Abstract
For simultaneous identification of members of the betaproteobacterial order “Rhodocyclales” in environmental samples, a 16S rRNA gene-targeted oligonucleotide microarray (RHC-PhyloChip) consisting of 79 probes was developed. Probe design was based on phylogenetic analysis of available 16S rRNA sequences from all cultured and as yet uncultured members of the “Rhodocyclales.” The multiple nested probe set was evaluated for microarray hybridization with 16S rRNA gene PCR amplicons from 29 reference organisms. Subsequently, the RHC-PhyloChip was successfully used for cultivation-independent “Rhodocyclales” diversity analysis in activated sludge from an industrial wastewater treatment plant. The implementation of a newly designed “Rhodocyclales”-selective PCR amplification system prior to microarray hybridization greatly enhanced the sensitivity of the RHC-PhyloChip and thus enabled the detection of “Rhodocyclales” populations with relative abundances of less than 1% of all bacteria (as determined by fluorescence in situ hybridization) in the activated sludge. The presence of as yet uncultured Zoogloea-, Ferribacterium/Dechloromonas-, and Sterolibacterium-related bacteria in the industrial activated sludge, as indicated by the RHC-PhyloChip analysis, was confirmed by retrieval of their 16S rRNA gene sequences and subsequent phylogenetic analysis, demonstrating the suitability of the RHC-PhyloChip as a novel monitoring tool for environmental microbiology.
Members of the provisional betaproteobacterial order “Rhodocyclales” comprise a physiologically versatile assemblage of bacteria, many of them responsible for the removal of anthropogenic compounds in the environment or in biotechnological systems. While most members of the genera Azoarcus and Thauera can couple the anaerobic reduction of nitrate with the degradation of aromatic hydrocarbons (7, 40) or halogenated compounds (50), other Azoarcus species are associated with grass roots, where they fix nitrogen (17, 44). Furthermore, it has been recognized only recently that the “Rhodocyclales” genera Dechloromonas and Azospira harbor the dominant (per)chlorate-reducing bacteria in the environment (1, 11). Another important bioremediation process which exploits bacteria of the order “Rhodocyclales” to ameliorate anthropogenic damage is sewage treatment. For example, an uncultured bacterium provisionally named Candidatus “Accumulibacter phosphatis” catalyzes enhanced biological phosphorous removal in wastewater treatment plants (WWTPs) (13, 25, 59). Other “Rhodocyclales” to date also recalcitrant to cultivation were the numerically dominant bacteria in activated sludge from a nitrifying-denitrifying WWTP, where they presumably contributed to denitrification (27).
Due to their importance for bioremediation and agriculture, several approaches for detection of members of the order “Rhodocyclales” have been developed. Besides traditional cultivation methods (48), molecular detection of members of this order has been based on taxon- or clone-selective 16S rRNA gene-targeted PCR primers or probes (4, 13, 24-27, 43, 45). While these molecular methods are well suited for the detection of a few selected subgroups or species within the order “Rhodocyclales,” tools for surveying the diversity of members of this order in parallel are missing. DNA microarrays, which have recently been introduced to microbial ecology (22) and generally fulfill all requirements for high-resolution monitoring of complex microbial communities (9, 10, 16, 32, 35, 41, 49, 55, 60-62), but a dedicated microarray for the order “Rhodocyclales” is not yet available.
In this study we have developed and applied a 16S rRNA gene-targeted oligonucleotide microarray consisting of 79 probes for the parallel detection of all bacteria of the order “Rhodocyclales” at different hierarchical or identical phylogenetic levels (RHC-PhyloChip). The use of three newly designed primer pairs for selective amplification of “Rhodocyclales” 16S rRNA genes prior to microarray hybridization allowed the detection of rare “Rhodocyclales” groups in activated sludge from an industrial sewage treatment plant. The microarray results were confirmed and extended by comparative 16S rRNA gene sequence and quantitative fluorescence in situ hybridization analyses.
MATERIALS AND METHODS
Reference organisms.
Tables 1 and 2 list the 12 pure cultures and the 17 16S rRNA gene-containing clones that were used to evaluate the RHC-PhyloChip.
TABLE 1.
“Rhodocyclales” reference strains
| Pure-culture species | Straina | 16S rRNA sequence accession no. |
|---|---|---|
| Azoarcus anaerobius | DSM 12081T | Y14701 |
| Azoarcus communis | DSM 12120 | AF011343 |
| Azoarcus evansii | DSM 6898T | X77679 |
| Azoarcus indigens | LMG 9092T | L15531 |
| Azoarcus sp. | LU1 | AJ007007 |
| Azonexus fungiphilus | LMG 19178T | AF011350 |
| Azospira oryzae (Dechlorosoma suillum) | DSM13638T | AF170348 |
| Dechloromonas agitata | DSM 13637T | AF047462 |
| Propionivibrio pelophilus | DSM 12018T | AF016690 |
| Rhodocyclus tenuis | DSM 109T | D16210 |
| Thauera mechernichensis | DSM 12266T | Y17590 |
| Thauera terpenica | DSM 12139T | AJ005817 |
Strains were obtained as lyophilized cells or active cultures. DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; LMG, Laboratorium voor Microbiologie, Universitat Gent, Belgium.
TABLE 2.
“Rhodocyclales” reference 16S rRNA gene clones
| 16S rRNA gene clone | 16S rRNA sequence accession no. or source | Insert region (E. coli numbering) | |
|---|---|---|---|
| Kraftisried WWTP clones | |||
| A13 | AF072927 | 0008-1545 | |
| A16 | AF234726 | 0008-1545 | |
| A33 | AF072925 | 0008-1545 | |
| H7 | AF234684 | 0008-1545 | |
| H23 | AF072926 | 0008-1545 | |
| S3 | AF072918 | 0008-1545 | |
| S21 | AF234738 | 0008-1545 | |
| S23 | AF072921 | 0008-1545 | |
| KRA34 | AY689089 | 0094-1439 | |
| KRR56 | AY689085 | 0175-1306 | |
| KRZ64 | AY689092 | 0066-1439 | |
| KRZ65 | AY689091 | 0066-1439 | |
| WWTP clones | |||
| BNP269 | N. Lee, unpublished data | 0008-1511 | |
| hBPR4 | N. Lee, unpublished data | 0107-1263 | |
| hBPR24 | N. Lee, unpublished data | 0107-1263 | |
| Wadi Gaza clones | |||
| WGAR24 | AY687927 | 0107-1263 | |
| WGAR25 | AY687928 | 0107-1263 |
Sampling of activated sludge.
Activated sludge samples were collected in June 2002 from the intermittently aerated nitrification-denitrification basin of an industrial WWTP. This treatment plant received its sewage from a rendering plant (Tierkörperbeseitigungsanstalt Kraftisried, Kraftisried, Germany). For DNA isolation, aliquots (4 ml) of the samples were pelleted by centrifugation (5,000 rpm for 2 min) at the treatment plant, immediately put on ice, and stored at −20°C upon arrival at the laboratory. For fluorescence in situ hybridization (FISH), an activated sludge sample was fixed at the WWTP with paraformaldehyde as outlined previously (15).
DNA extraction.
Genomic DNA was isolated from reference organisms by using the FastDNA kit (Bio101, Vista, Calif.). DNA from Kraftisried activated sludge was extracted by using a modification (35) of a previously described protocol (21).
PCR amplification of 16S rRNA genes.
For DNA microarray hybridization, 16S rRNA gene fragments from DNA of “Rhodocyclales” reference pure cultures were amplified by using the bacterial primer pair 616V and 630R (Table 3), whereas 16S rRNA gene inserts of reference clones were amplified with cloning vector-specific primers M13F(−20) (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′) (Invitrogen Corp., San Diego, Calif.). Amplification of 16S rRNA gene fragments from DNA of the activated sludge sample was performed by using the bacterial primer pairs 616V and 630R and 616V and 1492R or the newly designed primer pairs A, R, or Z, each targeting different “Rhodocyclales” subgroups (Table 3).
TABLE 3.
16S rRNA gene-targeted primers
| Short namea | Full nameb | Annealing temp (°C) | Sequence 5′-3′ | Specificityc | Reference |
|---|---|---|---|---|---|
| 616V | S-D-Bact-0008-a-S-18 | 52 | AGA GTT TGA TYM TGG CTC | Most Bacteria | 28 |
| 630R | S-D-Bact-1529-a-A-17 | 52 | CAK AAA GGA GGT GAT CC | Most Bacteria | 28 |
| 1492R | S-*-Proka-1492-a-A-19 | 52, 60c | GGY TAC CTT GTT ACG ACT T | Most Bacteria and Archaea | Modified from 29 |
| AT+94Fd | S-*-AT-0094-a-A-18 | 60 | GCC GGC GAG TGG CGA ACG | Genera Azoarcus, Thauera, and other bacteria | This study |
| ATD1420Rd | S-*AT-1420-a-S-20 | 60 | CCT ACT TCT GGT GAA ACC CA | Genera Azoarcus, Thauera, and Denitromonas | This study |
| RHC175Fe | S-*-Rhc-0175-a-A-19 | 60 | CCG CAT ATT CTG TGA GCA G | Genera Candidatus “Accumulibacter,” Rhodocyclus, Propionivibrio, Dechloromonas, Azospira, and Ferribacterium | This study |
| RHC+1289Re | S-*-Rhc-1289-a-S-18 | 60 | TCC GGA CTA CGA TCG GCT | Genera Candidatus “Accumulibacter,” Rhodocyclus, Propionivibrio, Dechloromonas, Azospira, Ferribacterium, and other bacteria | This study |
| ZOGLO66Ff | S-*-Zoglo-0066-a-A-18 | 60 | ACG GTA ACA GGG AGC TTG | Genus Zoogloea, not Z. resiniphila | This study |
| ZOGLO1421Rf | S-*Zoglo-1421-a-S-19 | 60 | CCT ACT TCT GGT AAA CCC C | Genus Zoogloea | This study |
The short name used in the reference or in this study.
