Abstract
Since virus concentrations in drinking waters are generally below the detection limit, the infectious risk from drinking water consumption requires assessment from the virus concentrations in source waters and removal efficiency of treatment processes. In this study, we estimated from reverse transcription-PCR on 10-fold serially diluted RNA that noroviruses, the most prevalent waterborne gastroenteritis agents, were present at 4 (0.2 to 38) to 4,900 (303 to 4.6 × 104) PCR-detectable units (PDU) per liter of river water (ranges are given in parentheses). These virus concentrations are still high compared with 896 to 7,499 PDU/liter of treated sewage and 5,111 to 850,000 PDU/liter in raw sewage. Sequencing analyses designated human norovirus GGII.4 Lordsdale as the most prevalent strain in the sampling period 1998 to 1999 in both sewage and surface waters. Other GGII strains were also very abundant, indicating that the majority of the virus contamination was derived from urban sewage, although very divergent strains and one animal strain were also detected in the surface and sewage waters. Rotaviruses were also detected in two large rivers (the Maas and the Waal) at 57 to 5,386 PDU/liter. The high virus concentrations determined by PCR may in part be explained by the detection of virus RNA instead of infectious particles. Indeed, reoviruses and enteroviruses that can be cultured were present at much lower levels, of 0.3 to 1 and 2 to 10 PFU/liter, respectively. Assuming 1% of the noroviruses and rotaviruses to be infectious, a much higher disease burden than for other viruses can be expected, not only because of the higher levels but also because of these viruses' higher infectivity and attack rates.
Noroviruses (NoV; previously known as Norwalk-like caliciviruses or small round-structured viruses) (35) belong to the most infectious group of causative agents of epidemic gastroenteritis (11, 44). Genetically, noroviruses can be divided into five genogroups (GGI, GGII, GGIII, GGIV, and GGV), which consist of different genotypes. A prototype virus represents each genotype: GGI includes Norwalk virus (GGI-1) and Southampton virus (GGI-2), GGII includes Hawaii virus (GGII-1) and others (2, 57), GGIII includes Jena virus, GGIV includes Alphatron, and GGV is found in mice (25, 53).
Numerous outbreaks of norovirus-associated gastroenteritis originating from contaminated drinking or recreational water have been ascribed to various causes (4, 6, 21, 29, 34, 37, 40). Contamination of drinking water by sewage through pump failure or blockage of a sewage system has been previously described (6, 34). Also, inadequate or failing treatment processes have led to the insufficient removal of viruses from source waters (4, 29, 37). Waterborne outbreaks may arise from direct exposure by ingestion of contaminated tap water or water-containing products, e.g., ice cubes, custard, and salads. Outbreaks associated with the consumption of contaminated drinking water may go unnoticed because of compliance with guidelines, including bacterial indicators and state-of-the-art treatment processes (17). In waterborne outbreaks, a very high proportion of the population can be affected, leading to several to hundreds of cases of gastroenteritis, followed by secondary spread and resulting in significant economic impact.
Noroviruses have been detected in raw urban sewage (32, 33). Primary and secondary sewage treatment processes do not efficiently reduce the virus concentration, in contrast with tertiary processes (13, 24, 46, 52). Therefore, depending on the applied processes, treated sewage discharged onto surface waters may significantly enhance the virus concentrations in the environment. Other sources for viral contamination of surface waters may occur during heavy rainfall and include discharges of raw sewage or washoff of animal manure. The latter may give rise to public health problems in the case of exposure to zoonotic pathogens, as was suggested occurs with caliciviruses and hepatitis E viruses (3, 18). Drinking water may be produced from groundwater or surface waters. Surface waters are more heavily contaminated with pathogenic microorganisms, although viruses have also been detected in groundwater (8, 14, 16, 46). The efficiency of advanced drinking water treatment processes may differ significantly with regard to removal or inactivation of microorganisms from source waters. Bacteria are more readily inactivated by chlorination than are protozoa (49). Viruses are effectively removed or inactivated by slow sand filtration and soil passage, but they are more resistant to UV and coagulation combined with sedimentation (9, 43, 50). However, a very low concentration of virus may suffice to cause infection, e.g., from 10 to <104 norovirus PCR-detectable units (PDU), leading to gastrointestinal disease in two-thirds of the individuals infected (30). Quantitative risk assessment based on source water quality and removal efficiency of treatment processes is required with regard to pathogenic viruses, since direct detection of pathogenic viruses in drinking water is not possible. This study focused on noroviruses, which are the most common waterborne gastroenteritis agents in The Netherlands. We estimated the numbers of particles containing norovirus and rotavirus RNA in two large rivers by molecular methods. We compared these estimates with the numbers of infectious enterovirus and reovirus particles and bacteriophages in these surface waters. Noroviruses in raw and treated sewage waters were quantified and typed by sequencing analysis and compared to strains detected in the surface waters to identify possible sources of contamination. Finally, norovirus sequences originating from water samples were compared to consensus strains circulating in the population.
MATERIALS AND METHODS
Viruses.
The positive control samples used in the cell culture method were virus isolates, reovirus type 3, and coxsackievirus type B4, originally obtained from the Voorns Canal and grown by buffalo green monkey kidney (BGM) cell culture. The positive control used in the detection of F-specific bacteriophages was MS2 (ATCC 15597-B1), and the positive control in the detection of somatic coliphages was ϕX174 (ATCC 13706-B1). Norovirus-positive stool specimens obtained from patients with gastroenteritis were used as a positive control (GGII.1; Hu/NV/6649/1994) for RNA extraction and reverse transcription-PCR (RT-PCR). Rotavirus WA was used as a positive control for RNA extraction and RT-PCR (Diagnostic Laboratory for Infectious Diseases, National Institute of Public Health and Environment).
Water samples.
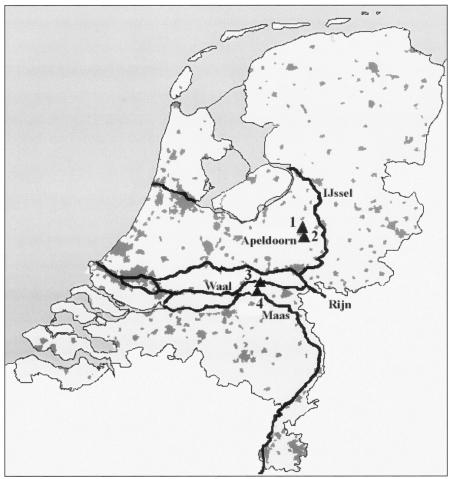
Ten liters of raw sewage and 200 to 400 liters of treated sewage were collected at a pumping-engine station in the city of Apeldoorn, The Netherlands (Fig. 1). The treatment process consists of primary settlement of the sewage, biological treatment (with activated sludge), and phosphorus removal. Large volumes of water (200 to 600 liters) were collected from two large Dutch rivers, the Maas and the Waal. Sampling at all locations was repeated at approximately 4-week intervals in the period from November 1998 to April 1999.
