Key Points
-
•
Adenine base editors efficiently correct the IVS1-110 (G>A) β-thalassemia–causing mutation.
-
•
Base editing efficiently corrects the β-thalassemic phenotype in vitro and in vivo.
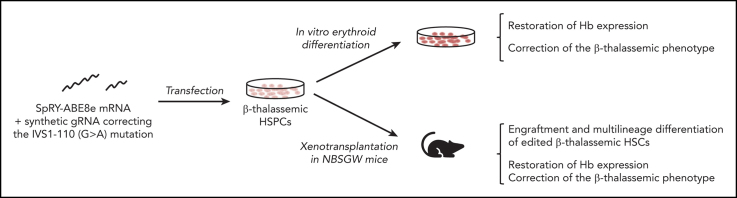
Visual Abstract
Abstract
β-Thalassemia (BT) is one of the most common genetic diseases worldwide and is caused by mutations affecting β-globin production. The only curative treatment is allogenic hematopoietic stem/progenitor cells (HSPCs) transplantation, an approach limited by compatible donor availability and immunological complications. Therefore, transplantation of autologous, genetically-modified HSPCs is an attractive therapeutic option. However, current gene therapy strategies based on the use of lentiviral vectors are not equally effective in all patients and CRISPR/Cas9 nuclease-based strategies raise safety concerns. Thus, base editing strategies aiming to correct the genetic defect in patients’ HSPCs could provide safe and effective treatment. Here, we developed a strategy to correct one of the most prevalent BT mutations (IVS1-110 [G>A]) using the SpRY-ABE8e base editor. RNA delivery of the base editing system was safe and led to ∼80% of gene correction in the HSPCs of patients with BT without causing dangerous double-strand DNA breaks. In HSPC-derived erythroid populations, this strategy was able to restore β-globin production and correct inefficient erythropoiesis typically observed in BT both in vitro and in vivo. In conclusion, this proof-of-concept study paves the way for the development of a safe and effective autologous gene therapy approach for BT.
β-thalassemia is a common recessive genetic disorder characterized by a reduction in β-globin production that results in mild to severe transfusion-dependent anemia. Existing gene therapies have high costs. Using adenine base editors that efficiently repair the mutation without causing the problematic double-stranded DNA breaks inherent with CRISPR/Cas9 systems, Hardouin and colleagues demonstrate in vitro correction of the common IVS1-110 mutation and in vivo resolution of the thalassemic phenotype in preclinical models. This sets the stage for new clinical approaches.
Introduction
β-Thalassemia (BT) is a highly prevalent1 recessive disorder caused by mutations in the β-globin gene (HBB) locus that reduce (β+) or eliminate (β0) β-globin production. Depending on the genotype, the clinical phenotype can vary from mild to transfusion-dependent anemia.
IVS1-110 (G>A) is one of the most common mutations in the Middle East and Mediterranean area.2, 3, 4 This mutation leads to the formation of a splice acceptor site in the first HBB intron causing partial intron retention and generation of a premature termination codon in 90% of the β-globin transcripts.5,6 Owing to the low levels of correctly spliced messenger RNAs (mRNAs), and the inhibition of their translation caused by the aberrant transcripts, this mutation leads to a strong β-globin downregulation.6 Thus, IVS1-110 is a severe β+ mutation, and homozygous patients or compound heterozygotes harboring this mutation and a β0 mutation have a clinical phenotype similar to patients with β0/β0.7
Transplantation of autologous, genetically-modified hematopoietic stem/progenitor cells (HSPCs) was investigated as a treatment option for patients with BT.7, 8, 9, 10, 11 Although a lentivirus-mediated gene addition therapy was approved for transfusion-dependent BT, this treatment was shown to be more efficacious in patients harboring a non-β0/β0 genotype.7,8
Editing approaches use nucleases, such as CRISPR/Cas9 that induce DNA double-strand breaks (DSBs) via a guide RNA (gRNA) complementary to the genomic target. The DSB can be repaired via homologous-directed repair (HDR) by providing a donor template containing the wild-type sequence, thus allowing gene correction. However, HDR is poorly efficient in hematopoietic stem cells (HSCs).12 Nuclease-based strategies are based on the disruption of the splice site generated by the IVS1-110 mutation through indel formation after nonhomologous end-joining–mediated DSB repair and are effective in restoring correct hemoglobin (Hb) expression.13,14 However, only a fraction of indels abolish the aberrant splice site.14 Furthermore, DSBs can induce cell toxicity15 and genomic rearrangements.16
Base editing allows precise DNA repair without generating DSBs. Adenine base editors (ABE) contain a Cas9 nickase and a deaminase, allowing A>G conversions.17 Here, we exploited ABEs to correct the IVS1-110 mutation in patients’ HSPCs and the pathological phenotype of HSPC-derived erythroid cells.
Methods
Base editor-expressing plasmids
Plasmids used in this study include NG-ABE8e (138491; Addgene, Watertown, MA), ABE8e (138489; Addgene), and pCMV-T7-ABEmax(7.10)-SpRY-P2A-EGFP (RTW5025) (140003; Addgene). The SpRY-ABE8e plasmid was created by replacing the Cas9 sequence of the ABE8e plasmid with the SpRY-P2A-EGFP from the pCMV-T7-ABEmax(7.10)-SpRY-P2A-EGFP (RTW5025) plasmid.
gRNA design
We manually designed gRNAs targeting the IVS1-110 (G>A) mutation (supplemental Table 1; available on Blood website). To evaluate the impact of bystander edits, we adapted the gRNA1 sequence to target the WT HBB gene (gRNA1_healthy donor [HD]). We used chemically modified synthetic gRNAs harboring 2′-O-methyl analogs and 3′-phosphorothioate nonhydrolyzable linkages at the first three 5′ and 3′ nucleotides (Synthego, Redwood City, CA).
mRNA in vitro transcription
NG-ABE8e, ABE8e, or SpRY-ABE8e plasmids were digested overnight with SapI (Thermo Fisher, Waltham, MA). The linearized plasmids were purified (28106; Qiagen, Venlo, Netherlands) and eluted in DNase/RNase-free water. The in vitro transcription protocol (AM1334; Thermo Fisher) was modified as follows. The GTP nucleotide solution was used at a final concentration of 3 mM and the antireverse cap analog N7-methyl-3'-o-methyl-guanosine-5'-triphosphate-5'-guanosine (N-7003; Trilink, San Diego, CA) was added at a final concentration of 12 mM. The incubation time was reduced to 30 minutes and followed by polyadenylation (AM1350; Thermo Fisher). mRNA was precipitated using LiCl.
Cell purification and culture
We obtained human peripheral blood non-mobilized CD34+ HSPCs from patients with BT (for in vitro experiments). HD CD34+ HSPCs samples were obtained from the bone marrow of healthy donors (for in vitro Granulocyte Colony Stimulating Factor [GCSF] experiments) or after GCSF mobilization (for in vivo experiments). Written informed consent was obtained from all patients. All experiments were performed in accordance with the Declaration of Helsinki. The study was approved by the regional investigational review board (reference, DC 2014-2272; CPP Ile-de-France II “Hôpital Necker-Enfants malades”). HSPCs were purified using the CD34 MicroBead Kit (Miltenyi Biotec, Bergisch Gladbach, Germany).
