's instructions (Gigapack III Gold; Stratagene). From each of the four cosmid libraries, 600 colonies were screened for isomaltulose production by an aniline-diphenylamine assay and capillary electrophoresis as described previously (27).
(ii) Subcloning and sequencing.
Cosmid inserts were partially digested with EcoRI, BamHI, or HindIII and ligated into the pZerO-2 vector (Invitrogen). For clones producing isomaltulose, the process was repeated by using the alternative restriction enzymes to obtain the smallest functional inserts for sequencing. Plasmid inserts were sequenced at the Australian Genome Research Facility by using an ABI PRISM Big Dye terminator reaction kit. Sequences were confirmed by using both strands of the DNA. Alignments to determine sequence similarity and identity were made with ANGIS by using the GCG program Gap with default parameters and the Dayhoff-Gribskov scoring matrix of evolutionary distances between amino acids.
(iii) Structure predictions and alignments.
Predicted protein sequences of sucrose isomerases (GenBank accession numbers A45846, A45854, A45856, A45858, AF279281, AY040843, AY223549, AY223550, and AY227804), glucosidases from the same organisms (GenBank accession numbers A45860, AF279283, AF279285), and the most similar enzymes with known crystal structures (PDB 1M53 A, 1UOK, and 1JGI A) were aligned by using tools in Cn3D (www.ncbi.nih.gov/Structure/CN3D/cn3d.shtml). Secondary-structure predictions were obtained by using the BMERC protein sequence analysis server (http://bmerc-www.bu.edu/psa/) with minimal assumptions. Three-dimensional (3D) structure predictions were obtained by using ESyPred3D (www.fundp.ac.be/urbm/bioinfo/esypred/) with PDB template 1M53 (sucrose isomerase) or 1UOK (glucosidase) and were aligned initially by using VAST Search (www.ncbi.nlm.nih.gov/Structure/VAST/vastsearch.html); then alignments were refined and viewed in Cn3D. For phylogenetic comparison, these sequences and additional glucosidase sequences were aligned by using Clustal W default/accurate parameters, followed by bootstrapping of the neighbor-joining tree (http://clustalw.genome.jp/).
Expression of the three SI genes in E. coli. (i) Cloning of three SI genes into E. coli expression vector pET24b.
Based on the sequences obtained as described above, PCR primers were designed for subcloning the SI genes (without noncoding regions and leader sequences) into expression vector pET24b (Novagen). For comparison we also cloned into the same expression system the SI gene from P. rubrum CBS574.77 (9), the organism currently used for industrial production of isomaltulose. Each forward primer included a BamHI restriction site and a start codon. Each reverse primer included a KpnI restriction site and a stop codon. The following primers were used: E. rhapontici forward primer 5′-GGA TCC AAC AAT GGC AAC CGT TCA GCA ATC AAA TG-3′, K. planticola forward primer 5′-GGA TCC AAC AAT GGC AAC CGT TCA CAA GGA AAG TG-3′, P. dispersa forward primer 5′-GGA TCC AAC AAT GGC AAC GAA TAT ACA AAA GTC C-3′, P. rubrum forward primer 5′-CGG GAT CCA ACA ATG GCA ACG AAA AGA GTA TCG AAC-3′, E. rhapontici reverse primer 5′-ATA GGT ACC TTA CTT AAA CGC GTG GAT G-3′, K. planticola reverse primer 5′-ATA GGT ACC TTA CCG CAG CTT ATA CAC ACC-3′, P. dispersa reverse primer 5′-ATA GGT ACC TCA GTT CAG CTT ATA GAT CCC-3′, and P. rubrum reverse primer 5′-GGG GTA CCT ATT TTG CGC TAA AAA AAC CGG-3′.
High-fidelity DNA polymerase Pfu (Stratagene) was used for PCR, with a 5-min extension of the final amplification cycle in the presence of Taq DNA polymerase to add TA tails. The PCR products were directly cloned into the pCR2.1 vector by using a TOPO TA cloning kit (Invitrogen). The SI genes were cloned from the pCR2.1 vector into pGEM-3Zf(+) (Promega) and then into the pET24b vector for expression in E. coli BL21(DE3) (Novagen).
(ii) Expression of SI genes in E. coli.
Fifteen cultures per construct were set up in 5 ml of Luria-Bertani medium with 50 μg of kanamycin per ml in 30-ml universal tubes. Cells were grown at 37°C with shaking at 225 rpm. Six to ten cultures per construct (optical density at 600 nm [OD600], 1.00) were selected for further induction. After a 0.5-ml sample was removed from each culture, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM, and incubation of the cultures was continued for another 3 h at 28°C. This allowed selection of three replicate cultures per construct with the same OD600 for sucrose conversion analysis and protein measurement. From each of the selected cultures, 1.5 ml was removed for protein quantification, 0.5 ml was removed for protein sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and 1.0 ml was removed for quantification of the efficiency of conversion from sucrose to isomaltulose. For SI protein purification, the culture volume was increased to 500 ml in a 2-liter flask.
(iii) Assay for SI in intact E. coli cells.
A 1.0-ml culture sample was centrifuged at 12,000 × g for 1 min and then resuspended in 5 ml of a 0.1 M citrate/phosphate (pH 6.0)-buffered 50% sucrose solution and incubated for 4 h at 37°C with shaking at 225 rpm. The reaction was terminated by boiling for 10 min, and this was followed by centrifugation at 15,000 × g for 20 min to remove the cell debris and denatured proteins. The conversion ratio was calculated from sucrose and isomaltulose peak areas normalized with standards whose concentrations were known by using a Beckman P/ACE 5000 capillary electrophoresis apparatus as described previously (27).
Protein extraction, purification, and determination. (i) Preparation of crude extracts.
Cells were harvested by centrifugation (3,000 × g, 4°C, 10 min), resuspended in 50 mM Tris-HCl (pH 8.0)-2 mM EDTA, and then recentrifuged. The cell pellet was immediately frozen in liquid nitrogen and stored at −75°C. Cells were suspended in extraction buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA, 1 mM azide, 10 mM β-mercaptoethanol) and then lysed by sonication (nine 15-s pulses at 50 W with a Branson Sonifier 450 microprobe), centrifuged (10,000 × g, 4°C, 10 min), and filtered through a 0.45-μm-pore-size membrane (Gelman Acrodisc).
(ii) SI protein purification.