The full name of the 16S rRNA gene-targeted oligonucleotide primer is based on the nomenclature of Alm et al. (2).
Target organisms with a perfectly matched primer target site.
Primer pair A.
Primer pair R.
Primer pair Z.
Positive controls containing purified DNA from suitable reference organisms were included in all of the PCR amplification experiments along with negative controls (no template DNA added). For 16S rRNA gene amplification, reaction mixtures (total volume, 50 μl) containing each primer at a concentration of 25 pM were prepared by using 10× Ex Taq reaction buffer and 2.5 U of Ex Taq polymerase (Takara Biomedicals, Otsu, Shiga, Japan). Additionally, 20 mM tetramethylammonium chloride (Sigma, Deisenhofen, Germany) was added to each amplification mixture to enhance the specificity of the PCR (31). Thermal cycling was carried out by using an initial denaturation step of 94°C for 1 min, followed by 30 cycles of denaturation at 94°C for 40 s, annealing at temperatures ranging from 52°C to 60°C (depending on the primer pair [Table 3]) for 40 s, and elongation at 72°C for 1.5 min. Cycling was completed by a final elongation step of 72°C for 10 min.
The presence and sizes of the amplification products were determined by agarose (1%) gel electrophoresis of the reaction product. Ethidium bromide-stained bands were digitally recorded with a video documentation system (Cybertech, Hamburg, Germany).
DNA microarray hybridization.
16S rRNA-targeted oligonucleotides were designed in silico by using the ARB probe design and probe match tools (37) and obtained from MWG Biotech (Ebersberg, Germany). Table 4 lists the sequence, specificity, and microarray position of all oligonucleotide probes. The theoretical melting temperatures (Tm) of the probes were calculated according to the nearest neighbor method by using the OligoAnalyzer 3.0 software with default settings (http://biotools.idtdna.com/analyzer/oligocalc.asp). For each probe and each reference organism, the free energies, ΔG, of the perfectly matched and the mismatched (up to 4.5 weighted mismatches; as determined by using the ARB probe match tool) probe-target hybrids were calculated. ΔG calculation was performed with the two-state hybridization server (concentration of Na+ and temperature were set to 0.829 M and 42°C, respectively) at the mfold website (http://www.bioinfo.rpi.edu/applications/mfold/) (63). Additional information on RHC-PhyloChip probes can be viewed at the probeBase website (http://www.microbial-ecology.net/probebase) (34).
TABLE 4.
16S rRNA-targeted probes used for microarray hybridzation.
| Original name | Name | Full namea | Primerb | Microarray position(s) | Sequence (5′-3′) | Tm (°C) | ΔG (kcal mol−1) | Specificity | Reference |
|---|---|---|---|---|---|---|---|---|---|
| CONT-COMP | CTT CCT TCC TTC CTT CCT | Complementary to control oligonucleotide | 35 | ||||||
| CONT | A1, A48, B1, B48, C1, C48, D1, D48 | AGG AAG GAA GGA AGG AAG | 51.3 | −18.8 | Control oligonucleotide | 35 | |||
| NONSENSE | A2, A25, A47, C25, D47 | AGA GAG AGA GAG AGA GAG | 48.5 | −17.7 | Nonbinding control | 35 | |||
| UNIV1390 | UNIV1389a | S-D-Univ-1389-a-A-18 | R | A27, C27, D5 | ACG GGC GGT GTG TAC AAG | 58.3 | −22.2 | Bacteria, not Epsilonproteobacteria | Modified from 51 |
| UNIV1390 | UNIV1389b | S-D-Univ-1389-b-A-18 | R | D6 | ACG GGC GGT GTG TAC AAA | 58 | −21.9 | Eucarya | Modified from 51 |
| UNIV1390 | UNIV1389c | S-D-Univ-1389-c-A-18 | R | D7 | ACG GGC GGT GTG TGC AAG | 62 | −23.9 | Archaea | Modified from 51 |
| EUB338 | EUB338 | S-D-Bact-0338-a-A-18 | A3, A46, C26, D2, D46 | GCT GCC TCC CGT AGG AGT | 59.5 | −22.4 | Most Bacteria | 5 | |
| EUB338II | EUB338II | S-*-BactP-0338-a-A-18 | D3 | GCA GCC ACC CGT AGG TGT | 60.7 | −23.0 | Planctomycetes | 14 | |
| EUB338III | EUB338III | S-*-BactV-0338-a-A-18 | D4 | GCT GCC ACC CGT AGG TGT | 60.7 | −23.0 | Verrucomirobia | 14 | |
| BTWO23a | BTWO663 | S-*-Btwo-0663-a-A-18 | A34, D9 | GGA ATT CCA CCC CCC TCT | 57 | −21.0 | Most Rhodocyclales, but not the Zoogloea and the Azospira lineages | Modified from 4 | |
| RHC143 | S-*-RHC-0143-a-A-18 | R | A4, A19, A44, D19 | TCG CTA CGT TAT CCC CCA | 56 | −21.2 | Most members of the Dechloromonas-Ferribacterium-Quadricoccus-Azonexus, the Azospira, and the Rhodocyclus-Propionivibrio-Accumulibacter lineages, Zoogloea spp., and few members of the Azoarcus-Thauera-Denitromonas lineage | This study | |
| S-G-Rhc-0175-a-A-18 | RHC175a | S-*-RHC-0175-a-A-18 | A42, D17 | TGC TCA CAG AAT ATG CGG | 53 | −20.0 | Most members of the Dechloromonas-Ferribacterium-Quadricoccus-Azonexus, the Azospira, and the Rhodocyclus-Propionivibrio-Accumulibacter lineages | 25 | |
| S-G-Rhc-0439-a-A-18 | RHC439 | S-*-RHC-0439-a-A-18 | A43, D18 | CNA TTT CTT CCC CGC CGA | 54.9-58.3 | Rhodocyclus spp., most members of the Candidatus “Accumulibacter” cluster, Azospira lineage | 25 | ||
| RHAC855 | S-*-RhAc-0855-a-A-18 | A7 | TCA CGC GTT AGC TAC GGC | 58.2 | −22.8 | Rhodocyclus spp., most members of the Candidatus “Accumulibacter” cluster | This study | ||
| PAO846 | ACCBA846 | S-G-Accba-0846-a-A-18 | A10 | AGC TAC GGC ACT AAA AGG | 52.6 | −19.7 | Most members of the Candidatus “Accumulibacter” cluster | Modified from 13 | |
| PAO651 | ACCBA651 | S-G-Accba-0651-a-A-18 | A6 | CCC TCT GCC AAA CTC CAG | 55.8 | −20.9 | Most members of the Candidatus “Accumulibacter” cluster | 13 | |
| ACCBA443 | S-*-Accba-0443-a-A-18 | A8 | CAA GCA ATT TCT TCC CCG | 52.2 | −19.6 | Some members of the Candidatus “Accumulibacter” cluster | This study | ||
| S-G-Rhx-0456-a-A-17 | ACCBA455 | S-*-Accba-0455-a-A-18 | A9 | AGG GTA TTA ACC CAA GCA | 51.4 | −18.8 | Some members of the Candidatus “Accumulibacter” cluster | 25 | |
| RHO828 | S-G-Rho-0828-a-A-18 | A12 | TTA ACC CCA CCA ACA CCT | 54.4 | −20.0 | Rhodocyclus spp. | This study | ||
| RHO842 | S-G-Rho-0842-a-A-18 | A13 | CGG CAC TAA TGG GTT TAA | 50.5 | −18.8 | Rhodocyclus spp. | This study | ||
| RHOTE1280 | S-S-Rho.te-1280-a-A-18 | A15 | CGA TCG GCT TTG CGG GAT | 58.9 | −22.6 | Rhodocyclus tenuis | This study | ||
| AZP471 | S-G-Azp-0471-a-A-18 | A29 | GTA CCG TCA TCA ACA ACG | 51.4 | −19.5 | Most Azospira spp. | This study | ||
| AZP456 | S-G-Azp-0456-a-A-18 | A30 | ACG GAT ATT AGC CGT TGC | 53.1 | −20.1 | Most Azospira spp. | This study | ||
| AZP737 | S-G-Azp-0737-a-A-18 | A28 | GTC AGT ACT AAC CCA GGG | 51.6 | −19.1 | Most Azospira spp. | This study | ||
| DAF1030 | S-*-D.A.F-1030-a-A-18 | A22, A45, D20 | TGT GTT CCA GCT CCC TTT | 54.7 | −20.2 | Some bacteria of the Dechloromonas-Ferribacterium-Quadricoccus-Azonexus lineage | This study | ||