Plerixafor/GCSF-mobilized peripheral blood CD34+ cells (used for in vivo experiments) were selected from a patient with BT on signed informed consent approved by the Ethical Committee of the San Raffaele Hospital (Milan, Italy). After the mobilization and cell collection with the Spectra Cobe or Spectra Optia apheresis system (Terumo BCT, Lakewood, CO), CD34+ cells were purified using immunomagnetic beads (CliniMACS, Miltenyi Biotec) by MolMed SpA (Milan, Italy).11
Forty-eight hours before electroporation, CD34+ cells were thawed and cultured at a concentration of 5 × 105 cells/mL in a “HSPC medium” containing StemSpan (STEMCELL Technologies, Vancouver, Canada) supplemented with penicillin/streptomycin (Thermo Fisher), StemRegenin1 (STEMCELL Technologies), and the following recombinant human cytokines (PeproTech, Cranbury, NJ): human stem cell factor, Fms-like tyrosine kinase receptor 3 ligand, thrombopoietin, and interleukin-3 (IL-3). Four days before electroporation, the CD34– fraction was thawed and cultured at a concentration of 5 × 106 cells/mL in a “T cell medium” containing RPMI 1640 + GlutaMAX (Thermo Fisher) supplemented with FBS (Thermo Fisher), penicillin/streptomycin (Thermo Fisher), and recombinant human IL-2 (PeproTech). Five hours after thawing, cells were transferred to a “T cell activation medium” containing RPMI 1640 + GlutaMAX (Thermo Fisher), which was supplemented with CD28 monoclonal antibody (Clone CD28.2; eBioscience, San Diego, CA) in plates coated with CD3 monoclonal antibody (Clone OKT3; eBioscience).
RNA electroporation
For in vitro experiments, 1 × 106 T cells or 1 × 104 to 5 × 105 HSPCs per condition were electroporated with 3.0 μg of the ABE-encoding mRNA and 3.2 μg of the synthetic gRNA using the P3 primary cell 4D-Nucleofector X Kit S (Lonza, Basel, Switzerland) and the EO-115 or CA-137 program (Nucleofector 4D), respectively. For T cells, when ABE was not fused to GFP (ABE8e, NG-ABE8e), 0.25 μg of GFP-encoding mRNA (L-7201, Trilink) was added to the electroporation mix. For in vivo experiments, 2.3 × 106 to 7.5 × 106 HSPCs per condition were electroporated with 15.0 μg of the ABE-encoding mRNA and 16.0 μg of the synthetic gRNA using the P3 primary cell 4D-Nucleofector X Kit L (Lonza) and the CA-137 program (Nucleofector 4D). Cells electroporated only with Tris-EDTA (TE) buffer or with ABE-encoding mRNA served as negative controls.
Colony-forming cell (CFC) assay
For in vitro and in vivo experiments, CD34+ HSPCs and human bone marrow CD45+ cells were plated at a concentration of 500 cells per mL (according to pre-nucleofection counting) or 200 000 cells per mL, respectively, in a methylcellulose-based medium (GFH4435; STEMCELL Technologies) under conditions supporting erythroid and granulo-monocytic differentiation. BFU-E and CFU-GM colonies were counted after 14 days. Single colonies were collected to evaluate base editing efficiency and indels.
HSPC differentiation
Electroporated BT and HD CD34+ HSPCs were differentiated into mature red blood cells (RBCs) using a three-phase erythroid differentiation protocol, as previously described.18,19
Evaluation of editing efficiency
Genomic DNA was extracted from control and edited cells using PURE LINK Genomic DNA Mini kit (Thermo Fisher) or Quick-DNA/RNA Miniprep (ZD7001; Zymo Research, Irvine, CA) following the manufacturer’s instructions. To evaluate base editing efficiency at gRNA target sites, we performed a nested polymerase chain reaction (PCR) using previously published primers,13 followed by Sanger sequencing and EditR analysis.20 Tracking of indels by decomposition (TIDE) analysis was performed to evaluate the percentage of indels in edited samples.21
On- and off-target regions were also PCR-amplified and subjected to next-generation sequencing (NGS). Off-targets were in silico predicted using COSMID.22 We selected the top10 predicted off-targets and assessed editing at day 9 or 13 of differentiation. On-target and off-target sites were PCR-amplified using the Phusion High-Fidelity polymerase (M0530; NEB, Ipswich, MA) and primers containing specific DNA stretches (MR3 for forward primers and MR4 for reverse primers; supplemental Table 3) located 5’ to the sequence recognizing the off-target. For the on-target site, a nested PCR was applied as described above. Amplicons were purified using Ampure XP beads (A63881; Beckman Coulter, Brea, CA). Illumina-compatible barcoded DNA amplicon libraries were prepared by a second PCR step using the Phusion High-Fidelity polymerase (M0530; NEB) and primers containing Unique Dual Index barcodes and annealing to MR3 and MR4 sequences. Libraries were pooled, purified using the High Pure PCR product purification kit (11732676001; Sigma-Aldrich, Saint Louis, MO), and sequenced using Illumina NovaSeq 6000 system (paired-end sequencing; 2×100-bp) to obtain a minimum of 100 000 reads per amplicon. Targeted NGS data were analyzed using CRISPResso2.23
RT-PCR and RT-qPCR
RNA was extracted from cells at day 13 of differentiation (74004; Qiagen; or ZD7001; Zymo Research) and retrotranscribed (18080051; Thermo Fisher). RT-PCR analysis of β-globin mRNAs was performed using previously described primers spanning the exon 1 to 2 junction.13 RT-qPCR was performed using previously described primers for the detection of correctly spliced β-globin mRNA13 and the following primers amplifying α-globin transcripts: α-globin-F 5’-CGGTCAACTTCAAGCTCCTAA-3’ and α-globin-R 5’- ACAGAAGCCAGGAACTTGTC-3’. For the detection of the effect of bystander edits on HBB expression, the following primers were used for RT-qPCR: β-globin-F 5’-GCAAGGTGAACGTGGATGAAGT-3’ and β-globin-R 5’-TAACAGCATCAGGAGTGGACAGA-3’.
Flow cytometry analysis
Flow cytometry analysis of erythroid surface markers on HSPC-derived erythroid cells was performed using anti-CD36 (561535; BD Horizon, Franklin Lakes, NJ), anti-CD71 (555536; BD Pharmingen), and anti-GPA (563666; BD Pharmingen) antibodies. Flow cytometry analysis of enucleated or viable cells was performed using DRAQ5 (65-0880-96; Thermo Fisher) and 7AAD (559925; BD Pharmingen) dyes, respectively. Apoptosis was evaluated using PE Annexin V Apoptosis Detection Kit I (BD Biosciences). Flow cytometry analysis of reactive oxygen species (ROS) was performed using H2DCFDA (D399; 65-0880-96). Flow cytometry analyses were performed using Gallios (Beckman coulter) or Novocyte (Agilent, Santa Clara, CA) flow cytometer. Data were analyzed using the FlowJo (BD Biosciences) software.
