Abstract
TRF-CUT, an ARB-implemented tool, was developed to predict in silico the terminal restriction fragments of aligned small-subunit rRNA gene or functional gene sequences. Application of this new tool to perform directed terminal restriction fragment length polymorphism analysis of pmoA products obtained from a forest soil revealed that novel cluster I methanotrophic bacteria were dominant.
T-RFLP (terminal restriction fragment length polymorphism) is a method commonly applied to study the structure of microbial communities (1, 2, 6, 14). PCR products of 16S rRNA genes or of functional genes end labeled with a fluorescent dye are cleaved with a site-specific restriction endonuclease to obtain genetic fingerprints of microbial communities. Recently some online tools, e.g., TAP-TRFLP (12; http://rdp.cme.msu.edu), torast (http://www.torast.de), and MiCA (http://mica.ibest.uidaho.edu/), have been developed to perform in silico hydrolysis of 16S rRNA gene sequences. However, analysis software predicting T-RFs from databases of functional marker genes (e.g., pmoA, nirK, and nifH), which are used to describe the structure of functional microbial groups, is not available.
The freely available software ARB (11; www.arb-home.de) is a graphics-oriented program including a database of small-subunit rRNA gene sequences and various sequence analysis tools. Since ARB is widely used for the phylogenetic analysis of strain-specific and environmental sequence data, it was considered worthwhile to integrate an in silico T-RFLP analysis tool (TRF-CUT) into this software package. The upgrade with TRF-CUT is simple and does not alter the ARB source code. TRF-CUT and a manual file can be obtained from http://www.uni-marburg.de/mpi/downloads/trfcut/trfcut.zip.
In addition to the small-subunit RNA database, TRF-CUT is applicable to any set of nucleic acid sequences aligned in ARB. Specification of parameters such as primer position and restriction enzyme allows the generation of individual T-RF commands that can be stored permanently. T-RFs are even predicted for sequences lacking a primer region, since the algorithm uses absolute alignment positions to determine the priming region instead of searching for individual primer sequences. This feature distinguishes TRF-CUT from any other T-RF prediction tool available to date and is of special interest because priming regions are frequently omitted from sequences submitted to public databases or may contain mismatches with respect to the primer. In these cases, these sequences would remain undetected by an in silico analysis, although they can be obtained by PCR. Furthermore, TRF-CUT indicates any species that have incomplete sequences between the primer region and the first restriction site, thus enabling the user to manually check or to exclude them from analysis if desired. T-RFs can be calculated with TRF-CUT for any restriction endonuclease with an unambiguous restriction site provided by the Rebase database (http://rebase.neb.com/rebase/rebase.html). Results of TRF-CUT are stored in ARB data fields and therefore can be used to perform searches and manipulations and can be combined for multiple-enzyme digests. Results can be exported as lists, e.g., for downstream analysis with the online tool PAT (8; http://trflp.limnology.wisc.edu) or the tRFLP Fragment Sorter (http://www.oardc.ohio-state.edu/trflpfragsort/) or can be displayed at the tips of the phylogenetic trees, thus allowing the direct assignment and visualization of T-RFs specific to phylogenetic groups (Fig. 1A). In conclusion, TRF-CUT is suitable for (i) T-RF prediction based on various enzymes to select enzymes with high phylogenetic resolution and (ii) assigning T-RFs from experimental T-RFLP data to potentially corresponding sequences in the database.
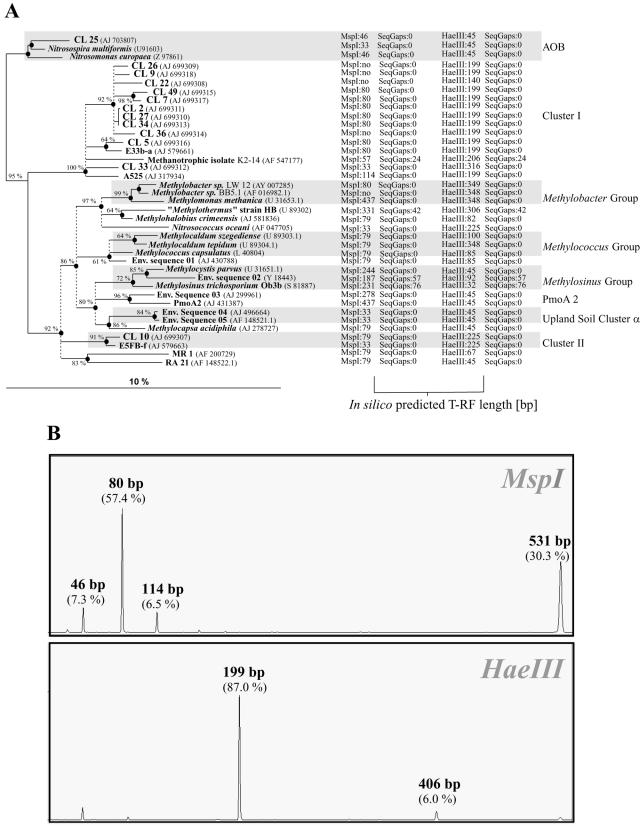
FIG. 1.
