Abstract
FOXO3 is a ubiquitous transcription factor expressed in response to cellular stress caused by nutrient deprivation, inflammatory cytokines, reactive oxygen species, radiation, hypoxia, and other factors. We showed previously that the association of inherited FOXO3 variants with longevity was the result of partial protection against mortality risk posed by aging-related life-long stressors, particularly cardiometabolic disease. We then referred to the longevity-associated genotypes as conferring “mortality resilience.” Serum proteins whose levels change with aging and are associated with mortality risk may be considered as “stress proteins.” They may serve as indirect measures of life-long stress. Our aims were to (1) identify stress proteins that increase with aging and are associated with an increased risk of mortality, and (2) to determine if FOXO3 longevity/resilience genotype dampens the expected increase in mortality risk they pose. A total of 4500 serum protein aptamers were quantified using the Somalogic SomaScan proteomics platform in the current study of 975 men aged 71–83 years. Stress proteins associated with mortality were identified. We then used age-adjusted multivariable Cox models to investigate the interaction of stress protein with FOXO3 longevity-associated rs12212067 genotypes. For all the analyses, the p values were corrected for multiple comparisons by false discovery rate. This led to the identification of 44 stress proteins influencing the association of FOXO3 genotype with reduced mortality. Biological pathways were identified for these proteins. Our results suggest that the FOXO3 resilience genotype functions by reducing mortality in pathways related to innate immunity, bone morphogenetic protein signaling, leukocyte migration, and growth factor response.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-023-00740-6.
Keywords: FOXO3, Genetics, Disease resilience, Cytokines, Inflammation, Cardiometabolic disease, Bone morphogenic protein signaling
Introduction
Considerable contemporary research is being devoted to increasing lifespan and, in particular, improving health-span, defined as the number of years free of disease. The number of years living with a chronic disease (YLCD) could therefore be referred to as “morbidity-span.” The latter is a relatively under-investigated area. YLCD has a major impact on public health and medical systems since these are the years of life that consume the most healthcare resources. At present, YLCD cannot be reliably anticipated. Being able to predict the length of YLCD would offer better allocation of resources to improve population health. This could include further resources devoted to strategies that increase biological resilience (currently under-leveraged), and spending more money treating or preventing disease in people predicted to have high YLCD, since this will have high impact at a population level. Understanding the biological pathways may permit therapies for those morbidities for which the patient is most at risk.
Aging is accompanied by deterioration of bodily structures and function. The capacity of organisms to withstand the impact of stress-induced damage is referred to as “resilience.” Possession of such an ability is likely to result in a greater number of years free of disease (i.e., improved health-span), but could also reduce mortality in those with disease (i.e., increase YLCD). Heritable genetic variants in only two genes, namely, APOE [1] and FOXO3 [2] (see review: [3]), have been consistently associated with longevity in multiple populations globally.
Please note that, herein, FOXO3 (in italics) refers to the gene, whereas FOXO3 (in plain text) refers to the protein, “FOXO” refers to both FOXO1 and FOXO3 proteins, Foxo3 is the mouse version of the gene, dFOXO is the Drosophila version of the gene, and daf16 is the Caenorhabditis elegans version of the gene.
In a previous study, we found that longevity associated FOXO3 single nucleotide polymorphisms (SNPs) consisted of a group of variants in close genetic proximity (within a haplotype block) [4]. In that study, we identified at least 14 putative functional sequence variants that were predicted to modify transcription factor binding efficiencies and therefore had the potential to modulate FOXO3 mRNA transcription. In the present study, we focus on the SNP rs12212067. FOXO3 longevity-associated SNPs were associated with mitigation of the lifespan-shortening effect of having a cardiometabolic disease (CMD, i.e., one or more of the conditions coronary heart disease (CHD), hypertension, and type 2 diabetes) in late life [5]. This was an example of an effect on cellular and organismal resilience, whereby FOXO3 genotypes did not prevent chronic disease, but, rather, extended the years of morbidity associated with having one or more of these conditions (YLCD).
FOXO proteins play important roles in many cellular processes, including glucose and lipid metabolism, apoptosis, autophagy, cell cycle inhibition, stress resistance, DNA repair, angiogenesis, inflammation, immune response, pluripotency, and differentiation. FOXO transcription factors are thought to protect cells from these insults and to assist in repair or elimination of damaged cells. These functions of FOXO proteins likely underlie their impact on longevity. In contrast, the deregulation of FOXO proteins has been shown to be involved in several diseases owing to their roles in autophagy [6]. FOXOs are, moreover, tumor suppressors and have other functions that may contribute to healthy aging (see review: [7]).
