Abstract
Despite considerable interest in studying Burkholderia cepacia complex in the environment, we still do not have efficient methods to detect, isolate, and screen large numbers of B. cepacia isolates. To better describe the ecology and diversity of B. cepacia complex, a colony hybridization assay was developed to detect specifically all species of the complex based on polymorphism of the variable V3 region of the 16S rRNA sequence. The sensitivity of the assay was dramatically enhanced by using a probe consisting of three repeats of a B. cepacia complex-specific probe, each separated by a phosphoramidite spacer. In addition, a duplex PCR targeting B. cepacia complex-specific recA and 16S rRNA sequences was developed to enable a fast and reliable diagnostic assay for members of the complex. When applied to maize rhizosphere samples, colony hybridization results were in good agreement with those of most-probable-number duplex PCR, both indicating a >100-fold fluctuation of abundance between individual plants. Using restriction analysis of recA for a total of 285 confirmed isolates of the B. cepacia complex, up to seven B. cepacia complex species were identified; however, their diversity and abundance were not evenly distributed among individual plants, and several allelic variants were commonly found from the same rhizosphere sample. These results indicate that not only complex communities of B. cepacia complex species and closely related strains of the same species may coexist at high population levels but also species composition and abundance may dramatically vary between individual plants.
In the genus Burkholderia of the β subdivision of the proteobacteria, the Burkholderia cepacia complex consists of at least nine closely related Burkholderia species, namely, B. cepacia (formerly genomovar I), B. multivorans (II), B. cenocepacia (III), B. stabilis (IV), B. vietnamiensis (V), B. dolosa (VI), B. ambifaria (VII), B. anthina (VIII), and B. pyrrocinia (IX) (10, 11, 22, 49-53). They share moderate levels of DNA-DNA hybridization (30 to 50%) but high sequence similarity at the level of 16S rRNA (98 to 99%) and recA (94 to 95%) genes. Moreover, they all possess an unusually complex and plastic genome that consists of two to four replicons (chromosomes), which harbor several insertion sequences that may increase the frequency of genetic mutations and recombinations (7, 30). This may favor their adaptation to different environments, and it is not surprising that strains of the complex are found in habitats as diverse as soil, plant, and animal surfaces, rhizosphere, and water (9, 42).
This remarkable versatility is further exemplified by the wide array of properties they possess. Although first described as plant pathogens causing sour skin of onion (6), strains of the B. cepacia complex have also been reported to be effective biological control agents against soilborne (5, 25), foliar (28), and postharvest (27) plant pathogens, to promote plant growth (16, 48), and to decontaminate polluted soil or groundwater (29, 39). Despite the useful properties those organisms may offer to bioremediation or as an alternative to pesticides in agriculture, commercial application has been hampered by the risk B. cepacia complex may pose to human health. Indeed, in the last decade, strains of the complex have been found associated with nosocomial infection of immunocompromised patients (26) and chronic or rapid decline in pulmonary function in patients with cystic fibrosis (31).
To date, no clear distinction has been established between clinical and environmental isolates on the basis of phenotypic or genotypic criteria (1, 3, 34), and all B. cepacia complex species have been isolated from sputum cultures of cystic fibrosis patients (9). Moreover, the emergence of new strains each year in cystic fibrosis patients suggests the ongoing acquisition of strains from the environment (8, 32, 34).
Although B. cepacia complex strains deposited in culture collections have been sampled from various habitats, information concerning major reservoirs of B. cepacia complex in the environment are still lacking. Indeed, there are few studies concerning the ecology of the B. cepacia complex in the environment, and they deal with a few strains isolated sporadically or were done when the description of the complex was limited to one species or a few species (9, 42). Recently, several studies have examined the rhizospheres of trees (43) and crop plants (1, 3, 14, 19) as potential reservoirs for B. cepacia complex. Although most of the latter studies dealt with 50 to 100 isolates, a more thorough description of B. cepacia complex populations in the environment is needed, despite the laborious work of isolation and screening that this entails. The main issue, however, is not B. cepacia complex identification because molecular identification of B. cepacia complex species is made possible using specific primers targeting recA (37) and 16S rRNA (2, 33) sequences. Furthermore, recA restriction analysis has been shown to be a convenient tool to distinguish each of the species in the complex (37). The greater difficulty is in the way those isolates are arbitrarily selected on dilution plates prior to identification. Indeed, Miller et al. (41) reported the presence of B. cepacia complex in more than 82% of 91 urban soils as detected by culture-independent methods, while B. cepacia complex colonies could be isolated in only 15% of all soils investigated. This underlines the need for better screening techniques to recover B. cepacia complex even when present in a sample at low population levels.
The low isolate yield may be attributed to the fact that semiselective media, e.g., trypan blue tetracycline medium (TB-T) (24) or Pseudomonas cepacia azelaic acid tryptamine medium (PCAT) (4), were developed when the taxonomic description of the B. cepacia complex was not yet fully established. Although unavoidable biases exist when recovering B. cepacia complex on selective or semiselective media (46), it is the only way to obtain isolates in pure cultures for further phenotypic and genotypic characterizations and to better describe their ecology and diversity.
The objectives of this study were to develop and assess the efficiencies of molecular assays to determine the population density and diversity of culturable organisms of the B. cepacia complex from environmental samples. The methods include colony hybridization (CH) and duplex PCR combined with most-probable-number PCR (MPN-PCR). The methods were first developed and tested using a representative collection of reference B. cepacia complex and non-B. cepacia complex species and further validated and compared with each other by thoroughly characterizing B. cepacia complex populations from a heterogeneous habitat, i.e., the maize rhizosphere.
MATERIALS AND METHODS
Bacterial cultures and storage.
All reference B. cepacia complex and non-B. cepacia complex strains were selected from the collection of the Burkholderia cepacia Research Laboratory and Repository (at the University of Michigan, Ann Arbor). Twenty-four strains representing the nine B. cepacia complex species were used to validate the molecular techniques. In the following list, a strain name followed by a T and/or P superscript indicates that the strain is a species type strain and/or a strain included in previously published experimental strain panels (12, 36), respectively. Their designations in the American Type Culture Collection (ATCC) and the Laboratorium voor Microbiologie (LMG) (Ghent University, Ghent, Belgium) are provided when available. The reference B. cepacia complex and non-B. cepacia complex strains were B. cepacia genomovar I PC783PT (ATCC 25416, LMG 1222), HI2140P, and HI2284; B. multivorans HI229P (ATCC 17616, LMG 17588), HI2132P, and HI2240P; B. cenocepacia J2315TP (ATCC BAA-245, LMG 16656), HI3240P, and HI3248P; B. stabilis HI2210TP (ATCC BAA-67, LMG 14294) and AU0244; B. vietnamiensis PC259P (LMG 18835, CEP40, PC259), AU1344, and HI2278P; B. dolosa AU0645TP (LMG 18943, R-5670), AU0158, and HI2238; B. ambifaria AMMDTP (ATCC BAA-244, LMG 19182), AU0212 (LMG 19466), and HI2474P; B. anthina AU1293P (LMG 21821) and HI2725P; and B. pyrrocinia BC011P (LMG 21823) and HI2710P. Negative controls consisted of six strains that were not B. cepacia complex strains but were closely related to them, i.e., Burkholderia gladioli AU3923, AU5002, AU5085, and HI2870; Burkholderia xenovorans LB400T (LMG 21463) (23); and Ralstonia pickettii PKO1 (21). Reference strains and rhizosphere isolates were routinely grown at 29°C on plate count agar (Difco Laboratories, Detroit, Mich.) and stored at −80°C in Luria-Bertani broth medium (Difco) containing 30% glycerol.