FIG. 1.
Sampling locations in The Netherlands. Grey fields indicate urbanization; thick lines indicate rivers. Numbers indicate sampling locations. 1, Sewage treatment plant at Apeldoorn, raw sewage; 2, sewage treatment plant at Apeldoorn, treated sewage; 3, river Waal; 4, river Maas.
Concentration of the water samples.
Water was first concentrated by a conventional filter adsorption-elution method. The average recovery of the membrane filtration step used in this study was 87 to 94% as determined by the use of F-specific bacteriophages (these phages have characteristics comparable to the NoVs with respect to size and isoelectric point) and 80% for coxsackie B4 virus. Briefly, for the concentration procedure, magnesium chloride was added to the water sample to a final concentration of 0.05 M to enable formation of a virus-magnesium chloride complex. By reducing the pH to 3.8, these positively charged complexes adsorb to a negatively charged cartridge filter (Nominal; 1.2 μm-pore-sized filter) (Millipore, Etten-Leur, The Netherlands). Viruses were eluted from the filter with a 3% beef extract (Difco, Amsterdam, The Netherlands) solution, to which Tris (Biosolve, Valkenswaard, The Netherlands) was added to a final concentration of 0.05 M to set the pH to 9.0. The typical retentate volume of 10-liter raw sewage water was approximately 650 ml, and the volume of large volumes of treated sewage and surface water (200 to 600 liters) was approximately 1,800 ml. The precipitate was dissolved and neutralized with a concentrated acetic acid buffer (pH 5.0) to a final pH of approximately 7.2. The resulting eluate in part was subjected further to the two-phase separation method and in part was analyzed by cell culture for enteric viruses and bacteriophages.
Concentration and purification of virus from the eluate by two-phase separation.
The two-phase separation method is based on the selective distribution of viruses into two incompatible phases (42), with slight modifications. Briefly, 650 ml of eluate, 1% (wt/vol) Dextran T40 (Pharmacia, Roosendaal, The Netherlands), 10% (wt/vol) polyethylene glycol 6000 (Merck, Amsterdam, The Netherlands), 0.2 M NaCl (Merck), and 10 mM phosphate buffer (pH 7.2) were mixed for 1 h at 4°C on a horizontal shaker. The suspension was then transferred to a separation funnel and left overnight at 4°C. After separation, the bottom phase and the interphase were harvested. Further purification was done by spin column gel chromatography with Sephadex G200 (ICN, Zoetermeer, The Netherlands) and by ultrafiltration with a Centricon 100 microconcentrator (100,000-molecular-weight cutoff; Amicon, Dronten, The Netherlands). The average retentate volumes were 1 to 5 ml.
RNA extraction.
RNA was extracted from the retentate obtained after two-phase separation by binding to silica beads in the presence of a high-molarity solution (5). This method was modified to allow larger volumes of retentate to be processed (32). Briefly, 2.7 M solid guanidinium isothiocyanate (Fluka, Zwijndrecht, The Netherlands), 25 mM EDTA (pH 8.0; ICN), 55 mM Tris-HCl (pH 6.4; Biosolve), and 20 μl of silica suspension were added to the retentate, resulting in a final volume of 1.7 to 8.7 ml. The sample was mixed on a rotary shaker, and then centrifuged briefly to pellet the silica particles. The pellet was washed twice with a guanidinium isothiocyanate-containing washing buffer (L2) (5), twice with ethanol 70% (vol/vol), and once with acetone. After removal of acetone by evaporation, the RNA was eluted in distilled water with RNAguard (200 U/ml; Pharmacia) and dithiothreitol (3 mM; Sigma, Zwijndrecht, The Netherlands) and was used directly in RT-PCR.
RT-PCR.
For norovirus detection, we used an RT-PCR assay that had been developed for the specific detection of norovirus in stool specimens from humans (56). The assay detects over 85% of a panel of different antigenic types of norovirus (27). Norovirus-specific primers were used, which anneal to a part of the norovirus genome within the viral RNA polymerase gene (open reading frame 1). The RT-PCR was done as described previously (54, 56). For rotaviruses, we used an RT-PCR assay developed for generic molecular detection, amplifying the VP6 gene fragment as described previously (55). RNA was diluted at 10-fold intervals to semiquantitatively determine the virus concentrations in the samples.
Gel electrophoresis, Southern blotting, and hybridization.
The amplification products were analyzed by electrophoresis with 2% agarose gels and visualized under UV illumination after being stained with SYBR Gold nucleic acid gel stain (Molecular Probes, Leiden, The Netherlands). The PCR products in the agarose gel were transferred to a positively charged nylon membrane (Amersham, ′s Hertogenbosch, The Netherlands) by a vacuum blotting system (Millipore) with 0.5 M NaOH (Merck) and 0.6 M NaCl (Merck) for 30 min. The confirmation of the specificity of the detected RT-PCR products of both viruses was done as described previously (54, 55).
Norovirus cloning, sequencing, and phylogenetic analysis.
Since water may contain multiple virus strains (32), each norovirus-positive RT-PCR product was cloned and selected for sequence analysis. Norovirus hybridization-positive RT-PCR products were excised from a 2% agarose gel and purified with a Qiaquick gel extraction kit (QIAGEN, Hilden, Germany). The purified RT-PCR products were cloned into the pGEM-T Easy Vector System II (Invitrogen, Leek, The Netherlands); after transformation, at least five positive colonies were selected. The pGEM-T Easy Vector system was checked for correct insertion size by direct PCR amplifications with M13 forward and M13 reverse primers supplied by the manufacturer. PCR products hybridizing with the generic norovirus probes (1, 56) were purified with a PCR purification kit (QIAGEN) and sequenced with the BigDye Terminator Cycle Sequencing Ready Reaction kit (Perkin-Elmer, Applied Biosystems, Foster City, Calif.). Nucleotide sequences were edited by SeqMan II (DNA-star) and aligned by Bionumerics, version 2.0 (Applied Maths, Kortrijk, Belgium) by the unweighted-pair group method using average linkages after multiple sequence alignment of a 145-nucleotide segment of the polymerase gene.
Cell culture.