The pET24b vector introduced a carboxy-terminal six-His tag into expressed proteins, which were purified by adsorption to Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) and elution with 25 mM NaH2PO4-150 mM NaCl-125 mM imidazole buffer (pH 8.0) by following the manufacturer's instructions. For storage, the eluted solution was diluted with 50% glycerol. The purity of SI proteins was determined by SDS-PAGE as described below. A batch procedure in which Ni-NTA agarose in suspension was used yielded predominantly (80 to 95%) SI along with a background of several other protein bands. A procedure in which Ni-NTA agarose was used in adsorption columns yielded preparations containing a single protein band. Unless otherwise specified, this was the form of the purified SI enzymes used for biochemical characterization.
For SDS-PAGE, samples were heated at 100°C for 5 min in loading buffer (25 mM Tris-HCl [pH 6.8], 1% [wt/vol] SDS, 5% [vol/vol] β-mercaptoethanol, 10% [vol/vol] glycerol, 0.005% [vol/vol] bromophenol blue) and then centrifuged at 12,000 × g for 1 min. To estimate the SI yield as a proportion of the total cellular protein, each sample was loaded into two adjacent lanes. After electrophoresis, one lane from the gel was stained in 0.025% (wt/vol) Coomassie blue R-250 and then destained in 30% (vol/vol) methanol with 10% (vol/vol) acetic acid. Then the SI was cut from the unstained lane at the position corresponding to the migration position in the stained lane. Proteins were eluted from gel slices by immersion in phosphate-buffered saline (pH 7.4) overnight at 4°C with gentle shaking and were quantified by the Bradford method by using bovine serum albumin as a standard.
Determination of SI activities. (i) Assay of isolated SI enzyme activity.
Enzyme activity was measured by incubating 5 μl of purified enzyme with 95 μl of a sucrose solution at a final concentration of 584 mM in 0.1 M citrate/phosphate buffer (pH 6.0) at 30°C. The sugar profiles in the reaction mixture were analyzed at intervals by high-performance liquid chromatography with electrochemical detection as described previously (27). Briefly, resolution and quantification of sucrose, isomaltulose, trehalulose, glucose, and fructose were achieved by isocratic high-performance liquid chromatography at high pH (100 mM KOH or 120 mM NaOH) by using a Dionex BioLC system with a PA20 analytical anion-exchange column and quad waveform pulsed electrochemical detection. All sugar quantification data presented below were obtained by this method, and calibration against a dilution series of sugar standards was performed for every sample batch. One unit of SI activity was defined as the amount of enzyme that released 1 μmol of isomaltulose or trehalulose in 1 min at the initial stage of the reaction.
(ii) Effects of pH and temperature on SI activity.
To determine the effect of pH on enzyme activity, the enzyme was equilibrated in reaction buffer at 35°C for 15 min before addition of the substrate sucrose. The buffer was 0.1 M citrate/phosphate (pH 3.0 to 7.0) or 0.1 M Tris-HCl (pH 8.0 to 9.0). The effects of temperatures from 20 to 60°C were determined after equilibration of the enzyme solution for 10 min at pH 6.0.
(iii) Effect of sucrose concentration in the absence or presence of glucose and fructose.
SI activity was measured by incubating the purified enzyme with different sucrose concentrations (2.92, 5.84, 14.6, 58.4, 146, 292, and 584 mM) under standard assay conditions and also with glucose added to a final concentration of 69, 139, or 277 mM, with fructose added to a final concentration of 277 mM, or with both glucose and fructose added to a final concentration of 277 mM. Data were analyzed by using double-reciprocal plots to calculate Km,Vmax, and Kcat values.
RESULTS
Three distinct SI genes were cloned and sequenced.
Functional screening of genomic cosmid libraries of E. rhapontici WAC2928, P. dispersa UQ68J, K. planticola UQ14S, and unidentified isolate UQ349J in E. coli yielded clones from each source that were able to convert sucrose to isomaltulose. After several cycles of subcloning and functional screening, the sizes of the smallest functional inserts in the pZerO-2 vector ranged from 3 to 5 kb.
The sequence from the E. rhapontici clone had an 1,899-bp open reading frame (ORF) encoding 632 amino acids (GenBank accession number AY223550). First-strand sequencing of the UQ349J clone revealed a gene with 99% identity to this E. rhapontici ORF, so sequencing of UQ349J was stopped. Mattes et al. (9) determined 1,305 bp of an SI gene from E. rhapontici NCPPB 1578. Börnke et al. (1) subsequently reported the complete sequence of a very similar gene (designated palI) from E. rhapontici DSM 4484 with 1,803 bp encoding 600 amino acids. The SI from E. rhapontici WAC2928 showed 96.6% nucleotide identity (92.8% with the 3′ extension) and 97.7% amino acid similarity (93.6% with the carboxy-terminal extension) to the gene described by Börnke et al. (1).
The sequence from K. planticola UQ14S revealed a 1,797-bp ORF encoding 598 amino acids (GenBank accession number AY227804). A database search with FASTA revealed 98.5% nucleotide identity and 98.9% amino acid similarity to an SI gene from Enterobacter strain SZ62 (9); the amino acid sequence varied by only three amino acids from the sequence encoded by the gene described from Klebsiella sp. strain LX3 (30).
The sequence obtained from P. dispersa UQ68J indicated that the SI gene was substantially different, with a 1,797-bp ORF encoding 598 amino acids (GenBank accession number AY223549). At the nucleotide level, the sequence exhibited less than 70% identity to known sequences encoding SIs, either with or without a leader fragment. At the amino acid level, the levels of identity to other SIs were between 63 and 71% with the leader and between 64 and 74% without the leader. The predicted mature SI gene product has an Mr of 65,819 and an isoelectric point of 7.0 due to 77 basic and 68 acidic amino acid residues. Throughout this paper, the numbering of amino acids in the UQ68J SI includes the predicted 33-amino-acid periplasm-targeting leader.
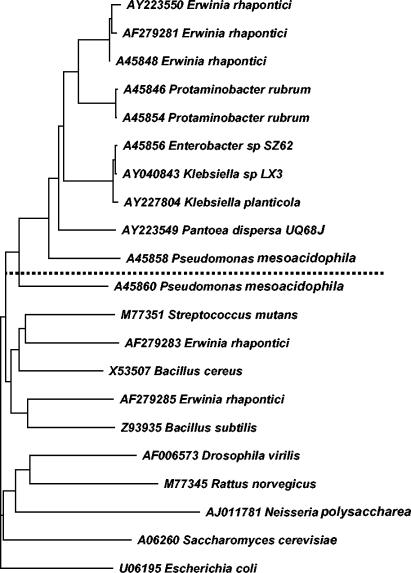
In a phylogenetic analysis of the amino acid sequences of representative hydrolases, glucosidases, and sucrose isomerases, the cloned SIs from E. rhapontici WAC2928 and K. planticola UQ14S fell into a cluster with other known sucrose isomerases. The SI from P. dispersa UQ68J diverged earlier from this cluster, along with the SI and hydrolase from the trehalulose-producing organism P. mesoacidophila and various glucosidases (Fig. 1). Because of its substantial differences in sequence and in enzymatic properties, as described below, here we use the designation sim (sucrose-isomaltulose isomerase) to distinguish the UQ68J SI gene from the gene in the Erwinia-Klebsiella-Enterobacter branch designated palI (1, 30), the gene in the Protaminobacter subbranch designated smu, and the P. mesoacidophila sucrose-trehalulose isomerase gene designated mutB (9).