Duplex PCR.
B. cepacia complex-specific primers targeting 16S rRNA and recA (Table 1) were synthesized by Integrated DNA Technologies (Coralville, Iowa). Using Primer3 software (44), a new forward primer (i.e., Burkf) was designed to amplify specifically 16S rRNA sequences of B. cepacia complex based on a CLUSTAL W (47) sequence alignment of B. cepacia complex and closely related genera and species.
TABLE 1.
B. cepacia complex-specific primers and probes
| Assay | Target | Primer or probea | Positionsb | Sequence (5′ → 3′)c | Length (no. of bases) | Reference |
|---|---|---|---|---|---|---|
| PCR | 16S rRNA gene | Burkf (f) | 133-153 | GGCGAAAGCCGGATTAATACC | 21 | This study |
| CeMuVi-16-2 (r) | 452-435 | CCGRCTGTATTAGAGCCA | 18 | 2 | ||
| recA | BCR1 (f) | 2-20 | TGACCGCCGAGAAGAGCAA | 19 | 37 | |
| BCR2 (r) | 1044-1024 | CTCTTCTTCGTCCATCGCCTC | 21 | 37 | ||
| CH | 16S rRNA gene | V3-30 | 460-431 | GTCATCCCCCGGCTGTATTAGAGCCAAGGA | 30 | This study |
| V3x2 | NA | GTCATCCCCCGGCTGTATTAGAGCCAAGGA/SpC3/GTCATCCCCCGGCTGTATTAGAGCCAAGGA | 60 | This study | ||
| V3x3 | NA | GTCATCCCCCGGCTGTATTAGAGCCAAGGA/SpC3/GTCATCCCCCGGCTGTATTAGAGCCAAGGA/SpC3/GTCATCCCCCGGCTGTATTAGAGCCAAGGA | 90 | This study |
(f), forward; (r), reverse.
For 16S rRNA gene-based assays, positions in the sequence of B. multivorans LMG 13010T (GenBank accession number Y18703) are shown. For recA-based assays, positions in the sequence of B. multivorans ATCC 17616 (U70431) are shown. The first coordinate corresponds to the 5′ end of the oligonucleotide sequence. NA, not applicable.
Wobble base, R = A or G; /SpC3/, hydrophilic C3 phosphoramidite spacer.
Each DNA preparation consisted of 1-μl loopful of a colony grown overnight that was lysed in 4 μl of lysis solution (50 mM KCl, 0.1% Tween 20, 10 mM Tris-HCl [pH 8.3]) at 99°C for 10 min. Sixteen microliters of PCR cocktail was subsequently added to the cell lysate. The PCR cocktail consisted of 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 9.0], 0.1% Triton X-100, 1.5 mM MgCl2; Promega Corp., Madison, Wis.), bovine serum albumin (0.5 g liter−1; Sigma, St. Louis, Mo.), 5% dimethyl sulfoxide (Sigma), 200 μM concentrations of the four deoxynucloside triphosphates (Promega), a 0.50 μM concentration of each primer targeting 16S rRNA (i.e., Burkf and CeMuVi-16-2), a 0.40 μM concentration of each primer targeting recA (i.e., BCR1 and BCR2), and 1.5 U of Taq DNA polymerase (Promega). After an initial denaturation step (3 min at 94°C), 30 PCR cycles (1 cycle consisting of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C) and a final extension step (10 min at 72°C) were used. PCR products were electrophoresed on 1.5% agarose gels in 1× Tris-acetate-EDTA (TAE) and stained with ethidium bromide by standard protocols (45). A 100-bp ladder (Invitrogen Corp., Carlsbad, Calif.) was used as molecular size markers.
recA RFLP.
PCR cocktail composition was as described above, except that only primers BCR1 and BCR2 (0.40 μM each) were used. After PCR amplification, 5 μl of amplified product was used for restriction analysis with 2 U of either HaeIII or MnlI (New England Biolabs, Inc., Beverly, Mass.) in a total volume of 15 μl. After 3-h digestion at 37°C, restriction fragments were separated by electrophoresis on 2.5% agarose gels by standard procedures (45). A 100-bp ladder and a 50-bp ladder (Invitrogen) were used as molecular size markers. Restriction analysis was generally done once and was repeated if restriction patterns could not be assigned to any of the reference patterns, i.e., those obtained by recA restriction fragment length polymorphism (RFLP) with reference B. cepacia complex strains of the nine B. cepacia complex species.
16S rRNA and recA sequencing.
To determine 16S rRNA sequences, PCR products were purified with the Qiaquick PCR purification kit (QIAGEN, Inc., Valencia, Calif.), and both strands were sequenced using universal eubacterial primers. Alternatively, when specific PCR products could not be obtained, PCR amplicons were first cloned with TOPO TA cloning kit for sequencing (Invitrogen) and then sequenced using Taq dye terminator chemistry and an ABI cycle sequencer (Genomic Technology Support Facility, Michigan State University). Both strands of recA sequences were sequenced using PCR primers BCR1, BCR2, BCR3, and BCR4 (37).
Phylogenetic reconstruction.
Sequences determined in this study were used to search for similarities among known sequences using the basic local alignment tool (BLAST) at the National Center for Biotechnology Information (Bethesda, Md.), and SEQUENCE MATCH of the Ribosomal Database Project. Multiple sequences yielding the highest similarity scores were aligned using CLUSTAL W, and sites presenting gaps or unknown bases were excluded from analysis.
Evolutionary distances were calculated according to the two-parameter Kimura model, and phylogenies were reconstructed on the basis of the neighbor-joining method of the Phylogeny Inference Package (PHYLIP) (17). Input order was randomized 5 to 10 times in all phylogenetic reconstructions. Nodal support of the inferred trees was assessed by 1,000 bootstrap replicates. DNApars in the PHYLIP package was also used to perform ordinary parsimony analyses using exhaustive search, and nodal support was assessed with 100 bootstrap replicates to confirm the phylogenetic positioning of the clades.
Isolation from rhizospheric soil.