RP-HPLC and CE-HPLC
High-performance liquid chromatography (HPLC) analysis was performed in HSPC-derived erythroid cells at day 16 of differentiation as previously described in the study by Locatelli et al.19
Quantitative phase image microscopy of RBCs
180 000 in vitro differentiated RBCs (at day 19 or 20) were resuspended in CellStab (005650; BioRad, Hercules, CA) and placed in an 8-well m-Slide (80826; Ibidi, Gräfelfing, Germany). We used the SID4 HR GE camera (Phasics, St Aubin, France) with an inverted microscope (Eclispe Ti-E; Nikon, Tokyo, Japan) to obtain quantitative phase images of label-free RBCs. The BIO-Data R&D software (version 2.7.1.46) was used to perform a segmentation procedure and isolate each RBC. We analyzed only enucleated RBCs, discarding cells with a surface density (dry mass/surface) >0.500 pg/μm2.
HSPC xenotransplantation in NBSGW mice
NOD.Cg-KitW-41JTyr +PrkdcscidIl2rgtm1Wjl/ThomJ (NBSGW) mice were housed in a pathogen-free facility. Control or edited mobilized healthy donor or β-thalassemic CD34+ cells (2.6 × 105 cells per mouse) were transplanted into non-irradiated NBSGW male and female mice of 6 to 9 weeks of age via retro-orbital sinus injection. NBSGW male and female mice were conditioned with busulfan (Sigma-Aldrich) injected intraperitoneally (15 mg/kg body weight) 24 hours before transplantation. Sixteen weeks after transplantation, NBSGW primary recipients were euthanized. Cells were harvested from bone marrow, thymus, spleen, and blood, and 105 cells from each organ were stained with antibodies against the following murine and human surface markers: murine CD45 (1/50 mCD45-VioBlue; Miltenyi Biotec), human CD45 (1/50 hCD45-APCvio770; Miltenyi Biotec), human CD3 (1/50 CD3-APC; Miltenyi Biotec), human CD14 (1/50 CD14-PECy7; BD Biosciences), human CD15 (1/50 CD15-PE; Miltenyi Biotec), human CD11b (1/100 CD11b-APC; Miltenyi Biotec), human CD19 (1/100 CD19-BV510; BD Biosciences), human CD235a (1/50 CD235a-PE; BD Biosciences), human CD71 (1/10 CD71-APC; BD Biosciences), CD36 (1/50 CD36-FITC; BD Biosciences), and CD34 (1/100 CD34-PE-Vio770; Miltenyi Biotec); and analyzed by flow cytometry using the Novocyte analyzer (Agilent) and the FlowJo software (BD Biosciences). Human bone marrow CD45+ cells were sorted by immunomagnetic selection with CD45 MicroBeads (Miltenyi Biotec). All experiments and procedures were performed in compliance with the French Ministry of Agriculture’s regulations on animal experiments and were approved by the regional Animal Care and Use Committee (APAFIS#2019061312202425_v4). Mice were housed in a temperature- (20°C-22°C) and humidity (40%-50%)-controlled environment with 12:12 hour light-dark cycle and fed ad libitum a standard diet.
Bone marrow cell sorting
Bone marrow (BM) cells were subjected to immunostaining with biotinylated antibodies that recognized the following surface markers: CD3 (dilution 1/25, clone HIT3a; BD), CD19 (dilution 1/25, clone HIB19; BD), B220 (dilution 1/50, clone RA3-6B2; BD), Ter119 (dilution 1/50, clone TER-119; BD), and mCD117 (clone 2B8; BD). BM cells were washed and incubated with 20 μL of Anti-Biotin beads (Miltenyi Biotec). After washing, the cells were magnetically purified using an LS column (Miltenyi Biotec) according to the manufacturer’s instructions. Cells from the negative fraction were immunostained with the following antibodies: CD235a-PE (dilution 1/5000, BD) and hCD45-BV510 (dilution 1/100, BD). The hCD45low/−/CD235ahigh cells were sorted using the MA900 cell sorter (Sony Biotechnology, San Jose, CA) and subjected to flow cytometry and reverse phase high performance liquid chromatography (RP-HPLC) analysis.
Macrophage depletion and human RBC sorting
Fifteen weeks after HSPC infusion, mice were injected intraperitoneally with 10 μl/g clodronate liposomes (5 mg/mL, Liposoma, Amsterdam, Netherlands). Four days after treatment, 10 μL of whole blood were stained with antibodies recognizing the following murine and human surface markers: murine CD45 (1/50 mCD45-VioBlue; Miltenyi Biotec), human CD45 (1/50 hCD45-APCvio770; Miltenyi Biotec), CD235a (1/5000, CD235a-PE; BD), and Ter119 (1/50, Ter119-FITC; BD). After 2 washings, human RBCs were sorted using an MA900 cell sorter (Sony Biotechnology). After gating on the mCD45−/hCD45low/− population, a minimum of 50 000 human RBCs (CD235ahigh/Ter119−) were sorted and lysed for RP-HPLC analysis.
Results
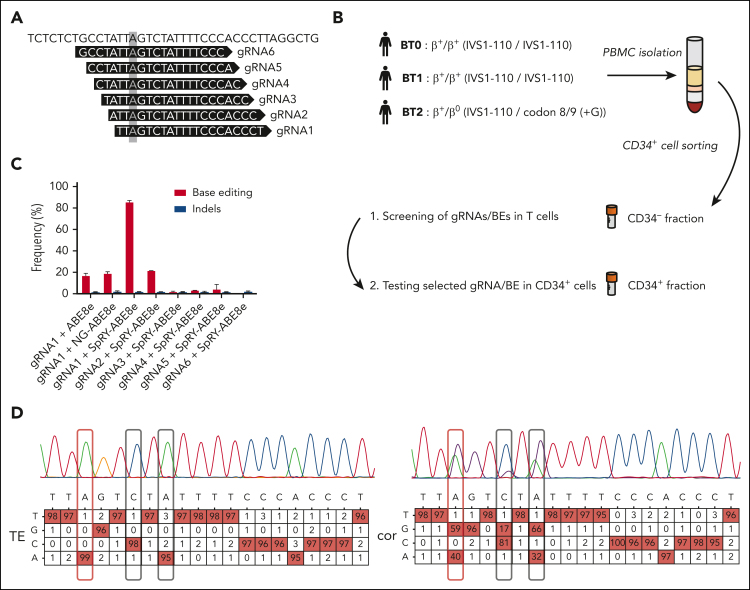
We designed 6 gRNAs (Figure 1A; supplemental Table 1) compatible with ABE8e and NG-ABE8e24 (recognizing NGG and NG PAM, respectively), and SpRY-ABE8e25 generated by combining the highly processive deaminase from ABE8e24 with the SpRY PAM-less–Cas9 nickase.26 We screened gRNA/base editor (BE) combinations in T cells obtained from patients with BT homozygous for the IVS1-110 mutation (BT0 and BT1, Figure 1B). gRNA1/SpRY-ABE8e was the most efficient combination that was able to correct the mutation in ∼90% of the HBB mutant alleles, as evaluated by Sanger sequencing (Figure 1C). Two bystander edits were observed at positions 6 and 8, including a C>G conversion occurring at a relatively low frequency (Figure 1D), as previously reported at other loci with ABEs.27,28 Importantly, these changes have likely no consequences on HBB expression as they occur in an intronic region, and single nucleotide polymorphisms described at these positions (rs777028217 and rs1480884739) are not associated with any clinical manifestations.29, 30, 31 Although bystander activity could be reduced by using precise ABE8e variants,32,33 their usage could also reduce the on-target correction efficiency. Thus, to confirm that these bystander edits have no impact on HBB expression, we generated these base conversions in HD HSPCs achieving frequencies similar to those detected in the BT samples (supplemental Figure 1A). We observed no decrease in HBB expression, neither at the mRNA level nor at the protein level in HSPC-derived erythroid cells with normal α-/non-α globin ratio and hemoglobin A expression (supplemental Figure 1B-D). Furthermore, the differentiation was not affected, as demonstrated by the normal expression of erythroid markers (GPA, CD71, and CD36) and enucleation rate along the differentiation (supplemental Figure 1E-F). Finally, the generation of the bystander edits did not induce apoptosis, confirming any impact of these base conversions on HBB expression and erythroid differentiation (supplemental Figure 1G).
