Abstract
Here we describe the rep gene, isolated from an environmental DNA library, which when transformed into Streptomyces species resulted in increased production of secondary metabolites and accelerated sporulation. We show that Streptomyces lividans strains bearing rep are particularly useful as expression hosts for heterologous antibiotic production.
Natural products from environmental microorganisms account for over 30% of all human therapeutics and over 60% of anti-infective and anticancer drugs. These include polyketides, produced by microbes such as Streptomyces, Saccharopolyspora, and Aspergillus species. Although the majority of microorganisms in the environment are still unknown, most are unculturable under standard laboratory conditions. If these uncultured species could be accessed, they could potentially provide a large, untapped pool of novel natural products. In a recent approach, microbial DNA was extracted directly from environmental samples, cloned into suitable vectors, and expressed in surrogate laboratory host strains (2, 7, 8, 10).
Because of their ease of use and ability to express polyketides, Streptomyces species are logical surrogate hosts for soil DNA libraries. Streptomyces lividans is usually preferred for genetic manipulation because it lacks methylation-dependent restriction, allowing efficient transfer of DNA from Escherichia coli cloning strains. However, S. lividans does not produce antibiotics as abundantly as does Streptomyces coelicolor, and its morphological development is delayed (6). It has been shown that overexpression of clpX (3) and mutations in the rpsL gene (11) and in glucose-6-phosphate dehydrogenase genes (1) result in increased endogenous actinorhodin synthesis in S. lividans, although it is not currently known whether these results extend to heterologous antibiotics. Therefore, a need remains for an expression host that combines the absence of methylation-dependent restriction systems with a high level of heterologous secondary metabolite production and that can be used conveniently for assaying soil DNA libraries for anti-infective and other activities. We show here that the rep gene, isolated from a soil environmental DNA library, confers to S. lividans both accelerated sporulation and overproduction of endogenous and heterologous secondary metabolites. Thus, S. lividans bearing this gene constitutes an improved expression host for heterologous DNA libraries.
Phenotypes conferred by cosmid 2A7.
Cosmid 2A7, previously isolated as part of a soil DNA library (2), contains approximately 40.4 kb of soil DNA cloned into the E. coli-Streptomyces shuttle vector pOSI700. When grown in glucose-containing medium (e.g., SMM or R5 [5]), S. lividans TK24 bearing this cosmid sporulated early (after 3 days in SMM versus 8 days for the control strain) and produced an enhanced level of actinorhodin, an endogenous pigment. To ascertain that this phenotype was encoded on the cosmid, 2A7 was retransformed into S. lividans TK24. Transformants again displayed both early sporulation and early, increased antibiotic production in SMM medium (Fig. 1). The 2A7 phenotypes were carbon source dependent; they were not observed when glucose was omitted or replaced by mannitol or glycerol (0.2 and 0.5%, respectively).
FIG. 1.
Cosmid 2A7 causes early antibiotic production and sporulation in S. lividans TK24. S. lividans TK24 containing cosmid 2A7 (left) or the empty vector pOSI700 (right) was grown in SMM for 6 days at 30°C. (Top) Agar plates; (Bottom) liquid cultures of the same strains.
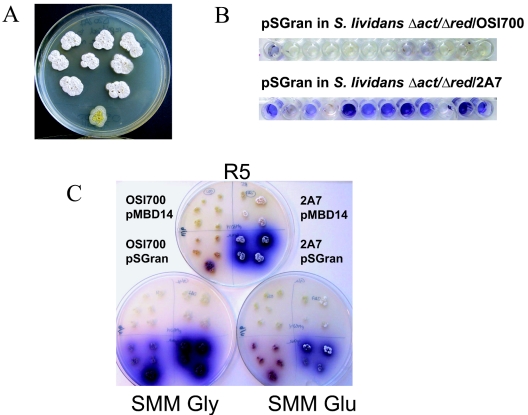
S. lividans Δact Δred (deleted for both endogenous antibiotics [8]) bearing cosmid 2A7 also displayed the early sporulation phenotype (Fig. 2A). To test whether 2A7 could increase heterologous antibiotic production, pSGran, an integrative bacterial artificial chromosome containing the granaticin biosynthetic cluster from Streptomyces violeacoruber Tu22 (8), and the corresponding empty BAC vector were conjugated into S. lividans Δact Δred containing cosmid 2A7. When grown in glucose-containing medium, 2A7 conferred significantly higher levels of granaticin production on both solid medium (Fig. 2C) and in a liquid 96-well format (Fig. 2B). As before, increased sporulation and secondary metabolite production were glucose dependent (not observed when glucose was replaced by glycerol or mannitol) and occurred even in the presence of a high copper concentration (R5 with 2 μM CuCl2), which has been shown to accelerate spore production in S. lividans (4). Thus, 2A7 appears to bypass glucose repression of sporulation and antibiotic biosynthesis in S. lividans (6).