Virus infectivity was determined by a monolayer plaque assay (7). Briefly, BGM cells were grown to confluent monolayers in 75-cm2 plastic flasks. The culture medium was removed, after which the eluate and antibiotic mixture were added to the flasks. The cultures were incubated at room temperature for 120 min to allow virus adsorption to the cells. The cells were overlaid with Medium 199 with Earl's salts (Life Technologies, Breda, The Netherlands) with 10% fetal bovine serum (Life Technologies), 0.9% Bacto agar (Difco, Amsterdam, The Netherlands), 0.2% bicarbonate, 100 IU of penicillin, and 100 μg of streptomycin (Life Technologies)/ml. After 9 days of incubation at 37°C, the cells were stained with 0.03% neutral red in 0.9% agar. After 24 h, the plaques were enumerated and the virus concentration in the original water sample was calculated from the test volume and the virus count.
Detection of bacteriophage.
The detection of F-specific bacteriophages and somatic coliphages was done according the ISO 10705-1 (1995) and ISO 10705-2 (2000) standards (www.iso.org), respectively. The host bacterial strains WG49 and WG5 were exposed to the eluates to culture the F-specific and somatic bacteriophages, respectively (20). In the presence of bacteriophages, plaques could be enumerated after overnight culture. The virus concentration in the original water sample was calculated from the test volume and the virus count.
Statistical methods.
The numbers of virus particles present in water were estimated by culture or by RT-PCR on 10-fold serially diluted RNA (end point dilution). Virus concentrations in the undiluted samples were estimated as most probable numbers by the use of the number of PFU or the presence or absence of virus genomes in the 10-fold RNA dilutions, under the assumption that negative samples do not contain virus or viral RNA. Application of the Poisson distribution was justified by the assumption that the infectious virus particles or viral RNA was dispersed randomly in the sample. The maximum likelihood method was used to estimate the number of virus particles in the undiluted sample (38). A negative binomial model gave the best fit for the distribution of virus particles in the original and diluted samples. The 95% confidence interval was estimated for each virus concentration.
RESULTS
Detection of viruses in surface water samples.
Virus concentrations in surface water samples were determined for samples from two large rivers in the centre of The Netherlands (Fig. 1). These rivers, the Maas and Waal Rivers, are influenced by (un)treated sewage originating from Belgium and France, and Germany, respectively. Each river water sample was positive for the presence of F-specific and somatic phages, noroviruses, rotaviruses, reoviruses, and enteroviruses (Table 1). In the Maas, the average concentrations of F-specific and somatic phages were 7 × 103 and 2 × 104 PFU/liter, respectively. The average reoviruses and enterovirus concentrations were much lower, varying from 1 to 6 PFU/liter. On average, noroviruses and rotaviruses were both detected at 2 × 102 PDU/liter in the Maas. In the Waal, the average concentrations of F-specific and somatic phages were 1 × 103 and 3 × 104 PFU/liter, respectively. As observed for the Maas, the concentrations of reoviruses and enteroviruses were much lower (0.3 to 10 PFU/liter), whereas noroviruses and rotavirus concentrations were higher (on average, 2 × 103 PDU/liter).
TABLE 1.
Detection of norovirus, rotavirus, reovirus, enterovirus, somatic phages, and F-specific phages in river water samples, Maas and Waal Rivers
| Date | Location | Concn (95% confidence interval)
|
|||||
|---|---|---|---|---|---|---|---|
| Somatic phages (PFU/liter) | F-specific phages (PFU/liter) | Enteroviruses (PFU/liter) | Reoviruses (PFU/liter) | Noroviruses (PDU/liter) | Rotaviruses (PDU/liter) | ||
| 10 December 1998 | Maas | 4.0 × 104 (3.1 × 104 to 4.3 × 104) | 1.5 × 104 (1.4 × 104 to 1.6 × 104) | 1 (0.7-2) | 6 (5-8) | 85 (5-756) | 57 (3-504) |
| 10 December 1998 | Waal | 6,963 (6,231-7,751) | 319 (279-362) | 0.6 (0.3-1) | 6 (5-7) | 776 (49-6,925) | 5,386 (333 to 5.1 × 104) |
| 28 December 1998 | Maas | 1.8 × 104 (1.2 × 104 to 2.7 × 104) | 1,572 (1,238-1,960) | 1 (0.9-2) | 5 (4-7) | 4 (0.2-38) | 285 (13-2,556) |
| 28 December 1998 | Waal | 7.7 × 104 (7.0 × 104 to 8.5 × 104) | 609 (541-682) | 0.3 (0.1-0.6) | 5 (4-6) | 4,900 (303 to 4.6 × 104) | 315 (20-2,823) |
| 28 January 1999 | Maas | 2.8 × 104 (2.5 × 104 to 3.1 × 104) | 4,460 (4,070-4,874) | 1 (0.7-2) | 4 (3-6) | 397 (19-3,554) | 264 (13-2,369) |
| 28 January 1999 | Waal | 2.1 × 104 (1.8 × 104 to 2.3 × 104) | 2,386 (2,141-2,648) | 0.3 (0.1-0.6) | 10 (9-12) | 29 (1-257) | 193 (12-1,730) |
| 11 March 1999 | Maas | 1.6 × 104 (1.3 × 104 to 1.9 × 104) | 6,054 (5,530-6,610) | 2 (1-3) | 3 (2-5) | 370 (18-3,304) | 248 (12-2,220) |
| 11 March 1999 | Waal | 5,305 (4,523-6,173) | 1,282 (1,159-1,413) | 0.8 (0.2-2) | 2 (0.8-4) | 540 (26-4,819) | 360 (17-3,213) |
Detection of norovirus variants in surface water.