FIG. 1.
Phylogram of SIs and representative glucosidases prepared by using Clustal W. The bootstrap values were more than 85% for the SI group. The dotted line separates SIs (above the line) from hydrolases. The P. mesoacidophila SI is unusual in producing predominantly trehalulose rather than isomaltulose.
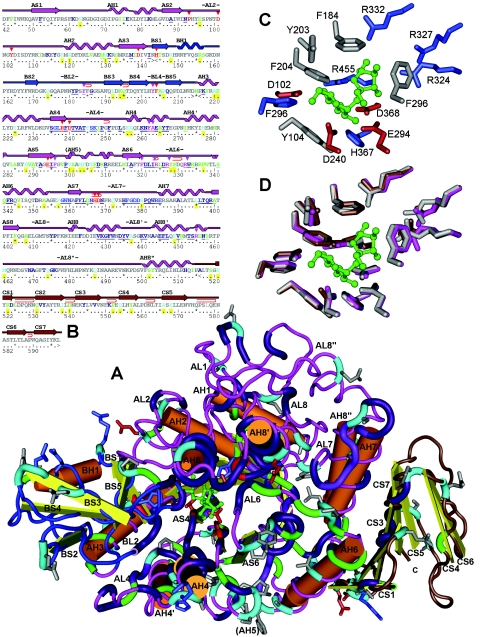
All of the sequenced SIs have predicted secondary structures comprising an N-terminal TIM barrel (β/α)8 A domain with a B subdomain inserted between β sheet 3 (AS3) and α helix 3 (AH3) and a C-terminal domain containing 7 to 10 β sheets. For convenience, we designated structure elements by domain (A, B, or C) and feature (S, β sheet; H, α helix; L, connecting loop). Features are numbered within domains from the N terminus, and loops are numbered the same as the preceding β sheet. Sequence alignment with the TIM glucosidases revealed that all the SIs have the 4/5/7 superfamily characteristic of catalytic acidic residues (D, E, D) at the C-terminal ends of AS4, AS5, and AS7 (14). The predicted 3D structures of the SIs superimposed closely on the 1M53 template (28). Several features described below are shown in a 3D structure diagram of the UQ68J gene product in Fig. 2.
FIG.2.
Predicted secondary and 3D structure of the P. dispersa UQ68J SI. The colors of the peptide backbone indicate domains (pink, A [TIM barrel] domain; blue, B subdomain; brown, C domain). (A) In the 3D structure, the core of the TIM barrel is under the loop marked AL6, and the eight beta sheets point upward. The substrate sucrose molecule (green) is located near the top of the barrel and is surrounded by conserved glycolytic residues (red). Other expanded sections of the peptide backbone represent residues that are conserved among sucrose isomerases (purple), unique to the Pseudomonas trehalulose synthase (green), or unique to the UQ68J isomaltulose synthase among residues functionally conserved across other sucrose isomerases (light blue). Amino acid side chains are shown only for the conserved active site and unique UQ68J residues (different colors indicate different charges). Prominent beta sheets (S), alpha helices (H), and connecting loops (L) are numbered by domain from the N terminus. (B) For the secondary structure the same numbering and color coding are used. Residues unique to UQ68J are indicated by yellow, conserved strings near the active site are underlined, residues closest to bound glucose and fructose moieties are indicated by red and pink arrows, glycolytic core residues are underlined with wavy lines, and tight folds in loop regions are indicated by hairpin symbols. (C) Amino acids around the UQ68J active site. (D) Overlaid active sites for SIs from strains UQ68J (pink), LX3 (brown), and WAC2928 (grey).
SI genes and glucosidases share conserved domains for sugar binding. As a result, conserved sequences and primers described previously (9) are not specific for SIs. For the genes described to date, there are conserved residues unique to sucrose isomerases flanking these catalytic residues and sequence strings specific for sucrose isomerases in AL4, AH4, AL6, AL7, AH4, AH8-8′, and BL2 (Table 1).
TABLE 1.
Sequence elements conserved among SIs
| Regiona | Residuesb | Amino acids |
|---|---|---|
| Conserved motifsc | ||
| AL4 | 241-251 | TVAT(Y/F)SP(I/T)PXF |
| AH4 | 260-267 | KNFAEXYT |
| AL6 | 321-327 | DLIR(L/Y)DR |
| AL7 | 380-385 | (R/S)PQWRE |
| AH7 | 395-400 | (T/L)LTQR(A/G) |
| AH8 | 428-436 | VKGFWXDYV |
| AH8′ | 440-454 | KVXAXEFL(D/Q)(N/E)V(R/A)XTS |
| Uniquely conserved residuesd | ||
| BL2 | 178-190 | PNNYPSFFGGSAW |
| BL4 | 198-213 | QYYLHYF(A/G)XQQPDLNW |
| AS4 | 233-240 | GVSGXRFD |
| AS5 | 290-300 | (A/V)TAGEIFGV/AP(L/V) |
| AS7 | 362-371 | GWNXFFL(D/G)NHDNPR |
Regions designated as shown in Fig. 2.
Numbering for the UQ68J SI sequence.
Motifs conserved among SIs but not among glucosidases.
Residues that are uniquely conserved in SIs within conserved loops near the substrate (underlined) or near the glycolytic core residues (boldface).
We examined the structure alignments for features that might contribute to the high efficiency and isomaltulose product specificity of the P. dispersa UQ68J SI or the trehalulose product specificity of the P. mesoacidophila MX-45 SI, allowing for a gap in the published sequence for the latter corresponding to residues 427 to 449 (AH8 to AH8′). Table 2 includes residues in these enzymes that vary from the residues conserved in other SIs, particularly near the C-terminal ends of the TIM barrel sheets or in other loops contributing to the substrate pocket.
TABLE 2.
