Abstract
Benzene dioxygenase and toluene dioxygenase from Pseudomonas putida have similar catalytic properties, structures, and gene organizations, but they differ in substrate specificity, with toluene dioxygenase having higher activity toward alkylbenzenes. The catalytic iron-sulfur proteins of these enzymes consist of two dissimilar subunits, α and β; the α subunit contains a [2Fe-2S] cluster involved in electron transfer, the catalytic nonheme iron center, and is also responsible for substrate specificity. The amino acid sequences of the α subunits of benzene and toluene dioxygenases differ at only 33 of 450 amino acids. Chimeric proteins and mutants of the benzene dioxygenase α subunit were constructed to determine which of these residues were primarily responsible for the change in specificity. The protein containing toluene dioxygenase C-terminal region residues 281 to 363 showed greater substrate preference for alkyl benzenes. In addition, we identified four amino acid substitutions in this region, I301V, T305S, I307L, and L309V, that particularly enhanced the preference for ethylbenzene. The positions of these amino acids in the α subunit structure were modeled by comparison with the crystal structure of naphthalene dioxygenase. They were not in the substrate-binding pocket but were adjacent to residues that lined the channel through which substrates were predicted to enter the active site. However, the quadruple mutant also showed a high uncoupled rate of electron transfer without product formation. Finally, the modified proteins showed altered patterns of products formed from toluene and ethylbenzene, including monohydroxylated side chains. We propose that these properties can be explained by a more facile diffusion of the substrate in and out of the substrate cavity.
The ring-hydroxylating dioxygenases of prokaryotes are nonheme iron enzymes that convert aromatic compounds to dihydrodiols in the presence of dioxygen and NADH (6). They act upon a wide range of compounds, which the organisms use as carbon and energy sources. The enzymes are important in the bioremediation of environmental pollutants. Although none of them is completely specific, the dioxygenases display selectivity for the compounds that they metabolize. Moreover, they convert these substrates into stereo- and regiospecific products. These properties make them valuable in the enantio-controlled synthesis of compounds (21).
The benzene 1,2-dioxygenase from Pseudomonas putida ML2 (EC 1.14.12.3) is an extensively studied enzyme system (6, 33, 35, 44) that catalyzes the dihydroxylation of benzene to (1R,2S)-cis-cyclohexa-3,5-diene-1,2-diol (benzene cis-dihydrodiol), the first step in biodegradation of this xenobiotic compound. The enzyme system has three components: a flavoprotein reductase and a ferredoxin, which transfer electrons from NADH, and a catalytic iron-sulfur protein (ISPBED). ISPBED contains a Rieske-type [2Fe-2S] cluster, a mononuclear nonheme iron oxygen activation center, and a substrate-binding site (6, 22, 25). The ISPBED component comprises two dissimilar subunits, ISPBEDα and ISPBEDβ (55). A closely related enzyme that preferentially metabolized ethylbenzene and toluene, toluene dioxygenase ISPTOD (EC 1.14.12.11), was isolated from P. putida F1 (13). Much evidence has accumulated that the α subunit of the ISP contains the region that confers substrate specificity (2, 3, 5, 8, 9, 17, 26, 34, 36-38, 47). The genes encoding benzene and toluene dioxygenase systems have been cloned, sequenced, and expressed separately and in combination in Escherichia coli (35, 49, 50, 57, 58), but the molecular structures of the enzymes have not been determined. However several structures are now available for the naphthalene dioxygenase ISP (25), which has a homologous amino acid sequence (35.2% identity with ISPBED) (24). The enzyme comprises a trimeric arrangement of two types of subunits, α3β3.
The manipulation of enzyme specificity, both for the substrates used and for the products formed, is important for biotechnology. It is necessary to minimize undesirable side reactions, such as the generation of products that cannot be metabolized or the direct oxidation of NADH without substrate oxidation (the uncoupling reaction). Enzymes that are isolated from the natural environment have been optimized by evolution to avoid these difficulties. To better design new enzymes, it is worthwhile to examine naturally occurring enzymes with different specificities in order to determine which features of the sequence are necessary for efficient and specific catalysis.
Alignment of the nucleotide and amino acid sequences of ISPBEDα (50) and ISPTODα (57) revealed that they differ by 33 amino acids, distributed through the sequence. The first objective of this study was to investigate which of these amino acid differences is necessary for conversion of the substrate preference of benzene dioxygenase from benzene to alkyl-substituted aromatic hydrocarbons, such as toluene and ethylbenzene. The N- and C-terminal domains of toluene and benzene dioxygenases were interchanged, and then individual amino acids were mutated. The recombinant proteins were subcloned and expressed in an isogenic background in E. coli. The properties of the variant enzymes were investigated by examining the activities with benzene, toluene, and ethylbenzene by an oxygen electrode assay and by performing product formation assays with radiolabeled substrates, TLC, and GC-MS.
MATERIALS AND METHODS
Abbreviations.
GC-MS, gas-chromatography-mass spectrometry; TLC, thin-layer chromatography; SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis; ISP, iron-sulfur protein of terminal dioxygenase; ISPBEDα and ISPTODα, iron-sulfur proteins of benzene and toluene dioxygenases, respectively; ISPBODα and ISPTEDα, chimeric α subunits with N-terminal regions from benzene and toluene dioxygenases and C-terminal regions from toluene and benzene dioxygenases, respectively; ISPBOD2α, ISPBEDα with residues 364 to 450 of ISPTODα; ISPBOD3α, ISPBEDα with residues 281 to 363 of ISPTODα.
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. E. coli JM109 was used for standard cloning and gene expression. E. coli CJ236 was used in the mutagenesis experiment with uracil-containing DNA. Transformations were carried out by the method of Hanahan (16).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics |
|---|---|
| Strains | |
| E. coli JM109a | endA1 recA1 gyrA96 thi hsdR17 relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] |
| E. coli CJ236b | dut-1 ung-1 thi-1 rela1 F′ pCJ105 (Cmr) |
| Plasmids | |
| pGEM 3Zf(+) | Plac lacZ Ampr |
| pKK223-3 | Plac Ampr |
| pCB1c | 1.6-kb DraI/EcoRI bedC1 insert in pGEM 3Zf(+) |
| pJRM502c | 2-kb EcoRI/Pst1 bedC1C2 insert in pKK223-3 |
| pJRM502-1 | 2-kb EcoRI/PstI todC1-bedC2 insert in pKK223-3 |
| pJRM502-2 | 2-kb EcoRI/PstI bodC1-bedC2 insert in pKK223-3 |
| pJRM503d | 1.7-kb EcoRI bedBA insert in pKK223-3 |
| pJRM504c | 1.6-kb EcoRI/HindIII bedC1 insert in pKK223-3 |
| pJRM505d | 0.7-kb EcoRI/PstI bedC2 insert in pKK223-3 |
| pJRM711 | 0.66-kb EcoRI todC2 insert in pKK223-3 |
| pSA109d | 1.6-kb EcoRI/PstI todC1 insert in pKK223-3 |
| pJRM5042 | 1.6-kb EcoRI/PstI todC1 modified insert in pKK223-3 |
| pJRM5041 | 1.6-kb EcoRI/PstI bodC1 insert in pKK223-3 |
| pJRM5041-1 | 1.6-kb EcoRI/HindIII bod2C1 insert in pKK223-3 |
| pJRM5041-2 | 1.6-kb EcoRI/HindIII bod3C1 insert in pKK223-3 |
| pCMB101 | 1.6-kb EcoRI/PstI tedC1 insert in pKK223-3 |
| pJRM5043 | 1.6-kb EcoRI/HindIII tedC1 modified insert in pKK223-3 |
| pCB1M1 | pCB1 (L285W) |
| pCB1M2 | pCB1 (A291S) |
| pCB1M3 | pCB1 (L285W/A291S) |
| pCB1M5 | pCB1 (V324I/I327V) |
| pCB1M7 | pCB1 (G404D) |
| pCB1M9 | pCB1 (K436R) |
| pJRM504M1 | pJRM504(L285W) |
| pJRM504M2 | pJRM504(A291S) |
| pJRM504M3 | pJRM504(L285W/A291S) |
| pJRM504M4e | pJRM504(I301V/T305S/I307L/L309V) |
| pJRM504M5 | pJRM504(V324I/I327V) |
| pJRM504M6e | pJRM504(L285W/A291S/G404D) |
| pJRM504M7 | pJRM504(G404D) |
| pJRM504M8e | pJRM504(I412V) |
| pJRM504M9 | pJRM504(K436R) |
| pJRM504M10e | pJRM504(E444D) |
See reference 54.