Figure 1.
A base editing strategy to efficiently correct the IVS1-110 (G>A) mutation. (A) gRNAs 1 to 6 were manually designed to place the IVS1-110 (G>A) mutation in positions 3 to 8 of the protospacer. The mutation is highlighted with a grey box. (B) Overview of the cell collection for testing the ability of gRNA/BE combinations to correct the IVS1-110 (G>A) mutation. (C) Frequency of corrected alleles (normalized to the frequency of GFP+ cells) and indels as assessed by Sanger sequencing in T cells electroporated with different combinations of synthetic gRNAs and ABE mRNAs. Data are expressed as mean ± standard error of the mean (SEM) (n = 3, 2 donors). (D) Representative percent composition of Sanger sequencing traces measured to be significantly different from noise (in red), as assessed by EditR following Sanger sequencing in T cells electroporated with gRNA1/SpRY-ABE8e (cor) or with TE buffer (TE). The target base position is outlined with a red box and the observed bystander edits with black boxes. Single nucleotide polymorphisms rs777028217 (G>A) and rs1480884739 (T>C) mapped to positions 6 and 8 are not associated with a clinical phenotype. PBMC, peripheral blood mononuclear cells; cor, corrected.
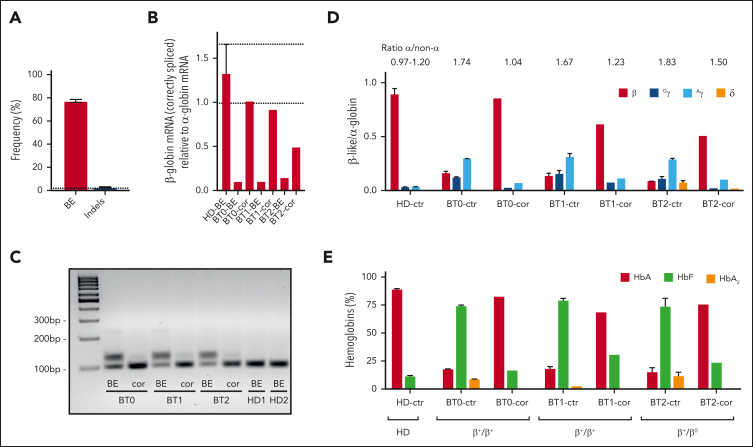
gRNA1 and SpRY-ABE8e mRNA were electroporated in HSPCs from 2 homozygous patients (BT0 and BT1) and 1 compound heterozygous patient harboring the IVS1-110 and a β0 mutation, respectively (codon 8/9 (+G); BT2) (Figure 1B; supplemental Figure 2A). Gene correction was high and similar between the 3 donors with little indel generation as evaluated by Sanger sequencing and NGS (Figure 2A; supplemental Figure 2B-D). The high percentage of bystander edit at position 8 in the heterozygous donor indicates that the gRNA also targets the allele containing the β0 mutation, likely because the mismatch is located at the 5’ end of the gRNA, where it is more easily tolerated34 (supplemental Figure 2B). NGS analysis confirmed a good product purity with most of the events being A>G conversions, and lower editing of cytosines leading mainly to C>G and more rarely to C>T conversions (supplemental Figure 2E).
Figure 2.
ABE-mediated correction of the IVS1-110 (G>A) mutation in BT HSPCs restores normal Hb production in their erythroid progeny. (A) The frequency of corrected alleles (BE) and indels as assessed by Sanger sequencing in BTHSPCs. Data are expressed as mean ± SEM (3 donors). The frequency of corrected alleles in the cells obtained from the compound heterozygous patient (BT2) was corrected considering only the alleles harboring the IVS1-110 (G>A) mutation. The dotted line represents the maximum background noise of indels calculated by TIDE. (B) RT-qPCR using primers detecting exclusively correctly spliced β-globin mRNAs in erythroid cells derived from BT HSPCs (cor). β-globin expression was normalized to α-globin. Data are expressed as mean ± SEM. The dotted lines indicate the maximum and minimum values observed in HD cells. (C) RT-PCR using primers amplifying a region spanning the HBB exon 1-exon 2 junctions. cDNA was obtained from erythroid cells derived from BT HSPCs (cor). (D) Expression of β-, Gγ-, Aγ- and δ- globin chains measured by RP-HPLC in BT and HD RBCs. β-like-globin expression was normalized to α-globin. The α-/non–α-globin ratio is reported on top of the graph. RBCs were obtained from BT HSPCs (cor). Data are expressed as mean ± SEM. (E) Analysis of HbA, HbF, and HbA2 by CE-HPLC in BT and HD RBCs. We calculated the percentage of each Hb type over the total Hb tetramers. RBCs were obtained from corrected BT HSPCs (cor). (B-C) As controls, we used erythroid cells derived from BT or HD HSPCs electroporated only with SpRY-ABE8e mRNA (BE) (3 patients with BT and 2 HDs). (D-E) As controls, we pooled data obtained in RBCs derived from BT or HD HSPCs electroporated with TE or with SpRY-ABE8e mRNA only (3 patients with BT and 2 HDs). Data are expressed as mean ± SEM. cor, corrected; ctr, control.
To evaluate the safety profile of our strategy, we performed NGS of the top-10 in silico predicted off-targets (supplemental Table 2). Base editing was observed only at 1 site that was mapped to a region devoid of known genes and did not impact cell viability and differentiation (supplemental Figure 2C). The off-target activity could be potentially mitigated using high-fidelity base editors or modulating DNA exposure time to the editing machinery. No indels were detected at off-target sites, thus minimizing the possibility of DSB-induced genomic rearrangements (eg, translocations).
We observed a similar number of progenitors before and after the treatment with no difference in the proportion of erythroid (BFU-E) and granulo-monocytic (CFU-GM) colonies, demonstrating the safety of our strategy (supplemental Figures 2A and 3A). Importantly, all the progenitors were edited with low to absent indels (supplemental Figure 3B).
Electroporated BT and control HD HSPCs were differentiated to the erythroid lineage (supplemental Figure 2A). Reverse transcription PCR (RT-PCR) showed restoration of the correct β-globin splicing and a substantial decrease of aberrant transcripts characterized by lower mobility owing to partial intron retention (Figure 2B-C). Wild-type–β-globin mRNA levels were similar to those observed in HD cells for homozygous donors while representing ∼50% of the HD β-globin transcripts for the compound heterozygote donor (Figure 2B). In untreated BT RBCs, HPLC showed elevated α-/non–α-globin ratios and low β-globin expression poorly compensated by fetal γ (γA+γG)-globins (Figure 2D; supplemental Figure 4). After treatment, RBCs exhibited higher levels of β-globin chain and hemoglobin A (Figure 2D-E; supplemental Figure 4). In edited cells from homozygous patients, the α-/non-α-globin ratio was comparable to the values measured in HD cells and substantially ameliorated in the erythroid cells from the compound heterozygote patient (Figure 2D).