Maize rhizosphere samples were obtained in mid-July, 2003, from the Michigan State University, W. K. Kellogg Biological Station Long-Term Ecological Research (KBS) site (Hickory Maizeers, Mich.) (http://lter.kbs.msu.edu). The maize field was managed under maize monoculture (one annual crop with no cover crop), with conventional tillage, and without application of fertilizers, herbicides, or insecticides. Entire root systems were sampled at 5, 10, 20, 120, 150, and 160 m from the first sample, and they were assigned sample number 2, 3, 4, 5, 6, and 7, respectively (Table 2). For each sample, 1 g of root system with closely adhering soil (rhizospheric soil) was transferred to a sterile tube containing 10 ml of 0.85% NaCl solution, and the tube was shaken at 350 rpm at room temperature for 30 min (Fig. 1A). The bacterial suspension was diluted 10-fold in 0.85% NaCl, and 100 μl of each dilution was plated in triplicate on B. cepacia complex semiselective medium trypan blue tetracycline medium (TB-T) (24), which contains (per liter) 2 g of glucose, 1 g of l-asparagine, 1 g of NaHCO3, 0.5 g of KH2PO4, 0.1 g of MgSO4 · 7H2O, 20 g of agar, 5 mg of crystal violet, and 50 mg of trypan blue. The pH was adjusted to 5.5, and 20 mg of tetracycline and 100 mg of cycloheximide were added after autoclaving. TB-T plates were incubated at 29°C for 4 days prior to counting the number of CFU. TB-T was chosen over other B. cepacia complex semiselective media, because all known B. cepacia complex species can grow on it, it yields more B. cepacia complex isolates than other media (41; our unpublished results), and it was formulated for environmental samples (24).
TABLE 2.
Population density, species abundance, and diversity of B. cepacia complex strains from maize rhizosphere samples
| Sample or parameter | Population level [log10 (CFU/g)]a
|
No. of isolates of B. cepacia complex speciesb:
|
Total no. of isolates | Sc | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | B. cepacia complex | I
|
II | III | IV | VI | VII
|
IX | ||||||||
| A | B | A | B | C | D | E | ||||||||||
| Plant 1 | 5.90 (0.07) | 5.41 | 2 | 19 | 3 | 6 | 2 | 30 | 2 | |||||||
| Plant 2 | 6.48 (0.14) | 5.81 | 20 | 1 | 3 | 3 | 1 | 2 | 30 | 3 | ||||||
| Plant 3 | 6.57 (0.08) | 6.51 | 1 | 35 | 3 | 5 | 1 | 2 | 1 | 1 | 1 | 1 | 51 | 6 | ||
| Plant 4 | 7.02 (0.08) | 6.72 | 13 | 6 | 6 | 3 | 28 | 1 | ||||||||
| Plant 5 | 5.86 (0.10) | 5.83 | 5 | 46 | 5 | 56 | 1 | |||||||||
| Plant 6 | 5.15 (0.04) | 4.58 | 2 | 7 | 8 | 1 | 18 | 3 | ||||||||
| Plant 7 | 4.76 (0.06) | 4.61 | 1 | 19 | 27 | 3 | 20 | 70 | 3 | |||||||
| Total abundance and richness | 22 | 36 | 3 | 9 | 1 | 26 | 75 | 10 | 60 | 36 | 2 | 3 | 285 | 7 | ||
| Mean (SD) | 5.96 (0.80) | 5.64 (0.84) | 40 (18.7) | 2.7 (1.7) | ||||||||||||
| CVd (%) | 14 | 15 | 47 | 63 | ||||||||||||
Total population levels were determined by plating on TB-T plates, and B. cepacia complex abundance was determined by CH, followed by confirmatory duplex PCR. Standard deviations of three replicate measurements per plant are indicated in parentheses.
The B. cepacia complex species found were B. cepacia (formerly genomovar I), B. multivorans (II), B. cenocepacia (III), B. stabilis (IV), B. dolosa (VI), B. ambifaria (VII), and B. pyrrocinia (IX). This classification is based on the results of recA RFLP analysis and was confirmed by sequencing. Different recA alleles are indicated by letters (A, B, C, and D) that correspond to the sequences of representative isolates KBC-1 (VII-E), KBC-2 (I-A), KBC-3 (I-B), KBC-4 (III), KBC-5 (VII-B), KBC-6 (VII-C), and KBC-7 (VII-D). The results of a phylogenetic analysis of those sequences are shown in Fig. 4. Amplicons yielding patterns that were similar to those of reference strains were not sequenced.
Observed species richness (S) was calculated by considering B. cepacia complex species and not the occurrence of multiple recA patterns within a species.
CV, coefficient of variation (standard deviation divided by the mean).
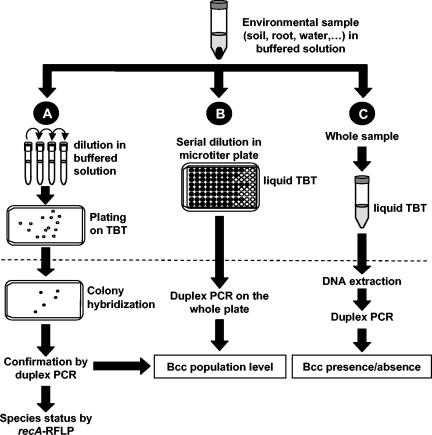
FIG. 1.
Determination of the presence of B. cepacia complex (Bcc) and B. cepacia complex population densities from environmental samples. (A) After the sample is shaken in buffered solution, it is serially diluted and plated on semiselective TB-T medium (TBT). Colonies are transferred to nylon membranes, and their DNA is hybridized with the probe V3x3 to localize B. cepacia complex candidates on dilution plates. The candidates are confirmed as B. cepacia complex members by performing duplex PCR targeting 16S rRNA gene and recA B. cepacia complex-specific sequences. (B) In parallel, the sample is diluted serially in liquid TB-T medium, and after 4 days of growth, an aliquot of each well is subjected to duplex PCR to detect whether B. cepacia complex cells are present at a given dilution. (C) When population levels are below the detection limit for strategies A and B [around 2 to 3 log (CFU/g)], the presence of B. cepacia complex in a sample can be assessed by enriching the sample for 4 days in liquid TB-T medium, extracting DNA from total cells, and testing by duplex B. cepacia complex-specific PCR. The dotted line represents the 4-day incubation period at 29°C for all methods.
DNA-DNA and colony hybridization.
A CLUSTAL W alignment of rRNA sequences of B. cepacia complex species and closely related organisms was inspected manually to identify putative B. cepacia complex-specific probes. Candidate probes were evaluated in silico using the Probe Match tool of the Ribosomal Database Project (38). A universal probe (for members of the domain Bacteria) served as a positive control for DNA fixation on the membranes. It consisted of a 510-bp fragment (produced with primers RHG-F and RHG-R) (33) that hybridized with all bacterial DNA spotted on the membrane. This probe was washed off the nylon membrane prior to hybridization with B. cepacia complex-specific probes.