Obtained from Bio-Rad.
Gift of C. S. Butler.
Gift of S. Anguravirutt.
Mutant ISPBEDα generated by PCR-based mutagenesis.
Cloning strategies.
To construct ISPBODα encoded by plasmid pJRM5041 (Fig. 1A), the EcoRI-BstEII fragment from pJRM504 encoding ISPBEDα (residues 1 to 280) was ligated to the BstEII-HindIII fragment (residues 281 to 450) from pSA109 encoding ISPTODα. ISPTEDα encoded by plasmid pCMB101 was constructed by ligating the EcoRI-BstEII fragment from pSA109 with the BstEII-HindIII fragment from pJRM504. The two constructs that had the 5′ end from plasmid pSA109 had a lower expression level due to a difference of nine nucleotides with a high adenosine content in the ribosome-binding domain (45). To increase the level of expression, this sequence (CCAAAAAGT) was introduced into the Shine-Dalgarno sequence by carrying out PCR with pSA109 by using forward primer KPIf, which contained the 9-nucleotide sequence and a KpnI restriction site, and reverse primer BEIIr, which contained a BstEII restriction site. The positions of the primers are shown in Table 2. PCRs were carried out for 25 cycles of denaturation at 94°C for 20 s, primer annealing at 55°C for 20 s, and primer extension at 74°C for 60 s. The amplified DNA was digested with KpnI and BstEII and ligated into the BstEII-KpnI fragment of pJRM504 to construct pJRM5043 (ISPTEDα modified). Plasmid pJRM5042, containing ISPTODα modified in the Shine-Dalgarno region, was constructed by ligation of the pJRM5043 HindIII-BstEII fragment with the BstEII-HindIII fragment of pSA109 (Fig. 1A).
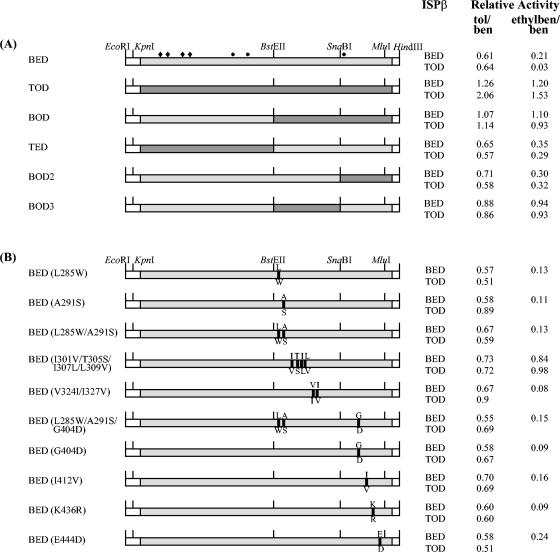
FIG. 1.
(A) Diagram of the genes encoding ISPBEDα, ISPTODα, ISPBODα, ISPTEDα, ISPBOD2α, and ISPBOD3α and relative activities of the corresponding cell extracts. (B) Diagram of the single and multiple mutations created in the ISPBEDα C-terminal region and the effects of the mutations on the relative activity. The bars indicate the open reading frames encoding ISP α subunit proteins. The nonconserved amino acids targeted for mutation are indicated by solid boxes. The letters above the bars indicate translated amino acids in ISPBEDα, and the letters below the bars indicate amino acids in ISPTODα. The numbering corresponds to the numbering for the amino acids in the ISPBEDα sequence. Solid diamonds indicate the conserved histidine (H98 and H119) and cysteine (C96 and C116) residues responsible for coordination of the [2Fe-2S] cluster. Solid circles indicate the conserved amino acids (two histidines [H222 and H228] and one aspartate [D376]) involved in coordination of the nonheme iron. The EcoRI, KpnI, BstEII, SnaBI, MluI, and HindIII sites are the restriction sites used for cloning the recombinant α subunit genes. The relative activities with toluene (tol) and ethylbenzene (ethylben) are compared to the activity with benzene (ben), and the data are based on the rate of O2 uptake measured polarographically by using ISPBED or ISPTOD β subunits, as described in Materials and Methods (means of three determinations).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotidea | Oligonucleotide sequenceb | Presence, loss, or gain of restriction site |
|---|---|---|
| Oligonucleotides for modification of the Shine-Dalgarno region | ||
| KPIfc | 5′ CTCGGTAC(CCAAAAAGT)GAGAAGACAATG 3′ | Gain of KpnI |
| BEIIrc | 5′ CAAGTAGCTGGTGACCTTCGGCCC 3′ | BstEII |
| Mutant oligonucleotidesd | ||
| L285W | 5′ CCAGCTACTGGACCGAAGGCCCGGCG 3′ | Loss of DraII |
| A291S | 5′ GAAGGCCCCGCGTCGGAAAAGGCGG 3′ | Loss of SacII |
| G404D | 5′ GCTGAGATGTCCATGGACCAAACCGTTG 3′ | Gain of NcoI |
| I301V/T305S/I307L/L309V | 5′ GCTCGACCATGAGTTTGGAGCCGCGCTCCACACTAC 3′ | Gain of BstXI |
| V324I/I327V | 5′ CCCAGGTATCAATACGGTCCGAACATGG 3′ | Gain of AvaII |
| I412V | 5′ CGACCCGGTATACCCTGGTCG 3′ | Gain of Bst1107I |
| K436R | 5′ GCACATTGGCTCCGGATGATGACC 3′ | Gain of BspEI |
| E444D | 5′ GACTGGGACGCATTAAAGGCGACG 3′ | Gain of BsmFI |
| Nonmutagenic oligonucleotides for PCR-based site-directed mutagenesis and/or cloning of bod2C1, bod3C1, todC1-bedC2, and bodC1-bedC2 genes | ||
| SBIfc | 5′ CGTCAGACACTACGTACCTTCTCTGCC 3′ | SnaBI |
| MUIrc | 5′ TTAGGTTTAACGCGTCGCCTTTAATGC 3′ | MluI |
| BEIIfc | 5′ GGGCCGAAGGTCACCAGCTACTTG 3′ | BstEII |
| SBIrc | 5′ GGCAGAGAAGGTACGTAGTGTCTGACG 3′ | SnaBI |
| HDIIIrc | 5′ GGCTGAAAATCTTCTCTCATCCGC 3′ | HindIII |
| PKKc | 5′ GCGGATAACAATTTCACACAGG 3′ | EcoRI |
| AGIr | 5′ CGGGCGCTACCGGTGCCGGC 3′ | AgeIe |
Most oligonucleotides were synthesized by Phil Marsh (King's College London); the exceptions were KPIf and BEIIr, which were synthesized by Pharmacia (UK) and Perkin-Elmer Ltd., respectively.
The base changes are indicated by boldface type. The underlined bases indicate loss or addition of the restriction site. Additional bases are in parentheses.
For the locations of nonmutagenic oligonucleotides see the positions of restriction sites in Fig. 1, sequences are within or close to the primer sequence.
In the mutant oligonucleotide designations the first letter is the amino acid in the ISPBEDα sequence, the number corresponds to the position in the amino acid sequence, and the second letter is the replacing amino acid in the ISPTODα sequence.
The AgeI restriction site is located between the bedC1 and bedC2 genes in the pJRM502 plasmid.
Two chimeric ISP α subunits in the C-terminal region, ISPBOD2α and ISPBOD3α, were generated by using an approach similar to the approach described above (Fig. 1A). ISPBOD2α contained the N-terminal region of ISPBEDα (residues 1 to 363) linked to the SnaBI-HindIII C-terminal region of ISPTODα (residues 364 to 450). ISPBOD3α was ISPBEDα with the BstEII-SnaBI region (residues 281 to 363) replaced with the BstEII-SnaBI region of ISPTODα.