| DEMFE455 | S-*-DemFe-0455-a-A-18 | A21 | AGG GTA TTA ACC CAT GCG | 52.5 | −19.5 | Ferribacterium limneticum, few Dechloromonas spp. | This study | ||
| QUACO135 | S-*-Quaco-0135-a-A-18 | D40 | TTA TCC CCC ACT CAA TGG | 51.8 | −19.0 | Quadricoccus australiensis, reactor clone PHOS-HE23 | This study | ||
| DCMAG455 | S-S-Dcm.ag-0455-a-A-18 | A20 | CAG GTA TTA GCT GAT GCG | 50.7 | −19.1 | Dechloromonas agitata | This study | ||
| A08KA458 | S-*-A08KA-0459-a-A-18 | D44 | ACA CCC CGT ATT AGA GAG | 50.7 | −18.7 | Oral strain A08KA lineage | This study | ||
| TH14 | RHC175b | S-*-RHC-0175-b-A-18 | A39, C4, C28, C40, D14 | CCC TCA GGA CGT ATG CGG | 57.9 | −22.1 | Some Thauera, Azoarcus, Zoogloea, Sterolibacterium, and Azovibrio spp. | Modified from 26 | |
| TH3 | RHC222 | S-*-RHC-0222-a-A-18 | A36, B2, C2, C41, D11 | ACA TCG GCC GCT CCA ATC | 58.6 | −22.4 | Some members of the Azoarcus-Thauera-Denitromonas, Zoogloea, and Azovibrio lineages | 26 | |
| AZV211 | S-G-Azv-0211-a-A-18 | C42 | TCC AAT CGC ACA AGG TCC | 55.7 | −21.2 | Azovibrio spp. | This study | ||
| AZVRE847 | S-*-Azv.re-0847-a-A-18 | C43 | TAG CTC CGT TAC TAA TAG | 45.1 | −17.1 | Azovibrio restrictus | This study | ||
| AT1458 | ATD1459 | S-*-ATD-1459-a-A-18 | A,R,Z | A35, D10 | TCT CAC CGT GGT AAG CGC | 58 | −22.4 | Most members of the Azoarcus-Thauera-Denitromonas lineage | Modified from 43 |
| RHC630 | S-*-RHC-0630-a-A-18 | A40, D15, D25 | TGC AGT CAC AAA CGC AGT | 56.1 | −21.2 | Most Thauera, Zoogloea, and Rhodocyclus spp. | This study | ||
| AZA1006 | S-*-Aza-1006-a-A-18 | A38, B4, D13 | TCC CTG ATC TCT CAA GGA | 52.1 | −19.5 | Most members of the Azoarcus cluster | This study | ||
| AZA483 | S-*-Aza-0483-a-A-18 | B6 | CTT CTT CTG ACA GTA CCG | 49.7 | −18.6 | Azoarcus cluster | This study | ||
| Azo644 | AZA645 | S-*-Aza-0645-a-A-18 | B5, B43 | GCC GTA CTC TAG CCG TGC | 58.3 | −22.5 | Most members of the Azoarcus cluster | 24 | |
| Azo1251 | AZA1252 | S-*-Aza-1252-a-A-18 | B7 | TCG CGC TTT GGC AGC CCT | 64.1 | −24.8 | Azoarcus evansii, Azoarcus toluvorans, Azoarcus tolulyticus, Azoarcus toluclasticus, and related Azoarcus spp. | Modified from 24 | |
| AZA444 | S-*-Aza-0444-a-A-18 | B9 | GGA AGC GTT TTC TTT CCG | 52.4 | −19.9 | Azoarcus evansii, Azoarcus tolulyticus str. Td-3, Azoarcus tolulyticus str.Td-19, Azoarcus sp. ToN1 | This study | ||
| AZTOLY452 | S-S-Az.toly-0452-a-A-18 | B10 | GTA TTG ACC CAC CCG ATT | 52.4 | −19.5 | Azoarcus tolulyticus | This study | ||
| AZANBU228 | S-*-Az.an.bu-0228-a-A-18 | B12 | AAT CCG ACA TCA GCC GCT | 57.7 | −21.8 | Azoarcus anaerobius, A. buckelli, and related Azoarcus spp. | This study | ||
| AZA844 | S-*-Aza-0844-a-A-18 | B20 | TGC GTC ACT CAG CGC GTT | 61.1 | −23.7 | Azoarcus spp. PH002 and CR23 | This study | ||
| AZA452 | S-*-Aza-0452-a-A-18 | B19 | CTA TTC ACG CAC CCG ATT | 53 | −20.0 | Azoarcus spp. PH002 and CR23 | This study | ||
| AZA463 | S-*-Aza-0463-a-A-18 | B15 | ATC CAG GCA CGC TAT TCA | 54.6 | −20.3 | Azoarcus spp. PbN1 and HxN1 | This study | ||
| AZA835 | S-*-Aza-0835-a-A-18 | B16 | CAG AAA GTT ACC TTC CCG | 50.5 | −18.8 | Azoarcus sp. PbN1 | This study | ||
| AZAN465 | S-S-Az.an-0465-a-A-18 | B14 | TCA TCC AGG CTC GCT ATT | 54 | −20.3 | Azoarcus anaerobius | This study | ||
| AZAN130 | S-S-Az.an-0130-a-A-18 | R | B13 | CCC CTC GAC TGG GTA CGT | 59 | −22.2 | Azoarcus anaerobius | This study | |
| S-*-OTU2-0132-a-A-18 | ATDe132 | S-*-ATDe-0132-a-A-18 | R | B24 | CCC CCA CAA CAT GGG TAC | 55.9 | −20.9 | Activated sludge clones A33, H25, H30, H35, S3, S10, and S23 of the Azoarcus-Thauera-Denitromonas lineage | 27 |
| Original name | Name | Full namea | Primerb | Microarray position(s) | Sequence (5′-3′) | Tm (°C) | ΔG (kcal mol−1) | Specificity | Reference |
| ATDe442 | S-*-ATDe-0442-a-A-18 | B25 | ACC CCG TTT CTT CCC AAC | 55.7 | −20.8 | Activated sludge clones A33, H25, H30, H35, S3, S10, and S23 of the Azoarcus-Thauera-Denitromonas lineage | This study | ||
| ATDe830 | S-*-ATDe-0830-a-A-18 | B26 | CGT TAC CGC TCC GAA CAA | 55.8 | −21.4 | Activated sludge clones A33, H25, H30, H35, S3, S10, and S23 of the Azoarcus-Thauera-Denitromonas lineage | This study | ||
| S-*-OTU2-0467-a-A-20 | ATDe467 | S-*-ATDe-0467-a-A-18 | B27 | CGT CAT TAG GAT CCT ATG | 46.6 | −17.2 | Activated sludge clones A33, H25, H30, H35, S3, S10, and S23 of the Azoarcus-Thauera-Denitromonas lineage | Modified from 27 | |
| S3-486 | S-*-S3-0486-a-A-18 | B31 | GTG CTT CTT CCG TCG GTA | 54.9 | −21.4 | Activated sludge clone S3 | This study | ||
| A33-587 | S-*-A33-0587-a-A-18 | B34 | CAC CTG TCT TAC CAA ACC | 50.7 | −18.9 | Activated sludge clone A33 | This study | ||
| DENAR176 | S-S-Denar-0176-a-A-18 | C16 | TCC CTC AGG AAA TAT GCG | 52.1 | −19.6 | Denitromonas aromaticus | This study | ||
| DENAR453 | S-S-Denar-0453-a-A-18 | C17 | CGT ATT CGG GGC GAT GAT | 55.3 | −21.0 | Denitromonas aromaticus | This study | ||
| DENAR845 | S-S-Denar-0845-a-A-18 | C18 | GCT GCG TTA CCC AGA AAG | 54.2 | −20.6 | Denitromonas aromaticus | This study | ||
| AZCOM447 | S-S-Az.com-0447-a-A-18 | B42 | AGC CCA CAC GTT TTC TTC | 53.8 | −20.2 | Azoarcus communis | This study | ||
| TH5 | AZIND1023 | S-S-Az.ind-1023-a-A-18 | A37, B3, B38, C3, D12 | CTG GTT CCC GAA GGC ACC | 58.9 | −22.3 | Azoarcus indigens, Azoarcus sp. BH72 | Modified from 26 | |
| AZIND433 | S-S-Az.ind-0433-a-A-18 | B39 | CTT TCC GTC CGA AAG AGC | 53.8 | −20.5 | Azoarcus indigens, Azoarcus sp. BH72 | This study | ||
| AZIND449 | S-St-Az.ind-0449-a-A-18 | B40 | TTA GCC CGC GCG ATT TCT | 58.2 | −22.2 | Azoarcus sp. BH72 | This study | ||
| AZIND455 | S-St-Az.ind-0455-a-A-18 | B41 | CGG GTA TTG GCC GAA GCG | 59.4 | −23.0 | Azoarcus indigens (T) | This study | ||
| THAU832 | S-G-Thau-0832-a-A-18 | C5 | TGC ATT GCT GCT CCG AAC | 57 | −21.7 | Thauera spp. | This study | ||
| THAU455a | S-*-Thau-0455-a-A-18 | C6 | ACT ATG TTA GAG TGC GCG | 52.6 | −19.9 | Thauera chlorobenzoica, Thauera mechernichensis | This study | ||
| THAU443 | S-*-Thau-0443-a-A-18 | C7 | AAC ACG ATT TCT TCC CGG | 53.2 | −20.0 | Thauera selenatis, Thauera phenylacetica, and related Thauera spp. | This study | ||
| THAU580 | S-*-Thau-0580-a-A-18 | C8 | CTT ACA AAA CCG GCC TCG | 54.2 | −20.6 | Thauera selenatis, soil clone AX39 | This study | ||
| THAU455b | S-*-Thau-0455-b-A-18 | C9 | ACT ATG TTA GAG TCG CCG | 51.6 | −19.4 | Thauera sp. PIV-1, TCB-transforming microbial consortium clone SJA-186 | This study | ||
| THAU468 | S-*-Thau-0468-a-A-18 | C10 | CCG TCA TCC AGC GAC TAT | 54.5 | −20.6 | Thauera sp. PIV-1, TCB-transforming microbial consortium clone SJA-186 | This study | ||