For dot blot hybridization, full-length PCR-amplified 16S rRNA was spotted onto Hybond-N+ membranes (Amersham, Piscataway, N.J.), and fixed by UV cross-linking (Stratalinker; Stratagene, La Jolla, Calif.). For CH, colonies were grown at 29°C either overnight on PCAT or for 4 days on TB-T dilution plates. Prior to applying colonies to membranes, it was generally necessary to incubate the colonies overnight at 4°C to increase colony adhesiveness. For TB-T dilution plates, larger plates (Omnitray; Nalge Nunc International, Rochester, N.Y.) were used instead of round petri dishes to maximize distance between colonies. DNA from colonies adhering to the membranes was denatured and neutralized following the manufacturer's recommendations (Amersham) and fixed by UV cross-linking.
Oligoprobes were synthesized by Integrated DNA Technologies. They were diluted to a concentration of 1.25 μM and labeled according to the instructions for the Alkphos direct labeling kit (Amersham) for 30 min for fragment probes and for at least 2 h for oligoprobes. Prehybridization, hybridization, and posthybridization stringency washes were done at 40°C using the manufacturer's recommended buffers and protocols (Amersham). After signal generation in the presence of CDP-Star chemiluminescent substrate (Amersham), light emission was detected by autoradiography using film sensitive to blue light (Hyperfilm ECL; Amersham).
Prior to hybridizing the same membranes with a new probe, probes were removed by dipping membranes into boiling 0.5% (wt/vol) sodium dodecyl sulfate and allowing them to cool down to room temperature with gentle agitation. The absence of signal was checked by autoradiography before further hybridization with other probes.
DE-PCR and MPN-PCR.
A dilution endpoint PCR (DE-PCR) was performed by the method of McSpadden Gardener et al. (40) with the following modifications. Ten microliters of the initial rhizospheric wash suspension was diluted 10-fold six times in a 96-well microtiter plate with liquid TB-T as the diluent (Fig. 1B). Four replicate samples were prepared, which enabled the simultaneous analysis of four rhizospheric samples in each 96-well plate. Plates were incubated at 29°C with gentle shaking (150 rpm) for 4 days. Five microliters from each well was transferred to 95 μl of lysis buffer using a 12-channel pipetman, and cell lysis was performed as described above. Two microliters of the DNA lysate was used in 10-μl screening duplex PCR with the PCR cocktail composition and cycling conditions described above. Population levels were estimated on the basis of the last dilution in which a PCR product could be observed after electrophoresis on standard agarose gels. Based on the same PCR results, estimates of B. cepacia complex population levels were also calculated using the MPN method and its corresponding 95% confidence intervals (20). DE-PCR and MPN-PCR differ only in the way PCR results are used to calculate final population levels.
Enrichment and detection at low population levels.
Samples (1 g) were shaken in liquid TB-T medium 4 days at 29°C (Fig. 1C). Total DNA was extracted and purified using the UltraClean soil DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, Calif.). Duplex PCR was performed as described above using 4 μl of undiluted or 10-fold-diluted soil extracts for a total volume of 20 μl.
Statistical analyses.
B. cepacia complex population levels were determined at least twice, and representative results of one experiment are presented. Restriction analysis to determine B. cepacia complex species was done for one of those experiments. The numbers of CFU expressed per gram of fresh roots were log10 transformed (35) prior to statistical analysis. Analysis of variance, regression analysis followed by Bonferroni's two-tailed significance test, and nonparametric tests were done using SYSTAT (version 8.0; SPSS, Inc., Chicago, Ill.), and a P value of <0.05 was considered statistically significant.
Nucleotide sequence accession numbers.
16S rRNA and recA sequences have been deposited in the GenBank and EMBL databases under accession numbers AY769902 to AY769913 for 16S rRNA sequences and AY769895 to AY769901 for recA sequences.
RESULTS AND DISCUSSION
Duplex PCR.
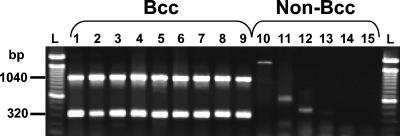
In order to determine quickly and reliably whether an isolate belongs to the B. cepacia complex, a duplex PCR assay was designed to specifically differentiate B. cepacia complex from closely related bacteria. Existing primers and a new forward primer targeting 16S rRNA sequences (Table 1) were combined to enable the simultaneous amplification of recA (1,040 bp) and 16S rRNA (320 bp) in only B. cepacia complex species (Fig. 2). All reference B. cepacia complex strains used in this study produced the double band, whereas closely related genera and species did not. The Burkf primer was designed to provide more specificity to the 16S rRNA gene-based PCR assay than the combination of eubacterial primer Eub-16-1 with the B. cepacia complex-specific CeMuVi-16-2 (2, 41). Initially, the two primer pairs were mixed at equal concentrations, but this sometimes produced a weaker signal for the 16S rRNA gene band than for the recA band. Comparable band intensities were obtained by using 25% more of the 16S rRNA gene primers than of the recA primers. As a result, a reliable and efficient method to identify all B. cepacia complex species was obtained, eliminating closely related isolates that could produce weak, unspecific fragments (e.g., Fig. 2, lanes 10 to 12). This was particularly valuable for high-throughput PCR screening and rapid identification of environmental isolates displaying the characteristic double band (data not shown).
FIG. 2.
B. cepacia complex-specific duplex PCR. The B. cepacia complex-specific PCR yielded fragment sizes of 1,040 and 320 bp, corresponding to 16S rRNA and recA amplicons, respectively. Representative strains of the nine B. cepacia complex (Bcc) species are shown in lanes 1 to 9. Lanes 1 to 9, B. cepacia PC783, B. multivorans HI229, B. cenocepacia J2315, B. stabilis HI2210, B. vietnamiensis PC259, B. dolosa AU0645, B. ambifaria AMMD, B. anthina AU1293, and B. pyrrocinia BC011, respectively. Lanes 10 to 13, B. gladioli strains AU3923, AU5002, AU5085, and HI2870, respectively; lane 14, B. xenovorans LB400; lane 15, R. pickettii PKO1. L, 100-bp ladder.
Specificity and sensitivity of B. cepacia complex-specific probes.