To construct the coexpression vectors encoding ISPTODα-ISPBEDβ and ISPBODα-ISPBEDβ, PCRs were carried out with plasmids pJRM5042 (encoding ISPTODα) and pJRM5041 (encoding ISPBODα), respectively, by using primers PKKf and AGIr (Table 2). The PCR products were digested with EcoRI and AgeI and ligated into pJRM502 (encoding ISPBEDα-ISPBEDβ) previously digested with the same enzymes to construct plasmids pJRM502-1 and pJRM502-2, respectively. The correct clones were selected by restriction analysis and DNA sequencing.
Mutagenesis.
Site-directed mutagenesis was performed by using both the uracil-containing DNA-based mutagenesis method developed by Kunkel (27) and a PCR-based mutagenesis method adapted from the method of Kammann et al. (23). For uracil-containing DNA-based mutagenesis, plasmid pCB1 containing an F1 origin and encoding ISPBEDα was transformed into E. coli strain CJ236 (ung dut). After production of single-stranded DNA with the VCM13 helper phage (Stratagene), mutagenesis was carried out with a Muta-gene kit (Bio-Rad). The mutated DNA was further cloned into expression vector pJRM504 (ISPBEDα). For PCR-based mutagenesis, the first PCR was carried out with the wild-type ISPBEDα gene (plasmid pJRM504) by using the mutant oligonucleotide and a nonmutagenic oligonucleotide facing in the opposite direction at a distance of about 100 bp. The product was then used as a megaoligonucleotide in a second PCR with another nonmutagenic oligonucleotide facing in the opposite direction at a distance of about 250 bp. The final product was digested and ligated into the wild-type ISPBEDα gene. The mutagenic oligonucleotides were designed with a silent mutation that generated a new restriction site (Table 2) to facilitate identification of clones carrying the desired mutation. Successful mutations were identified by restriction analysis and DNA sequencing.
Expression of the genes in E. coli JM109 and preparation of cell extracts.
Cultures of E. coli JM109 cells carrying the different plasmids were grown at 37°C in L broth containing ampicillin (100 μg ml−1) to an optical density at 660 nm of 0.7 and then were grown at 30°C after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were harvested in the late exponential phase, washed once in 50 mM potassium phosphate (pH 7.2), and resuspended in 25 mM potassium phosphate (pH 7.2)-1 mM dithiothreitol (2 ml/g [wet weight]). The cells were disrupted by three passages through a chilled French pressure cell (Aminco, Silver Spring, Md.) at 27.5 MPa. After centrifugation at 17,400 × g for 1 h, the supernatant was used for enzyme activity measurements.
Protein determination, SDS-PAGE, and Western blot analysis.
Protein concentrations were determined by the modified Lowry method of Hess et al. (19). Proteins were resolved by SDS-PAGE (28). Western blot analysis with antibodies raised to purified ISPα was performed as described by Zamanian and Mason (55). To determine the ISPα concentration expressed per milligram of cell extract protein, samples were resolved by SDS-PAGE, stained with Coomassie brilliant blue R250, and subjected to quantitative image analysis with the EASYWin program (Herolab, Wiesloch, Germany). Duplicate samples (5 and 10 μg of cell extract protein) were taken, and a calibration curve was constructed by using a range of concentrations of pure ISPBEDα (1 to 15 μg).
Oxygen uptake measurement.
Dioxygenase activities of cell extracts were determined polarographically by measuring O2 consumption with a Clark-type oxygen electrode as previously described (11). The assay mixture contained 0.42 mM NADH, 0.5% (vol/vol) Triton X-100, 0.1 mM ferrous ammonium sulfate, and 1.5 mg of cell extract protein from the strain expressing reductaseBED and ferredoxinBED (plasmid pJRM503). The following enzymes were also added to the assay mixture: for the coexpressed enzymes, 1.5 mg of cell extract protein from the strains expressing ISPBEDα-ISPBEDβ (plasmid pJRM502), ISPTODα-ISPBEDβ (plasmid pJRM502-1), or ISPBODα-ISPBEDβ (plasmid pJRM502-2); and when the α and β subunits were expressed separately, 0.25 mg of ISP α subunits present in the cell extracts from the different strains and 0.5 mg of either ISPBEDβ present in the cell extract from the strain containing plasmid pJRM505 or ISPTODβ present in the cell extract from the strain containing plasmid pJRM711. The reaction was initiated by addition of 300 μM benzene, toluene, or ethylbenzene dissolved in buffer. Enzyme activity was expressed in nanomoles of O2 per minute per milligram of ISPα.
Enzyme activity.
Benzene dioxygenase activity was determined by using 14C-labeled substrates by measuring the accumulation of nonvolatile products on TLC squares by using a modification of the method described by Jiang et al. (22). Each reaction mixture (0.5 ml) was similar to the reaction mixture described for the polarographic assay, except that after 5 min of preincubation at room temperature, 1 mM NADH and either 20 μl of l-[U-14C]benzene, 20 μl of l-[U-14C]toluene (Sigma) in methanol (22.2 × 104 dpm μl−1; final concentration, 0.8 mM), or 20 μl of l-[U-14C]ethylbenzene (Sigma) in methanol (4.8 × 104 dpm μl−1; final concentration, 0.8 mM) were added. The enzyme specific activity was expressed in nanomoles of total product formed per minute per milligram of ISPα.
Detection and quantification of reaction products.
The oxidation products of the recombinant proteins from the different substrates were determined by using a modified method described by Ensley et al. (7). The assay with a radiolabeled substrate was allowed to proceed for 45 min. Aliquots (8 μl for experiments in which [14C]benzene and [14C]toluene were used and 16 μl for experiments in which [14C]ethylbenzene was used) were applied to the origin of a TLC plate (Silica Gel 60 F254; thickness, 0.2 mm). Standard samples of cis-3,5-cyclohexadiene-1,2-diol, catechol, 3-methylcatechol, o-cresol, m-cresol, p-cresol, and benzyl alcohol were also applied to the chromatograms, which were air dried for 30 min and developed with chloroform-acetone (80:20, vol/vol). Radioactive spots were located by autoradiography with X-ray film exposed at −70°C for 6 days before development. The area containing the radioactive product was scraped from the TLC plate, and the amount of each nonvolatile 14C-labeled product was determined by scintillation counting. Nonradioactive compounds were detected by spraying the plate with 2% (wt/vol) 2,6-dichloroquinone-4-chloroimide (13).
The identities of the reaction products were further established by GC-MS. Assays with unlabeled substrates were performed as described above, and the reaction products were extracted with 2 volumes of ethyl acetate. The organic phase was dried over anhydrous sodium sulfate, and the samples were concentrated to 100 μl under a flow of nitrogen. Samples were injected into a Hewlett-Packard 6890 GC coupled to a 5970 mass selective detector operating in splitless mode (injector temperature, 300°C) with a fused silica capillary column (inside diameter, 0.25 mm; length, 25 m) coated with a 0.1-μm phase of DB-5 (5% phenyl, 95% dimethylpolysiloxane; J& W Scientific, Folsom, Calif.). The column was kept at 50°C for 3 min, and then the temperature was increased to 300°C at a rate of 15°C/min and finally kept at 300°C for 10 min. The detector was operated in the scan mode with electron impact ionization at 70 eV and with scanning from 550 to 50 atomic mass units with a scan time of 0.86 s. The transfer line temperature was 280°C. All products were identified either by comparison of their GC-MS properties (retention times and mass fragments) with those of reference compounds used in the TLC experiments or by interpretation of the mass spectra. Relative yields of products were determined by integration of total ion current peak areas.