FOXOs impact cardiovascular disease through the maintenance of cardiomyocyte function in health and in pathological conditions such as hyperglycemic and ischemic stress [8]. FOXO proteins have been shown to inhibit and reverse cardiac hypertrophy, as observed in heart failure, through maintenance of a quiescent state and the promotion of apoptosis after mechanical stress-induced hypertrophy [9–12]. FOXO proteins are involved in the pathogenesis of type 2 diabetes. The insulin pathway senses the nutritional status of an organism and FOXO transcription factors relay this information to specific transcriptional targets. Therefore, FOXO proteins are considered as metabolic master regulators that control the response to nutrient availability [13].
FOXO proteins play critical roles in dementia as they are responsible for the maintenance of quiescence of neuronal stem cells and the clearance of reactive oxygen species (ROS) [14]. In a cellular model of Huntington disease, the co-expression of wild-type FOXO1 with mutant huntingtin protein promoted autophagy and clearance of the aberrant protein [15]. In addition, levels of nuclear FOXO3 were found to be increased in cells homozygous for Huntington disease mutation [14].
A more complete understanding of the pathways by which FOXO3 exerts its resilience effects to help promote longevity should provide better targets for aging intervention and a reduction in late-life morbidity. Multiple stress signaling pathways converge on the protein homeostasis network of cells. Activation of the integrated stress response during aging involves increased expression of stress response genes whose encoded proteins affect pathways capable of slowing the aging processes [16]. Deregulation in aging-related diseases leads to earlier mortality. From mechanistic, therapeutic, and financial perspectives, it would be valuable to discern pathways that differentiate “healthy” from “unhealthy” aging, namely, avoidance of, as compared to resilience against, chronic disease. There is abundant literature now that shows changes in serum concentration of particular proteins with aging, and that some of these are proteins associated with increased mortality. A prime aim of geroscience is to identify biomarkers of aging and implement this knowledge to reduce the burden of aging-related diseases, slow functional decline, and promote healthy aging [17]. We believe that proteins associated with increased mortality may be used as surrogates/biological markers for life-long stress, permitting us to examine and perhaps separate the effects of longevity/resilience genotypes of FOXO3, for example, on mortality in individuals experiencing chronic disease–related stress.
The present analysis was conducted as part of the Kuakini Hawaii Lifespan Study (KHHP) [18] and the Kuakini Hawaii Asia Aging Study (KHAAS) [2, 18–20], an embedded cohort study of healthy aging drawn from the original KHHP-KHAAS population. The KHHP cohort is robust for phenotype–genotype associations since the data collection was exceptionally accurate and involved cross-validation utilizing an expert Morbidity and Mortality Committee. The Japanese population in Hawaii is of Japanese origin, with little outbreeding and, based on the authors’ unpublished data, exhibits a smaller degree of genetic diversity than the overall population of Japan. The relatively high degree of genetic homogeneity made our cohort ideal for phenotype–genotype discovery.
In the present study, we use proteomic analyses in a population, followed for over five decades, to ascertain changes in concentrations of serum proteins with aging. In an attempt to better understand the mechanisms of action of the FOXO3 stress-response protein, we were particularly interested in those proteins that increase with biological age and have a propensity to increase risk of mortality. We refer to these as “stress proteins.” Our aims were (1) to identify proteins whose serum levels increase in old age and that are associated with increased risk of mortality and (2) to determine the association, if any, of these proteins with FOXO3 genotype and mortality in those at increased risk (i.e., in the upper tertile of high-risk protein levels). We hypothesized that this study would identify pathways in which FOXO3 genotype may influence not only longevity and healthy aging (i.e., avoidance), but also unhealthy aging (i.e., resilience) imposed by chronic diseases (YLCD). Such information should prove valuable to public health and provide more accurate estimates for the allocation of funds for late-life resource needs, such as Medicaid, and long-term care. Identification of druggable pathways may provide more focused therapeutic relief to those at the greatest risk.
Methods
Study cohort
Participants were American men of Japanese ancestry living on the island of Oahu, Hawaii. The men were recruited in 1965–1968 from World War II Selective Service records for the Kuakini Honolulu Heart Program (KHHP) [18], which continued from 1991 onwards as the Kuakini Honolulu-Asia Aging Study (KHAAS) [2, 18–20]. The present analysis was conducted as part of the Kuakini Hawaii Lifespan Study and the Kuakini Hawaii Health-span Study, an embedded cohort study of healthy aging drawn from the original KHHP-KHAAS population. Subjects had parents who were almost all from a limited geographic area of Japan, mostly the western, central, and southern regions [18, 21]. Recruitment took place at the same time and place (Oahu), meaning there was no apparent reason why genetic background should be substantially different among subjects. The KHHP cohort is robust for phenotype–genotype associations since the data collection was exceptionally accurate and involved cross-validation utilizing an expert Morbidity and Mortality Committee. The Japanese population in Hawaii is of Japanese origin, with little outbreeding and, based on the authors’ unpublished data, exhibits a smaller degree of genetic diversity than the overall population of Japan.