We identified the variable V3 region of the 16S rRNA sequence (positions 430 to 457 in B. multivorans LMG 13010T; accession number Y18703) as the best candidate to differentiate B. cepacia complex species from closely related species and genera, such as B. gladioli, Ralstonia spp., Chryseobacterium meningosepticum, Stenotrophomonas maltophilia, Comamonas acidovorans, and Pseudomonas spp. These bacteria are often found in mixed cultures with B. cepacia complex isolates from either environmental or clinical samples, often have similar growth requirements as B. cepacia complex species, and thus, may often be misidentified as B. cepacia complex species (9). In the B. cepacia complex, sequence polymorphism in the V3 region consisted of two or three mismatches distantly located from each other, whereas more than seven mismatches (i.e., with B. gladioli) were found for non-B. cepacia complex species. Generally, about 10 mismatches were obtained in the genus Burkholderia, with the exception of Burkholderia andropogonis, which displayed 15 mismatches. Other closely related genera listed above displayed either 15 to 20 mismatches or complete deletions (e.g., Chryseobacterium meningosepticum with gaps at seven sites [data not shown]).
Several probes ranging from 30 to 150 bases long, spanning the V3 region of the 16S rRNA, were tested for specificity using PCR-amplified DNA from B. cepacia complex and closely related organisms. Non-B. cepacia complex strains were chosen on the basis of the high similarities of their rRNA gene sequences, since they would be more difficult to differentiate from the B. cepacia complex, and consisted of B. gladioli, B. xenovorans, and Ralstonia spp.
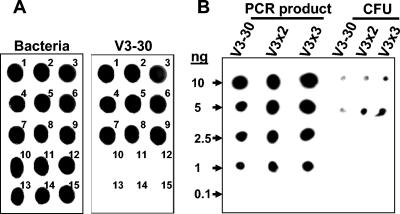
Only probe V3-30 specifically detected all B. cepacia complex species (Fig. 3A), while longer probes lacked specificity (data not shown). However, the hybridization assay using probe V3-30 was not sensitive enough to satisfactorily detect small colonies growing on dilution plates (Fig. 3B), for which a weak signal was often difficult to distinguish from the low level of background hybridization signal.
FIG. 3.
Specificity and sensitivity of the CH assay. (A) The specificity of the V3-30 probe was tested against all reference strains. 16S rRNA was amplified by PCR and fixed to the membranes as described in Materials and Methods. DNA fixation on the membranes was controlled by using a universal probe (for members of the domain Bacteria) that was subsequently washed off the membrane before hybridization with different B. cepacia complex-specific probes. Representative strains of each of the nine B. cepacia complex species are numbered 1 to 9, with the numbers 1 to 9 corresponding to B. cepacia PC783, B. multivorans HI229, B. cenocepacia J2315, B. stabilis HI2210, B. vietnamiensis PC259, B. dolosa AU0645, B. ambifaria AMMD, B. anthina AU1293, and B. pyrrocinia BC011, respectively. The numbers 10 to 13 indicate B. gladioli strains AU3923, AU5002, AU5085, and HI2870, respectively. The numbers 14 and 15 indicate B. xenovorans LB400 and Ralstonia pickettii PKO1, respectively. (B) Comparison of the sensitivities of probes V3-30 and its corresponding dimer (V3x2) and trimer (V3x3 [Table 1]) for different amounts of PCR-amplified 16S rRNA of B. cepacia LMG 1222 strain (left) and for a minimal number of colonies (CFU) deposited on the membrane with a toothpick (right).
Longer probes were designed by linking together several copies of the V3-30 probe using one or two phosphoramidite spacers (Table 1), because signal intensity was directly related to probe length due to probe labeling with alkaline phosphatase, but probes of increased sequence length were found to lack specificity. The sensitivities of dimer (V3x2) and trimer (V3x3) probes (Table 1) were substantially higher than that of V3-30, especially when working with colonies (Fig. 3B), without losing the specificity of the monomer (data not shown). Generally, the limit of detection of the CH assay using V3x3 ranged from 0.1 to 1 ng, and the limit of detection of the assay using V3-30 or V3x2 was >1 ng (Fig. 3B; our unpublished data).
The difference in the sensitivities of the V3-30 and V3x3 probes was further confirmed by a preliminary experiment in which 1,100 isolates from a maize rhizosphere sample were randomly selected from TB-T dilution plates, transferred to PCAT plates, and submitted to CH with either V3-30 or V3x3. Among the 700 isolates that were identified as B. cepacia complex strains by using V3-30, only 78% were confirmed by duplex PCR. In comparison, 99% of the 548 colonies identified as B. cepacia complex with V3x3 were duplex PCR positive. This can be explained by the weaker hybridization signals that are generated with V3-30 compared with those of V3x3; hence, when using V3-30, more doubtful colonies had to be picked on the plates to be screened by duplex PCR. In conclusion, probe V3x3 was the best probe in terms of both specificity and sensitivity and was used in subsequent studies on B. cepacia complex population densities in maize rhizospheres.
Population levels in the maize rhizosphere.
Seven individual rhizosphere samples were used to determine the B. cepacia complex population densities, species abundance, and diversity in the same field. The number of total bacteria growing on TB-T plates was high, with an average of 6.01 ± 0.83 log (CFU/g of root) (mean ± standard deviation) and with noticeable differences between maize plants (e.g., plants 4 and 7 [Table 2]). CH revealed that B. cepacia complex population levels were generally found within 0.5 to 1 log unit below the total number of bacteria growing on TB-T plates, with an average of 5.64 ± 0.84 log (CFU/g of root), very close to the estimates previously found in Italy on maize with 6.20 ± 0.20 log (CFU/g of root) (15). As for total counts on TB-T, B. cepacia complex colonization was found to vary by several orders of magnitude between individual rhizospheres (e.g., plants 4 and 6, with a 140-fold difference). Experimental repeatability that consisted of sampling the same root system at different times ranged from 0.5 to 1.5 log units, depending on the rhizosphere sample considered (data not shown). The percentage of B. cepacia complex isolates recovered from TB-T plates ranged from 17 to 81, which led to the isolation of about 40 ± 18 isolates per rhizosphere for a total of 285 isolates (Table 2).
Restriction analysis of recA amplicons with two restriction enzymes (HaeIII and MnlI [37]) for all B. cepacia complex strains identified by CH was used to determine species abundance and richness. Samples giving patterns that were not identical to those of reference strains were sequenced further to ascertain the correct species (Table 2 and Fig. 4).
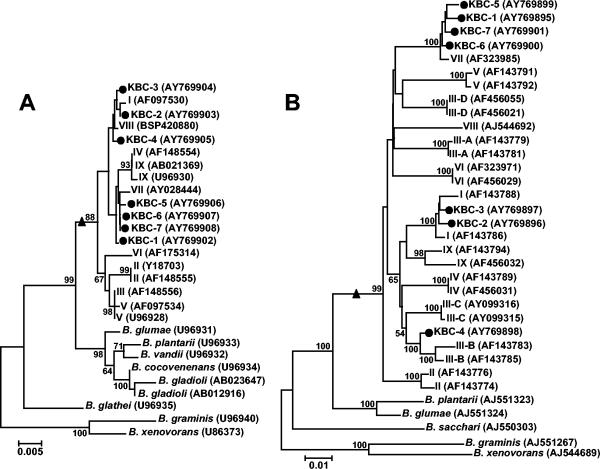
FIG. 4.