RESULTS
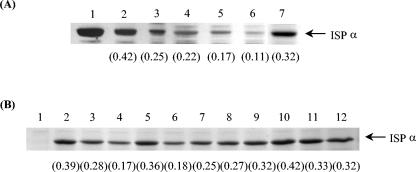
Chimeric proteins were constructed with the sequences containing residues 1 to 280 and 281 to 450 of ISPBEDα and ISPTODα, which yielded proteins designated ISPBODα and ISPTEDα (Fig. 1A). Two additional chimeric α subunits in the C-terminal region were constructed. ISPBOD2α contained the N-terminal region of ISPBEDα (residues 1 to 363) linked to the C-terminal region of ISPTODα (residues 364 to 450). ISPBOD3α was ISPBEDα with residues 281 to 363 replaced with the residues of ISPTODα. The cell extracts from each of the recombinant strains contained a protein that immunocross-reacted with a band whose relative mobility was identical to that of purified ISPBEDα (data not shown). The level of protein production was determined by quantitative SDS-PAGE (Fig. 2A). Despite the presence of identical promoter and ribosome-binding sites, the level of expression for the recombinant plasmids generally was not as high as that for the ISPBEDα protein (Fig. 2A, lane 2).
FIG. 2.
SDS-PAGE-resolved E. coli cell extracts expressing recombinant ISP α subunits. The numbers in parentheses indicate the levels of expression of the soluble ISP α subunit in the cell extract (in milligrams per milligram of total protein; means of two determinations). (A) Wild-type and chimeric ISP α-subunit cell extracts. Lane 1, purified ISPBEDα (2 μg); lanes 2 to 7, cell extracts (5 μg of protein per lane) from E. coli strains containing plasmids pJRM504 (ISPBEDα), pJRM5041 (ISPBODα), pJRM5042 (ISPTODα), pJRM5043 (ISPTEDα), pJRM5041-2 (ISPBOD3α), and pJRM5041-1 (ISPBOD2α), respectively. (B) Wild-type and mutant ISP α subunit cell extracts. Lanes 1 to 12, cell extracts (5 μg of protein per lane) from E. coli strains containing plasmids pKK223-3, pJRM504 (ISPBEDα), pJRM504M1 [ISPBEDα(L285W)], pJRM504M2 [ISPBEDα(A291S)], pJRM504M7 [ISPBEDα(G404D)], pJRM504M3 [ISPBEDα(L285W/A291S)], pJRM504M6 [ISPBEDα(L285W/A291S/G404D)], pJRM504M4 [ISPBEDα(I301V/T305S/I307L/L309V)], pJRM504M5 [ISPBEDα(V324I/I327V)], pJRM504M8 [ISPBEDα(I412V)], pJRM504M9 [ISPBEDα(K436R)], and pJRM504M10 [ISPBEDα(E444D)], respectively.
When the O2 uptake assay was used to measure activity, all the recombinant ISP α subunits reconstituted with ISPBEDβ or ISPTODβ were active with the three substrates (Fig. 1 and Table 3). The ratios of the oxygen uptake rates measured with toluene or ethylbenzene to the oxygen uptake rates measured with benzene (Fig. 1) gave a first measure of the substrate preferences of the enzymes. The chimeric ISPBODα and ISPBOD3α proteins combined with ISPBEDβ had a pattern of substrate specificity more like that of ISPTODα; the toluene/benzene activity ratios of the three proteins were 1.07, 0.88, and 1.26, respectively, and the ethylbenzene/benzene activity ratios were 1.10, 0.94, and 1.20, respectively. Furthermore, ISPTEDα and ISPBOD2α were more similar to ISPBEDα, and the ethylbenzene/benzene activity ratios of the three proteins were 0.35, 0.30, and 0.21, respectively (Fig. 1). The nature of the β subunit did not have a consistent effect on the substrate specificity. It should be noted that the specific activities of ISPTODα, ISPBODα, ISPTEDα, and ISPBOD3α combined with ISPBEDβ were lower than the specific activity of ISPBEDα (Table 3). However, this had no effect on the substrate preference since it was the relative activity with all three substrates that was important.
TABLE 3.
Comparison of the rates of oxygen uptake and nonvolatile product formation by different ISP α subunits combined with ISPBED β subunita
| Source of ISP α subunit in assay (plasmid) | Type of assay | Sp act (nmol of O2 min−1 mg−1 or nmol of product min−1 mg−1)
|
% of uncoupling
|
||||
|---|---|---|---|---|---|---|---|
| Benzene | Toluene | Ethyl-benzene | Benzene | Toluene | Ethylbenzene | ||
| ISPBED (pJRM504) | Polarographic | 115 ± 9.6b | 70.5 ± 5.5 | 25 ± 1.5 | 0 | 22 | 0 |
| Radiolabeled | 120 ± 2.9 | 54.9 ± 1.9 | 30.7 ± 1.7 | ||||
| ISPTOD (pJRM5042) | Polarographic | 44.9 ± 2.2 | 56.4 ± 3.9 | 54.1 ± 3.4 | 0 | 0 | 21 |
| Radiolabeled | 45.5 ± 0.7 | 61.3 ± 2.5 | 42.5 ± 3.4 | ||||
| ISPBOD (pJRM5041) | Polarographic | 44.5 ± 3.1 | 47.5 ± 4.1 | 49.1 ± 5.6 | 0 | 0 | 45 |
| Radiolabeled | 47.6 ± 2.6 | 46.2 ± 0.7 | 26.8 ± 2.3 | ||||
| ISPTED (pJRM5043) | Polarographic | 39.1 ± 2.3 | 25.3 ± 3.0 | 13.8 ± 0.5 | 0 | 0 | 48 |
| Radiolabeled | 42.1 ± 2.9 | 25.5 ± 1.7 | 8.8 ± 0.7 | ||||
| ISPBOD2 (pJRM5041-1) | Polarographic | 112 ± 8.5 | 79.7 ± 8.1 | 33.7 ± 4.1 | 0 | 37 | 24 |
| Radiolabeled | 115 ± 2.9 | 49.6 ± 2.2 | 25.7 ± 2.6 | ||||
| ISPBOD3 (pJRM5041-2) | Polarographic | 36.8 ± 4.6 | 32.2 ± 6.1 | 34.5 ± 4.0 | 0 | 0 | 31 |
| Radiolabeled | 39.1 ± 1.5 | 32.2 ± 0.6 | 23.6 ± 2.5 | ||||
| I301V/T305S/I307L/L309V (pJRM504M4) | Polarographic | 84.5 ± 4.4 | 62.1 ± 8.9 | 70.7 ± 9.9 | 0 | 0 | 44 |
| Radiolabeled | 85.1 ± 1.9 | 70.8 ± 6.1 | 39.9 ± 1.4 | ||||
Activities were determined with ISPBED β subunit, ferredoxinBED, and reductaseBED cell extracts. The specific activity is expressed as the rate of O2 uptake measured polarographically (three determinations) or as the amount of nonvolatile 14C-labeled product formed from oxidation of [14C]benzene, [14C]toluene, and [14C]ethylbenzene (nine determinations).
Mean ± standard deviation.
The results for the chimeric α subunits (Fig. 1A) revealed that a region of the protein responsible for specificity was between residues 281 and 450. In this region, benzene and toluene dioxygenases differ at 12 amino acid positions. Single and multiple amino acid mutations were generated, in which the nonconserved residues of ISPBEDα were replaced by the corresponding residues of ISPTODα (Fig. 1B). The ISPα concentration was estimated by quantitative SDS-PAGE (Fig. 2B) to be between 0.17 and 0.42 mg mg of protein−1. When the activity was measured polarographically, the majority of the mutant proteins had a substrate preference similar to that of the wild type. However, ISPBEDα(I301V/T305S/I307V/L309V) showed an increased preference for ethylbenzene, with an ethylbenzene/benzene activity ratio of 0.84 (Fig. 1B).