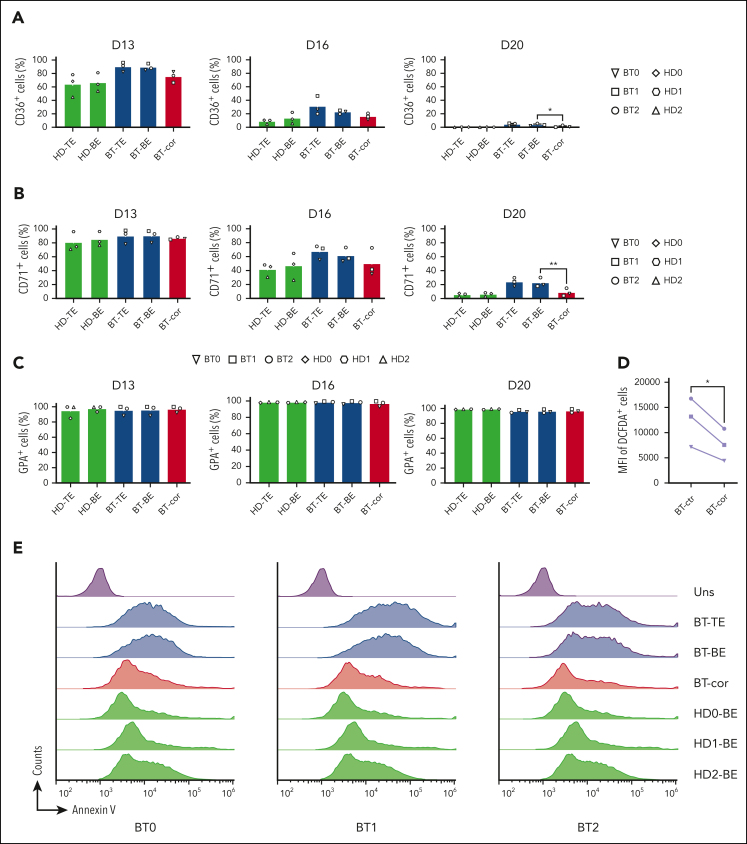
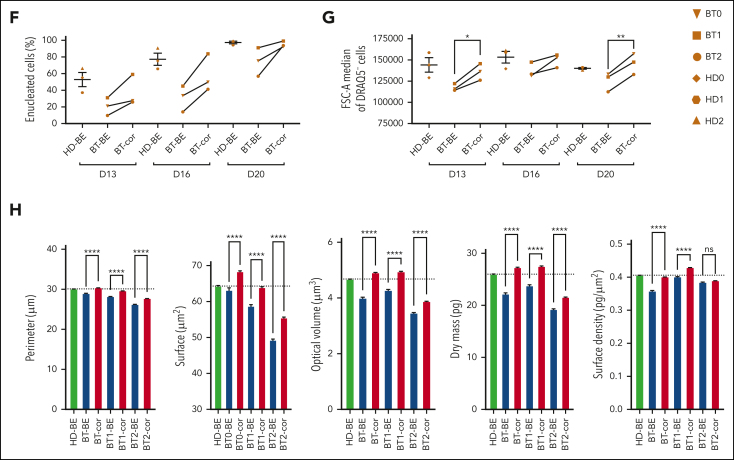
In BT, the imbalance between α- and β-globin production leads to the precipitation of α-globins causing apoptosis of erythroid precursors and ineffective erythropoiesis.35 Flow cytometry analysis of CD36 and CD71, known early erythroid markers, showed a proper downregulation in treated samples, demonstrating that the typically delayed erythroid differentiation of BT cells was efficiently corrected (Figure 3A-C). The production of ROS observed in BT control erythroid cells was significantly decreased in treated samples (Figure 3D). Importantly, flow cytometry showed the rescue of edited BT erythroid precursors from apoptosis (Figure 3E).
Figure 3.
Efficient reversion of the IVS1-110 (G>A) mutation in BT HSPCs corrects ineffective erythropoiesis. (A-C) Frequency of CD36+ (A), CD71+ (B), and GPA+ (C) cells at days 13, 16, and 20 of erythroid differentiation, as measured by flow cytometry analysis. Control BT or HD cells were either electroporated with TE buffer (TE), or only with SpRY-ABE8e mRNA (BE). Data are expressed as mean ± SEM (3 patients with BT and 3 HDs). ∗P ≤ .05; ∗∗P ≤ .01 (paired t test; BT-BE vs BT-cor). (D) Mean Fluorescence Intensity (MFI) in ROS-containing cells (DCFDA+) for control and edited (cor) BT samples. For the control, we pooled data obtained in RBCs derived from BT HSPCs electroporated with TE or with SpRY-ABE8e mRNA only (3 patients with BT). ∗P ≤ .05 (paired t test). (E) Flow cytometry histograms showing the frequency of apoptotic cells (annexin V+ cells) in the 7AAD− cell population in unstained (Uns), BT and HD samples at day 13 of erythroid differentiation (3 patients with BT and 3 HDs). (F) Frequency of enucleated cells at days 13, 16, and 20 of erythroid differentiation, as measured by flow cytometry analysis of cells stained with the DRAQ5 nuclear dye (3 patients with BT and 3 HDs). Data for HD samples are expressed as mean ± SEM. (G) Cell size of enucleated cells at days 13, 16, and 20 of erythroid differentiation, as measured by flow cytometry analysis of the median FSC-A intensity (3 patients with BT and 3 HDs). Data for HD samples are expressed as mean ± SEM. ∗P ≤ .05; ∗∗P ≤ .01 (paired t test; BT-BE vs BT-cor). (H) Single RBC parameters (perimeter, surface, optical volume, dry mass, and surface density) evaluated by quantitative phase image microscopy in RBCs obtained from BT HSPCs (cor). As controls, we used RBCs derived from BT or HD HSPCs electroporated only with SpRY-ABE8e mRNA (BE). Data are expressed as mean ± SEM (3 patients with BT and 2 HDs). ∗∗∗∗P ≤ .0001 (Ordinary one-way ANOVA). cor, corrected; ctr, control; D, day.
In the treated samples, we observed an increased enucleation rate along the differentiation, reaching levels similar to those observed in HD at the end of the differentiation (Figure 3F). Flow cytometry showed that the size of enucleated cells (typically reduced in BT cell cultures, namely microcytosis) was increased in the treated samples (Figure 3G). Finally, we used phase microscopy to evaluate several physical parameters in a quantitative manner (Figure 3H; supplemental Figure 5A-E). BT RBCs were characterized by a reduced perimeter, surface, and volume owing to the microcytosis and decreased dry mass and surface density owing to the low Hb content. These parameters were normalized or substantially improved in treated RBCs.