The demographic characteristics of subjects were as described previously [2, 22]. After recruitment in 1965–1968, subjects in the cohort had been followed with regular examinations and blood work until 2020, or death up to the end of 2021. Of 8006 men, 7965 had died (mean age at death 89.0 ± 6.2 SD years; range 72–108 years), and 36 (1%) were still alive (mean age 101.6 ± 1.9 SD years; range 100–108 years). Archived phenotypic data and blood samples from Examination 4 of the KHHP (1991–1993) were used. For the present study, we randomly selected 1000 subjects from among those men in KHHP examination 4 who were aged 71–83, and who also participated in KHHP examination 3 (in 1971–1974), and were, at that time, free from prevalent chronic diseases such as diabetes, kidney disease, liver disease, CHD, stroke, and chronic obstructive pulmonary disease.
Procedures performed were in accord with institutional guidelines and were approved by the Institutional Review Board of Kuakini Medical Center. Written informed consent was obtained at each examination from all study participants or from family representatives, if participants could not provide consent.
Genotyping
Leukocyte DNA obtained from participants underwent genotyping of FOXO3 SNP rs12212067 by allelic discrimination assays using TaqMan (Applied Biosystems, Inc.) and a Life Technologies QuantStudio 12K Flex OpenArray system. The longevity-associated minor (G) allele of rs12212067 creates a myeloid zinc finger 1 transcription factor binding site [22, 23]. Details of other SNPs in linkage disequilibrium with rs12212067 and their predicted effect on transcription factor binding appear in Fig. S1.
Proteomics methods
Protein concentrations in fasting serum were measured using the SOMAScan assay platform (SomaLogic Inc.). SOMAScan is an aptamer-based assay allowing for the simultaneous measurement and quantification of thousands of individual proteins. The assay uses modified aptamers, which are DNA strands created to have a 3D conformation so that each selectively recognizes a specific protein, to measure relative protein abundance via fluorescence readings from microarrays [24].
Samples selected for proteomic profiling were distributed across 12 assay plates, each of which accommodates 96 samples. Each plate includes, in addition to the cohort samples, special standardized samples for calibration and quality control as follows: three with buffer only (no sample added), five calibrator samples, and three quality control samples. Buffer-only samples are used to establish a baseline level of signal for each aptamer representing the absence of protein. Calibrator samples are standardized samples added to all plates and used for correction of systematic differences across plates.
Raw data were normalized using a probabilistic model taking into account sample-specific effects, dilution group effects, and plate effects, and which uses hybridization control sequences and calibrator samples [24] to estimate these effects. Modeling was performed using Gaussian distributions on log-transformed data, with inference using Markov-Chain Monte Carlo method.
Statistical analyses
Proteins that were increased with age were determined using a linear regression model. The false discovery rate (FDR) method was applied for multiple testing [25]. A protein that increases with age was classified according to stress status, namely, “stressed” if the protein level was greater than or equal to its upper tertile and “non-stressed” if otherwise. Then, the proteins (“stresses”) were screened for their effects, adjusting for age effect on mortality in Cox proportional hazard models. Those proteins that showed as a risk factor for mortality adjusting for age were classified as stress proteins. To assess the FOXO3 gene resilience effect of a stress protein, a Cox proportional hazard model was applied. All statistical analyses were performed using the Statistical Analysis System (SAS) version 9.4 [26]. Figures were generated using STATA 16 [27].
Pathway analysis
An online pathway analysis package (GeneMANIA; http://genemania.org) was used to scan for significant FOXO3 interacting protein/gene pathways [28]. A brief description of their data sources can be found in Mostafavi et al. [28]. By default, the GeneMANIA prediction server uses one of two different adaptive network weighting methods. For longer gene lists, such as those used in the present study, GeneMANIA adopts the basic weighting method (called GeneMANIAEntry-1 in [28] and termed “assigned based on query genes” on its web site) and weights each network so that after the networks are combined, the query genes interact as much as possible with each other while interacting as little as possible with genes not in the list. GeneMANIA “learns” from longer gene lists, allowing a gene list–specific network weighting to be calculated.