(A) Phylogeny of pmoA sequences and corresponding T-RFs predicted by in silico analysis through TRF-CUT. The tree is slightly modified from the ARB display. The tree was calculated from 127 deduced amino acid positions by the tree puzzle, maximum-likelihood, and neighbor-joining methods. Ten thousand puzzling steps were performed, and probability estimates (≥60%) are given as percentages at nodes. Accession numbers are in parentheses. Thickened nodes were common to both maximum-likelihood and neighbor-joining analyses. Dashed vertical lines (polytomies) indicate unresolved branching, which was inferred from conflicting results of the phylogenetic analyses and low (≤60%) tree puzzle support values. The scale bar indicates 10% sequence divergence. T-RF lengths after MspI and HaeIII in silico hydrolysis are given by numbers following the enzyme names; no, no restriction site within the analyzed pmoA fragment; SeqGaps 0, no gap; another number following SeqGaps indicates the size of the gap between the priming site and the start of the actual sequence. (B) T-RFs retrieved from a pH-neutral temperate forest soil by T-RFLP with MspI and HaeIII. Lengths of T-RFs represented by pmoA clones are indicated. Values in parentheses are the relative contributions of the T-RFs to the total sample fluorescence. x axis, size of T-RFs in base pairs; y axis, relative fluorescence units.
As an example, we applied TRF-CUT to evaluate the community structure of methanotrophic bacteria (MB) in a pH-neutral upland soil located near Göttingen, Germany. MB play a crucial role in the global carbon cycle. The initial oxidation of methane to methanol is catalyzed by either a particulate methane monooxygenase (pMMO) or a soluble methane monooxygenase. The membrane-bound form, pMMO, occurs in almost all known MB and is homologous to the ammonium monooxygenase of ammonia-oxidizing bacteria. The pmoA gene encodes the catalytic center of pMMO and is widely used as a functional marker in environmental studies (4, 9, 10). Its application to study MB communities in a variety of methane-consuming upland soils indicated that two novel pmoA sequence clusters, USCα and USCγ, were dominant (7, 9). Recently, novel MB belonging to the α subgroup of the class Proteobacteria but harboring pmoA sequences only distantly related to known pmoA genes (here referred to as cluster I) were isolated from a tundra soil (13). However, in contrast to USCα and USCγ, information on the distribution of this cluster is very limited.
Community analyses of MB by T-RFLP have been performed mainly by MspI hydrolysis of 6-carboxyfluorescein-labeled pmoA PCR products (6), which were amplified with the primers A189f and A682r (5). The application of this primer-enzyme combination as described by Horz et al. (6) to the Göttingen forest soil sample resulted in four major peaks with one dominant T-RF of 80 bp (57%) (Fig. 1B). Application of TRF-CUT to a database including all public-domain pmoA and amoA sequences (June 2004) revealed that the 80-bp T-RF was indicative for an environmental sequence from an upland soil (E33b-a, cluster I) and for Methylobacter sp. LW12. A 79-bp T-RF, which is experimentally difficult to distinguish from the 80-bp T-RF, corresponded to several taxa. Since the MspI hydrolysis did not resolve the MB community structure in this soil properly, we cloned PCR products and sequenced 13 environmental pmoA genes. Reconstruction of the gene phylogeny showed that the majority of sequences (11) were affiliated with cluster I sequences (Fig. 1A). Only two clones clustered with amoA genes (clone 25) and environmental cluster II sequence E5FB-f (clone 10 [9]), respectively. The lack of clones affiliated with any other group of MB suggested a dominance of cluster I pmoA sequences in this pH-neutral temperate forest soil.