| THATE461 | S-S-Tha.te-0461-a-A-18 | C11 | CCA CAC CCT ATG TTA GAG | 49.2 | −18.2 | Thauera terpenica | This study | ||
| ZOGLO828 | S-G-Zoglo-0828-a-A-18 | C30 | TCT CCT CAC CGA ACA ACT | 53.6 | −20.1 | Zoogloea spp. | This study | ||
| S-*-OTU1-1415-a-A-20 | ZOGLO1416 | S-G-Zoglo-1416-a-A-18 | C29 | TCT GGT AAA CCC CAC TCC | 54.1 | −20.2 | Zoogloea spp. | Modified from 27 | |
| ZRA (ZRA23a) | ZOGLO647 | S-*-Zoglo-0647-a-A-18 | C31 | CTG CCG TAC TCT AGT TAT | 48.2 | −17.8 | Most members of the Zoogloea lineage, not Z. resiniphila | 45 | |
| ZOGLO455 | S-*-Zoglo-0455-a-A-18 | C32 | AGA GTA TTA TCC TGC GCG | 52.1 | −19.6 | Some members of the Zoogloea lineage (activated sludge clones H13, H11, H22, H10, H40, H27, and S21), not Z. ramigera and Z. resiniphila | This study | ||
| S-*-H7-1014-a-A-18 | H7-1014 | S-*-H7-1014-a-A-18 | C33 | TCG GGC ACC CCT CAA TCT | 59.4 | −22.3 | Activated sludge clone H7 | 27 | |
| ZORAM211 | S-S-Zo.ram-0211-a-A-18 | C34 | TCG TAT AAC GTG AGG CCT | 53.4 | −20.2 | Zoogloea ramigera | This study | ||
| ZORAM441 | S-S-Zo.ram-0441-a-A-18 | C35 | TGC GAT TTC TTT CCA CCT | 52.6 | −19.5 | Zoogloea ramigera | This study | ||
| S-*-OTU3-1426-a-A-18 | STEBA1426 | S-*-Steba-1426-a-A-18 | A, R, Z | D26 | ACT ACC TAC TTC TGG TGG | 50.6 | −18.5 | Some members of the Sterolibacterium lineage | 27 |
| STEBA468 | S-*-Steba-0468-a-A-18 | D28 | CCG TCA TTA GTA GCC CGT | 54.5 | −20.5 | Some members of the Sterolibacterium lineage (activated sludge clones S28, A13, S40, H12, H23, and H20) | This study | ||
| S-*-OTU3-0445-a-A-20 | STEBA448 | S-*-Steba-0448-a-A-18 | D27 | TAG GGG CCA CCG TTT CGT | 59.8 | −23.1 | Some members of the Sterolibacterium lineage (activated sludge clones S28, A13, S40, H12, H23, and H20) | Modified from 27 | |
| STEBA635 | S-*-Steba-0635-a-A-18 | D29 | AGT CCT ACA GTC ACA AAC | 49.3 | −18.2 | Few members of the Sterolibacterium lineage (activated sludge clones S28 and A13) | This study | ||
| STEBA643 | S-*-Steba-0643-a-A-18 | D30 | CAC ACT CGA GTT ATG CAG | 50.8 | −19.2 | Few members of the Sterolibacterium lineage (activated sludge clones S40, H12, H23, and H20) | This study | ||
| STEBA214 | S-*-Steba-0214-a-A-18 | D32 | CGC TCC TCT CGC GCG AGG | 63.5 | −25.1 | Few members of the Sterolibacterium lineage (clones SBR1001, SBR2080, and GC24) | This study | ||
| BONE23a | BONE663c | S-*-Bone-0663-a-A-18 | A33, D8 | GGA ATT CCA TCC CCC TCT | 54.1 | −19.9 | Beta1 group of Betaproteobacteria | Modified from 4 | |
| RHOTE206c | S-S-Rho.te-0206-a-A-18 | A14 | AAG CGC AAG GTC CTA AGA | 54.4 | −20.5 | Rhodocyclus tenuis | This study | ||
| PPV1239c | S-*-Ppv-1239-a-A-18 | A11 | ACC CTC TGA ACC GAC CAT | 56.4 | −20.9 | Propionivibrio spp., few members of the Candidatus “Accumulibacter” cluster | This study | ||
| AZA1269c | S-*-Aza-1269-a-A-18 | B8 | AAG GGA TTG GCT CCA GCT | 57.2 | −21.1 | Azoarcus evansii, Azoarcus toluvorans, Azoarcus tolulyticus, Azoarcus toluclasticus, and related Azoarcus spp. | This study | ||
| AZA829c | S-*-Aza-0829-a-A-18 | B18 | GTT ACC GCA CCG AAC AAC | 54.7 | −21.0 | Azoarcus spp. EbN1 and pCyN1 | This study | ||
| AZA234c | S-*-Aza-0234-a-A-18 | B17 | CCA GCT AAT CCG ACA TCA | 52.1 | −19.4 | Azoarcus spp. EbN1 and pCyN1 | This study | ||
| AZA221c | S-S-Aza-0221-a-A18 | B45 | CAT CGG CCA CTC CAA TCA | 55.5 | −20.9 | Azoarcus sp. LU1 | This study |
Name of 16S rRNA gene-targeted oligonucleotide probe based on the nomenclature of Alm et al. (2).
When “Rhodocyclales” subgroup-selective primer pair A, R, or Z was used for microarray analysis, this probe has its target site outside the amplified 16S rRNA gene fragments and must be ignored during interpretation of the hybridization pattern.
Probe was removed from the RHC-PhyloChip because it hybridized nonspecifically to many reference organisms that have mismatches in the 16S rRNA gene target site.
Each oligonucleotide probe contained a spacer element consisting of 15 dTTPs at the 5′ end and was aminated at the 5′-terminal nucleotide to allow covalent coupling to aldehyde group-coated CSS-100 glass slides (CEL Associates, Houston, Tex.). Fluorescence labeling of PCR amplicons, manufacturing and processing of microarrays, and reverse hybridization on microarrays were performed as outlined previously (35). The concentration of oligonucleotide probes before printing was adjusted to 50 pmol μl−1 in 50% dimethyl sulfoxide to prevent evaporation during the printing procedure. RHC-PhyloChips with triplicate spots for each probe were printed by using a GMS 417 contact arrayer (Affymetrix, Santa Clara, Calif.). Spotted DNA microarrays were dried overnight at room temperature in the dark to allow efficient cross-linking. Free aldehyde groups at the slide surface were reduced with sodium borohydride solution (35). For each reference organism, a separate microarray was hybridized, washed, and scanned under identical conditions and settings.
Scanning of microarrays and image analysis.
Fluorescence images of the RHC-PhyloChips were recorded by scanning the slides with a GMS 418 array scanner (Affymetrix). The fluorescence signals were quantified by using the ImaGene 4.0 software (BioDiscovery, Inc., Los Angeles, Calif.). A grid of individual circles defining the location of each spot on the array was superimposed on the image to designate each fluorescent spot to be quantified. The mean signal intensity of each spot and the local background area surrounding each spot was determined. Subsequently, for each probe the signal-to-noise ratio (SNR) was calculated according to the following formula:
SNR = [IP − (IN − INLB)] × IPLB−1
where IP is the mean pixel intensity of all replicate probe spots, IN is the mean pixel intensity of all nonsense probe spots, INLB is the mean pixel intensity of the local background area around all nonsense probe spots (note that IN − INLB must always have a lower value than IP), and IPLB is the mean pixel intensity of the local background area around all replicate probe spots. Probes for which the SNR was equal to or greater than 2.0 were considered positive (35). Furthermore, in the reference strain evaluation experiments the SNR of each probe was normalized against the SNR of the bacterial EUB338 probe, recorded on the same microarray, according to the following formula:
nSNR = SNR × {[IEUB − (IN − INLB)] × IEUBLB−1}−1
where nSNR is the normalized SNR of the specific probe, IEUB is the mean pixel intensity of all EUB338 probe spots, and IEUBLB is the mean pixel intensity of the local background area around all EUB338 probe spots.
Cloning and sequencing.