Phylogenetic positions of representative isolates of the maize field based on their 16S rRNA gene sequences (A) and recA sequences (B). Phylogenetic trees were generated by the neighbor-joining method based on the two-parameter Kimura correction of evolutionary distances. For both phylogenies, nodal support was assessed by 1,000 bootstrap replicates. Support values lower than 50% are not indicated. The number of substitutions per site is indicated by the bar at the bottom left of each phylogeny. Sequences determined in this study are indicated by a solid black circle (i.e., KBC-1 to -7). For presentation purposes, roman numbers were used to designate B. cepacia complex species and correspond to B. cepacia (formerly genomovar I), B. multivorans (II), B. cenocepacia (III), B. stabilis (IV), B. vietnamiensis (V), B. dolosa (VI), B. ambifaria (VII), B. anthina (VIII), and B. pyrrocinia (IX). Different recA lineages in B. cenocepacia (genomovar III) (37, 48) are indicated by capital letters A, B, C, and D after a hyphen in the genomovar designation. GenBank accession numbers of the sequences are given in parentheses. For each phylogeny, the B. cepacia complex monophyletic clade is indicated by a solid black triangle.
Sequencing of the recA and 16S rRNA gene amplicons was performed for seven randomly selected isolates belonging to the main recA types and allelic variants to confirm that they were B. cepacia complex isolates and determine their phylogenetic positions. The 16S rRNA gene-based phylogeny indicated that all selected isolates were indeed B. cepacia complex isolates (Fig. 4A). They clustered with B. ambifaria (KBC-1, -5, -6, and -7), B. cepacia (formerly genomovar I) (KBC-2 and -3) or close to but separately from B. anthina (genomovar VIII) (KBC-4) (Fig. 4A).
The phylogenetic positions of the isolates in the B. cepacia complex were further confirmed by the analysis of recA sequences (Fig. 4B). Noticeably, while all selected isolates clustered similarly when recA or 16S rRNA gene sequences were considered, KBC-4 was found between B. cenocepacia III-B and III-C clusters in the recA-based phylogeny, with significant nodal support (Fig. 4B) for this topology. The existence of low bootstrap values between B. cepacia complex species (e.g., between clusters III, V, VI, and VII [Fig. 4B]) may suggest that nucleotide substitutions at this locus could display some phylogenetic saturation (our unpublished data).
As a whole, one to seven B. cepacia complex species were recovered on each plant rhizosphere with B. ambifaria (formerly genomovar VII) found in all samples (i.e., 64% of all isolates), followed by B. cepacia (genomovar I) with 58 isolates (i.e., 20% of the total number) found on three plants (Table 2). The other species consisted of B. multivorans (genomovar II) with three isolates found on one plant, B. cenocepacia (genomovar III) with nine isolates found on three plants, B. stabilis (genomovar IV) with a unique isolate, B. dolosa (genomovar VI) found on two plants for a total of 26 isolates, and B. pyrrocinia (genomovar IX) found on two plants for a total of three isolates. To our knowledge, this is the first report of B. stabilis and B. dolosa isolated from the environment. Noteworthy, B. multivorans and B. cenocepacia, although rarely evidenced in our study, are the most frequent species isolated in patients with cystic fibrosis. Indeed, those two species were found in up to 38 and 50%, respectively, of patients with cystic fibrosis in the United States, accounting for mortality rates of up to 15 and 45%, respectively, in cystic fibrosis patients in Canada (42).
Interestingly, in the dominant species, substantial allelic polymorphism was evidenced for recA with two and four alleles for B. cepacia (genomovar I) and B. ambifaria, respectively. This allelic polymorphism was also observed for populations from the same sample, e.g., two recA variants found in genomovar I and three to four recA variants of B. ambifaria on plants 2 and 3 (Table 2).
If the abundance of each species is expressed in log10 so as to identify significant difference in species abundance in a given sample, it becomes clear that although up to six B. cepacia complex species may coexist on a given maize rhizosphere (e.g., plant 3 [Table 2]), each plant is colonized by a dominant species and/or one variant type. For instance, plant 1 is dominated by B. ambifaria variant A, while other recA variants were three to six times less numerous (Table 2). The only case where abundance was similar between species in a given sample was for plant 6 with seven to nine isolates for both B. dolosa and B. ambifaria. Note that no direct relationship was found between the location of the sample and the total population density of B. cepacia complex strains, species distribution, and abundance (P > 0.05 by regression [Table 2]).
In summary, our results indicate that in each maize rhizosphere, a complex community of B. cepacia complex species and variants of the same species coexist at high population densities. Since recA restriction analysis is rather a coarse method to resolve intraspecific diversity, further studies using DNA fingerprinting may prove very valuable to reveal the level of genetic differentiation of B. cepacia complex species on each individual plant.
Previous studies that examined B. cepacia complex population levels in maize rhizospheres reported that B. cenocepacia and B. ambifaria were predominant, while B. cepacia and B. pyrrocinia were found less frequently (3, 13, 19). This was partly corroborated here because B. ambifaria and B. cepacia were the most dominant in our study, whereas B. cenocepacia and B. pyrrocinia were less frequently isolated (Table 2). Furthermore, species not yet encountered on maize, such as B. multivorans and B. stabilis, were also found at significant population densities at the KBS site (Table 2). Therefore, without higher capacity methods, such as proposed here, sampling bias may distort our understanding of the diversity and ecology of B. cepacia complex strains, which may underestimate species distribution and abundance, and hence could potentially miss the identification of true environmental reservoirs.
rRNA sequencing was also done for five randomly chosen isolates that were not identified as B. cepacia complex isolates by any of the methods used in this study but that were often present at high densities on TB-T dilution plates. These organisms belonged to the α, β, and γ subdivisions of the proteobacteria. In the α subdivision, one isolate (S8 [accession number AY769909]) showed 70 to 90% identity of its rRNA sequence with Rhizobium or Agrobacterium. In the β subdivision, isolates S9 (accession number AY769910) and S12 (AY769913) displayed 98% sequence similarity with Burkholderia spp. and Variovorax spp., respectively. Two isolates belonged to the γ subdivision (S10 [accession number AY769911] and S11 [AY769912]) and shared more than 97% identity of their 16S rRNA sequences with Lysobacter spp. and Xanthomonas spp., respectively. The diversity of organisms growing on semiselective media in company with B. cepacia complex strains further underlines the necessity of using an efficient and specific screening strategy to recover B. cepacia complex organisms from environmental samples.
Determination of B. cepacia complex population levels by DE- and MPN-PCR.