On the basis of these findings, we applied TRF-CUT to test various restriction endonucleases in order to select one that allowed the discrimination of cluster I sequence types from all other pmoA and amoA genes. In silico digestion with HaeIII revealed a 199-bp fragment unique to the majority of cluster I sequences (Fig. 1A). For two cluster I sequences, TRF-CUT indicated equally unique T-RFs of 140 and 316 bp, respectively. Subsequent T-RFLP analysis of pmoA amplified from the soil DNA extract indeed showed a predominance of a 199-bp T-RF when HaeIII was used for digestion (87.0%), thus confirming the predicted result. An additional 406-bp T-RF corresponded exclusively to the pseudo-T-RF of cluster I sequences (3). Summing up the relative abundance of these two peaks, cluster I MB comprised 93.0% of the detected MB community. Peaks of the predicted sizes of 140 and 316 bp for cluster I, of 225 bp for cluster II, and of 45 bp for clones affiliated with ammonia-oxidizing bacteria were all relatively small (<1.0%) or undetected, indicating that organisms with these genotypes were only minor constituents of the methanotrophic guild in this forest soil.
In summary, application of TRF-CUT prior to T-RFLP analysis revealed cluster I as the predominant MB in this pH-neutral methane-consuming soil and indicated that the distribution of this novel gene cluster is more widespread than previously known. In this study, a clone library was necessary because of the limited availability of pmoA sequences from different habitats. However, with the prerequisite of an extensive data set, in silico predictions of multiple-enzyme digests should allow assignment of T-RFs to potentially corresponding organisms without extensive prior cloning efforts.
Acknowledgments
We acknowledge Peter F. Dunfield for critically reading the manuscript.
This work was financially supported by the German Federal Ministry of Education and Research within the Competence Network Göttingen project “Genome Research on Bacteria” (GenoMik) and the BIOLOG Biodiversity Program (01LC0021) and by grants from the Max Planck Society (Munich).
REFERENCES
- 1.Avrahami, S., R. Conrad, and G. Braker. 2002. Effect of soil ammonium concentration on N2O release and on the community structure of ammonia oxidizers and denitrifiers. Appl. Environ. Microbiol. 68:5685-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, bacteria, and archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egert, M., and M. W. Friedrich. 2003. Formation of pseudo-terminal restriction fragments, a PCR-related bias affecting terminal restriction fragment length polymorphism analysis of microbial community structure. Appl. Environ. Microbiol. 5:2555-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henckel, T., U. Jäckel, S. Schnell, and R. Conrad. 2000. Molecular analysis of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 66:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmes, A. J., A. Costello, M. E. Lidstrom, and J. C. Murrell. 1995. Evidence that particulate methane monooxygenase and ammonia monooxygenase may be evolutionary related. FEMS Microbiol. Lett. 132:203-208. [DOI] [PubMed] [Google Scholar]
- 6.Horz, H.-P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen, S., A. J. Holmes, R. A. Olsen, and J. C. Murrell. 2000. Detection of methane oxidizing bacteria in forest soil by monooygenase PCR amplification. Microbiol. Ecol. 39:282-289. [PubMed] [Google Scholar]
- 8.Kent, A. D., D. J. Smith, B. J. Benson, and E. W. Triplett. 2003. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 69:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knief, C., A. Lipski, and P. F. Dunfield. 2003. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 69:6703-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, K. Yadhu, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsh, T. L., P. R. Saxman, J. R. Cole, and J. M. Tiedje. 2000. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacheco-Oliver, M., I. R. McDonald, D. Groleau, J. C. Murrell, and C. B. Miguez. 2002. Detection of methanotrophs with highly divergent pmoA genes from Arctic soils. FEMS Microbiol. Lett. 209:313-319. [DOI] [PubMed] [Google Scholar]
- 14.Yeager, C. M., J. L. Kornosky, D. C. Housman, E. E. Grote, J. Belnap, and C. R. Kuske. 2004. Diazotrophic community structure and function in two successional stages of biological soil crusts from the Colorado Plateau and Chihuahuan Desert. Appl. Environ. Microbiol. 70:973-983. [DOI] [PMC free article] [PubMed] [Google Scholar]