Prior to cloning, the PCR amplification products were purified by low-melting-point agarose (1.5%) gel electrophoresis (NuSieve 3:1; FMC Bioproducts, Biozym Diagnostics GmbH, Oldendorf, Germany) and stained in SYBR Green I solution (10 μl of 10,000× SYBR Green I stain in 100 ml of TAE buffer [40 mM Tris, 10 mM sodium acetate, 1 mM EDTA, pH 8.0]; Biozym Diagnostics GmbH) for 45 min. Bands of the expected size were excised from the agarose gel with a glass capillary and melted with 80 μl of double-distilled water for 10 min at 80°C (this procedure was also done for the amplicon obtained with primer pair A, although no band was visible). Four microliters of each solution was ligated as recommended by the manufacturer (Invitrogen Corp.) into the cloning vector pCR2.1 of the TOPO TA cloning kit. Nucleotide sequences were determined by the dideoxynucleotide method (46) following a previously published protocol (42).
Phylogeny inference.
All phylogenetic analyses were performed by using the alignment and treeing tools implemented in the ARB program package (37). All almost-full-length 16S rRNA sequences (>1,300 bases) which have been assigned to the order “Rhodocyclales” in the preview release of the RDP II database (version September 2003) (12) and all 16S rRNA sequences obtained from the activated sludge samples in this study were added to an ARB alignment of about 20,000 small-subunit rRNA sequences by using the alignment tool ARB_EDIT. Alignments were refined by visual inspection. Chimeric “Rhodocyclales” sequences were identified by independently subjecting base positions 1 to 513, 514 to 1026, and 1027 to 1539 (Escherichia coli numbering) of the 16S rRNA sequence to phylogenetic analysis. Inconsistent affiliation of the gene fragments in the phylogenetic trees was interpreted as being caused by a chimeric sequence. In addition, the CHECK_CHIMERA program of the RDP II was used for confirmation. Ambiguous base positions were excluded during calculation of 16S rRNA sequence similarities.
16S rRNA phylogenetic analyses were performed by applying distance-matrix, maximum-parsimony, and maximum-likelihood methods (36). A representative assortment of type strain sequences of different orders of the Beta- and Gammaproteobacteria was used as the outgroup for treeing. Variability of the individual alignment positions was determined by using the ARB_SAI tools and used as a criterion to remove or include variable positions for phylogenetic analyses. The neighbor-joining method combined with a Jukes-Cantor correction was used to infer distance-matrix trees. Maximum-likelihood trees were calculated by Tree-puzzle (54) and by applying the new A(x)ccelerated Maximum-Likelihood (AxML) algorithm (52). Maximum-parsimony analyses (treeing and bootstrapping) were performed with the Phylogeny Inference Package (PHYLIP, version 3.57c, J. Felsenstein, Department of Genetics, University of Washington, Seattle). For parsimony bootstrap analysis, 100 resamplings were used. All phylogenetic consensus trees were drawn according to recommendations outlined previously (36).
FISH.
The abundance of selected “Rhodocyclales” groups in the activated sludge sample was determined by FISH combined with subsequent image analysis (14, 47). Fluorescently labeled oligonucleotide probes (Table 5) (34) were purchased from Thermo Hybaid (Ulm, Germany). Hybridization under optimal conditions was performed as described previously (27, 38).
TABLE 5.
16S rRNA-targeted probes used for FISH
| Name | Sequence, 5′-3′ | Formamide concn (%) | Specificity | Reference |
|---|---|---|---|---|
| EUB338a | GCT GCC TCC CGT AGG AGT | 0-50 | Most Bacteria | 6 |
| EUB338IIa | GCA GCC ACC CGT AGG TGT | 0-50 | Planctomycetes | 14 |
| EUB338IIIa | GCT GCC ACC CGT AGG TGT | 0-50 | Verrucomicrobia | 14 |
| GAM42a | GCC TTC CCA CAT CGT TT | 35 | “Gammaproteobacteria” | 38 |
| BET42a | GCC TTC CCA CTT CGT TT | 35 | “Betaproteobacteria” | 38 |
| AT1458 | GAA TCT CAC CGT GGT AAG CGC | 50 | Most members of the Azoarcus-Thauera-Denitromonas lineage | 43 |
| S-*-OTU1-1415-a-A-20 | TTC TGG TAA ACC CCA CTC CC | 25 | Zoogloea lineage (including all KRZ Kraftisried clones from this study) | 27 |
| S-*-OTU3-0445-a-A-20 | TTA GGG GCC ACC GTT TCG TT | 30 | Kraftisried activated sludge clones of the Sterolibacterium lineage | 27 |
EUB338, EUB338II, and EUB338III were applied simultaneously to target most Bacteria (14).
Bacterial nomenclature.
The names of the bacterial taxa were used in accordance with the prokaryotic nomenclature proposed in the latest taxonomic outline of the second edition of Bergey′s Manual of Systematic Bacteriology (http://dx.doi.org/10.1007/bergeysoutline200310) (20).
Nucleotide sequence accession numbers.
The sequences determined in this study were deposited at GenBank under accession numbers AY689085 to AY689093.
RESULTS AND DISCUSSION
16S rRNA-based phylogeny of “Rhodocyclales.”
The latest taxonomic outline of Bergey′s Manual of Systematic Bacteriology lists 30 validly published species assigned into the 12 recognized genera of the “Rhodocyclaceae” (Azoarcus, Thauera, Zoogloea, Azovibrio, Azospira, Rhodocyclus, Propionivibrio, Dechloromonas, Ferribacterium, Quadricoccus, Azonexus, and Sterolibacterium), the only family within the betaproteobacterial order “Rhodocyclales.” The order additionally encompasses the species “Denitromonas aromaticus” and the as yet uncultured Candidatus species “Accumulibacter phosphatis” (25), both of which still await valid description. In addition, 92 isolates and 104 environmentally retrieved 16S rRNA sequences affiliated to this order were included in the analysis.
To establish a robust and detailed phylogenetic backbone for subsequent design of microarray probes and for future taxonomic and environmental studies, an evaluation of the phylogeny of cultivated and yet uncultivated “Rhodocyclales” was performed. Initially, 10 sequences from environmental 16S rRNA clones (GenBank accession numbers AJ009452, AF245350, AF280861, AF281119, AY118150, AF529340, AY082472, AB089100, AB089101, and AF204249) were identified as chimeras and omitted from all subsequent analyses. The remaining 218 almost-full-length 16S rRNA sequences of “Rhodocyclales” were phylogenetically analyzed by calculating similarities and applying distance-matrix, maximum-parsimony, and maximum-likelihood methods for treeing.
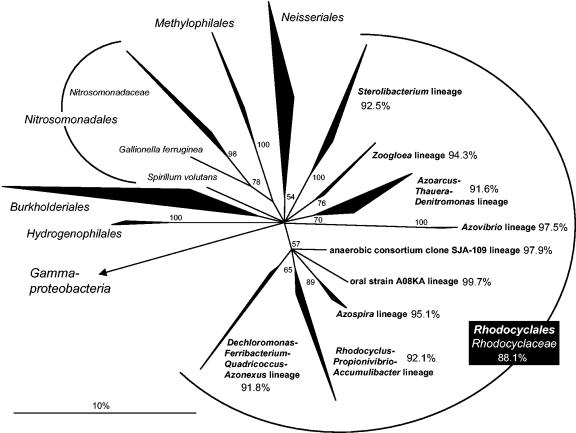
The minimum 16S rRNA sequence similarity of two members of the “Rhodocyclales” was 88.1%. This is in the range of minimal similarities previously reported for other bacterial families e.g., 83% each for “Desulfobacteraceae” and “Desulfovibrionaceae” (33), 89% for “Nitrosomonadaceae” (30), and 90% for Chlamydiaceae (18), and therefore legitimates subclassification of all “Rhodocyclales” into the single family “Rhodocyclaceae” (http://dx.doi.org/10.1007/bergeysoutline200310) (20) from an rRNA-based point of view.
Independently of the phylogeny inference method applied, members of the “Rhodocyclaceae” could be subdivided into nine different monophyletic lineages (Fig. 1). The phylogenetic positions of these lineages to other betaproteobacterial orders could not be unambiguously determined, as shown by a polytomic tree topology (Fig. 1). With the exception of the anaerobic consortium clone SJA-109 lineage, designated according to an environmental clone sequence from an anaerobic, trichlorobenzene-transforming microbial consortium (58), each lineage was represented by at least one validly described or cultured species, indicating that lineage-level biodiversity of “Rhodocyclaceae” is well reflected by available cultured strains. Detailed phylogenetic trees showing the affiliation of all members of each “Rhodocyclaceae” lineage can be downloaded at http://www.microbial-ecology.net/supplements.asp (supplemental Fig. S1).
FIG. 1.
16S rRNA-based phylogenetic tree of the “Rhodocyclales” and selected type strains of other betaproteobacterial orders. The consensus tree is based on maximum-likelihood analysis (AxML) performed with a 50% conservation filter for the “Betaproteobacteria.” The bar indicates 10% estimated sequence divergence. Polytomic nodes connect branches for which a relative order could not be determined unambiguously by applying neighbor-joining, maximum-parsimony, and maximum-likelihood treeing methods. Numbers at branches indicate percent parsimony bootstrap values. Branches without numbers had bootstrap values of less than 50%. The minimum 16S rRNA sequence similarity for each “Rhodocyclales” lineage is shown.
Probe design and microarray format.
In general, the same strategies for in silico development and the same technical set-up for fabrication and hybridization of the RHC-PhyloChip were used as for the development of a 16S rRNA-targeted oligonucleotide microarray for detection of all lineages of recognized sulfate-reducing prokaryotes (SRP-PhyloChip) (35).