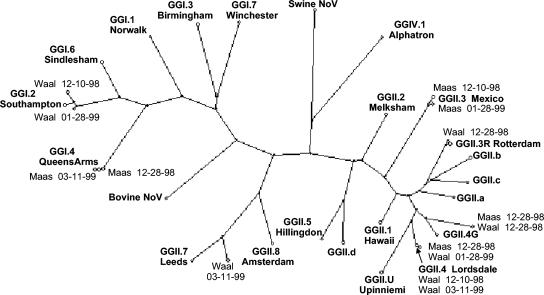
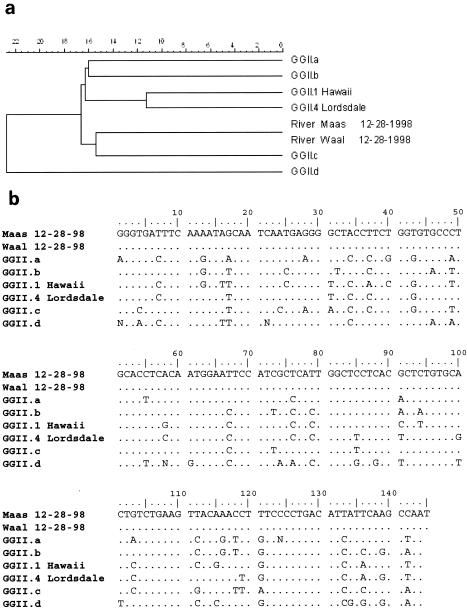
Strains belonging to seven different norovirus genotype strains (GGI.2 Southampton, GGI.4 QueensArms, GGII.3 Mexico, GGII.3R Rotterdam, GGII.4 Lordsdale, GGII.7 Leeds, and an unclassified GGII strain) were found in the surface water samples out of a total of 38 clones. Figure 2 shows these different norovirus strains detected in surface water and the relation of these strains to the consensus of the prototypes found in the population (28). As can be seen from Fig. 2, most norovirus strains from the water samples were closely related to the respective prototypes, differing from 0% (GGII.4 Lordsdale) to 12% (GGII.7 Leeds). However, one strain was found to differ by over 15% in two clones isolated from either the Maas or Waal River sampled on the same day (28 December 1998). The aberrant strain (designated Maas/Waal) was compared to consensus strains of variants found in fecal samples from patients in the population. Aberrant or other variants were circulating in the population years later and varied over 15% with the strains found in the surface water samples (Fig. 3a). To determine the recombination or mutation rate of these strains, a comparison was made between the sequences found in the two surface waters and the sequences of these consensus strains. Figure 3b shows the differences in nucleotides between these strains. In each surface water sample, strains were detected which belonged to at least one norovirus genotype strain, but sometimes two or three strains. In all four Maas samples, strains were found grouped with four different norovirus genotypes (QueensArms, Mexico, Lordsdale, and the Maas/Waal strain) and in the river Waal samples, five different norovirus genotypes (Southampton, Rotterdam, Lordsdale, Leeds, and the Maas/Waal strain) were found. In samples from both rivers, strains were present from the Lordsdale and the aberrant GGII strain genotypes. The additional strains were found in several samples and several sequenced clones. On two of four sampling dates in either river, strains genotyped as Southampton, QueenArms, and Mexico were found. The strains belonging to the three additional genotypes (Rotterdam, Leeds, and the Maas/Waal strain) were found each in only one out of the four sampling dates and were found scattered over the sampling period.
FIG. 2.
Phylogenetic tree illustrating the genetic relationship of norovirus isolates from river water samples from the Maas and Waal Rivers and the consensus sequences of norovirus prototypes (GG) found in the population (28). The genetic relationship is based on a 145-nt fragment of the polymerase gene.
FIG. 3.
(a) Phylogenetic analysis of the polymerase gene (145-bp region) showing the aberrant Maas/Waal strain (River Maas 12-28-98; River Waal 12-28-98) detected in the river water, Maas and Waal Rivers, and different GGII consensus prototype strains. (b) Alignment of the polymerase gene (145-bp region) showing the aberrant Maas/Waal strain (Maas 12-28-98; Waal 12-28-98) detected in the river water, Maas and Waal Rivers, and different GGII consensus prototype strains.
Detection of viruses in sewage samples.
Raw and treated sewage samples were collected every 3 to 4 weeks during the winter of 1998 to 1999. F-specific and somatic phages, enteroviruses, reoviruses, rotaviruses, and noroviruses were detected in all sewage samples (Table 2).
TABLE 2.
Detection of norovirus, rotavirus, reovirus, enterovirus, somatic phages, and F-specific phages in sewage samples (both raw and treated sewage) and the virus removal at the sewage treatment plant
| Date | Sewage | Concn (95% confidence interval)
|
|||||
|---|---|---|---|---|---|---|---|
| Somatic phages (PFU/liter) | F-specific phages (PFU/liter) | Enteroviruses (PFU/liter) | Reoviruses (PFU/liter) | Noroviruses (PDU/liter) | Rotaviruses (PDU/liter) | ||
| 16 November 1998 | Raw | 3.5 × 106 (2.9 × 106 to 4.2 × 106) | 4.0 × 106 (3.4 × 106 to 4.7 × 106) | 191 (140-253) | 1,253 (1,114-1,403) | 7,035 (444 to 6 .3 × 104) | 4,690 (296 to 4.2 × 104) |
| 16 November 1998 | Treated | 3.6 × 105 (2.9 × 105 to 4.4 × 105) | 1.0 × 105 (9.2 × 104 to 1.2 × 105) | 39 (31-49) | 48 (39-59) | 896 (57-7,999) | 598 (38-5,333) |
| Calculated log 10 removal | 1.0 | 1.6 | 0.7 | 1.4 | 0.9 | 0.9 | |
| 21 December 1998 | Raw | 6.8 × 106 (6.3 × 106 to 7.3 × 106) | 6.8 × 106 (6.9 × 105 to 7.7 × 105) | 288 (226-361) | 2,143 (1,964-2,332) | 1.2 × 105 (7,359 to 1.1 × 106) | 763 (36-8,804) |
| 21 December 1998 | Treated | 3.0 × 105 (2.5 × 105 to 3.4 × 105) | 6.2 × 103 (5.3 × 103 to 7.2 × 103) | 7 (4-10) | 92 (83-102) | 1,367 (88 to 1.2 × 104) | 908 (57-8,106) |
| Calculated log 10 removal | 1.4 | 2.0 | 1.6 | 1.4 | 1.9 | −0.1 | |
| 11 January 1999 | Raw | 6.4 × 106 (5.0 × 106 to 8.1 × 104) | 1.2 × 106 (1.1 × 106 to 1.3 × 106) | 296 (186-424) | 555 (414-725) | 8.5 × 105 (5.3 × 104 to 8.0 × 106) | 5.5 × 104 (2,592 to 4.9 × 105) |
| 11 January 1999 | Treated | 1.0 × 106 (9.3 × 105 to 1.1 × 106) | 6.7 × 104 (5.9 × 104 to 7.6 × 104) | 10 (7-13) | 76 (68-85) | 7,499 (463 to 7.1 × 104) | 4,819 (228 to 4.3 × 104) |
| Calculated log 10 removal | 0.8 | 1.3 | 1.5 | 0.9 | 2.1 | 1.1 | |
| 4 February 1999 | Raw | 3.3 × 106 (3.0 × 106 to 4.0 × 106) | 7.5 × 105 (6.7 × 105 to 8.4 × 106) | 140 (99-190) | 416 (343-499) | 4.6 × 104 (2,896 to 4.1 × 104) | 3.1 × 104 (1,931 to 2.7 × 105) |
| 4 February 1999 | Treated | 1.5 × 105 (1.3 × 106 to 1.7 × 105) | 1.3 × 104 (1.2 × 104 to 1.4 × 104) | 5 (3-7) | 23 (19-28) | 4,283 (265 to 4.0 × 1 04) | 2.9 × 104 (1,763 to 2.7 × 105) |
| Calculated log 10 removal | 1.4 | 1.8 | 1.5 | 1.3 | 1.0 | 0.03 | |
| 25 February 1999 | Raw | 5.2 × 106 (4.5 × 106 to 6.0 × 106) | 3.5 × 106 (3.2 × 106 to 4.0 × 106) | 833 (587-1,141) | 111 (40-239) | 5,111 (322 to 4.6 × 104) | 339 (16-3,027) |
| 25 February 1999 | Treated | 1.9 × 105 (1.8 × 108 to 2.1 × 105) | 5.4 × 104 (4.8 × 104 to 6.2 × 104) | 12 (9-18) | 8 (5-12) | 3,582 (221 to 3.4 × 104) | 2.4 × 104 (1,475 to 2.2 × 105) |
| Calculated log 10 removal | 1.4 | 1.8 | 1.8 | 1.1 | 0.2 | −1.8 | |
In raw sewage, the concentrations of F-specific and somatic phages were higher than that found in river water (average, 106 PFU/liter). On average, reoviruses and enteroviruses were present at 102 PFU/liter, whereas noroviruses and rotaviruses, respectively, were detected on average at 2 × 105 and 2 × 104 PDU/liter of raw sewage. The average virus concentrations in treated sewage were lower than in raw sewage, except for rotaviruses (Table 2).