Residues near the substrate pocket or in key loops of the enzyme structure that are different in SIs with different product specificities
| Regiona | Residueb | Residue in:
|
||
|---|---|---|---|---|
| UQ68J isomaltulose synthase | Other sucrose isomerases | MX-45 trehalulose synthase | ||
| AL2 | 96 | E | D | A |
| BL3 | 192 | L | K | K |
| BL3 | 194 | K | D/E/A | P |
| BL3 | 195 | Q | K | V |
| BL3 | 197 | D | G/N | G |
| BL4 | 206 | P | R/K | R |
| AL4 | 245 | F | Y | Y |
| AL4 | 248 | I | I | T |
| AL4 | 256 | K | P/Q | P |
| AL4 | 257 | A | E/Q | E |
| AL6 | 325 | L | L | Y |
| AL6 | 328 | Y | D | A |
| AL6 | 329 | P | S/A | L |
| AL6 | 337 | P | S/D | P |
| AL6 | 341-342 | SQ | SQ | AD |
| AL7 | 365 | D | D | G |
| AL7 | 372 | Q | A | A |
| AL7 | 380 | S | R | R |
| AS8 | 405 | F | Y | F |
| AL8 | 408 | A | S | D |
| AH8′ | 456 | D | D | A |
| AH8′ | 458 | S | S | A |
Regions designated as shown in Fig. 2.
Numbering for the UQ68J SI sequence.
SI from P. dispersa UQ68J showed the highest conversion efficiency among the isomerases expressed in E. coli cells that were tested.
When the SI genes were arranged for expression by using the same promoter, start codon context, and termination sequences in vector pET24b, the efficiency of conversion from sucrose to isomaltulose by intact E. coli cells expressing the cloned P. dispersa UQ68J gene was 10 times that of the cells expressing the cloned gene from E. rhapontici and 18 times that of the cells expressing the cloned gene from K. planticola.
There was no significant difference in the total protein content or in the expressed SI content (around 10% of the total protein), but there were substantial differences in the soluble SI contents (estimated by recovery of the His-tagged protein) and particularly in the apparent SI enzyme efficiencies in the intact cell assay. The Protaminobacter SI cloned for comparison proved to be the SI most susceptible to inactivation by misfolding or precipitation. No soluble enzyme was recovered from the Ni-NTA agarose step, and SI activity was barely detectable in the E. coli cells expressing this SI (<0.1% of the activity in E. coli cells expressing the Pantoea strain UQ68J SI). For the enzymes produced in an active form, the efficiency of UQ68J SI was estimated to be 6 and 38 times the efficiency of the Klebsiella and Erwinia enzymes, respectively, per unit of soluble SI in the transformed E. coli cells (Table 3).
TABLE 3.
Total protein contents and estimated SI protein contents in E. coli cells expressing cloned SI genesa
| SI gene source | Total protein (%, dry wt) | Total SI (% of cellular proteins)b | Soluble His-tagged SI
|
Amt of isomaltulose produced (μmol)d | Cellular soluble SI efficiency (mol of isomaltulose g of SI−1) | |
|---|---|---|---|---|---|---|
| % of total SI | Concn (μg ml−1)c | |||||
| E. rhapontici WAC2928 | 16.0 ± 1.6 | 10.2 ± 1.5 | 26.5 | 129.3 | 511 ± 65 | 4 |
| K. planticola UQ14S | 15.8 ± 1.4 | 9.8 ± 0.5 | 2.7 | 12.2 | 292 ± 67 | 24 |
| P. dispersa UQ68J | 16.1 ± 1.8 | 10.4 ± 1.2 | 8.1 | 36.4 | 5552 ± 212 | 152 |
| Control (pET24b) | 14.4 ± 2.0 | 0 | 0 | 0 | 0 | |
The data are means ± standard errors for three replicates.
Corrected for 2% proteins in the PAGE SI zone of the control crude lysates.
Amount of SI recovered from batch adsorption to Ni-NTA agarose per milliliter of IPTG-induced culture.
Amount of isomaltulose produced in the cellular assay per milliliter of IPTG-induced culture.
The cells expressing P. dispersa UQ68J SI produced less contaminating glucose, fructose, and trehalulose in the SI assay (10% of the supplied sucrose) than the cells expressing SIs from E. rhapontici (15%) or K. planticola (16%) produced. These results indicate the high potential of the cloned SI from P. dispersa UQ68J in industrial applications for isomaltulose production.
The efficiencies of conversion by cells of E. coli expressing the cloned SI genes were lower than the efficiencies of conversion by cells of the corresponding native SI-producing strains in the same assay (27). In the present work, the cloned sequences encoded the mature SI enzymes without the leader sequences involved in transport to the periplasmic space. The overexpressed cytosolic recombinant SIs may not have been in a fully active conformation, and the cytosolic location may have imposed additional barriers to diffusion of the substrate during the assay. For industrial applications with intact bacterial cells, the periplasmic form of the enzymes may be preferable, whereas clones encoding the mature form of the enzyme may be preferable for applications in which purified enzyme or nonbacterial cells are used.
Purified SI proteins varied greatly in specific activity and stability.
We obtained the maximum yield of soluble SI from E. coli strain BL21(DE3) expressing SI genes in vector pET24b under the following conditions: (i) cultures were grown at 37°C to an OD600 of 1.0 before IPTG induction; (ii) the IPTG concentration was 0.5 mM for induction; and (iii) growth for SI production occurred at 28°C.
After purification of His-tagged proteins on Ni-NTA agarose columns, a single band from each of the three constructs was revealed by Coomassie blue R-250 staining after SDS-PAGE. The purified fresh enzymes from the Ni-NTA agarose columns had the following specific activities for isomaltulose production: E. rhapontici, 35 U mg of protein−1; K. planticola, 351 U mg of protein−1; and P. dispersa UQ68J, 562 U mg of protein−1. The purified SI from E. rhapontici WAC2928 lost function during overnight storage at −20°C in elution buffer diluted with 50% glycerol. In contrast, the purified enzymes from K. planticola UQ14S and P. dispersa UQ68J retained 100% of the fresh enzyme efficiency for isomaltulose production after 6 months of storage at −20°C or 60% of the fresh enzyme efficiency after 15 days of storage at room temperature in the same buffer. Because of instability, the E. rhapontici SI was not characterized further in purified form. Cheetham (2) found substantial differences between E. rhapontici strains for SI stability in immobilized cells, and the enzyme from strain NCPPB 1578 was stable in purified form (2). The partial sequence of the SI from strain NCPPB 1578 (9) exhibits a high level of similarity to the WAC2928 sequence. The different stabilities could reflect sequence differences in the COOH region that have not been revealed yet, the use of His-tagged enzyme versus native enzyme, or the greater enzyme purity in the present study.
Purified SI from P. dispersa UQ68J converted sucrose faster.