Initially, the specificities of all previously published 16S rRNA-targeted probes and primers for “Rhodocyclales” (13, 24-27, 43, 45) were reevaluated with the updated 16S rRNA database. Eighteen probes were found to target “Rhodocyclales” only and were therefore included on the RHC-PhyloChip, although not all of them target monophyletic “Rhodocyclales” groups (Table 4 and supplemental Fig. S2). In addition, 60 oligonucleotide probes were designed according to the “multiple probe concept” (3, 8) to specifically target “Rhodocyclales” at hierarchical and identical phylogenetic levels (Table 4 and Fig. S2). Because multiple nested probes can at least partly compensate for unspecificities of individual probes (32, 35), this probe design strategy is particularly valuable if microarray formats are used which only allow hybridization or washing at a single stringency. In summary, the RHC-PhyloChip probe set consists of 78 specific probes covering the complete diversity of “Rhodocyclales” known so far (see above), two probes targeting betaproteobacterial groups at a broader specificity (BONE663 and BTWO663) (4), six bacterial or universal probes, and two probes as positive and negative hybridization controls (CONT and NONSENSE) (Table 4). All probes were designed to have the same length (18 bases, excluding the T-spacer), but the G+C contents of the probes varied between 38.9 and 77.8%. To attenuate the influence of differing G+C contents of probe-target duplexes on duplex stabilities, 3M tetramethylammonium chloride was added to the washing buffer (8, 35, 39).
RHC-PhyloChip evaluation with reference strains.
The specificity of the individual probes was tested under monostringent conditions (i.e., the same hybridization and washing conditions for all probes and microarrays). Cy5-labeled 16S rRNA gene amplicons of each pure culture and each 16S rRNA gene-containing clone (n = 29) were hybridized with a separate RHC-PhyloChip. For 60 “Rhodocyclales”-specific probes, this set of reference 16S rRNA genes contained at least one sequence with a perfectly matched target site. None of these probes showed false-negative signals. Out of the 18 probes for which no perfectly matched reference sequence was available, 10 yielded positive signals with mismatching reference 16S rRNA gene amplicons, indicating that the respective probe-target sites were accessible for hybridization. A detailed list of the individual hybridization results can be downloaded at http://www.microbial-ecology.net/supplements.asp (supplemental Table S1). Seven of the probes hybridized nonspecifically with many reference organisms not having fully complementary probe target sites and were therefore excluded from the final version of the RHC-PhyloChip (listed separately in Table 4 and supplemental Table S1).
In order to compare the hybridization efficiency of the different RHC-PhyloChip probes, the signal-to-noise ratios (SNRs) of the probes were normalized against the SNR of the bacterial probe EUB338 recorded on the same microarray. The resulting nSNRs for perfectly matched probe-target duplexes ranged from 0.2 (for probe A33-587 with Kraftisried WWTP clone A33) to 48.0 (for probe BTWO663 with Azonexus fungiphilus), demonstrating that the signal intensities of individual probes vary strongly if excess target DNA is added. It has been observed previously that the duplex yield of different rRNA gene-targeted microarray probes can differ considerably (32, 35, 41), and on the RHC-PhyloChip the duplex yield was significantly positively correlated with the theoretical Tm of the probe (Spearman nonparametric correlation test: R = 0.342, P = 0.013). The latter point demonstrated that the addition of tetramethylammonium chloride did not completely abolish the influence of the G+C content on the duplex yield of different probes.
Of the 2,291 different hybridizations (each reference DNA with each probe) which were performed in total, 208 (9.1%) were false-positive (positive probe signal with a nontarget organism having one or more mismatches in the probe target site), while no hybridizations were false-negatives (supplemental Table S1). The occurrence of some false-positive results is almost impossible to avoid with a monostringent microarray hybridization approach, because the stability of mismatched probe-target hybrids is difficult to predict in silico and influenced by many factors, such as (i) the number of mismatches, (ii) the nature of the mismatching nucleotides, (iii) the position of the mismatches in the probe target site, and (iv) possible stacking interactions of nucleotides adjacent to the mismatches (19, 53, 56, 57). However, specific identification of target organisms is still possible with the RHC-PhyloChip because of the multiple probe concept (the theoretical specificities of the nested probes are depicted in supplemental Fig. S2). Nevertheless, future microarray probe design would be further improved if oligonucleotide probe parameters were available which allow estimation of the hybridization behavior of each probe in silico (53).
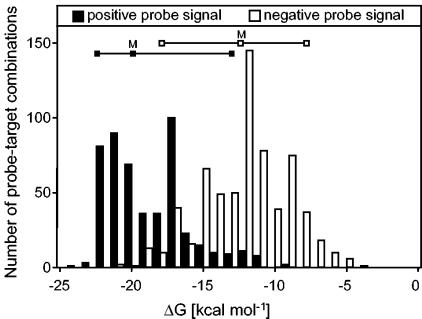
One hybridization parameter that might be a suitable candidate is the free energy, ΔG, of a given perfectly matching or mismatching probe-target hybrid (23, 55). On the RHC-PhyloChip, the ΔG values of most (88%) of the false-positive probe-target hybrids with one or two mismatches were in a similar range (−22 to −16 kcal mol−1) to ΔG values of all perfectly matched probe-target pairs (−25 to −17 kcal mol−1), providing an explanation of why discrimination was not successful under monostringent hybridization conditions. As can be inferred from Fig. 2, most of the positive probe-target hybrids (84%) (including false-positive signals) had a ΔG below −16 kcal mol−1. Additionally, only 3% of all probe-target combinations that yielded no hybridization signal had also a ΔG below −16 kcal mol−1. Therefore, a ΔG threshold of −16 kcal mol−1 could provide useful guidance for future preselection of appropriate probes in silico (Fig. 2) but does not abolish the need for extensive empirical testing of the hybridization behavior of microarray probes. It should be further stressed that the ΔG threshold of −16 kcal mol−1 might only apply to the microarray set-up and the hybridization conditions used in this study.
FIG. 2.
Frequency distribution of ΔG values for positive (black bars) and negative (white bars) probe-target combinations having up to five mismatches. The horizontal lines indicate the 5th and 95th percentiles and the median value (M). The difference between the ΔG values of positive and negative probe-target combinations was highly significant (analysis of variance, P < 0.001).
RHC-PhyloChip application in activated sludge.
To demonstrate the applicability of the RHC-PhyloChip for rapid screening of “Rhodocyclales” diversity in environmental samples, activated sludge from the nitrifying-denitrifying WWTP Kraftisried was analyzed. Kraftisried was chosen as a model system because in 1996 members of diverse lineages of “Rhodocyclales” comprised more than one third of the entire bacterial biovolume in this WWTP (27).
Initially, 16S rRNA genes were amplified from Kraftisried DNA by using standard bacterial primers, fluorescently labeled, and hybridized with the RHC-PhyloChip. Surprisingly, the hybridization pattern obtained did not indicate the presence of members of the “Rhodocyclales” below the lineage level [some probes (BTWO663, ATD1459, RHC630, RHC175a, RHC143, RHC222, and RHAC855) targeting “Rhodocyclales” at broader phylogenetic levels showed positive signals but almost all probes of higher specificity were negative] (Fig. 3A). To find an explanation for this unexpected result, the relative abundance of “Rhodocyclales” in this WWTP was analyzed quantitatively by FISH. Compared to 1996, the relative abundance of “Betaproteobacteria” in the activated sludge from 2002 decreased from 47 to 18% of all bacteria detectable by FISH (Table 5). Similarly, the abundance of members of the “Rhodocyclales” decreased dramatically between the two samples. While the activated sludge from 1996 contained significant amounts of “Rhodocyclales” detectable by probes AT1458, S-*-OTU1-1415-a-A-20, and S-*-OTU3-0445-a-A-20, [each targeting a “Rhodocyclales” group found previously in this WWTP (27)] (Table 5), less than 1% of the cells hybridized with these probes in the WWTP sample from 2002.
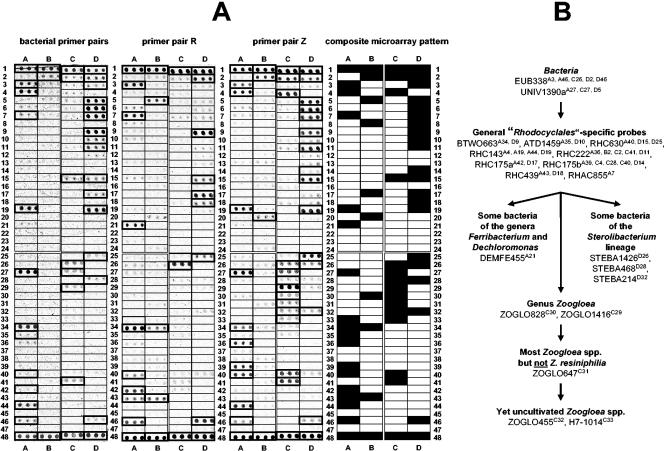
FIG. 3.