Virus removal at the sewage treatment plant was on average 1.6 and 1.1 log10 units for F-specific and somatic phages, respectively. For enteroviruses and reoviruses, removal was similar at 1.4 and 1.3 log10 units on average, respectively. For noroviruses, the average removal was 1.8 log10 units, whereas for rotaviruses the removal was much lower, approximately 0.2 log10 units (Table 2).
Typing of norovirus variants in sewage waters.
To distinguish between different norovirus strains present in water, we determined the genetic variability of these noroviruses by sequencing the RT-PCR products after cloning. The sequences of 145 nucleotides were aligned and subjected to phylogenetic analysis, with 86 clones sequenced in all. The sequences amplified from water samples were compared to consensus strains found in samples from the human population.
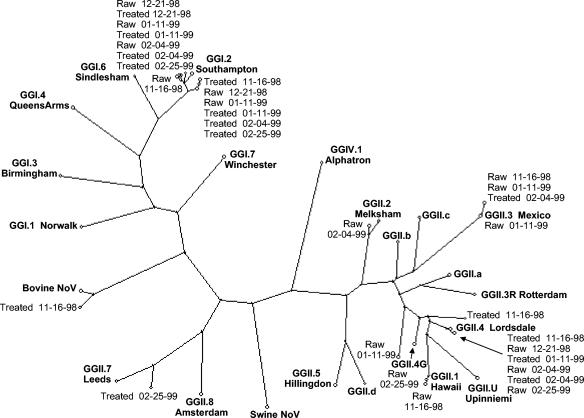
Strains belonging to eight different norovirus genotype strains (GGI.2 Southampton, GGII.1 Hawaii, GII.2 Melksham, GGII.3 Mexico, GGII.4 Lordsdale, GGII.7 Leeds, an unclassified GGII strain [Apeldoorn RS01-11-99], and bovine NoV) were found in 10 sewage samples (Fig. 4). As can be seen from Fig. 4, most norovirus strains from the sewage samples are closely related to the respective prototypes differing from 0% (GGII.4 Lordsdale and GGII.3 Mexico) to 12% (GGII.7 Leeds), except one strain differing by over 15% (Apeldoorn RS01-11-99). In each sewage sample, strains were detected which belonged to at least one norovirus genotype strain, but sometimes two or three different strains. In the raw sewage samples, strains were found belonging to six different norovirus genotypes strains in all (GGI.2 Southampton, GGII.1 Hawaii, GII.2 Melksham, GGII.3 Mexico, GGII.4 Lordsdale, and the Apeldoorn RS01-11-99 strain). In the treated sewage samples, strains were found belonging to five different norovirus genotype strains (GGI.2 Southampton, GGII.3 Mexico, GGII.4 Lordsdale, GGII.7 Leeds, and bovine NoV). Strains belonging to the genotypes Southampton, Mexico, and Lordsdale were predominantly present in both raw and treated sewage. The strains belonging to the other genotypes were retrieved from 1 or 2 of the 86 sequenced clones. On all sampling dates, strains belonging to the genotypes Southampton and Lordsdale were present in either raw or treated sewage or both. On three of five sampling dates, strains belonging to the genotype Mexico were found in raw sewage or treated sewage. The strains belonging to the five additional genotypes (Hawaii, Melksham, Leeds, the Apeldoorn RS01-11-99 strain, and bovine NoV) were found scattered over the sampling period.
FIG. 4.
Phylogenetic tree illustrating the genetic relationship of norovirus isolates from sewage samples (both raw and treated sewage) and the consensus sequences of norovirus prototypes (GG) found in the population (28). The genetic relationship is based on a 145-nt fragment of the polymerase gene.
DISCUSSION
Surface waters may be directly or indirectly contaminated with human pathogenic viruses by (un)treated sewage or washoff of animal manure. We found noroviruses, rotaviruses, and enteroviruses in each of the samples taken from two large rivers in The Netherlands. In the summer of the same year, Schvoerer et al. (45) did not detect rotaviruses, adenoviruses, or hepatitis A viruses in fresh waters in France by PCR, but in 1 of 10 samples, noroviruses were detected when 5 ml was analyzed; in 3 of 10 samples, enteroviruses were detected. In another French study, viruses were detected by PCR and culture methods in concentrates of 20 liters of river water (23). From the 68 samples tested, only 2 were positive for infectious enterovirus but 60 were positive by enterovirus RT-PCR. Also other viruses were detected with PCR, one sample was positive for hepatitis A virus, one sample was positive for norovirus genogroup II, and three samples were positive for astrovirus. From 11 of 22 800-ml river water samples in Japan, 100 reovirus strains were identified by electropherotyping (36). In comparison, we detected reoviruses by cell culture in each of the eight river water samples tested. By RT-PCR, three of eight 1-liter water samples from three rivers in Germany were positive for the presence of enteroviruses in 1993 (15). Two years later, each of six 1-liter river water samples tested positive for the presence of GGI noroviruses, rotaviruses, and enteroviruses; in two of six samples, GGII noroviruses were detected, but no hepatitis A viruses were detected (16). In a recent Finnish study, 1-liter surface water samples were concentrated, of which 13 of 139 samples tested positive for the presence of noroviruses and 47 of 139 samples tested positive for the presence of F-RNA phages (22). These few papers document presence-absence data; however, to be able to estimate the infectious risk by consumption of drinking water, quantitative data are necessary (19). We determined the virus concentrations varying from 0.3 to 10 PFU of enteroviruses and reoviruses/liter of river water and varying from 319 to 7.7 × 104 PFU of F-specific and somatic phages/liter. Since noroviruses and rotaviruses are hard or impossible to culture (12), these viruses were detected by molecular methods resulting in much higher concentrations, varying from 4 to 5,386 PDU/liter of river water. No sensitive cell lines are available; alternatively, the presence of the opposite strand of these single-strand viruses could be determined to establish if replication took place. This has been described for adenoviruses but yet has to be developed for noroviruses (26). In absence of such methods, we estimated the virus concentrations by RT-PCR on 10-fold serial dilutions of the extracted RNA as a semiquantitative approach. A negative binomial model was used to estimate the virus concentration in the original sample because this model gives the best fit for the distribution of virus particles in the samples (51).