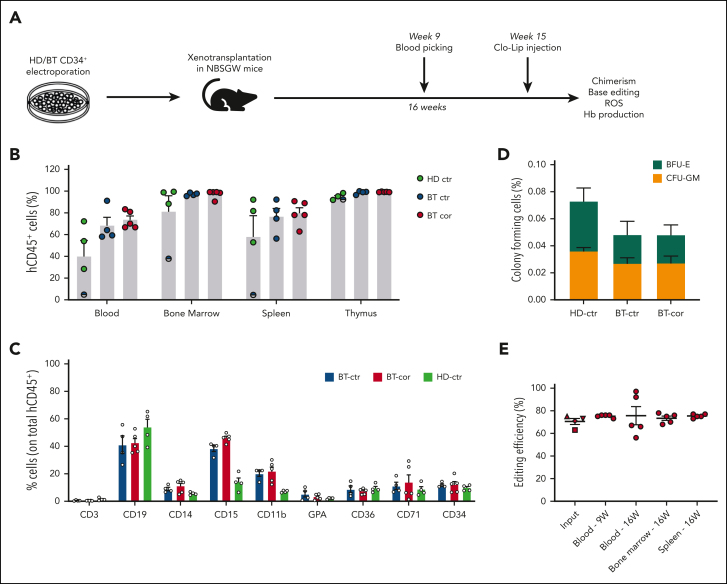
To evaluate the ability of SpRY-ABE8e to correct HBB in repopulating HSCs, we transplanted human BT HSPCs homozygous for the IVS1-110 mutation after electroporation with SpRY-ABE8e mRNA and gRNA1 into immunodeficient NBSGW mice (Figure 4A). HD HSPCs and unedited BT HSPCs served as controls. Sixteen weeks after transplantation, no differences were observed between edited and control HSPCs in terms of engraftment and differentiation potential, as measured by the frequency of human CD45+ cells in the hematopoietic tissues and the expression of lineage-specific markers (Figure 4B-C). Human CD45+ bone marrow cells were isolated and subjected to a CFC assay. Control and edited BT samples showed a similar number of erythroid and granulo-monocytic colonies, demonstrating no impact of base editing on the clonogenic potential of engrafted human cells (Figure 4D). Importantly, 16 weeks after transplantation, on-target base editing efficiency was similar in input HSPCs (70.5% ± 2.6) and bone marrow (73.4% ± 1.9), blood (75.6% ± 8.0) and spleen (75.4% ± 0.7), demonstrating successful engraftment of base-edited HSCs (Figure 4E). Surprisingly, though editing at bystander 2 position (A>G) was similar in input HSPCs and in the progeny of repopulating HSCs, we observed an increase in the frequency of bystander edit 1 (C>G) after engraftment (supplemental Figure 6). These data suggest that C>G conversions resulting from ABE editing might be favored in HSCs vs HSPCs, a population containing mostly progenitor cells.
Figure 4.
Correction of the IVS1-110 (G>A) mutation in repopulating HSCs. (A) Overview of the experimental protocol of HSPC xenotransplantation. BT HSPCs were subjected to RNA-mediated base editing and xenotransplanted into NBSGW immunodeficient mice. HD and BT HSPCs electroporated with TE buffer or only with SpRY-ABE8e mRNA were injected as controls. Peripheral blood analysis was performed at weeks 9 and 16. Mice were euthanized 16 weeks after engraftment, after Clo-Lip injection, and their hematopoietic tissues and organs were collected and analyzed. (B) Engraftment of human cells in NBSGW mice transplanted with HD or BT control (HD-ctr; BT-ctr) or corrected (BT-cor) HSPCs 16 weeks post-transplantation (HD-ctr, n = 4; BT-ctr, n = 4; BT-cor, n = 5). Engraftment is represented as the percentage of human CD45+ cells in the total murine and human CD45+ cell population in peripheral blood, bone marrow, spleen, and thymus. Each data point represents an individual mouse. The mouse with the lowest chimerism is indicated with the symbol ◓. Data are expressed as mean ± SEM. (C) Frequency of human T (CD3) and B (CD19) lymphoid, myeloid (CD14, CD15, and CD11b), erythroid (GPA, CD36, CD71), and HSPC (CD34) cells in BM 16 weeks after the transplantation (HD-ctr, n = 4; BT-ctr, n = 4; BT-cor, n = 5). Each data point represents an individual mouse. Data are expressed as mean ± SEM. (D) Human hematopoietic progenitor content in BM human CD45+ cells derived from mice transplanted with control and edited HSPCs (HD-ctr, n = 4; BT-ctr, n = 4; BT-cor, n = 5). We plotted the percentage of human CD45+ cells giving rise to BFU-E and CFU-GM. Data are expressed as mean ± SEM. (E) Base editing efficiency, calculated by the EditR software, in input, peripheral blood-, bone marrow- and spleen-derived HD, and BT human samples subjected to Sanger sequencing. Data are expressed as mean ± SEM (BT-cor, n = 5). The frequency of base editing in the input was calculated in cells cultured in the HSPC medium (▲), in liquid erythroid cultures (▼), BFU-E (■) and CFU-GM (◆) colonies. Each data point represents an individual mouse. Data are expressed as mean ± SEM. ctr, control. BFU-E, burst forming units-erythroid; CFU-GM, colony forming units-erythroid.
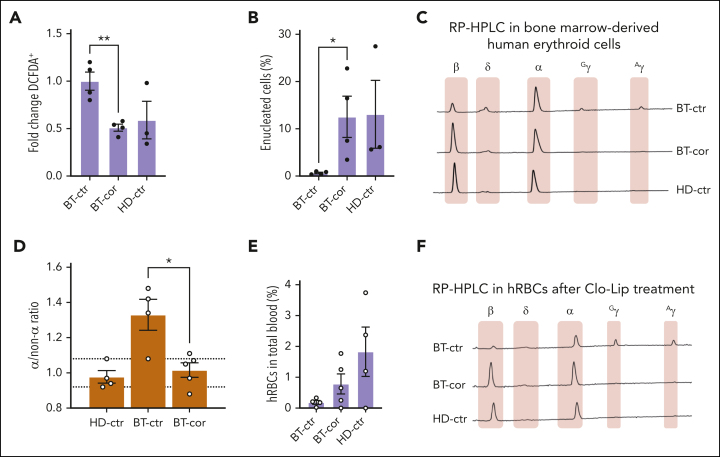
To assess the correction of the BT phenotype in the erythroid progeny of HSCs, we sorted human GPA+ erythroid cells from the bone marrow (72.0% ± 0.9; Figure 4E). Interestingly, ROS levels and enucleation rate were normalized in erythroid cells derived from edited HSPCs (Figure 5A-B). Moreover, HPLC analysis showed the expression of WT β-globin in treated samples and restoration of the α-/non–α-globin ratio (Figure 5C-D).
Figure 5.