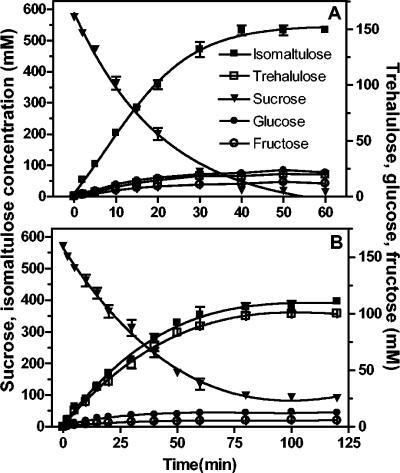
For UQ68J SI, 97% of the sucrose was converted into isomaltulose, fructose, and glucose within 45 min (Fig. 3A). Isomaltulose accounted for 91% of the sucrose consumed, and trehalulose accounted for 3%; glucose and fructose accounted for the remainder.
FIG. 3.
Sucrose conversion by the purified sucrose isomerases cloned from P. dispersa UQ68J (A) and K. planticola UQ14S (B). Note the difference in the time scales between panels A and B. The values are means ± standard errors for three replicates.
Sucrose conversion by K. planticola UQ14S SI was much slower. The same enzyme concentration depleted only 84% of the sucrose within 2 h; 66% was converted to isomaltulose, 15% was converted to trehalulose, and a small amount was converted to glucose and fructose (Fig. 3B).
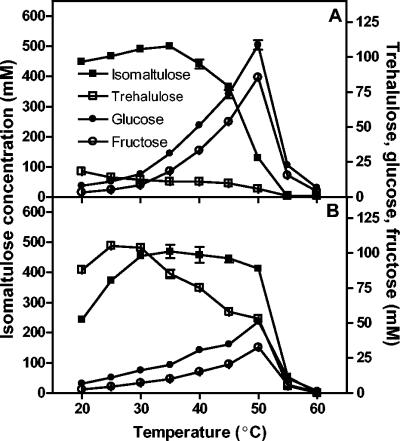
Purified SI from P. dispersa UQ68J produced less trehalulose at all temperatures and pHs.
The optimal temperature for isomaltulose production by the purified enzyme was 30 to 35°C for P. dispersa UQ68J SI and 30 to 45°C for K. planticola UQ14S SI (Fig. 4). Between 20 and 40°C, the molar ratio of isomaltulose to other conversion products (glucose, fructose, and trehalulose) was in the range from 21:1 to 8:1 for the UQ68J SI but never exceeded 5:1 for the UQ14S SI (Table 4). The UQ68J SI produced less trehalulose across the temperature range tested. Although both SIs generated more trehalulose as the temperature decreased, this effect was far more pronounced for the UQ14S SI (Fig. 4 and Table 4). Invertase activity increased at temperatures above 40°C, particularly for the UQ68J SI (Fig. 4).
FIG. 4.
Effects of temperature on activity and product specificity of the purified SIs from P. dispersa UQ68J (A) and K. planticola UQ14S (B). The values are means ± standard errors for three replicates.
TABLE 4.
Comparison of isomaltulose/trehalulose ratios for the purified SIs from P. dispersa UQ68J and K. planticola UQ14S
| Temp (°C) | Isomaltulose/trehalulose ratio
|
Isomaltulose/(trehalulose + glucose + fructose) ratio
|
||
|---|---|---|---|---|
| UQ68J | UQ14S | UQ68J | UQ14S | |
| 20 | 24.5 ± 0.1a | 2.8 ± 0.1 | 18.8 ± 0.1 | 2.6 ± 0.1 |
| 25 | 33.4 ± 0.4 | 3.6 ± 0.1 | 21.2 ± 0.3 | 3.3 ± 0.1 |
| 30 | 40.2 ± 1.1 | 4.4 ± 0.1 | 20.0 ± 0.4 | 4.0 ± 0.1 |
| 35 | 45.3 ± 1.4 | 5.5 ± 0.1 | 14.0 ± 0.3 | 4.7 ± 0.1 |
| 40 | 40.6 ± 1.1 | 6.1 ± 0.2 | 8.3 ± 0.4 | 4.7 ± 0.2 |
| 45 | 37.7 ± 2.4 | 7.7 ± 0.1 | 5.0 ± 0.1 | 5.2 ± 0.1 |
| 50 | 22.4 ± 0.7 | 7.8 ± 0.2 | 1.3 ± 0.1 | 4.4 ± 0.1 |
The data are means ± standard errors for three replicates.
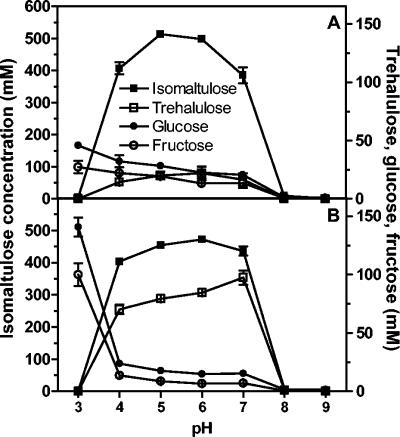
The optimal pH range for isomaltulose production was 4 to 7 for both purified enzymes. UQ14S SI generated more trehalulose than UQ68J SI generated across the active pH range, and there was a more pronounced increase in undesired products at high pH (trehalulose) and at low pH (monosaccharides) (Fig. 5).
FIG. 5.
Effects of pH on activity and product specificity of the purified SIs from P. dispersa UQ68J (A) and K. planticola UQ14S (B). The values are means ± standard errors for three replicates.
P. dispersa UQ68J SI had a lower Km and higher Vmax for isomaltulose production.
The purified P. dispersa UQ68J SI had a 48% lower Km and a 50% higher Vmax than the K. planticola UQ14S SI (Table 5). The Kcat/Km value for UQ68J SI was 2.9 times that for UQ14S SI (1.79 × 104 M−1 s−1 versus 6.19 × 103 M−1 s−1). For trehalulose production the Km values were similar, but UQ14S SI had a 220% higher Vmax than UQ68J SI (Table 5), and both Kcat/Km values were lower than those for isomaltulose (8.78 × 102 M−1 s−1 for UQ14S SI versus 5.63 × 102 M−1 s−1 for UQ68J SI).
TABLE 5.