The Maas is a river fed by rainwater. In total, 8 million people live in the Maas catchment area, with over 5 million people depending on it for drinking water, compared with 30 million people who drink water directly or indirectly produced from the river Rhine. The Rhine is a river fed by both rain and meltwater. Our results generally showed higher concentrations of pathogenic viruses in the Waal, a tributary of the Rhine, than in the Maas. The Rhine catchment is five times as large as the Maas, collecting more microbiological contamination from wastewater sources. Since part of the population in the catchments is not yet connected to sewage treatment and part of the sewage is discharged without treatment with (for instance) heavy rainfall, this would indicate that high virus loads may reach the surface waters. Indeed, we found loads varying from 111 to 2,143 PFU of enterovirus and reovirus/liter of raw sewage, 6.8 × 105 to 6.8 × 106 PFU of F-specific and somatic phages/liter, and 339 to 8.5 × 105 PDU of rotaviruses and noroviruses /liter. In treated sewage, we determined virus concentrations varying from 5 to 92 enterovirus and reovirus PFU/liter, 6.2 × 103 to 1.5 × 105 PFU of F-specific and somatic phages/liter, and 598 to 2.4 × 104 PDU of rotaviruses and noroviruses/liter. The contribution of the sewage treatment processes to virus removal varied between 0.7 and 2.1 log10 units. This indicates that discharge of both raw and treated sewage will considerably contribute to the virus burden of the rivers.
Noroviruses are genetically very diverse because of accumulating mutations during replication (57). Sequencing analysis of norovirus PCR products showed the presence of 10 different genotypes in sewage and surface waters. Half of these genotypes were detected in both sewage and surface waters, indicating that these virus genotypes were very prevalent in The Netherlands. In addition, the diverging genotypes that were detected either in sewage at the same plant or in rivers several tens of kilometers away pointed at regional differences between genotypes circulating in smaller groups of the population or sporadic cases.
Recently, the circulation of viruses in the population and environment was shown for enteroviruses (47). In 1998 to 1999, mainly GGII strains were found in fecal samples taken from humans in The Netherlands (27). In both sewage and surface waters, GGII strains were also predominantly detected. More specifically, GGII.4 Lordsdale was the main genotype in both the Dutch human population and waters, followed by GGII.3 Mexico, GGII.7 Leeds, and GGII.1 Hawaii. The predominant GGI strain in sewage, surface waters, and the population was GGI.2 Southampton. Remarkably, a bovine NoV strain was detected in urban sewage, known as the Jena variant (31). However, in both sewage and surface waters aberrant strains were found which were not represented in clinical samples. One diverging variant (Apeldoorn RS01-11-99 strain) was detected in raw sewage in January 1999, which differed significantly from any variant identified to that point. The highest identity of 85% in the 145-nucleotide segment of the polymerase gene was found for GGIIU Upinniemi. Another divergent strain (Maas/Waal) detected in both the rivers Maas and Waal showed the highest homology (86%) with GGIIc Den Haag. At least four different GGII.4 Lordsdale strains emerged in 1998 to 1999, causing gastroenteritis in the population.
Noroviruses are generally more prevalent in winter (39). Here, we screened sewage and surface waters sampled in the cold season in which noroviruses were present. However, the screening period was not very extensive, ranging only from November to April. We have also detected noroviruses in water samples taken in warmer periods of the year at other locations, which is relevant when surface waters are used for recreational purposes (10, 48). We have also previously shown that viruses occur at peak concentrations, suggesting that the risk of infections may also differ greatly between exposures (52, 58). Moreover, consumption of drinking water may also pose a health risk if it coincides with failed drinking water treatment. Exposure to high levels in surface waters by either recreation, shellfish culture, or drinking water production leads to public health hazards. Although a low dose of only 10 PDU may lead to infection (30), not all of these virus units making up the high-virus concentrations found in surface waters will lead to infection, because of host factors. But even if only 1% of noroviruses detected in surface waters are infectious, this would lead to a much higher disease burden in case of noroviruses (attack rate, 65% gastroenteritis cases independent of blood group) (41) than for enteroviruses or reoviruses.
Acknowledgments
This work was funded by the Environmental Inspectorate under project number 330000, Health Related Water Microbiology.
We thank the employees of the sewage treatment plant in Apeldoorn. Our colleages from the RIVM, Microbiological Laboratory for Health Protection, Olaf Nijst, Harold van den Berg, Sylvain Skraber, and Saskia Rutjes are gratefully acknowledged for technical assistance and critical reading of the manuscript.
We gratefully acknowledge the help of Harry Vennema with regard to the comparison of the norovirus sequences and Peter Teunis from the Computerization and Methodological Consultancy Unit for statiistical analyses.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181:S336-S348. [DOI] [PubMed] [Google Scholar]
- 3.Balayan, M. S. 1997. Epidemiology of hepatitis E virus infection. J. Viral Hepat. 4:155-165. [DOI] [PubMed] [Google Scholar]
- 4.Boccia, D., A. E. Tozzi, B. Cotter, C. Rizzo, T. Russo, G. Buttinelli, A. Caprioli, M. L. Marziano, and F. M. Ruggeri. 2002. Waterborne outbreak of Norwalk-like virus gastroenteritis at a tourist resort, Italy. Emerg. Infect. Dis. 8:563-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim van Dillen, and J. S. O. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, C. M., J. W. Cann, G. Simons, R. L. Fankhauser, W. Thomas, U. D. Parashar, and M. J. Lewis. 2001. Outbreak of Norwalk virus in a Caribbean island resort: application of molecular diagnostics to ascertain the vehicle of infection. Epidemiol. Infect. 126:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahling, D. R. 2002. An improved filter elution and cell culture assay procedure for evaluating public groundwater systems for culturable enteroviruses. Water Environ. Res. 74:564-568. [DOI] [PubMed] [Google Scholar]
- 8.Dahling, D. R., and B. A. Wright. 1986. Optimization of the BGM cell line culture and viral assay procedures for monitoring viruses in the environment. Appl. Environ. Microbiol. 51:790-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Roda Husman, A. M., P. Bijkerk, W. Lodder, H. van den Berg, W. Pribil, A. Cabaj, P. Gehringer, R. Sommer, and E. Duizer. 2004. Calicivirus inactivation by nonionizing (253.7-nanometer-wavelength [UV]) and ionizing (gamma) radiation. Appl. Environ. Microbiol. 70:5089-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Roda Husman, A. M., E. J. M. Penders, A. P. Krom, G. L. Bakker, and W. Hoogenboezem. 2004. Viruses in the Rhine and source waters for drinking water production. RIVM report 330200 003/2004. National Institute of Public Health and the Environment, Bilthoven, The Netherlands.