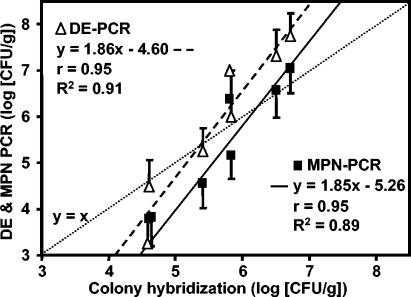
In parallel to the detection by CH, a liquid culture screening assay was developed to determine B. cepacia complex population levels (Fig. 1B). CH gave the best estimate, since all dilution plates are screened, and positive isolates are further confirmed as B. cepacia complex by duplex PCR, leading to an accurate counting of B. cepacia complex at each dilution level. Liquid culture PCR screening assays gave results that showed good agreement with the results of CH (Fig. 5). Both DE-PCR and MPN-PCR indicated a strong linear relationship (r > 0.90, P < 0.001, R2 = 0.90) with CH estimates of population levels (Fig. 5). DE-PCR and CH gave the same results, ranging from 4.5 to 6.5 log (CFU/g), whereas above 6.5 log (CFU/g), DE-PCR overestimated the abundance of B. cepacia complex. For MPN-PCR, good estimates of population levels were obtained over the range 5.5 to 7 log (CFU/g) with an underestimation below 5.5 log (CFU/g). As a consequence, the three methods agreed with each other in the range 5 to 5.5 log (CFU/g) to 6.5 to 7 log (CFU/g). Below 4.5 to 5 log (CFU/g of sample), liquid culture PCR screening methods gave lower estimates of B. cepacia complex abundance than those of CH. These results are supported by previous studies reporting that MPN counts generally underestimate the actual number of bacteria in a sample (18). Therefore, a quick estimation of B. cepacia complex population abundance can be obtained using liquid culture screening methods in order to decide whether isolation should be performed by randomly picking isolates or by CH in the case of low B. cepacia complex population levels.
FIG. 5.
Comparison of DE-PCR, MPN-PCR, and CH methods to estimate the densities of environmental B. cepacia complex populations isolated from seven independent maize rhizospheres in the same field. CH was done using probe V3x3 on TB-T dilution plates. All positive colonies were confirmed as B. cepacia complex by duplex PCR and sequencing for representative isolates (Fig. 4 and Table 2). B. cepacia complex population levels were estimated by DE-PCR and MPN-PCR as described in Materials and Methods. Error bars indicate standard deviations for DE-PCR and 95% confidence intervals for MPN-PCR (both based on four measurements per sample). Regression equations, Pearson correlation coefficients (r), and the coefficients of determination (R2) are indicated for each PCR-based method. The theoretical line representing the same population estimate by PCR-based methods and CH is depicted by a dotted line (y = x).
The actual detection limit of liquid culture PCR screening is 1,000 CFU/g, given that only 10 μl of the initial 10 ml of sample is inoculated into the wells of the dilution plate (Fig. 1B). For CH, the detection limit is 10-fold lower (i.e., 100 CFU/g), since it is possible to detect one B. cepacia complex colony on the semiselective medium plate corresponding to the lowest dilution of the sample suspension. Because not finding B. cepacia complex by these methods cannot definitely rule out their presence at population levels below 100 CFU/g (i.e., what is not detected by CH), incubation of the sample in liquid TB-T medium for 4 days prior to plating on TB-T plates (Fig. 1C) can be used to enrich as few as 1 to 10 cells in the original sample to levels detectable by duplex PCR after DNA extraction of the whole sample (our unpublished results). If the sample gave positive results by PCR, screening of dilution plates by CH may then be successfully used to isolate B. cepacia complex. However, due to the enrichment step, a reliable estimate of the actual population level in the sample is no longer possible. This enrichment step can prove valuable in the case of bulk soil in which B. cepacia complex abundance may range from 102 to 104 CFU/g of soil (24) or even lower (our unpublished results).
In conclusion, a combination of the methods presented here offers efficiency and flexibility, depending on the specific aims of each study. For instance, enrichment and duplex PCR may be used to screen numerous samples in parallel for just the presence or absence of the B. cepacia complex, liquid culture PCR screening assay may estimate B. cepacia complex abundance, and CH provides the best means of isolating the target organisms in pure cultures. Another variation would be to perform CH on the bacterial suspension from each well of liquid culture plates (Fig. 1B). Indeed, by directly spotting liquid cultures from several plates on hybridization membranes, determining B. cepacia complex population densities in tens of samples in parallel will no long be an obstacle. This will undoubtedly yield more data about the ecology and diversity of the B. cepacia complex in various habitats and environments.
Acknowledgments
We thank Melissa S. Meyers for excellent technical assistance and Andrew T. Corbin for help in sampling at the KBS site.
This work was supported in part by NSF grant no. DEB-00755564, a postdoctoral fellowship from the Swiss National Science Foundation to A.R., and a grant from the Cystic Fibrosis Foundation to J.J.L.
REFERENCES
- 1.Balandreau, J., V. Viallard, B. Cournoyer, T. Coenye, S. Laevens, and P. Vandamme. 2001. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 67:982-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1999. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J. Clin. Microbiol. 37:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevivino, A., C. Dalmastri, S. Tabacchioni, L. Chiarini, M. L. Belli, S. Piana, A. Materazzo, P. Vandamme, and G. Manno. 2002. Burkholderia cepacia complex bacteria from clinical and environmental sources in Italy: genomovar status and distribution of traits related to virulence and transmissibility. J. Clin. Microbiol. 40:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burbage, D. A., and M. Sasser. 1982. A medium selective for Pseudomonas cepacia. Phythopathology 72:706. (abstract) [Google Scholar]
- 5.Burkhead, K. D., D. A. Schisler, and P. J. Slininger. 1994. Pyrrolnitrin production by biological control agent Pseudomonas cepacia B37w in culture and in colonized wounds of potatoes. Appl. Environ. Microbiol. 60:2031-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 7.Cheng, H. P., and T. G. Lessie. 1994. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J. Bacteriol. 176:4034-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coenye, T., and J. J. LiPuma. 2003. Molecular epidemiology of Burkholderia species. Front. Biosci. 8:e55-e67. [DOI] [PubMed] [Google Scholar]
- 9.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 10.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Bacteriol. 5:1481-1490. [DOI] [PubMed] [Google Scholar]
- 11.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroucke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 12.Coenye, T., P. Vandamme, J. J. LiPuma, J. R. Govan, and E. Mahenthiralingam. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 41:2797-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalmastri, C., A. Fiore, C. Alisi, A. Bevivino, S. Tabacchioni, G. Giuliano, A. R. Sprocati, L. Segre, E. Mahenthiralingam, L. Chiarini, and P. Vandamme. 2003. A rhizospheric Burkholderia cepacia complex population: genotypic and phenotypic diversity of Burkholderia cenocepacia and Burkholderia ambifaria. FEMS Microbiol. Ecol. 46:179-187. [DOI] [PubMed] [Google Scholar]
- 14.Dalmastri, C., L. Chiarini, C. Cantale, A. Bevivino, and S. Tabacchioni. 1999. Soil type and maize cultivar affect the genetic diversity of maize root-associated Burkholderia cepacia populations. Microb. Ecol. 38:274-283. [DOI] [PubMed] [Google Scholar]
- 15.Di Cello, F., A. Bevivino, L. Chiarini, R. Fani, D. Paffetti, S. Tabacchioni, and C. Dalmastri. 1997. Biodiversity of a Burkholderia cepacia population isolated from the maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 63:4485-4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estrada-De Los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1993. PHYLIP (Phylogenetic Inference Package) and manual, version 3.5c. Department of Genetics, University of Washington, Seattle.