Km and Vmax values for conversion of sucrose into isomaltulose and trehalulose by purified SIs from P. dispersa UQ68J and K. planticola UQ14Sa
| Parameter | UQ685
|
UQ145
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sucrose | Sucrose + 69 mM glucose | Sucrose + 139 mM glucose | Sucrose + 277 mM glucose | Sucrose + 277 mM fructose | Sucrose + 277 mM glucose + 277 mM fructose | Sucrose | Sucrose + 69 mM glucose | Sucrose + 139 mM glucose | Sucrose + 277 mM glucose | Sucrose + 277 mM fructose | Sucrose + 277 mM glucose + 277 mM fructose | |
| Isomaltulose production | ||||||||||||
| Km (mM) | 40 | 56 | 76 | 164 | 72 | 3,017 | 76 | 102 | 138 | 329 | 170 | 709 |
| Vmax (U mg−1) | 638 | 623 | 627 | 635 | 624 | 636 | 423 | 423 | 416 | 431 | 430 | 403 |
| Trehalulose production | ||||||||||||
| Km (mM) | 71 | 140 | 244 | 1,009 | 168 | 670 | 147 | 241 | 360 | 687 | 368 | 3,012 |
| Vmax (U mg−1) | 36 | 36 | 35 | 35 | 39 | 35 | 116 | 115 | 115 | 111 | 117 | 118 |
The data are means for three enzyme reaction replicates. The kinetic constants were obtained from Lineweaver-Burk plots with sucrose concentrations of 2.92, 5.84, 14.6, 58.4, and 146 mM.
Glucose and fructose acted as competitive inhibitors for both purified SIs, as indicated by higher Km values but similar Vmax values in the presence of the inhibitor (Table 5). The slope of the Lineweaver-Burk plots increased in linear proportion to the glucose concentration up to 139 mM glucose for both isomaltulose and trehalulose production. Over this linear range, the inhibitor dissociation factors (Ki) of glucose for isomaltulose and trehalulose production were 146.2 and 50.1 mM for UQ68J SI, which were similar to the values for UQ14S SI (159.0 and 55.0 mM). Fructose at a concentration of 277 mM was about as inhibitory as glucose at a concentration of 139 mM. The two monosaccharides at a concentration of 277 mM were synergistic for inhibition of isomaltulose production for both UQ68J SI and UQ14S SI and for inhibition of trehalulose production by UQ14S SI. Glucose at a concentration of 277 mM was most inhibitory to trehalulose production by UQ68J SI; there was apparent interference between the monosaccharides in this case.
SI from P. dispersa UQ68J neither hydrolyzed isomaltulose nor produced isomaltulose from glucose and fructose.
The isomaltulase activity of the purified SI from P. dispersa UQ68J was investigated by incubating it with 50 mM isomaltulose at 30°C for 30 min at pH 3.0, 4.0, 5.0, 6.0, and 7.0. Glucose and fructose were tested as substrates under the same reaction conditions. The purified UQ68J SI did not hydrolyze isomaltulose, and no product was detected after incubation of the enzyme with these monosaccharides.
DISCUSSION
Comparative structure of the novel UQ68J sucrose isomerase.
P. dispersa UQ68J, which was previously shown to have exceptional efficiency and specificity for production of isomaltulose from sucrose (27), had a sucrose isomerase gene that was substantially different from those characterized previously (less than 70% nucleotide identity or 71% amino acid identity when the leader sequence for export to the periplasm was included). SI genes cloned from other bacteria isolated by screening for SI activity proved to be very similar to genes characterized previously (1, 9, 30). Phylogenetic analysis of SI and glucosidase amino acid sequences showed that the P. dispersa UQ68J SI is divergent from both the trehalulose-producing P. mesoacidophila SI and the cluster of predominantly isomaltulose-producing enzymes of P. rubrum, E. rhapontici, Klebsiella sp., and Enterobacter sp. (Fig. 1).
Homology modeling indicated that all sequenced SIs have the modified TIM (β/a)8 barrel scaffold typified by the Bacillus cereus oligo-1,6-glucosidase (26) and confirmed by crystallography for the Klebsiella strain LX3 SI (28). Because the SIs share substantial conserved regions with TIM glucosidases, the conserved SI sequences proposed previously (9) are not specific, and corresponding primers yield many non-SI genes from different organisms in PCR amplifications. Members of this superfamily with substrate specificities ranging from starch to glycogen, dextrin, isomaltose, isomaltulose, maltodextrin, pullulan, and sucrose have now been sequenced (14). Our analysis revealed several conserved regions that appear to be more diagnostic of SIs (Table 1).
There are indications that substrate specificity may be influenced particularly by the subdomain that protrudes from the TIM barrel core between AS3 and AH3 (5). This subdomain includes loops in the vicinity of the substrate, which contain sequences that are broadly conserved across glucosidases interspersed with residues that appear to be uniquely conserved among SIs (Table 1). Conserved SI-specific residues flanking the glycolytic core residues near the C-terminal ends of TIM barrel sheets AS4, AS5, and AS7 were also identified along with conserved sequence strings in loops AL4 and AL6 near the substrate and in several of the surrounding α helices (Table 1 and Fig. 2).
Differences in residues around the active site (Table 2) may not fully explain the different efficiencies and specificities of SIs. These properties are likely to be influenced not only by amino acid side chains in the vicinity of the substrate but also by remote sequence differences that alter the shape or stability of the substrate pocket. SI product specificity depends substantially on the opportunity for tautomerization of fructose during the reaction (6). Therefore, remote prolines or other residues involved in salt bridges or even hydrogen bonds that alter the rigidity of loops enclosing the substrate could be important, as recently found for the structurally related enzyme amylosucrase (23). Loops AL4, AL6, BL3, and BL4 are all interesting in this context.
For example, the highly efficient UQ68J SI differs from other isomaltulose synthases in AL6, with 324RLDRYP329 rather than RLDRDS/A and with P337 replacing S or D in this loop. Zhang et al. (29) found that mutations in any of the charged amino acids in the RLDRDS motif increased the ratio of trehalulose to isomaltulose produced by Klebsiella strain LX3 SI. However, the changes adversely affected kinetic properties, and the trehalulose specificity was generally less than that of the native Pseudomonas strain MX-45 SI (with RYDRAL). Two proline substitutions designed to increase thermostability in Klebsiella strain LX3 SI (one of which corresponds to P497 in the native UQ68J SI) had little effect on catalytic efficiency or product specificity (30), but deletions in the C domain showed that interdomain interactions are essential for the function of the catalytic A domain (28).
Enzymatic properties.