- 11.De Wit, M. A. S., M. P. G. Koopmans, L. M. Kortbeek, W. J. B. Wannet, J. Vinje, F. van Leusden, A. I. M. Bartelds, and Y. T. H. P. van Duynhoven. 2001. Sensor, a population-based cohort study on gastro-enteritis in the Netherlands, incidence and etiology. Am. J. Epidemiol. 154:666-674. [DOI] [PubMed] [Google Scholar]
- 12.Duizer, E., K. J. Schwab, F. H. Neill, R. L. Atmar, M. P. Koopmans, and M. K. Estes. 2004. Laboratory efforts to cultivate noroviruses. J. Gen. Virol. 85:79-87. [DOI] [PubMed] [Google Scholar]
- 13.Fleischer, J., K. Schlafmann, R. Otchwemah, and K. Botzenhart. 2000. Elimination of enteroviruses, other enteric viruses, F-specific coliphages, somatic coliphages and E. coli in four sewage treatment plants of Southern Germany. J. Water Supply Res. Technol. 49:127-137. [Google Scholar]
- 14.Fout, S. H., B. C. Martinson, M. W. N. Moyer, and D. R. Dahling. 2003. A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 69:3158-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilgen, M., B. Wegmuller, P. Burkhalter, H. P. Buhler, U. Muller, J. Luthy, U. Candrian. 1995. Reverse transcription PCR to detect enteroviruses in surface water. Appl. Environ. Microbiol. 61:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilgen, M., D. Germann, J. Luthy, and P. Hubner. 1997. Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int. J. Food Microbiol. 37:189-199. [DOI] [PubMed] [Google Scholar]
- 17.Goldstein, S. T., D. D. Juranek, O. Ravenholt, A. W. Hightower, D. G. Martin, J. L. Mesnik, S. D. Griffiths, A. J. Bryant, R. R. Reich, and B. L. Herwaldt. 1996. Cryptosporidiosis: an outbreak associated with drinking water despite state-of-the-art water treatment. Ann. Intern. Med. 124:459-468. [DOI] [PubMed] [Google Scholar]
- 18.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H.-J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181:S322-S330. [DOI] [PubMed] [Google Scholar]
- 19.Haas, C. N. 1983. Estimation of risk due to low doses of microorganisms: a comparison of alternative methodologies. Am. J. Epidemiol. 118:573-582. [DOI] [PubMed] [Google Scholar]
- 20.Havelaar, A. H., M. van Olphen, and J. F. Schijven. 1995. Removal and inactivation of viruses by drinking water treatment processes under full scale conditions. Water Sci. Technol. 31:55-62. [Google Scholar]
- 21.Hoebe, C. J. P. A., H. Vennema, A. M. de Roda Husman, and Y. T. H. P. van Duynhoven. 2004. Norovirus outbreak among primary school children who had played in a recreational water fountain. J. Infect. Dis. 189:699-705. [DOI] [PubMed] [Google Scholar]
- 22.Hörman, A., R. Rimhanen-Finne, L. Maunula, C. H. von Bonsdorff, N. Torvela, A. Heikinheimo, and M. L. Hänninen. 2004. Campylobacter spp., Giardia spp., Cryptosporidium spp., noroviruses, and indicator organisms in surface water in southwestern Finland, 2000-2001. Appl. Environ. Microbiol. 70:87-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hot, D., O. Legeay, J. Jacques, C. Gantzer, Y. Caudrelier, K. Guyard, M. Lange, and L. Andreoletti. 2003. Detection of somatic phages, infectious enteroviruses and enterovirus genomes as indicators of human enteric viral pollution in surface water. Water Res. 37:4703-4710. [DOI] [PubMed] [Google Scholar]
- 24.Hovi, T., M. Stenvik, H. Partanen, and A. Kangas. 2001. Poliovirus surveillance by examining sewage specimens. Quantitative recovery of virus after introduction into sewerage at remote upstream location. Epidemiol. Infect. 127:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karst, S. M., C. E. Wobus, M. Lay, J. Davidson, and H. W. Virgin IV. 2003. STAT1-dependent innate immunity to a Norwalk-like virus. Science 299:1575-1578. [DOI] [PubMed] [Google Scholar]
- 26.Ko, G., T. L. Cromeans, and M. D. Sobsey. 2003. Detection of infectious adenovirus in cell culture by mRNA reverse transcription-PCR. Appl. Environ. Microbiol. 69:7377-7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koopmans, M. P. G., J. Vinje, M. A. S. de Wit, E. J. T. M. Leenen, W. H. M. van der Poel, and Y. T. H. P. van Duynhoven. 2000. Human enteric caliciviruses in The Netherlands. J. Infect. Dis. 181:S262-S269. [DOI] [PubMed] [Google Scholar]
- 28.Koopmans, M., E. van Strien, and H. Vennema. 2002. Molecular epidemiology of human caliciviruses, p. 509-539. In U. Desselberger and J. Gray (ed.), Viral gastroenteritis. Elsevier Science B.V., Amsterdam, The Netherlands.