Correction of the IVS1-110 (G>A) mutation in xenotransplanted BT HSPCs rescues the ineffective erythropoiesis in vivo. (A) Frequency of ROS-containing (DCFDA+) human GPA+ erythroid cells derived from the bone marrow of mice transplanted with HD or BT control (HD-ctr; BT-ctr) or corrected (BT-cor) HSPCs 16 weeks after the transplantation (HD-ctr, n = 3; BT-ctr, n = 4; BT-cor, n = 4). ∗∗P ≤ .01 (unpaired t test; BT-ctr vs BT-cor). We plotted the fold change relative to BT-ctr samples. (B) Frequency of enucleated cells as measured by the flow cytometry analysis of cells stained with the DRAQ5 nuclear dye in human GPA+ erythroid populations from the bone marrow of mice transplanted with HD or BT control (HD-ctr; BT-ctr) or corrected (BT-cor) HSPCs 16 weeks after the transplantation (HD-ctr, n = 3; BT-ctr, n = 4; BT-cor, n = 4). ∗P ≤ .05 (unpaired t test; BT-ctr vs BT-cor). (C) Representative RP-HPLC chromatograms from sorted human GPA+ bone marrow erythroid cells 16 weeks posttransplantation. (D) α/non–α ratio calculated based on RP-HPLC data from sorted human GPA+ bone marrow erythroid cells obtained from mice transplanted with HD or BT control (HD-ctr; BT-ctr) or corrected (BT-cor) HSPCs 16 weeks after the transplantation (HD-ctr, n = 4; BT-ctr, n = 4; BT-cor, n=5). The dotted lines indicate minimum and maximum values observed in HD-ctr samples. ∗P ≤ .05 (unpaired t test; BT-ctr vs BT-cor). (E) Frequency of human RBCs in total peripheral blood 4 days after Clo-Lip injection in mice transplanted with HD or BT control (HD-ctr; BT-ctr) or corrected (BT-cor) HSPCs 16 weeks after the transplantation (HD-ctr, n = 4; BT-ctr, n = 4; BT-cor, n = 5). (F) Representative RP-HPLC chromatograms from sorted human circulating RBCs 16 weeks after the transplantation (BT-ctr, n = 4 pooled samples; BT-cor, n = 1 representative graph; HD-ctr, n = 1 representative graph). cor, corrected; ctr, control.
Before euthanization, the mice were subjected to clodronate liposomes (Clo-Lip) treatment to evaluate the egression of mature human RBCs (hRBCs) from the bone marrow to the blood.36,37 Interestingly, we observed an increased frequency of hRBCs in mice engrafted with corrected BT cells as compared with the levels observed in the control group of mice engrafted with unedited BT cells (Figure 5E). HPLC showed the correction of the BT phenotype in circulating hRBCs obtained from the treated group, with a high expression of β-globin and a globin expression profile overlapping with that observed in the HD control group (Figure 5F).
Discussion
Our results demonstrate that ABEs can efficiently correct the IVS1-110 mutation in patients’ HSPCs without causing DSBs and restore a normal to nearly normal Hb expression profile in HSPC-derived RBCs, thus correcting the BT cell phenotype in vitro and in vivo. Targeting only the βIVS1-110(G>A) allele in the compound heterozygote largely corrects ineffective erythropoiesis but is less effective in ameliorating the α-/non–α-globin ratio than correcting both mutations in homozygous patients’ cells. However, the observed α-/non–α-globin ratios might be close to those observed in erythroid cells from asymptomatic heterozygous carriers of BT mutations. If necessary, to fully correct the BT phenotype, we envision that the strategy proposed in this study could be combined with an approach either correcting the second mutation or reducing α-globin expression.38
Other strategies were proposed to specifically target the highly prevalent IVS1-110 mutation.6,13,14 Short hairpin RNA-mediated targeting of the aberrant mRNA increases β-globin chain production probably by alleviating translational inhibition. However, our strategy not only reduces the abnormally spliced transcripts but also increases the levels of correctly spliced mRNAs. Compared with nuclease-based strategies disrupting the abnormal splice acceptor site, our approach is more predictable and accurate as all the events lead to the correction of the splicing defect. Furthermore, compared with nuclease-based strategies, base editing should limit DSB-caused toxicity such as the activation of p53-mediated–DNA damage response16 and large genomic rearrangements (ie, large deletions, translocations, and chromothripsis). However, base editing retains a lack of precision because of bystander editing, which can cause unwanted events (particularly when targeting coding sequences), although, specific to our study, these mutations had no impact on the target gene expression. Prime editing, a new CRISPR/Cas9- based technology,39 could theoretically allow the development of more precise gene correction strategies. However, its efficiency in primary hematopoietic cells is still limited (A. Miccio, unpublished results), and further improvements are required to use this tool for developing gene therapy approaches for hematopoietic disorders.
Other gene therapy strategies for BT aim at inducing fetal Hb (HbF).40 These strategies have the advantage of being mutation-independent and can be potentially applied to all patients with BT, although it is still debated if high HbF levels are developmentally or physiologically harmful .41 Furthermore, for prevalent mutations such as IVS1-110, development of a mutation-specific correction strategy could allow the production of higher levels of therapeutic hemoglobin, as HBB promoter activity and HBB gene expression are favored in adult cells.42 Finally, our mutation-specific approach counteracts the partially dominant causative effect of aberrant HBB mRNAs that is likely still present after HbF induction or expression of a therapeutic globin in lentiviral vector-based approaches.6
Overall, our study provides in vitro and in vivo proof of efficacy of a base editing approach to treat patients with prevalent and severe BT mutations. The clinical development of our approach will require the establishment of a large-scale electroporation protocol with clinical-grade reagents and toxicology studies to demonstrate the safety of the base-edited drug product.
Conflict-of-interest disclosure: G.H. and A.M. are the inventors of a patent describing base editing approaches for β-Thalassemia (EP22305075.8: Base editing approaches for the treatment of BT). The remaining authors declare no competing financial interests.
Acknowledgments
The authors thank Christine Bole and the Imagine genomic facility for the generation of the next-generation sequencing (NGS) data. The authors also thank the patients for their participation.
This work was supported by state funding from the Agence Nationale de la Recherche under “Investissements d’avenir” program (ANR-10-IAHU-01), the Fondation Bettencourt Schueller, the Paris Ile de France Region under “DIM Thérapie génique” initiative, and the European Research Council (865797 DITSB).
Authorship
Contribution: G.H. designed, conducted experiments, analyzed data, and wrote the paper; P.A. and P.M. conducted experiments and analyzed data; S.M., L.J., M.C., S.S., and G.F. contributed to the design of the experimental strategy and provided patient samples; C.M. analyzed the NGS data; A.M. conceived the study, designed experiments, and wrote the paper.