The enzyme purified from expression of the cloned SI gene from P. dispersa UQ68J showed remarkable efficiency and product specificity, rapidly converting 91% of the sucrose to isomaltulose; the remainder was glucose, fructose, and trehalulose. All isomaltulose synthases previously tested in purified form produce a higher proportion of trehalulose. In the case of the best-characterized enzymes from S. plymuthica ATCC 15928 and Klebsiella strain LX3, as well as the trehalulose synthase from P. mesoacidophila MX-45, the product ratio varies with the assay temperature and pH, and trehalulose is favored particularly at lower temperatures (12, 24, 30). These effects were also evident for the purified SI from K. planticola UQ14S. In contrast, the enzyme from P. dispersa UQ68J maintained a high isomaltulose/trehalulose ratio across its activity range (Fig. 4 and 5 and Table 4). A temperature close to 30°C may be optimal for industrial use of UQ68J SI, because invertase activity increases at higher temperatures. This is also a suitable maximum temperature for use in plants engineered as biofactories, where the substantial activity at pH 4 is potentially useful in the acidic sucrose storage vacuoles (4). At temperatures from 40 to 50°C other enzymes, such as the UQ14S enzyme, may be preferable, depending on the relative interference from trehalulose versus glucose and fructose in the downstream industrial processes.
P. dispersa UQ68J SI showed the lowest Km (40 mM) and the highest Vmax (638 μmol of isomaltulose mg of protein−1 min−1) reported for purified isomaltulose synthases (Table 6). Also, in contrast to the unidirectional conversion by the UQ68J enzyme, other SIs use isomaltulose as a substrate to produce trehalulose, glucose, and fructose (24). This reverse activity can cause a gradual increase in the trehalulose/isomaltulose ratio during incubation, which is undesirable for industrial production of isomaltulose (3). Differences in reaction kinetics and specificity most likely reflect differences at the active site of the SI (6, 25). The possibilities include a higher rate constant for the conversion of the enzyme-glucose-fructofuranose complex to enzyme plus isomaltulose product, resulting in less opportunity for tautomerization to fructopyranose (consistent with the higher Vmax observed for the UQ68J enzyme) and a higher specificity of the transferase reaction for fructofuranose as the acceptor (consistent with the observed retention of isomaltulose product specificity at low temperatures and in the presence of added fructose despite inhibition of isomaltulose production).
TABLE 6.
General characteristics of purified sucrose isomerasesa
| Organism | Maximum yield (%)b
|
Optimal temp (temp range, °C) | Optimal pH (pH range) | Km (mM) | Vmax (sp act) (U mg−1) | Reference | |
|---|---|---|---|---|---|---|---|
| Isomaltulose | Trehalulose | ||||||
| P. dispersa UQ68J | 91 | 3 | 30-35 (10-55) | 5 (3-8) | 40 | 638 (562) | This study |
| K. planticola UQ14S | 66 | 15 | 35 (10-60) | 6 (4-7) | 76 | 423 (351) | This study |
| Klebsiella sp. strain LX3 | 83 | 21 | 35 (15-40) | 6 (5-6.5) | 55 | NS (328)c | 30 |
| Klebsiella sp. | 86 | NS | 35 | 6.0-6.5 | 120 | 110 (NS) | 16 |
| S. plymuthica ATCC 15928 | 73 | 9 | 30 | 6.2 | 65 | 120 (NS) | 24 |
| E. rhapontici NCPPB 1578 | 85 | 15 | 30 | 7.0 | 280 | NS (4) | 2 |
| P. rubrum CBS574.77d | 86 | 9 | 30 | 5.5 | 140 | NS (NS) | 15 |
| P. mesoacidophila MX-45 | 8 | 91 | 40 (20-40) | 5.8 (4-8) | 19 | NS (14) | 12 |
| Bemisia argentifolii | 0 | NS | NS | 6 | 110 | NS (245) | 18 |
The results are the results for the isomaltulose product, except for MX-45 and Bemisia (trehalulose product).
Ultimate percentage of conversion of sucrose into isomaltulose or trehalulose.
NS, not specified.
Immobilized cells (not purified enzyme) were used to obtain the data for P. rubrum CBS574.77.
Fructose increases the ratio of trehalulose production but does not inhibit the SI activity for the purified SIs from S. plymuthica, P. rubrum, and E. rhapontici (12, 25). In contrast, fructose acts as a competitive inhibitor for the SIs from P. dispersa UQ68J and K. planticola UQ14S (Table 5), which implies that there is tighter binding of fructose to the active site in these enzymes (although the binding is still weaker than that of glucose). Further comparisons of the kinetics of these SIs in the presence of different monosaccharides, along with mutagenesis studies to test the functional significance of apparent key amino acid differences (Table 2), are likely to be revealing.
Practical implications.
It has been speculated that the high Km values for many SIs have a functional benefit, allowing cells to consume sucrose that is in limited supply and to convert only excess sucrose into isomaltulose reserves (1). Under this hypothesis for SI function and evolution, the highly efficient isomaltulose synthase in P. dispersa UQ68J is initially surprising. However, isomaltulose is an inhibitor of microbial glucosyltransferases and invertase (1, 22). Production of an efficient isomaltulose synthase could therefore be an advantage under conditions in which there is intense competition for abundant sucrose, where the rapid release of isomaltulose could inhibit development of competing microbial populations. Consistent with this interpretation are the substantial constitutive production of isomaltulose, the inducibility of SI activity by sucrose or fructose, and the low propensity to use isomaltulose for growth in P. dispersa UQ68J (27).
Whatever the drivers for its evolution, the characteristics of the unusual isomaltulose synthase from P. dispersa UQ68J are advantageous for use in cell- or enzyme-based bioreactors or potentially in engineered plants for isomaltulose production. The key advantages revealed here are the low Km, the high Vmax, the high Kcat/Km, the high stability in purified form, the high substrate use efficiency, the high ratio of isomaltulose to trehalulose across wide pH and temperature activity ranges (optimum pH, pH 5; optimum temperature, 30°C), and the absence of a reverse reaction converting isomaltulose to glucose, fructose, and/or trehalulose. Together, these characteristics result in highly efficient conversion of sucrose into isomaltulose.
Further investigation of the unique structural features of the P. dispersa UQ68J SI compared with those of the less efficient and specific isomerases from other species should help to elucidate the mechanisms of isomerase action and indicate opportunities to further increase stability and activity under conditions used for industrial biosynthesis of isomaltulose.
Acknowledgments
We thank S. Hansom and Pan Yunrong for expert technical assistance.
This investigation was supported in part by grants from the Sugar Research and Development Corporation, CSR Sugar Limited, and the Australian Research Council and by a UQ Early Career Enabling Grant to L.W.