Table 3 shows the rates of product formation measured by 14C assays for a representative range of recombinant proteins combined with ISPBEDβ, and the data were compared with the rates of oxygen uptake under the same conditions, as shown in Fig. 1. The difference between the rate of product formation and the rate of oxygen consumption represented the rate of uncoupled oxidation of NADH. With benzene as the substrate, the rate of O2 consumption for ISPBEDα was equal to the amount of products formed, which was indicative of tight coupling, but for the chimeric proteins uncoupling was observed with toluene and, more particularly, with ethylbenzene. For example, the percentage of uncoupling (percentage of O2 consumed in excess of the amount of product formed) was 45% for ISPBODα and 44% for ISPBEDα(I301V/T305S/I307L/L309V), both with ethylbenzene (Table 3). Similar uncoupling reactions have been observed with naphthalene dioxygenase, in which the oxidation of benzene, toluene, and ethylbenzene was partially uncoupled from O2 consumption (29, 31). However, the ethylbenzene/benzene activity ratios as determined by the radiolabel assay were 0.67 for ISPBODα and 0.71 for ISPBOD3α, compared to 0.23 for ISPBEDα (Table 3), indicating that these enzymes had a higher substrate preference for ethylbenzene than the wild type. This phenomenon was less pronounced for ISPBEDα(I301V/T305S/I307L/L309V) which had a ratio of 0.47.
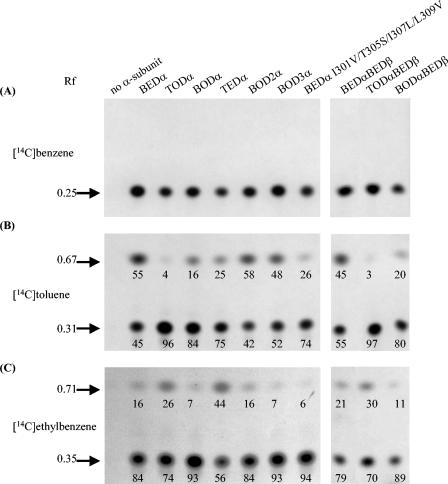
The products formed from the three substrates were investigated. Samples from 14C assays of the recombinant proteins, either expressed separately or coexpressed (ISPBEDα-ISPBEDβ, ISPTODα-ISPBEDβ, and ISPBODα-ISPBEDβ), were subjected to TLC (Fig. 3). With benzene as the substrate, a single compound with an Rf of 0.25, corresponding to the reference compound benzene cis-dihydrodiol, was detected in every case (Fig. 3A). This result is consistent with previous studies (12). With toluene as the substrate, two compounds (Rf, 0.31 and 0.67) were detected (Fig. 3B). By analogy to the benzene dioxygenase product, the compound with an Rf of 0.31 was provisionally identified as the expected cis-2,3-dihydroxy-1-methyl-cyclohexa-4,6-diene (toluene cis-dihydrodiol) (12). The Rf of 0.67 corresponded to the Rf of benzyl alcohol. Similarly, with all the dioxygenases that oxidized ethylbenzene, two compounds with Rf values of 0.35 and 0.71 were detected (Fig. 3C), and these compounds were provisionally identified as cis-2,3-dihydroxy-1-ethyl-cyclohexa-4,6-diene (ethylbenzene cis-dihydrodiol) and 1-phenethyl alcohol, respectively.
FIG. 3.
Autoradiograms of the products formed by using [14C]benzene (A), [14C]toluene (B), and [14C]ethylbenzene (C) by wild-type, chimeric, and mutant ISPBEDα subunits combined with the ISPBEDβ subunit and coexpressed ISPBEDα-ISPBEDβ, ISPTODα-ISPBEDβ, and ISPBODα-ISPBEDβ in the dioxygenase assay. The Rf values of the radiolabeled products are indicated on the left. The numbers on the autoradiograms indicate the percentages of the chain monohydroxylated and cis-diol products. The values are averages of two determinations. A repeat experiment gave similar results. Details of the experiment are described in Materials and Methods.
The products of the reactions described above, but without the use of radiolabeled substrates, were further analyzed by GC-MS (Tables 4 and 5). Since some compounds are known to undergo thermal decomposition during analysis, the phenol and catechols that were detected by GC-MC and not by other methods were interpreted as being the products of thermal dehydration of the cis-diols. Thus, with benzene as the substrate, peaks with masses corresponding to those of phenol, benzene cis-dihydrodiol, and catechol were detected, and the sum of the masses of these peaks corresponded to the mass of the single product, presumed to be benzene cis-dihydrodiol, observed in the autoradiogram (Fig. 3A).
TABLE 4.
Gas chromatography-mass spectrometric results for reaction products
| Iona | Retention time (min) | Product [ion mass (relative abundance)] |
|---|---|---|
| Phenol | 5.0 | 94 (100), 66 (38), 65 (28), 63 (9), 55 (9), 51 (7) |
| Benzene cis-dihydrodiol | 5.3 | 12 (15), 94 (23), 83 (36), 68 (26), 66 (100), 65 (32), 55 (40) |
| Catechol | 7.8 | 110 (100), 92 (12), 81 (19), 64 (47), 63 (23), 53 (18) |
| Benzaldehyde | 4.5 | 106 (82), 105 (82), 77 (100), 51 (53) |
| Benzyl alcohol | 5.7 | 108 (73), 107 (49), 79 (100), 77 (59), 51 (31) |
| o-Cresol | 6.0 | 108 (100), 107 (90), 90 (25), 79 (54), 77 (51), 51 (25) |
| m-Cresol | 6.3 | 108 (100), 107 (94), 79 (47), 77 (46), 53 (16), 51 (19) |
| Toluene cis-dihydrodiol | 6.4 | 126 (7), 108 (60), 97 (21), 80 (82), 79 (100), 77 (36), 65 (23), 55 (22) 53 (22) 51 (20) |
| Phenethyl alcohol | 6.0 | 122 (20), 107 (77), 79 (100), 77 (57), 51 (29) |
| 2-Hydroxyethylbenzene | 7.0 | 122 (39), 107 (100), 79 (17), 77 (34), 51 (10) |
| 3-Hydroxyethylbenzene | 7.4 | 122 (41), 107 (100), 79 (10), 77 (32), 51 (8) |
| Ethylbenzene cis-dihydrodiol | 7.5 | 140 (8), 122 (8), 111 (19), 94 (40), 84 (49), 83 (86), 81 (30), 79 (38), 68 (49), 67 (31), 57 (46), 55 (100), 53 (28), 51 (17) |
All the products formed by dioxygenase-catalyzed reactions with ISPBEDα, ISPTODα, or ISPBEDα(I301V/T305S/I307L/L309V) combined with the ISPBED β subunit by using benzene, toluene, or ethylbenzene as the substrate.
TABLE 5.
Percentages of products formed by dioxygenase-catalyzed reactions with ISPBEDα, ISPTODα, or ISPBEDα(I301V/T305S/I307L/L309V) combined with the ISPBED β subunit by using benzene, toluene, or ethylbenzene as the substrate
| Substrate | Product | % of product formeda
|
||
|---|---|---|---|---|
| ISPBEDα | ISPTODα | ISPBEDα mutant | ||
| Benzene | Benzene cis-dihydrodiol | 46 | 37 | 42 |
| Phenolb | 35 | 30 | 37 | |
| Catecholb | 19 | 33 | 21 | |
| Toluene | Toluene cis-dihydrodiol | 3 | 45 | 12 |
| Benzaldehyde | 18 | 1 | 8 | |
| Benzyl alcohol | 53 | 6 | 31 | |
| o-Cresolb | 22 | 38 | 42 | |
| m-Cresolb | 4 | 10 | 7 | |
| Ethylbenzene | Ethylbenzene cis-dihydrodiol | 5 | 24 | 16 |
| 1-Phenethyl alcohol | 24 | 39 | 5 | |
| 2-Hydroxyethylbenzeneb | 58 | 30 | 65 | |
| 3-Hydroxyethylbenzeneb | 13 | 7 | 14 | |
The values are averages of two determinations. A repeat experiment gave similar results.
Product expected from thermal decomposition. See text for details.