Footnotes
Data are available on request from the corresponding author, Annarita Miccio (annarita.miccio@institutimagine.org).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Supplementary Material
References
- 1.Modell B, Darlison M. Global epidemiology of haemoglobin disorders and derived service indicators. Bull World Health Organ. 2008;86(6):480–487. doi: 10.2471/BLT.06.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thein SL. The Molecular basis of β-thalassemia. Cold Spring Harb. Perspect. Med. 2013;3(5) doi: 10.1101/cshperspect.a011700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao A, Gossens M, Pirastu M. Beta thalassaemia mutations in Mediterranean populations. Br J Haematol. 1989;71(3):309–312. doi: 10.1111/j.1365-2141.1989.tb04285.x. [DOI] [PubMed] [Google Scholar]
- 4.IthaGenes Ithanet.eu. IthaID: 113. IthaGenes IthaID. 2022;113 [Google Scholar]
- 5.Busslinger M, Moschonas N, Flavell RA. Beta + thalassemia: aberrant splicing results from a single point mutation in an intron. Cell. 1981;27(2 Pt 1):289–298. doi: 10.1016/0092-8674(81)90412-8. [DOI] [PubMed] [Google Scholar]
- 6.Patsali P, Papasavva P, Stephanou C, et al. Short-hairpin RNA against aberrant HBBIVSI-110(G>A) mRNA restores β-globin levels in a novel cell model and acts as mono- and combination therapy for β-thalassemia in primary hematopoietic stem cells. Haematologica. 2018;103(9):e419–e423. doi: 10.3324/haematol.2018.189357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson AA, Walters MC, Kwiatkowski J, et al. Gene therapy in patients with transfusion-dependent β-thalassemia. N Engl J Med. 2018;378(16):1479–1493. doi: 10.1056/NEJMoa1705342. [DOI] [PubMed] [Google Scholar]
- 8.Magrin E, Semeraro M, Hebert N, et al. Long-term outcomes of lentiviral gene therapy for the β-hemoglobinopathies: the HGB-205 trial. Nat Med. 2022;28(1):81–88. doi: 10.1038/s41591-021-01650-w. [DOI] [PubMed] [Google Scholar]
- 9.Locatelli F, Thompson AA, Kwiatkowski JL, et al. Betibeglogene autotemcel gene therapy for non–β0/β0 genotype β-thalassemia. N Engl J Med. 2022;386(5):415–427. doi: 10.1056/NEJMoa2113206. [DOI] [PubMed] [Google Scholar]
- 10.Cavazzana M, Antoniani C, Miccio A. Gene therapy for β-hemoglobinopathies. Mol Ther. 2017;25(5):1142–1154. doi: 10.1016/j.ymthe.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marktel S, Scaramuzza S, Cicalese MP, et al. Intrabone hematopoietic stem cell gene therapy for adult and pediatric patients affected by transfusion-dependent ß-thalassemia. Nat Med. 2019;25(2):234–241. doi: 10.1038/s41591-018-0301-6. [DOI] [PubMed] [Google Scholar]
- 12.Park SH, Lee CM, Dever DP, et al. Highly efficient editing of the β-globin gene in patient-derived hematopoietic stem and progenitor cells to treat sickle cell disease. Nucleic Acids Res. 2019;47(15):7955–7972. doi: 10.1093/nar/gkz475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu S, Luk K, Yao Q, et al. Editing aberrant splice sites efficiently restores β-globin expression in β-thalassemia. Blood. 2019;133(21):2255–2262. doi: 10.1182/blood-2019-01-895094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patsali P, Turchiano G, Papasavva P, et al. Correction of IVS I-110(G>A) β-thalassemia by CRISPR/Cas-and TALEN-mediated disruption of aberrant regulatory elements in human hematopoietic stem and progenitor cells. Haematologica. 2019;104(11):e497–e501. doi: 10.3324/haematol.2018.215178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med. 2018;24(7):927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 16.Boutin J, Cappellen D, Rosier J, et al. ON-target adverse events of CRISPR-Cas9 nuclease: more chaotic than expected. CRISPR J. 2022;5(1):19–30. doi: 10.1089/crispr.2021.0120. [DOI] [PubMed] [Google Scholar]
- 17.Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet. 2018;19(12):770–788. doi: 10.1038/s41576-018-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giarratana M-C, Kobari L, Lapillonne H, et al. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotechnol. 2005;23(1):69–74. doi: 10.1038/nbt1047. [DOI] [PubMed] [Google Scholar]
- 19.Weber L, Frati G, Felix T, et al. Editing a γ-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci Adv. 2020;6(7) doi: 10.1126/sciadv.aay9392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kluesner MG, Nedveck DA, Lahr WS, et al. EditR: A method to quantify base editing from sanger sequencing. CRISPR J. 2018;1(3):239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42(22):e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: A web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 2014;3(12):e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clement K, Rees H, Canver MC, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37(3):224–226. doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter MF, Zhao KT, Eton E, et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol. 2020;38(7):883–891. doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren Q, Sretenovic S, Liu S, et al. PAM-less plant genome editing using a CRISPR-SpRY toolbox. Nat Plants. 2021;7(1):25–33. doi: 10.1038/s41477-020-00827-4. [DOI] [PubMed] [Google Scholar]
- 26.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368(6488):290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurt IC, Zhou R, Iyer S, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol. 2021;39(1):41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HS, Jeong YK, Hur JK, Kim J-S, Bae S. Adenine base editors catalyze cytosine conversions in human cells. Nat Biotechnol. 2019;37(10):1145–1148. doi: 10.1038/s41587-019-0254-4. [DOI] [PubMed] [Google Scholar]
- 29.RefSNP Report - dbSNP (NCBI). rs777028217. https://www.ncbi.nlm.nih.gov/snp/rs777028217#clinical_significance
- 30.RefSNP Report - dbSNP (NCBI).rs1480884739. https://www.ncbi.nlm.nih.gov/snp/rs1480884739
- 31.ClinVar Genomic variation as it relates to human health(NCBI).VCV000795005.6. https://www.ncbi.nlm.nih.gov/clinvar/variation/795005/?oq=((787776[AlleleID]))&m=NM_000518.5(HBB):c.93-18C%3ET
- 32.Chen L, Zhang S, Xue N, et al. Engineering a precise adenine base editor with minimal bystander editing. Nat Chem Biol. 2023;19(1):101–110. doi: 10.1038/s41589-022-01163-8. [DOI] [PubMed] [Google Scholar]
- 33.Tu T, Song Z, Liu X, et al. A precise and efficient adenine base editor. Mol Ther. 2022;30(9):2933–2941. doi: 10.1016/j.ymthe.2022.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musallam KM, Rivella S, Vichinsky E, Rachmilewitz EA. Non-transfusion-dependent thalassemias. Haematologica. 2013;98(6):833–844. doi: 10.3324/haematol.2012.066845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adigbli G, Hua P, Uchiyama M, et al. Development of LT-HSC-reconstituted non-irradiated NBSGW mice for the study of human hematopoiesis in vivo. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.642198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahmig S, Kronstein-Wiedemann R, Fohgrub J, et al. Improved human erythropoiesis and platelet formation in humanized NSGW41 mice. Stem Cell Rep. 2016;7(4):591–601. doi: 10.1016/j.stemcr.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng J, Wu Y, Ren C, et al. Therapeutic base editing of human hematopoietic stem cells. Nat Med. 2020;26(4):535–541. doi: 10.1038/s41591-020-0790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frangoul H, Altshuler D, Cappellini MD, et al. CRISPR-Cas9 gene editing for sickle cell disease and β-Thalassemia. N Engl J Med. 2021;384(3):252–260. doi: 10.1056/NEJMoa2031054. [DOI] [PubMed] [Google Scholar]
- 41.Steinberg MH. Fetal hemoglobin in β hemoglobinopathies: Is enough too much? Am J Hematol. 2022;97(6):676–678. doi: 10.1002/ajh.26518. [DOI] [PubMed] [Google Scholar]
- 42.Topfer S, Feng R, Huang P, et al. Disrupting the adult-globin promoter alleviates promoter competition and reactivates foetal-globin gene expression. Blood. 2022;139(14):2107–2118. doi: 10.1182/blood.2021014205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.