REFERENCES
- 1.Börnke, F., M. Hajirezaei, and U. Sonnewald. 2001. Cloning and characterization of the gene cluster for palatinose metabolism from the phytopathogenic bacterium Erwinia rhapontici. J. Bacteriol. 183:2425-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheetham, P. S. J. 1984. The extraction and mechanism of a novel isomaltulose-synthesizing enzyme from Erwinia rhapontici. Biochem. J. 220:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheetham, P. S. J., C. E. Imber, and J. Isherwood. 1982. The formation of isomaltulose by immobilized Erwinia rhapontici. Nature 299:628-631. [Google Scholar]
- 4.Gnanasambandam, A., and R. G. Birch. 2004. Efficient developmental mis-targeting by the sporamin NTPP vacuolar signal to plastids in young leaves of sugarcane and Arabidopsis. Plant Cell Rep. 23:435-447. [DOI] [PubMed]
- 5.Janecek, S., B. Svensson, and B. Henrissat. 1997. Domain evolution in the alpha-amylase family. J. Mol. Evol. 45:322-331. [DOI] [PubMed] [Google Scholar]
- 6.Kakinuma, H., H. Yuasa, and H. Hashimoto. 1998. Glycosyltransfer mechanism of alpha-glucosyltransferase from Protaminobacter rubrum. Carbohydr. Res. 312:103-115. [Google Scholar]
- 7.Lichtenthaler, F. W. 2002. Unsaturated O- and N-heterocycles from carbohydrate feedstocks. Accounts Chem. Res. 35:728-737. [DOI] [PubMed] [Google Scholar]
- 8.Lina, B. A. R., D. Jonker, and G. Kozianowski. 2002. Isomaltulose (Palatinose): a review of biological and toxicological studies. Food Chem. Toxicol. 40:1375-1381. [DOI] [PubMed] [Google Scholar]
- 9.Mattes, R., K. Klein, H. Schiweck, M. Kunz, and M. Munir. July. 1995. Preparation of acariogenic sugar substitutes. International patent WO 95/20047 A3.
- 10.McAllister, M., C. T. Kelly, E. Doyle, and W. M. Fogarty. 1990. The isomaltulose synthesizing enzyme of Serratia plymuthica. Biotechnol. Lett. 12:667-672. [Google Scholar]
- 11.Munir, M., B. Schneider, and H. Schiweck. 1987. 1-O-Alpha-d-glucopyranosyl-d-fructose: preparation from saccharose and reduction to 1-O-alpha-d-glucopyranosyl-d-glucitol. Carbohydr. Res. 164:477-485. [Google Scholar]
- 12.Nagai, Y., T. Sugitani, and K. Tsuyuki. 1994. Characterization of alpha-glucosyltransferase from Pseudomonas mesoacidophila MX-45. Biosci. Biotechnol. Biochem. 58:1789-1793. [DOI] [PubMed] [Google Scholar]
- 13.Nagai-Miyata, Y., K. Tsuyuki, T. Sugitani, T. Ebashi, and Y. Nakajima. 1993. Isolation and characterization of a trehalulose-producing strain of Agrobacterium. Biosci. Biotechnol. Biochem. 57:2049-2053. [Google Scholar]
- 14.Nagano, N., C. A. Orengo, and J. M. Thornton. 2002. One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321:741-765. [DOI] [PubMed] [Google Scholar]
- 15.Nakajima, Y. 1984. Palatinose production by immobilized α-glucosyl transferase. Proc. Res. Soc. Jpn. Sugar Refineries Technol. 33:55-63. [Google Scholar]
- 16.Park, Y. K., R. T. Uekane, and H. H. Sato. 1996. Biochemical characterization of a microbial glucosyltransferase that converts sucrose to palatinose. Rev. Microbiol. 27:131-136. [Google Scholar]
- 17.Priefer, U., R. Simon, and A. Pühler. 1984. Cloning with cosmids, p. 190-201. In A. Pühler and K. N. Timmis (ed.), Advanced molecular genetics. Springer-Verlag, Berlin, Germany.
- 18.Salvucci, M. E. 2003. Distinct sucrose isomerases catalyze trehalulose synthesis in whiteflies, Bemisia argentifolii, and Erwinia rhapontici. Comp. Biochem. Physiol. B 135:385-395. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Schiweck, H., M. Munir, K. M. Rapp, B. Schneider, and M. Vogel. 1990. New developments in the use of sucrose as an industrial bulk chemical. Zuckerindustrie 115:555-565. [Google Scholar]
- 21.Schiweck, H., M. Munir, K. M. Rapp, B. Schneider, and M. Vogel. 1991. New developments in the use of sucrose as an industrial bulk chemical, p. 57-94. In F. W. Lichtenthaler (ed.), Carbohydrates as organic raw materials. Wiley-VCH, Weinheim, Germany.
- 22.Takazoe, I. 1989. Palatinose—an isomeric alternative to sucrose, p. 143-167. In T. H. Grenby (ed.), Progress in sweeteners. Elsevier, Barking, United Kingdom.
- 23.van der Veen, B. A., G. Potocki-Veronese, C. Albenne, G. Joucla, P. Monsan, and M. Remaud-Simeon. 2004. Combinatorial engineering to enhance amylosucrase performance: construction, selection, and screening of variant libraries for increased activity. FEBS Lett. 560:91-97. [DOI] [PubMed] [Google Scholar]
- 24.Veronese, T., and P. Perlot. 1999. Mechanism of sucrose conversion by the sucrose isomerase of Serratia plymuthica ATCC 15928. Enzyme Microb. Technol. 24:263-269. [Google Scholar]
- 25.Veronese, T., and P. Perlot. 1998. Proposition for the biochemical mechanism occurring in the sucrose isomerase active site. FEBS Lett. 441:348-352. [DOI] [PubMed] [Google Scholar]
- 26.Watanabe, K., K. Chishiro, K. Kitamura, and Y. Suzuki. 1997. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0A resolution: structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 269:142-153. [DOI] [PubMed] [Google Scholar]
- 27.Wu, L., and R. G. Birch. 2004. Characterisation of Pantoea dispersa UQ68J: producer of a highly efficient sucrose isomerase for isomaltulose biosynthesis. J. Appl. Microbiol. 97:93-103. [DOI] [PubMed] [Google Scholar]
- 28.Zhang, D. H., N. Li, S. M. Lok, L. H. Zhang, and K. Swaminathan. 2003. Isomaltulose synthase (PalI) of Klebsiella sp. LX3—crystal structure and implication of mechanism. J. Biol. Chem. 278:35428-35434. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, D. H., N. Li, K. Swaminathan, and L. H. Zhang. 2003. A motif rich in charged residues determines product specificity in isomaltulose synthase. FEBS Lett. 534:151-155. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, D. H., X. Z. Li, and L. H. Zhang. 2002. Isomaltulose synthase from Klebsiella sp. strain LX3: gene cloning and characterization and engineering of thermostability. Appl. Environ. Microbiol. 68:2676-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]