FIG. 2.
Cosmid 2A7 causes early sporulation and enhanced polyketide production in S. lividans Δact Δred. (A) Independent transformants bearing cosmid 2A7 or the control vector pOSI700 (bottom colony) grown on R5 solid medium for 6 days at 30°C. (B and C) S. lividans Δact Δred bearing cosmid 2A7 or the corresponding empty shuttle vector (pOSI700), combined with the granaticin BAC vector (pSGran) or the corresponding empty shuttle BAC vector (pMBD14), was grown for 7 days in liquid R5 medium (B) or on solid medium for 5 days (C) as follows: top plate, R5; bottom left, SMM with glycerol (Gly); bottom right, SMM with glucose (Glu). Plasmid combinations are indicated.
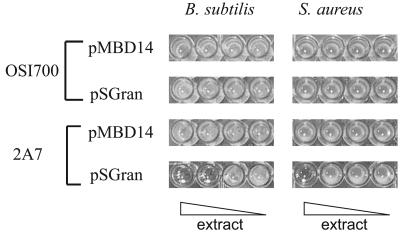
Extracts of S. lividans Δact Δred/2A7 with and without pSGran were prepared and tested for antibacterial activity. Cells were grown in shallow 96-well plates (non-tissue-culture treated) containing 150 μl of a modified R5 liquid medium (5), omitting sucrose and agar, and incubated at 30°C for 5 to 9 days in a sealed plastic bag containing a wet paper towel. Water was then evaporated, and the residue was extracted with 150 μl of 100% methanol. After a 5-min centrifugation at 4,000 rpm (1,500 × g), 100 μl of the methanol extract was removed into a clean 96-well plate, the methanol was evaporated, and dried extracts were stored at 4°C. Antibacterial assays were then carried out in the 96-well plates by resuspending the extracts in 150 μl of Luria-Bertani medium that had been inoculated with a tester strain (Bacillus subtilis BR151/pPL608 or Staphylococcus aureus Smith). After overnight incubation at 35°C, wells containing antibacterial compounds were clear as compared with the turbid control. S. lividans Δact Δred containing both cosmid 2A7 and pSGran showed a 10- to 100-fold increase in antibacterial activity compared with the same strain in the absence of 2A7 (Fig. 3), demonstrating that 2A7 can increase heterologous polyketide production from single-copy gene clusters to levels that are sufficient for biological screening in small culture volumes. Importantly, S. lividans Δact Δred containing 2A7 showed no background antibacterial or antifungal activity, and no new compounds were detectable by high-pressure liquid chromatography analysis in extracts of this strain (data not shown). Therefore, the presence of cosmid 2A7 did not itself result in significant levels of new molecules that could interfere with screening efforts.
FIG. 3.
Increased granaticin production conferred by 2A7 results in increased antibacterial activity. S. lividans Δact Δred/2A7 containing cosmid 2A7 or the corresponding empty shuttle vector (pOSI700), combined with pSGran or the corresponding empty shuttle vector (pMBD14), were grown for 9 days and extracted (see the text). Extracts were used undiluted and diluted 10-, 100-, and 1,000-fold and tested for antibacterial activity.
A single open reading frame (ORF), rep, is necessary and sufficient to confer the 2A7 phenotypes.