- 18.Feray, C., B. Volat, V. Degrange, A. Clays Josserand, and B. Montuelle. 1999. Assessment of three methods for detection and quantification of nitrite-oxidizing bacteria and Nitrobacter in freshwater sediments (MPN-PCR, MPN-Griess, immunofluorescence). Microb. Ecol. 37:208-217. [DOI] [PubMed] [Google Scholar]
- 19.Fiore, A., S. Laevens, A. Bevivino, C. Dalmastri, S. Tabbacchioni, P. Vandamme, and L. Chiarini. 2001. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 3:137-143. [DOI] [PubMed] [Google Scholar]
- 20.Garthright, W. G., and R. J. Blodgett. 2003. FDA's preferred MPN methods for standard, large or unusual tests, with a spreadsheet. Food Microbiol. 20:439-445. [Google Scholar]
- 21.Gibson, T. L., A. S. Abdul, and R. H. Olsen. 1988. Microbial degradation of aromatic hydrocarbons in hydrogeologic material: microcosm studies, p. 53-69. In Proceedings of the Second National Outdoor Action Conference on Aquifer Restoration: Groundwater and Geophysical Methods, vol. 1. National Water Well Association, Dublin, Ohio.
- 22.Gillis, M., V. Tran Van, R. Bardin, M. Goor, P. Hebbar, A. Willems, P. Segers, K. Kersters, T. Heulin, and M. P. Fernandez. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45:274-289. [Google Scholar]
- 23.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 24.Hagedorn, C., W. D. Gould, T. R. Bardinelli, and D. R. Gustavson. 1987. A selective medium for enumeration and recovery of Pseudomonas cepacia biotypes from soil. Appl. Environ. Microbiol. 53:2265-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heungens, K., and J. L. Parke. 2000. Zoospore homing and infection events: effects of the biocontrol bacterium Burkholderia cepacia AMMDRI on two oomycete pathogens of pea (Pisum sativum L.). Appl. Environ. Microbiol. 66:5192-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes, A., R. Nolan, R. Taylor, R. Finley, M. Riley, R. Z. Jiang, S. Steinbach, and R. Goldstein. 1999. An epidemic of Burkholderia cepacia transmitted between patients with and without cystic fibrosis. J. Infect. Dis. 179:1197-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janisiewicz, W. J., and J. Roitman. 1988. Biological control of blue mold and gray mold on apple and pear with Pseudomonas cepacia. Phytopathology 78:1697-1700. [Google Scholar]
- 28.Knudsen, G. R., and H. W. Spurr, Jr. 1987. Field persistence and efficacy of five bacterial preparations for control of peanut leaf spot. Plant Dis. 71:442-445. [Google Scholar]
- 29.Leahy, J. G., A. M. Byrne, and R. H. Olsen. 1996. Comparison of factors influencing trichloroethylene degradation by toluene-oxidizing bacteria. Appl. Environ. Microbiol. 62:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lessie, T. G., W. Hendrickson, B. D. Manning, and R. Devereux. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol. Lett. 144:117-128. [DOI] [PubMed] [Google Scholar]
- 31.LiPuma, J. J. 2003. Burkholderia and emerging pathogens in cystic fibrosis. Semin. Respir. Crit. Care Med. 24:681-692. [DOI] [PubMed] [Google Scholar]
- 32.LiPuma, J. J. 1998. Burkholderia cepacia: management issues and new insights. Clin. Chest Med. 19:473-486. [DOI] [PubMed] [Google Scholar]
- 33.LiPuma, J. J., B. J. Dulaney, J. D. McMenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LiPuma, J. J., T. Spilker, T. Coenye, and C. F. Gonzalez. 2002. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 359:2002-2003. [DOI] [PubMed] [Google Scholar]
- 35.Loper, J. E., T. V. Suslow, and M. N. Schroth. 1984. Lognormal distribution of bacterial subpopulations in the rhizosphere. Phytopathology 74:1454-1460. [Google Scholar]
- 36.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massol-Deya, A., R. Weller, L. Rios-Hernandez, J. Z. Zhou, R. F. Hickey, and J. M. Tiedje. 1997. Succession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl. Environ. Microbiol. 63:270-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McSpadden Gardener, B. B., D. V. Mavrodi, L. S. Thomashow, and D. M. Weller. 2001. A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-DAPG-producing bacteria. Phytopathology 91:44-54. [DOI] [PubMed] [Google Scholar]
- 41.Miller, S. C., J. J. LiPuma, and J. L. Parke. 2002. Culture-based and non-growth-dependent detection of the Burkholderia cepacia complex in soil environments. Appl. Environ. Microbiol. 68:3750-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parke, J. L., and D. Gurian-Sherman. 2001. Diversity of the Burkholderia cepacia complex and implications for risk assessment of biological control strains. Annu. Rev. Phytopathol. 39:225-258. [DOI] [PubMed] [Google Scholar]
- 43.Richardson, J., D. E. Stead, J. G. Elphinstone, and R. H. Coutts. 2002. Diversity of Burkholderia isolates from woodland rhizosphere environments. J. Appl. Microbiol. 93:616-630. [DOI] [PubMed] [Google Scholar]
- 44.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Tabacchioni, S., L. Chiarini, A. Bevivino, C. Cantale, and C. Dalmastri. 2000. Bias caused by using different isolation media for assessing the genetic diversity of a natural microbial population. Microb. Ecol. 40:169-176. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tran Van, V., O. Berge, S. Ngo Ke, J. Balandreau, and T. Heulin. 2000. Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield components in low fertility sulfate acid soils of Vietnam. Plant Soil 218:273-284. [Google Scholar]
- 49.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov., a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 50.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 51.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 52.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Viallard, V., I. Poirier, B. Cournoyer, J. Haurat, S. Wiebkin, K. Ophel-Keller, and J. Balandreau. 1998. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of [Pseudomonas] phenazinium, [Pseudomonas] pyrrocinia and [Pseudomonas] glathei as Burkholderia. Int. J. Syst. Bacteriol. 48:549-563. [DOI] [PubMed] [Google Scholar]