When toluene was used, toluene cis-dihydrodiol and o- and m-cresols, the expected dehydration products, were detected (Table 5); again, the sum of the levels of these products was taken to represent the level of cis-diols produced. In addition, benzyl alcohol and benzaldehyde were observed only in trace amounts with ISPTOD, but the amounts were larger with ISPBED. In principle, the benzyl alcohol could be formed either by direct monohydroxylation of toluene by benzene dioxygenase or by production of toluene cis-dihydrodiol, followed by dehydration and isomerization to benzyl alcohol. This possibility was eliminated by mixing ISPTODα and ISPBEDβ in a dioxygenase assay mixture and allowing them to react with [14C]toluene to produce the cis-dihydrodiol. The preparation was then further incubated with a reaction mixture containing ISPBEDα and ISPBEDβ, and the results were analyzed by TLC. No further formation of benzyl alcohol was observed (data not shown). Therefore, the benzyl alcohol was the result of chain monohydroxylation of the substrate by the enzyme system; benzaldehyde was most probably the direct product of further oxidation of benzyl alcohol by the dioxygenase. Analogous reactions have been observed during oxidation of toluene by naphthalene dioxygenase (30) and xylene monooxygenase (18). However, the possibility that a nonspecific host alcohol dehydrogenase caused the conversion of benzyl alcohol to benzaldehyde cannot be ruled out.
Similarly, for ethylbenzene, in addition to the cis-dihydrodiol detected in the autoradiogram analysis, the dehydration products 2- and 3-hydroxyethylbenzene were observed by GC-MS (Table 5). As observed for the oxidation of toluene, ethylbenzene was monohydroxylated on its side chain to form 1-phenethyl alcohol. Lee and Gibson (31) reported that naphthalene dioxygenase oxidized ethylbenzene to 1-phenethyl alcohol, followed by slower oxidation of the latter compound to acetophenone. This rate-determining step (31) may explain why acetophenone was not observed.
Table 5 shows that all of the enzymes tested catalyzed the side chain monohydroxylation of toluene and ethylbenzene. The products were quantified by TLC (Fig. 3), and in general, toluene dioxygenase was more efficient than benzene dioxygenase in the dihydroxylation of toluene and produced less of the monohydroxylation products benzyl alcohol and benzaldehyde. ISPBEDα(I301V/T305S/I307L/L309V) was intermediate between these two proteins. As expected for benzene, which has no side chains to be oxidized, no products other than the dihydrodiol were detected.
DISCUSSION
In this work, sections and individual amino acids of the toluene dioxygenase sequence were replaced in the benzene dioxygenase α subunit in order to probe the regions responsible for substrate preference for toluene and ethylbenzene. All of the ISP α subunits were active when they were combined with β subunits in vitro, in contrast to some other dioxygenases, in which the α and β subunits must be coexpressed to form a functional enzyme (10, 20, 46, 48). The ISPBOD3α chimera narrows the region for substrate preference for alkylbenzenes between residues 281 and 363. In this region, the amino acid substitutions I301V, T305S, I307L, and L309V resulted in a recombinant enzyme with a rate of O2 uptake with ethylbenzene that was substantially higher than that of the wild type (Table 3). The dioxygen data combined with the amount of nonvolatile products showed that for some of the ISP α subunits tested, partial uncoupling of O2 consumption was observed with toluene and, more particularly, with ethylbenzene. The first product of uncoupled reduction of O2 is expected to be H2O2 from the protonation of iron-bound peroxide species, as proposed for cytochrome P450 (14). However, formation of H2O2 is not observed as a result of uncoupling in assays of benzene dioxygenase (1). As suggested by Lee (29), this could be due to iron-catalyzed Fenton chemistry with H2O2 and, ultimately, water as the final product. ISPBODα and ISPBOD3α in which partial uncoupling reactions were observed did have higher substrate specificity for ethylbenzene than the wild-type ISPBEDα. However, the amino acids I301, T305, I307, and L309 are not the only amino acids involved in substrate specificity because although they increased the substrate preference for ethylbenzene, the high uncoupling reaction (44% with ethylbenzene) made them less efficient than the wild-type ISPTOD.
There was no discernible correlation between the degree of uncoupling and the type of ISP α subunit in the assay; for example, ISPBEDα showed partial uncoupling of oxygen consumption with toluene (22%), while ISPTEDα, which contained the same C-terminal region, showed no significant uncoupling with the same substrate. However, ethylbenzene, which had the largest substituent, induced uncoupling of O2 uptake in most cases, indicating that substrate shape or size is a factor. The fact that the specific activities of some of the chimeric α subunits combined with ISPBEDβ were lower with all three substrates (Table 3) than the specific activity of wild-type ISPBEDα could be explained by some mismatch at the subunit interface in the heterologous α-β pair, although, as indicated above, this would not have any effect on the relative activity with the substrates. The source of the β subunit had no systematic effect on substrate specificity (Fig. 1).
Analysis of product formation indicated that both dihydroxylation and chain monohydroxylation occurred in all the recombinant ISP α subunits combined with ISP β subunits. Several studies have reported that dioxygenases, such as toluene dioxygenase (4, 32, 40-42, 51), catalyze monohydroxylation reactions. However, to our knowledge, this study is the first study to report the chain monohydroxylation of toluene and ethylbenzene by benzene dioxygenase to form benzyl alcohol and 1-phenethyl alcohol, respectively. The amounts of cis-diols and chain monohydroxylated products were similar whether the ISP α and β subunits were expressed separately or not (Fig. 3), confirming that the two subunits can combine with each other to form a functional enzyme. For example, around 50% of toluene cis-dihydrodiol and benzyl alcohol was produced with ISPBEDα and coexpressed ISPBEDα-ISPBEDβ when toluene was the substrate. This also showed that the amount of cis-diols and chain monohydroxylated compounds depended upon the nature of the α subunit in the assay. Furthermore, ISPTODα, ISPBODα, and ISPBOD3α, all of which had the C-terminal region of ISPTOD, had different amounts of chain monohydroxylated products (for example, 4, 16, and 48% benzyl alcohol, respectively), indicating that no region of the ISP α subunit seemed to correlate with the amount of the benzyl and 1-phenethyl alcohols.
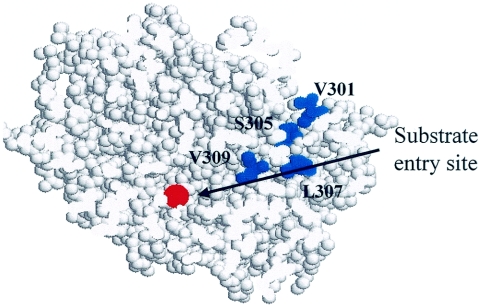
We modeled the sequence of the ISPBEDα(I301V/T305S/I307L/L309V) by homology with the known structure of naphthalene dioxygenase (25) (Fig. 4). As determined by sequence comparisons, I301V, T305S, I307L, and L309V correspond to V287, I291, R293, and H295 in naphthalene dioxygenase. Surprisingly, these residues are not in the active site cavity, nor do they line the channel leading from this site to the surface. H295 is located behind F352, and modification of the latter amino acid has been shown to alter the regiospecificity of naphthalene dioxygenase (39). L307 in toluene dioxygenase corresponds to the residue which was shown by Zhang et al. (56) (by using substitution in a proline) to change the proportion of 1-indenol, 1-indanone, and cis-indandiol produced. The other amino acids are aligned adjacent to residues lining the substrate channel. Replacement of I301 and L309 in benzene dioxygenase by the less bulky amino acid valine should facilitate access to the larger substrates, such as toluene and ethylbenzene, either by decreasing steric hindrance or by increasing flexibility in the channel lining. Although the amino acids surrounding the binding site for the aromatic ring are unchanged, the effects of the mutations on the conformational flexibility of the substrate cavity are difficult to predict. The consequences of the combination are a higher rate of oxygen uptake for toluene and ethylbenzene but a high rate of uncoupling, especially with ethylbenzene. Some monohydroxylated products were observed, but the amount was intermediate between the amounts for all the novel enzymes. The uncoupling is not directly associated with the formation of the chain monohydroxylated product because ISPBEDα(I301V/T305S/I307L/L309V) had a high level of uncoupling with ethylbenzene yet produced the largest proportion of cis-diols.
FIG. 4.
Structural representation of the putative substrate entry channel in ISPBEDα(I301V/T305S/I307L/L309V). The protein is represented in spacefill. Iron is indicated by red, and the four amino acid substitutions (I301V, T305S, I307L, and L309V) are indicated by blue spheres. The view is a section through the substrate channel, and the arrow indicates the putative route that the substrate takes to get to the iron catalytic site. The model was generated with the SWISS-MODEL program (15; http://expasy.hcuge.ch/swissmod/SWISS-MODEL.html) and was displayed by using RasWin Molecular Graphics Window, version 2.6 (43).