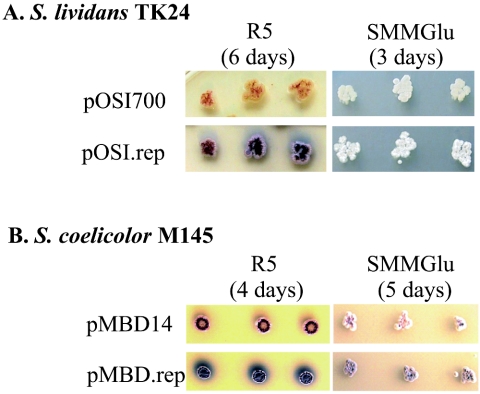
The insert in cosmid 2A7 was GC rich (74%) and showed no identities with existing GenBank sequences. To identify the gene(s) responsible for the 2A7 phenotype, we used transposon GPSApra, a modified form of transposon GPS1, used for genome sequencing (New England Biolabs, Beverly, Mass.), to create insertions in 35 of the 36 predicted ORFs. We transformed each into S. lividans TK24 act+ red+ and found that disruption of a single ORF, rep, resulted in complete loss of glucose-dependent early sporulation, indicating that rep is necessary for this phenotype (data not shown). To test whether rep was sufficient to confer the 2A7 phenotypes, a 1.5-kb fragment of cosmid 2A7 containing the rep ORF and 290 bp of upstream sequence was PCR amplified, using Vent polymerase (New England Biolabs) and primers 2A7.repS.5′ (GCAAGCTTGGTGCGTCGCCGGCACGTCGAGCTCGTGCA) and 2A7.rep.3′ (GCAAGCTTGTAGACGCCGCAGTCCACGATGACACGG) as recommended by the manufacturer in the presence of 10% dimethyl sulfoxide, with the following conditions: 98°C for 10 min; 30 cycles of 98°C for 1 min, 60°C for 1 min, and 72°C for 2 min; and 72°C for 10 min. The PCR product was cloned into pBlunt-TOPO (Invitrogen), yielding pTOPO.rep, and then rep was reisolated by HindIII digestion and cloned into the HindIII site of pOSI700, yielding pOSI.rep. The phenotype of S. lividans TK24 bearing pOSI.rep and grown on glucose-containing medium (Fig. 4A) showed that the rep gene was sufficient to confer early sporulation and increased antibiotic production. rep was also tested in S. coelicolor to assess its effect in other Streptomyces species. BstXI/EcoRI adapters (Invitrogen) were first ligated to the rep PCR product, using a standard blunt-end ligation procedure. The resulting rep fragment was gel purified and ligated to 20 ng of BstXI-cut pMBD13 vector (10:1, vector to insert molar ratio) at 16°C for 6 h. The resulting plasmid, pMBD.rep, was transformed into E. coli ET12567/pUB307 and conjugated into S. coelicolor M145 by standard procedures (5). S. coelicolor M145 transformants containing pMBD.rep overproduced antibiotics in glucose-containing medium compared to control S. coelicolor strains transformed with the empty vector (Fig. 4B). Therefore, rep can increase antibiotic production in at least one additional Streptomyces species and thus may be of value for other Streptomyces strains of commercial interest.
FIG. 4.
(A) rep is sufficient to confer the 2A7 phenotype. S. lividans TK24 transformed with pOSI700 (top) or pOSI.rep (bottom) was inoculated onto R5 or SMM glucose (Glu) agar and grown at 30°C for 6 or 3 days, respectively. (B) rep increases antibiotic production in S. coelicolor M145. S. coelicolor M145 containing the control vector pMBD14 (top) or pMBD.rep (bottom) was inoculated onto R5 or SMM glucose agar and grown for 4 or 5 days, respectively.
Sequence analyses indicated that rep encodes a putative 389-amino-acid transcriptional repressor of the ROK family (for repressor, ORF, kinase; Pfam motif PF00480 [12]). The transcriptional repressors of this family, such as B. subtilis xylR and E. coli nagC, encompass a DNA-binding helix-turn-helix motif at the N terminus followed by a ROK sugar-binding domain. In the absence of the appropriate inducing sugar, these repressors bind to their operator sequences and regulate gene expression. Binding of the inducing sugars to the ROK domain results in dissociation of the repressor-operator complex (9). Among the members of the ROK family, Rep is most similar (48% identity, 68% similarity) to NP630122, a putative transcriptional repressor of unknown function from S. coelicolor A3 (2); NP823424, also a putative repressor from Streptomyces avermitilis; and NagR, the putative repressor of an ABC transporter system that mediates uptake of N-acetylglucosamine in Streptomyces olivaceoviridis (13). Rep also exhibits 27% identity and 49% similarity to XylR of B. subtilis. An alignment of Rep to its closest homologs is shown in Fig. 5.
FIG. 5.