- 29.Kukkula, M., L. Maunula, E. Silvennoinen, and C.-H. von Bonsdorff. 1999. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J. Infect. Dis. 180:1771-1776. [DOI] [PubMed] [Google Scholar]
- 30.Lindesmith, L., C. Moe, S. Marionneau, N. Ruvoen, X. Jiang, L. Lindblad, P. Stewart, J. Lependu, and R. Baric. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548-553. [DOI] [PubMed] [Google Scholar]
- 31.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lodder, W. J., J. Vinje, R. van de Heide, A. M. de Roda Husman, E. J. T. M. Leenen, and M. P. G. Koopmans. 1999. Molecular detection of Norwalk-like caliciviruses in sewage. Appl. Environ. Microbiol. 65:5624-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loisy, F., P. Le Cann, M. Pommepuy, and F. LeGuyader. 2000. An improved method for the detection of Norwalk-like caliciviruses in environmental samples. Lett. Appl. Microbiol. 31:411-415. [DOI] [PubMed] [Google Scholar]
- 34.Maurer, A. M., and D. Sturchler. 2000. A waterborne outbreak of small round structured virus, campylobacter and shigella co-infections in La Neuville, Switzerland, 1998. Epidemiol. Infect. 125:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matson, D. O., and G. Szucs. 2003. Calicivirus infections in children. Curr. Opin. Infect. Dis. 16:241-246. [DOI] [PubMed] [Google Scholar]
- 36.Matsuura, K., M. Ishikura, T. Nakayama, S. Hasegawa, O. Morita, K. Katori, and H. Uetake. 1993. Ecological studies on reovirus pollution of rivers in Toyama Prefecture. II. Molecular epidemiological study of reoviruses isolated from river water. Microbiol. Immunol. 37:305-310. [DOI] [PubMed] [Google Scholar]
- 37.McCarthy, N., B. de Jong, T. Ziese, R. Sjolund, C. A. Hjalt, and J. Giesecke. 1998. Epidemiological investigation of an outbreak of gastroenteritis in Sweden in the absence of detailed microbiological information. Eur. J. Epidemiol. 14:711-718. [DOI] [PubMed] [Google Scholar]
- 38.Mood, M., F. A. Graybill, and D. C. Boes. 1974. Introduction to the theory of statistics. McGraw-Hill Book Company, Singapore, Republic of Singapore.
- 39.Mounts, A. W., T. Ando, M. Koopmans, J. S. Bresee, J. Noel, and R. I. Glass. 2000. Cold weather seasonality of gastroenteritis associated with Norwalk-like viruses. J. Infect. Dis. 181:S284-287. [DOI] [PubMed] [Google Scholar]
- 40.Nygard, K., M. Torven, C. Ancker, S. B. Knauth, K. O. Hedlund, J. Giesecke, Y. Andersson, and L. Svensson. 2003. Emerging genotype (GGIIb) of norovirus in drinking water, Sweden. Emerg. Infect. Dis. 9:1548-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nygard, K., L. Vold, E. Halvorsen, E. Bringeland, J. A. Rottingen, and P. Aavitsland. 2004. Waterborne outbreak of gastroenteritis in a religious summer camp in Norway, 2002. Epidemiol. Infect. 132:223-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pöyry, T., M. Stenvik, and T. Hovi. 1988. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl. Environ. Microbiol. 54:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schijven, J. F., and S. M. Hassanizadeh. 2000. Removal of viruses by soil passage: overview of modeling, processes, and parameters. Crit. Rev. Environ. Sci. Technol. 30:49-127. [Google Scholar]
- 44.Schreier, E., F. Doring, and U. Kunkel. 2000. Molecular epidemiology of outbreaks of gastroenteritis associated with small round structured viruses in Germany in 1997/98. Arch. Virol. 145:443-453. [DOI] [PubMed] [Google Scholar]
- 45.Schvoerer, E., F. Bonnet, V. Dubois, G. Cazaux, R. Serceau, H. J. Fleury, and M. E. Lafon. 2000. PCR detection of human enteric viruses in bathing areas, waste waters and human stools in southwestern France. Res. Microbiol. 151:693-701. [DOI] [PubMed] [Google Scholar]
- 46.Schvoerer, E., M. Ventura, O. Dubos, G. Cazaux, R. Serceau, N. Gournier, V. Dubois, P. Caminade, H. J. Fleury, and M. E. Lafon. 2001. Qualitative and quantitative molecular detection of enteroviruses in water from bathing areas and from a sewage treatment plant. Res. Microbiol. 152:179-186. [DOI] [PubMed] [Google Scholar]
- 47.Sedmak, G., D. Bina, and J. MacDonald. 2003. Assessment of an enterovirus sewage surveillance system by comparison of clinical isolates with sewage isolates from Milwaukee, Wisconsin, collected August 1994 to December 2002. Appl. Environ. Microbiol. 69:7181-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skraber, S., R. Italiaander, W. Lodder, and A. M. de Roda Husman. Noroviruses in archival samples. Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 49.Sobsey, M. D. 1989. Inactivation of health-related microorganisms in water by desinfection processes. Water Sci. Technol. 21:179-195. [Google Scholar]
- 50.Sommer, R., W. Pribil, S. Appelt, P. Gehringer, H. Eschweiler, H. Leth, A. Cabaj, and T. Haider. 2001. Inactivation of bacteriophages in water by means of non-ionizing (UV-253.7 nm) and ionizing (gamma) radiation: a comparative approach. Water Res. 35:3109-3116. [DOI] [PubMed] [Google Scholar]
- 51.Teunis, P. F., and A. H. Havelaar. 2000. The Beta Poisson dose-response model is not a single-hit model. Risk Anal. 20:513-520. [DOI] [PubMed] [Google Scholar]
- 52.Van den Berg, H., W. Lodder, W. van der Poel, H. Vennema, and A. M. de Roda Husman. Genetic diversity of noroviruses in raw and treated sewage water. Res. Microbiol., in press. [DOI] [PubMed]
- 53.Van der Poel, W. H., J. Vinje, R. van der Heide, M. I. Herrera, A. Vivo, and M. P. Koopmans. 2000. Norwalk-like calicivirus genes in farm animals. Emerg. Infect. Dis. 6:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vennema, H., E. de Bruin, and M. Koopmans. 2002. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 25:233-235. [DOI] [PubMed] [Google Scholar]
- 55.Villena, C., W. M. El-Senousy, F. X. Abad, R. M. Pinto, and A. Bosch. 2003. Group A rotavirus in sewage samples from Barcelona and Cairo: emergence of unusual genotypes. Appl. Environ. Microbiol. 69:3919-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vinje, J., S. A. Altena, and M. P. G. Koopmans. 1997. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 176:1374-1378. [DOI] [PubMed] [Google Scholar]
- 57.Vinje, J., R. A. Hamidjaja, and M. D. Sobsey. 2004. Development and application of a capsid VP1 (region D) based reverse transcription PCR assay for genotyping of genogroup I and II noroviruses. J. Virol. Methods 116:109-117. [DOI] [PubMed] [Google Scholar]
- 58.Westrell, T., P. Teunis, H. van den Berg, W. Lodder, H. Ketelaars, T. A. Stenström, and A. M. de Roda Husman. Short and long term fluctuations of norovirus concentration in surface water and their implications for public health. Submitted for publication.