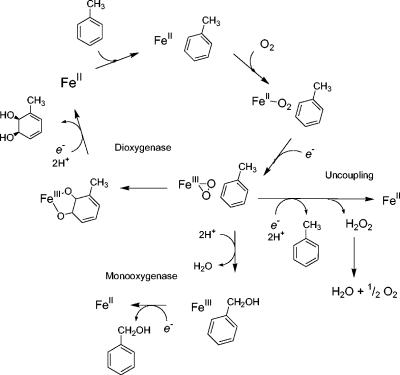
One must assume that the native enzymes evolved to maximize the dihydroxylation of the preferred substrates to the required stereospecific products and to avoid the side reactions. From the crystal structures of naphthalene dioxygenase it is clear that the position of the substrate relative to the Fe-bound O2 is favorable for the dioxygenase reaction (24). The three different reactions, dihydroxylation, monohydroxylation, and uncoupled reduction of O2 to H2O2, may be considered to be the result of competing pathways in the oxidoreduction reactions of iron with oxygen (Fig. 5). Wolfe et al. (52, 53) have shown that there is strict control of the redox events within the enzyme. To avoid uncontrolled reduction of O2, electron transfer from the Rieske cluster to Fe(III) does not occur until substrate is present in the active site, and the product is not released until the iron is reduced to Fe(II) for the next cycle. The dihydroxylation of the substrate by the adjacent side-on peroxo complex is the dominant reaction for native benzene and toluene dioxygenases. However, in the chimeric enzymes the shape of the substrate channel is distorted, and it is possible that the substrate is less tightly bound than it is in the native enzyme. Movement of the substrate away from the binding pocket should allow the side chain hydroxylation, possibly by repositioning of the substrate in the active site, and the uncoupling reaction with reduction to water. Thus, the different product ratios can be understood in terms of the tightness of binding of the substrate to the active site. Following the course of directed evolution seems to be a promising strategy for the engineering of enzymes to metabolize novel substrates.
FIG. 5.
Alternative pathways for dioxygenase and monooxygenase activities and uncoupling. Dihydroxylation results from the addition of O2 and two-electron reduction to the peroxo derivative. In the structure of naphthalene dioxygenase with O2 and substrate in the active site (24), the oxygen is bound side-on to the iron and next to the aromatic ring. Addition of both oxygen atoms, with two protons, would lead to addition of two —OH groups. The monooxygenation reactions are concerted reactions in which one oxygen atom is protonated and reduced to H2O and the other oxygen atom is inserted into the C—H bond of the side chain. The uncoupling reaction is shown as a protonation of the Fe-peroxo species, releasing H2O2.
Acknowledgments
This work was supported by a grant from the United Kingdom Natural Environment Research Council (grant GR3/R9674) and by a TMR Network grant from the European Commission (grant CERB FMRX CT 980174, NOHEMIP).
We thank S. Anguravirutt and C. Butler for providing recombinant strains and S. Jickells for assistance with the GC-MS experiments.
REFERENCES
- 1.Axcell, B. C., and P. J. Geary. 1975. Purification and some properties of a soluble benzene-oxidizing system from a strain of Pseudomonas. Biochem. J. 146:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barriault, D., M. M. Plante, and M. Sylvestre. 2002. Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J. Bacteriol. 184:3794-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barriault, D., C. Simard, H. Chatel, and M. Sylvestre. 2001. Characterisation of hybrid biphenyl dioxygenases obtained by combining Burkholderia sp. strain LB400 bphA with the homologous gene of Comamonas testosteroni B-356. Can. J. Microbiol. 47:1025-1032. [PubMed] [Google Scholar]
- 4.Brand, J. M., D. L. Cruden, G. J. Zylstra, and D. T. Gibson. 1992. Stereospecific hydroxylation of indan by Escherichia coli containing the cloned toluene dioxygenase genes from Pseudomonas putida F1. Appl. Environ. Microbiol. 58:3407-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruhlmann, F., and W. Chen. 1999. Tuning biphenyl dioxygenase for extended substrate specificity. Biotechnol. Bioeng. 63:544-551. [DOI] [PubMed] [Google Scholar]
- 6.Butler, C. S., and J. R. Mason. 1997. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv. Microb. Physiol. 38:47-84. [DOI] [PubMed] [Google Scholar]
- 7.Ensley, B. D., D. T. Gibson, and A. L. Laborde. 1982. Oxidation of naphthalene by a multicomponent enzyme system from Pseudomonas sp. strain NCIB 9816. J. Bacteriol. 149:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson, B. D., and F. J. Mondello. 1993. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl. Environ. Microbiol. 59:3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furukawa, K., J. Hirose, S. Hayashida, and K. Nakamura. 1994. Efficient degradation of trichloroethylene by a hybrid aromatic ring dioxygenase. J. Bacteriol. 176:2121-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa, K., J. Hirose, A. Suyama, T. Zaiki, and S. Hayashida. 1993. Gene components responsible for discrete substrate-specificity in the metabolism of biphenyl (Bph operon) and toluene (Tod operon). J. Bacteriol. 175:5224-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geary, P. J., J. R. Mason, and C. L. Joannou. 1990. Benzene dioxygenase from Pseudomonas putida (NCIB 12190). Methods Enzymol. 188:52-60. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, D. T., G. E. Cardini, F. C. Maseles, and R. E. Kallio. 1970. Incorporation of oxygen-18 into benzene by Pseudomonas putida. Biochemistry 9:1631-1635. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, D. T., M. Hensley, H. Yoshioka, and T. J. Mabry. 1970. Formation of (+)-cis-2,3-dihydroxy-1-methylcyclohexa-4,6-diene from toluene by Pseudomonas putida. Biochemistry 9:1626-1630. [DOI] [PubMed] [Google Scholar]
- 14.Guengerich, F. P. 1991. Reactions and significance of cytochrome p-450 enzymes. J. Biol. Chem. 266:10019-10022. [PubMed] [Google Scholar]
- 15.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modelling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 17.Harayama, S., M. Kok, and E. L. Neidle. 1992. Functional and evolutionary relationships among diverse oxygenases. Annu. Rev. Microbiol. 46:565-601. [DOI] [PubMed] [Google Scholar]
- 18.Harayama, S., R. A. Leppik, M. Rekik, N. Mermod, P. R. Lehrbach, W. Reineke, and K. N. Timmis. 1989. Gene order of the TOL catabolic upper pathway operon and oxidation of both toluene and benzylalcohol by the XylA product. J. Bacteriol. 167:455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess, H. H., M. B. Lees, and J. E. Derr. 1978. A linear Lowry-Folin assay for both water-soluble and sodium dodecyl sulphate-solubilised proteins. Anal. Biochem. 85:295-300. [DOI] [PubMed] [Google Scholar]
- 20.Hirose, J., A., S. Suyama, K. Hayashida, and K. Furukawa. 1994. Construction of hybrid biphenyl (bph) and toluene (tod) genes for functional analysis of aromatic ring dioxygenases. Gene 138:27-33. [DOI] [PubMed] [Google Scholar]
- 21.Hudlicky, T., M., J. Mandel, R. S. Rouden, B. B. Lee, T. Dudding, K. J. Yost, and J. S. Merola. 1994. Microbial oxidation of aromatics in enantiocontrolled synthesis. Part 1. Expedient and general asymmetric synthesis of inositols and carbohydrates via an unusual oxidation of a polarized diene with potassium permanganate. J. Chem. Soc. Perkin Trans. 1:1553-1567. [Google Scholar]
- 22.Jiang, H. Y., R. E. Parales, N. A. Lynch, and D. T. Gibson. 1996. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J. Bacteriol. 178:3133-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kammann, M., J. Laufs, J. Schell, and B. Gronenborn. 1989. Rapid insertional mutagenesis of DNA by polymerase chain reaction (PCR). Nucleic Acids Res. 17:5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson, A., J. V. Parales, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 2003. Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron. Science 299:1039-1042. [DOI] [PubMed] [Google Scholar]