Multiple sequence alignment showing the deduced sequence of the Rep protein encoded by 2A7 (Rep) with its closest homologs: NP630122 of S. coelicolor (Sc NP630122), NP823424 of S. avermitilis (Sa NP823424), and NgcR of S. olivaceoviridis (So NgcR).
Summary.
The rep gene, derived from an environmental library, was shown both to accelerate sporulation and to enhance production levels of secondary metabolites, specifically endogenous and heterologous antibiotics. Streptomyces strains bearing this sequence can thus be used not only as improved production hosts for endogenous secondary metabolites but also for screening libraries of soil DNA samples for heterologous secondary metabolites, thereby enhancing our ability to detect novel compounds produced in environmental libraries.
Nucleotide sequence accession number.
The sequence of the rep gene has been deposited in GenBank (accession number AY684299).
Acknowledgments
We thank our colleagues Ian MacNeil and Kara Brown for helpful discussions during the course of this work.
Footnotes
This paper is dedicated to the memory of Steven J. Kolvek, our colleague and friend.
REFERENCES
- 1.Butler, M. J., P. Bruheim, S. Jovetic, F. Marinelli, P. Postma, and M. J. Bibb. 2002. Engineering of primary carbon metabolism for improved antibiotic production in Streptomyces lividans. Appl. Environ. Microbiol. 68:4731-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courtois, S., C. M. Cappellano, M. Ball, F. X. Francou, P. Normand, G. Helynck, A. Martinez, S. J. Kolvek, J. Hopke, M. S. Osburne, P. R. August, R. Nalin, M. Guerineau, P. Jeannin, P. Simonet, and J. L. Pernodet. 2003. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl. Environ. Microbiol. 69:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Crecy-Lagard, V., P. Servant-Moisson, J. Viala, C. Grandvalet, and P. Mazodier. 1999. Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol. Microbiol. 32:505-517. [DOI] [PubMed] [Google Scholar]
- 4.Keijser, J. F., G. P. van Wezel, G. W. Canters, T. Kieser, and E. Vijgenboom. 2000. The ram-dependence of Streptomyces lividans differentiation is bypassed by copper. J. Mol. Microbiol. Biotechnol. 2:565-574. [PubMed] [Google Scholar]
- 5.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 6.Kim, E. S., H. J. Hong, C. Y. Choi, and S. Y. Cohen. 2001. Modulation of actinorhodin biosynthesis in Streptomyces lividans by glucose repression of afsR2 gene transcription. J. Bacteriol. 183:2198-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacNeil, I. A., C. L. Tiong, C. Minor, P. R. August, T. H. Grossman, K. A. Loiacono, B. A. Lynch, T. Phillips, S. Narula, R. Sundaramoorthi, A. Tyler, T. Aldredge, H. Long, M. Gilman, D. Holt, and M. S. Osburne. 2001. Expression and isolation of antimicrobial small molecules from soil DNA libraries. J. Mol. Microbiol. Biotechnol. 3:301-308. [PubMed] [Google Scholar]
- 8.Martinez, A., S. J. Kolvek, C. L. Tiong Yip, J. Hopke, K. A. Brown, I. A. MacNeil, and M. S. Osburne. 2004. Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple bacterial hosts. Appl. Environ. Microbiol. 70:2452-2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plumbridge, J. 2001. Regulation of PTS gene expression by the homologous transcriptional regulators, Mlc and NagC, in Escherichia coli (or how two similar repressors can behave differently). J. Mol. Microbiol. Biotechnol. 3:371-380. [PubMed] [Google Scholar]
- 10.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, I. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shima, J., A. Hesketh, S. Okamoto, S. Kawamoto, and K. Ochi. 1996. Induction of actinorhodin production by rpsL (encoding ribosomal protein S12) mutations that confer streptomycin resistance in Streptomyces lividans and Streptomyces coelicolor A3(2). J. Bacteriol. 179:7276-7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Titgemeyer, F., J. Reizer, A. Reizer, and M. H. Saier, Jr. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349-2354. [DOI] [PubMed] [Google Scholar]
- 13.Xiao, X., F. Wang, A. Saito, J. Majka, A. Schlosser, and H. Schrempf. 2002. The novel Streptomyces olivaceoviridis ABC transporter Ngc mediates uptake of N-acetylglucosamine and N,N′-diacetylchitobiose. Mol. Genet. Genomics 267:429-439. [DOI] [PubMed] [Google Scholar]