(A) DNA microarray diversity analysis of “Rhodocyclales” in activated sludge from the industrial WWTP Kraftisried. Three RHC-PhyloChips were hybridized separately with fluorescently labeled 16S rRNA gene PCR amplicons that were retrieved from the activated sludge sample by using either bacterial or “Rhodocyclales” subgroup-selective (R or Z) primer pairs (Table 3). Each probe was spotted in triplicate. For each microarray position, the probe sequence and specificity are depicted in Table 4. Probe spots having a signal-to-noise ratio (SNR) equal to or greater than 2.0 are indicated by boldface boxes and were considered positive. In the composite microarray pattern, probes which were positive in any of the three individual RHC-PhyloChip hybridizations are indicated by black boxes. (B) Flow chart illustrating the presence of distinct “Rhodocyclales” groups in the activated sludge from Kraftisried as inferred from the composite microarray pattern. For each probe, the position on the microarray is indicated in the superscript text.
To increase the sensitivity of the RHC-PhyloChip, three “Rhodocyclales”-subgroup-selective primer pairs called A, R, and Z (together targeting almost all “Rhodocyclales”) (Table 3) were designed and applied for amplification of 16S rRNA genes prior to microarray hybridization. Although these new primers were selected to amplify 16S rRNA gene fragments of the maximum possible length, the target sites of some RHC-PhyloChip probes are outside the amplified 16S rRNA gene region (Table 4), and these probes must thus be ignored during interpretation of hybridization patterns.
Each “Rhodocyclales”-subgroup-selective primer pair was used separately for amplification of Kraftisried activated sludge DNA at low stringency (Table 3) to allow potential primer binding to 16S rRNA genes of “Rhodocyclales” having mismatches in the primer target sites. PCR products of the expected length were obtained for primer pairs R and Z, but no primer pair A PCR product was observed after gel electrophoresis. The “Rhodocyclales” subgroup-selective PCR amplicons obtained were fluorescently labeled and hybridized with two separate RHC-PhyloChips. The RHC-PhyloChip hybridization patterns of the R and Z amplicons differed from each other and from the pattern obtained by using general bacterial primer pairs (Fig. 3A). In more detail, the hybridization pattern obtained with primer pair R indicated the presence of bacteria related to the genera Ferribacterium and Dechloromonas, whereas the hybridization pattern obtained with primer pair Z pointed to the presence of Zoogloea species.
A composite microarray fingerprint of the “Rhodocyclales” community present in activated sludge from Kraftisried was created by merging the separate microarray hybridization patterns obtained with the “Rhodocyclales”-subgroup-selective and the common bacterial 16S rRNA gene amplicons (Fig. 3A). Besides Ferribacterium/Dechloromonas-related bacteria and Zoogloea species, this composite microarray fingerprint additionally indicated the presence of members of the Sterolibacterium lineage (Fig. 3B).
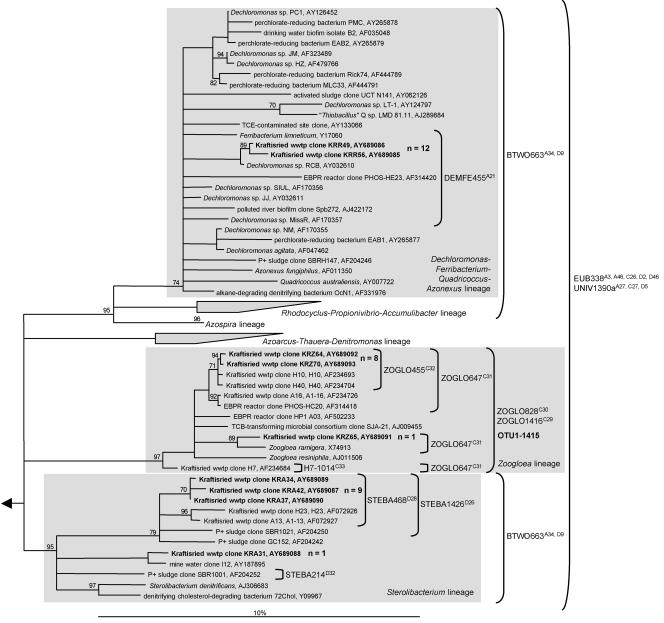
The microarray results were confirmed independently by cloning and sequencing of the 16S rRNA gene PCR products obtained with the three “Rhodocyclales” subgroup-selective primers. It should be noted that cloning of PCR products amplified from Kraftisried DNA with primer pair A was successful, although only small amounts of PCR product could be retrieved (see above). All 16S rRNA gene clones obtained with primer pairs Z and A were closely related to clones already found in the Kraftisried WWTP in 1996 (27) and belonged to the genus Zoogloea and the Sterolibacterium lineage, respectively (Fig. 4). In contrast, all 16S rRNA gene sequences obtained by using primer pair R clustered with members of the genera Dechloromonas and Ferribacterium (Fig. 4), which were not detected by the 16S rRNA full-cycle approach in Kraftisried WWTP samples from 1996 (27). The phylogeny of all retrieved 16S rRNA gene sequences was in perfect agreement with the microarray results. Furthermore, the sequenced 16S rRNA genes have perfectly matched target sites for the probes that showed a positive signal in the RHC-PhyloChip analyses (Figs. 3 and 4).
FIG. 4.
16S rRNA gene phylogenetic consensus tree based on maximum-likelihood analysis (Tree-puzzle) performed with a 50% conservation filter for the “Betaproteobacteria.” The tree shows the affiliation of clone sequences (boldface type) retrieved from the sewage treatment plant Kraftisried by using “Rhodocyclales” subgroup-selective primer pairs A (KRA clones), R (KRR clones), and Z (KRZ clones) for PCR. The grey box shows affiliation to a “Rhodocyclales” lineage. The bar indicates 10% estimated sequence divergence. Polytomic nodes connect branches for which a relative order could not be determined unambiguously by applying neighbor-joining, maximum-parsimony, and maximum-likelihood treeing methods. The percent reliability value of each internal branch indicates how often the corresponding cluster was found among 50,000 intermediate trees during quartet puzzling. Values below 70% are not shown. Parentheses indicate the perfect-match target organisms of the probes. Probe S-*-OTU1-1415-a-A-20 (OTU1-1415) (Table 5) is depicted in bold and was used for quantitative FISH analysis. The microarray position is depicted after the probe name. Probes RHC630, RHC143, RHC222, RHC175a, and RHC175b, perfectly matching some of the Kraftisried clones, are not shown to enhance clarity.
The microarray hybridizations, the retrieved 16S rRNA sequences, and the quantitative FISH data collected in this study provide corroborating evidence that substantial changes have occurred within the “Rhodocyclales” community in Kraftisried since the first bacterial community analysis of this WWTP (27). The dramatic decline in “Rhodocyclales,” assumed to be the major denitrifiers in this system (27), from 35% of the total bacterial biovolume in 1996 to less than 1% in 2002 may have been caused by the seasonal implementation of a partial ammonium stripping step prior to biological nitrogen removal in 1999. This physical sewage treatment step reduces the ammonia concentration and increases the salt concentration in the sewage and probably had dramatic consequences for the population structure of nitrifiers (data not shown) and potentially denitrifying heterotrophs in the activated sludge.
Acknowledgments
We thank Barbara Reinhold-Hurek (Universität Bremen, Bremen, Germany) for kindly providing Azoarcus anaerobius, A. communis, and A. evansii, Denis Rho (Biotechnology Research Institute, Montreal, Canada) for Azoarcus sp. strain LU1; and Natuschka Lee (Technische Universität München, Munich, Germany) for clones BNP269, hBPR24, hBPR4, WGAR24, and WGAR25. The excellent technical assistance of Sibylle Schadhauser and Helga Gaenge and critical review of the manuscript by Michael Taylor are acknowledged.
This research was supported by grants from the Deutsche Forschungsgemeinschaft (Wa1558/1-1), the Bayerische Forschungsstiftung (Development of Oligonucleotide-DNA-Chips in cooperation with MWG Biotech, Ebersberg; project 368/99), the bmb+f (project 01 LC 0021A-TP2 in the framework of the BIOLOG II program) to M.W., and by a Marie Curie Intra-European Fellowship (VENTSULFURMICDIV) within the 6th European Community Framework Programme to A.L.
REFERENCES
- 1.Achenbach, L. A., U. Michaelidou, R. A. Bruce, J. Fryman, and J. D. Coates. 2001. Dechloromonas agitata gen. nov., sp. nov. and Dechlorosoma suillum gen. nov., sp. nov., two novel environmentally dominant (per)chlorate-reducing bacteria and their phylogenetic position. Int. J. Syst. Evol. Microbiol. 51:527-533. [DOI] [PubMed] [Google Scholar]
- 2.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., and K.-H. Schleifer. 2001. Nucleic acid probes and their application in environmental microbiology, p. 67-82. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y. [Google Scholar]
- 4.Amann, R., J. Snaidr, M. Wagner, W. Ludwig, and K. H. Schleifer. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders, H. J., A. Kaetzke, P. Kämpfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 45:327-333. [DOI] [PubMed] [Google Scholar]
- 8.Behr, T., C. Koob, M. Schedl, A. Mehlen, H. Meier, D. Knopp, E. Frahm, U. Obst, K. Schleifer, R. Niessner, and W. Ludwig. 2000. A nested array of rRNA targeted probes for the detection and identification of enterococci by reverse hybridization. Syst. Appl. Microbiol. 23:563-572. [DOI] [PubMed] [Google Scholar]
- 9.Bodrossy, L., and A. Sessitsch. 2004. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 7:245-254. [DOI] [PubMed] [Google Scholar]
- 10.Bodrossy, L., N. Stralis-Pavese, J. C. Murrell, S. Radajewski, A. Weilharter, and A. Sessitsch. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566-582. [DOI] [PubMed] [Google Scholar]
- 11.Coates, J. D., U. Michaelidou, R. A. Bruce, S. M. O'Connor, J. N. Crespi, and L. A. Achenbach. 1999. Ubiquity and diversity of dissimilatory (per)chlorate-reducing bacteria. Appl. Environ. Microbiol. 65:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crocetti, G. R., P. Hugenholtz, P. L. Bond, A. Schuler, J. Keller, D. Jenkins, and L. L. Blackall. 2000. Identification of polyphosphate-accumulating organisms and design of 16S rRNA-directed probes for their detection and quantitation. Appl. Environ. Microbiol. 66:1175-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daims, H., A. Brühl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 15.Daims, H., K. Stoecker, and M. Wagner. Fluorescence in situ hybridization for the detection of prokaryotes. In Advanced methods in molecular microbial ecology, in press. BIOS Scientific Publishers, Abingdon, United Kingdom.