- 25.Kauppi, B., K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy. 1998. Structure of an aromatic-ring-hydroxylating dioxygenase-naphthalene 1,2-dioxygenase. Structure 6:571-586. [DOI] [PubMed] [Google Scholar]
- 26.Kimura, N., A. Nishi, M. Goto, and K. Furukawa. 1997. Functional analyses of a variety of chimeric dioxygenases constructed from two biphenyl dioxygenases that are similar structurally but different functionally. J. Bacteriol. 179:3936-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Lee, K. 1999. Benzene-induced uncoupling of naphthalene dioxygenase activity and enzyme inactivation by production of hydrogen peroxide. J. Bacteriol. 181:2719-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, K., and D. T. Gibson. 1996. Stereospecific dihydroxylation of the styrene vinyl group by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. J. Bacteriol. 178:3353-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, K., and D. T. Gibson. 1996. Toluene and ethylbenzene oxidation by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl. Environ. Microbiol. 62:3101-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehning, A., U. Fock, R. M. Wittich, K. N. Timmis, and D. H. Pieper. 1997. Metabolism of chlorotoluenes by Burkholderia sp. strain PS12 and toluene dioxygenase of Pseudomonas putida F1: evidence for monooxygenation by toluene and chlorobenzene dioxygenases. Appl. Environ. Microbiol. 63:1974-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Link, T. A., O. M. Hatzfeld, P. Unalkat, J. K. Shergill, R. Cammack, and J. R. Mason. 1996. Comparison of the “Rieske” [2Fe-2S] center in the bc1 complex and in bacterial dioxygenases by circular dichroism spectroscopy and cyclic voltammetry. Biochemistry 35:7546-7552. [DOI] [PubMed] [Google Scholar]
- 34.Maeda, T., Y. Takahashi, H. Suenaga, A. Suyama, M. Goto, and K. Furukawa. 2001. Functional analyses of Bph-Tod hybrid dioxygenase, which exhibits high degradation activity toward trichloroethylene. J. Biol. Chem. 276:29833-29838. [DOI] [PubMed] [Google Scholar]
- 35.Mason, J. R., C. S. Butler, R. Cammack, and J. K. Shergill. 1997. Structural studies on the catalytic component of benzene dioxygenase from Pseudomonas putida. Biochem. Soc. Trans. 25:90-95. [DOI] [PubMed] [Google Scholar]
- 36.Mondello, F. J., M. P. Turcich, J. H. Lobos, and B. D. Erickson. 1997. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl. Environ. Microbiol. 63:3096-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parales, J. V., R. E. Parales, S. M. Resnick, and D. T. Gibson. 1998. Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the C-terminal region of the alpha subunit of the oxygenase component. J. Bacteriol. 180:1194-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parales, R. E., M. D. Emig, N. A. Lynch, and D. T. Gibson. 1998. Substrate specificities of hybrid naphthalene and 2,1-dinitrotoluene dioxygenase enzyme systems. J. Bacteriol. 180:2337-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parales, R. E., K. Lee, S. M. Resnick, H. Y. Jiang, D. J. Lessner, and D. T. Gibson. 2000. Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol. 182:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phumathon, P., and G. M. Stephens. 1999. Production of toluene cis-glycol using recombinant Escherichia coli strains in glucose-limited fed batch culture. Enzyme Microb. Technol. 25:810-819. [Google Scholar]
- 41.Robertson, J. B., J. C. Spain, J. D. Haddock, and D. T. Gibson. 1992. Oxidation of nitrotoluenes by toluene dioxygenase: evidence for a monooxygenase reaction. Appl. Environ. Microbiol. 58:2643-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakamoto, T., J. M. Joern, A. Arisawa, and F. H. Arnold. 2001. Laboratory evolution of toluene dioxygenase to accept 4-picoline as a substrate. Appl. Environ. Microbiol. 67:3882-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sayle, R. A., and E. J. Miller-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374. [DOI] [PubMed] [Google Scholar]
- 44.Shergill, J. K., C. L. Joannou, J. R. Mason, and R. Cammack. 1995. Coordination of the Rieske-type [2Fe-2S] cluster of the terminal iron-sulfur protein of Pseudomonas putida benzene 1,2-dioxygenase, studied by one- and two-dimensional electron spin-echo envelope modulation spectroscopy. Biochemistry 34:16533-16542. [DOI] [PubMed] [Google Scholar]
- 45.Singer, M., and P. Berg. 1991. mRNA translation in prokaryotes, p. 167-173. In Genes and genomes: a changing perspective. University Sciences Books, Sausalito, Calif.
- 46.Suen, W.-C., and D. T. Gibson. 1993. Isolation and preliminary characterization of the subunits of the terminal component of naphthalene dioxygenase from Pseudomonas putida NCIB 9816-4. J. Bacteriol. 175:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suenaga, H., M. Mitsuoka, Y. Ura, T. Watanabe, and K. Furukawa. 2001. Directed evolution of biphenyl dioxygenase: emergence of enhanced degradation capacity for benzene, toluene, and alkyl benzenes. J. Bacteriol. 183:5441-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan, H. M., and C. M. Cheong. 1994. Substitution of the ISP alpha subunit of biphenyl dioxygenase from Pseudomonas results in a modification of the enzyme activity. Biochem. Biophys. Res. Commun. 204:912-917. [DOI] [PubMed] [Google Scholar]
- 49.Tan, H.-M., and J. R. Mason. 1990. Cloning and expression of the plasmid-encoded benzene dioxygenase genes from Pseudomonas putida ML2. FEMS Microbiol. Lett. 72:259-264. [DOI] [PubMed] [Google Scholar]
- 50.Tan, H. M., H. Y. Tang, C. L. Joannou, N. H. Abdel-Wahab, and J. R. Mason. 1993. The Pseudomonas putida Ml2 plasmid-encoded genes for benzene dioxygenase are unusual in codon usage and low in G+C content. Gene 130:33-39. [DOI] [PubMed] [Google Scholar]
- 51.Wackett, L. P., L. D. Kwart, and D. T. Gibson. 1988. Benzylic monooxygenation catalyzed by toluene dioxygenase from Pseudomonas putida. Biochemistry 27:1360-1367. [DOI] [PubMed] [Google Scholar]
- 52.Wolfe, M. D., D. J. Altier, A. Stubna, C. V. Popescu, E. Munck, and J. D. Lipscomb. 2002. Benzoate 1,2-dioxygenase from Pseudomonas putida: single turnover kinetics and regulation of a two-component Rieske dioxygenase. Biochemistry 41:9611-9626. [DOI] [PubMed] [Google Scholar]
- 53.Wolfe, M. D., J. V. Parales, D. T. Gibson, and J. D. Lipscomb. 2001. Single turnover chemistry and regulation of O-2 activation by the oxygenase component of naphthalene 1,2-dioxygenase. J. Biol. Chem. 276:1945-1953. [DOI] [PubMed] [Google Scholar]
- 54.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and Puc19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 55.Zamanian, M., and J. R. Mason. 1987. Benzene dioxygenase in Pseudomonas putida. Subunit composition and immuno-cross-reactivity with other aromatic dioxygenases. Biochem. J. 244:611-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, N., B. G. Stewart, J. C. Moore, R. L. Greasham, D. K. Robinson, B. C. Buckland, and C. Lee. 2000. Directed evolution of toluene dioxygenase from Pseudomonas putida for improved selectivity towards cis-indandiol during indene bioconversion. Metab. Eng. 2:339-348. [DOI] [PubMed] [Google Scholar]
- 57.Zylstra, G. J., and D. T. Gibson. 1989. Toluene degradation by Pseudomonas putida F1: nucleotide sequence of the Todc1C2BADE genes and their expression in Escherichia coli. J. Biol. Chem. 264:14940-14946. [PubMed] [Google Scholar]
- 58.Zylstra, G. J., W. R. McCoombie, D. T. Gibson, and B. A. Finette. 1988. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl. Environ. Microbiol. 54:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]