- 16.El Fantroussi, S., H. Urakawa, A. E. Bernhard, J. J. Kelly, P. A. Noble, H. Smidt, G. M. Yershov, and D. A. Stahl. 2003. Direct profiling of environmental microbial populations by thermal dissociation analysis of native rRNAs hybridized to oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2377-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelhard, M., T. Hurek, and B. Reinhold-Hurek. 2000. Preferential occurrence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ. Microbiol. 2:131-141. [DOI] [PubMed] [Google Scholar]
- 18.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 19.Fotin, A. V., A. L. Drobyshev, D. Y. Proudnikov, A. N. Perov, and A. D. Mirzabekov. 1998. Parallel thermodynamic analysis of duplexes on oligodeoxyribonucleotide microchips. Nucleic Acids Res. 26:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrity, G. M., and J. G. Holt. 2001. The road map to the manual, p. 119-166. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y. [Google Scholar]
- 21.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Held, G. A., G. Grinstein, and Y. Tu. 2003. Modeling of DNA microarray data by using physical properties of hybridization. Proc. Natl. Acad. Sci. USA 100:7575-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hess, A., B. Zarda, D. Hahn, A. Haner, D. Stax, P. Hohener, and J. Zeyer. 1997. In situ analysis of denitrifying toluene- and m-xylene-degrading bacteria in a diesel fuel-contaminated laboratory aquifer column. Appl. Environ. Microbiol. 63:2136-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hesselmann, R. P. X., C. Werlen, D. Hahn, J. R. van der Meer, and A. J. B. Zehnder. 1999. Enrichment, phylogenetic analysis and detection of a bacterium that performs enhanced biological phosphate removal in activated sludge. Syst. Appl. Microbiol. 22:454-465. [DOI] [PubMed] [Google Scholar]
- 26.Hurek, T., and B. Reinhold-Hurek. 1995. Identification of grass-associated and toluene-degrading diazotrophs, Azoarcus spp., by analyses of partial 16S ribosomal DNA sequences. Appl. Environ. Microbiol. 61:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 28.Juretschko, S., G. Timmermann, M. Schmid, K. H. Schleifer, A. Pommerening-Roser, H. P. Koops, and M. Wagner. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koops, H.-P., U. Purkhold, A. Pommerening-Röser, G. Timmermann, and M. Wagner. 2003. The lithoautotrophic ammonia-oxidizing bacteria. .In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.13, March 2003. Springer-Verlag, New York, N.Y.
- 31.Kovárová, M., and P. Dráber. 2000. New specificity and yield enhancer of polymerase chain reactions. Nucleic Acids Res. 28:E70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, W. T., A. D. Mirzabekov, and D. A. Stahl. 2001. Optimization of an oligonucleotide microchip for microbial identification studies: a non-equilibrium dissociation approach. Environ. Microbiol. 3:619-629. [DOI] [PubMed] [Google Scholar]
- 33.Loy, A. 2003. DNA microarray technology for biodiversity inventories of sulfate-reducing prokaryotes. Ph.D. thesis. (http://tumb1.biblio.tu-muenchen.de/publ/diss/ww/2003/loy.pdf.) Technische Universität München, Munich, Germany.
- 34.Loy, A., M. Horn, and M. Wagner. 2003. probeBase: an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 39.Maskos, U., and E. M. Southern. 1992. Parallel analysis of oligodeoxyribonucleotide (oligonucleotide) interactions. I. Analysis of factors influencing oligonucleotide duplex formation. Nucleic Acids Res. 20:1675-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mechichi, T., E. Stackebrandt, N. Gad'on, and G. Fuchs. 2002. Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch. Microbiol. 178:26-35. [DOI] [PubMed] [Google Scholar]
- 41.Peplies, J., F. O. Glöckner, and R. Amann. 2003. Optimization strategies for DNA microarray-based detection of bacteria with 16S rRNA-targeting oligonucleotide probes. Appl. Environ. Microbiol. 69:1397-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Purkhold, U., A. Pommering-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rabus, R., H. Wilkes, A. Schramm, G. Harms, A. Behrends, R. Amann, and F. Widdel. 1999. Anaerobic utilization of alkylbenzenes and n-alkanes from crude oil in an enrichment culture of denitrifying bacteria affiliating with the beta-subclass of Proteobacteria. Environ. Microbiol. 1:145-157. [DOI] [PubMed] [Google Scholar]
- 44.Reinhold-Hurek, B., T. Hurek, M. Gillis, M. Hoste, M. Vancanneyt, K. Kersters, and J. De Ley. 1993. Azoarcus gen. nov., nitrogen fixing Proteobacteria associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth), and description of two species, Azoarcus indigens sp. nov. and Azoarcus communis sp. nov. Int. J. Syst. Bacteriol. 43:574-584. [Google Scholar]
- 45.Rosselló-Mora, R. A., M. Wagner, R. Amann, and K.-H. Schleifer. 1995. The abundance of Zoogloea ramigera in sewage treatment plants. Appl. Environ. Microbiol. 61:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. Jetten, J. W. Metzger, K. H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 48.Shinoda, Y., Y. Sakai, H. Uenishi, Y. Uchihashi, A. Hiraishi, H. Yukawa, H. Yurimoto, and N. Kato. 2004. Aerobic and anaerobic toluene degradation by a newly isolated denitrifying bacterium, Thauera sp. strain DNT-1. Appl. Environ. Microbiol. 70:1385-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song, B., N. J. Palleroni, L. J. Kerkhof, and M. M. Häggblom. 2001. Characterization of halobenzoate-degrading, denitrifying Azoarcus and Thauera isolates and description of Thauera chlorobenzoica sp. nov. Int. J. Syst. Evol. Microbiol. 51:589-602. [DOI] [PubMed] [Google Scholar]
- 51.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 52.Stamatakis, A. P., T. Ludwig, H. Meier, and M. J. Wolf. 2002. AxML: a fast program for sequential and parallel phylogenetic tree computations based on the maximum likelihood method, p. 21-28. In Proceedings of the 1st IEEE Bioinformatics Conference (CSB2002). IEEE Press, Piscataway, N.J. [PubMed]
- 53.Stralis-Pavese, N., A. Sessitsch, A. Weilharter, T. Reichenauer, J. Riesing, J. Csontos, J. C. Murrell, and L. Bodrossy. 2004. Optimization of diagnostic microarray for application in analysing landfill methanotroph communities under different plant covers. Environ. Microbiol. 6:347-363. [DOI] [PubMed] [Google Scholar]
- 54.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 55.Taroncher-Oldenburg, G., E. M. Griner, C. A. Francis, and B. B. Ward. 2003. Oligonucleotide microarray for the study of functional gene diversity in the nitrogen cycle in the environment. Appl. Environ. Microbiol. 69:1159-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urakawa, H., S. El Fantroussi, H. Smidt, J. C. Smoot, E. H. Tribou, J. J. Kelly, P. A. Noble, and D. A. Stahl. 2003. Optimization of single-base-pair mismatch discrimination in oligonucleotide microarrays. Appl. Environ. Microbiol. 69:2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vasiliskov, V. A., D. V. Prokopenko, and A. D. Mirzabekov. 2001. Parallel multiplex thermodynamic analysis of coaxial base stacking in DNA duplexes by oligodeoxyribonucleotide microchips. Nucleic Acids Res. 29:2303-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.von Wintzingerode, F., B. Selent, W. Hegemann, and U. B. Göbel. 1999. Phylogenetic analysis of an anaerobic, trichlorobenzene-transforming microbial consortium. Appl. Environ. Microbiol. 65:283-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wagner, M., and A. Loy. 2002. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 13:218-227. [DOI] [PubMed] [Google Scholar]
- 60.Wilson, K. H., W. J. Wilson, J. L. Radosevich, T. Z. DeSantis, V. S. Viswanathan, T. A. Kuczmarski, and G. L. Andersen. 2002. High-density microarray of small-subunit ribosomal DNA probes. Appl. Environ. Microbiol. 68:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu, L., D. K. Thompson, G. Li, R. A. Hurt, J. M. Tiedje, and J. Zhou. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou, J. 2003. Microarrays for bacterial detection and microbial community analysis. Curr. Opin. Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 63.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]