Results
Final sample size and baseline variables
Of the 1000 men selected, 5 were excluded since we did not have complete proteomics data for these, and 20 were excluded since we did not have genotyping data for them in the final dataset. The sample size for the present study was thus 975. Baseline variables for FOXO3 rs12212067 TT and TG/GG genotypes are shown in Table S1. During 28 years of follow-up, 956 (98%) men died. The median follow-up time was 12.7 years.
Modification of mortality risk by FOXO3 genotype on mortality-increased protein levels
We screened 4575 serum protein aptamers using the Somalogic SomaScan v4.1 proteomics platform. Of these, 317 proteins, passed FDR adjustment, showed increases with age, and 184 proteins classified as stress proteins were, after FDR adjustment, associated with increased mortality.
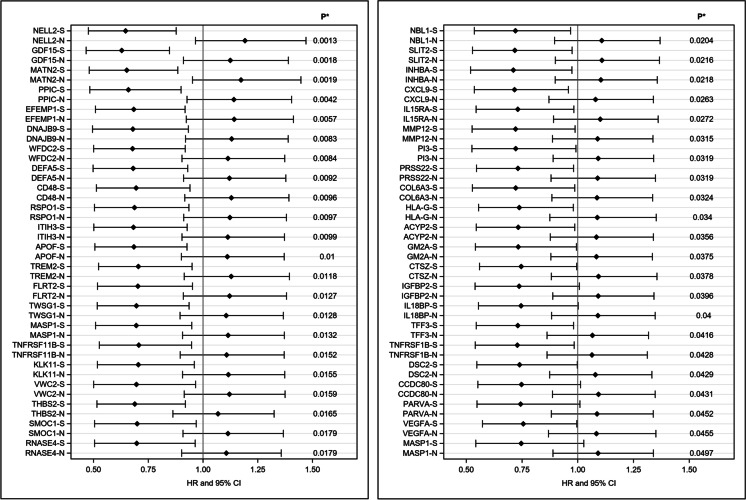
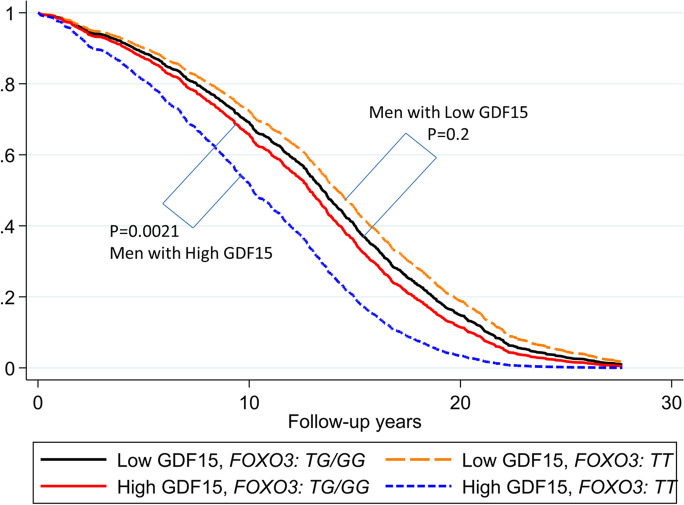
We then assessed FOXO3 genotypes for potential reduction in mortality for these stress proteins in a multivariate Cox model adjusting for age, BMI, fasting glucose, smoking (pack-years), alcohol consumption (oz/month), and physical activity index. We refer to this reduction in expected mortality under conditions of stress versus no stress as “resilience,” and the proteins were termed as “FOXO3 resilience proteins.” We included the interaction term of FOXO3 genotype with stress on mortality in the Cox model. When the interaction term was significant, it meant the effect of FOXO3 genotype on mortality differed between “stressed subjects” compared with “non-stressed subjects.” To investigate the effect of FOXO3-associated resilience proteins, we compared FOXO3 rs12212067 resilience genotypes (TG and GG) with the major allele homozygote, TT. In this way, we identified 44 FOXO3 resilience proteins (Table 1, Fig. 1). Due to the nature of low power of the test for interaction, we did not perform multiple testing adjustment for the interaction terms. Table 1 and Fig. 1 show the hazard ratios and p values of the interaction between FOXO3 genotype (TG/GG vs. TT) and stress status (stressed vs. non-stressed) for mortality.
Table 1.
Effect of FOXO3 genotype (TG/GG vs TT) on risk of mortality for aging-related proteins
| Protein/gene name | Gene symbol | UniProt ID | Stress | p** | HR*** | p for interaction |
|---|---|---|---|---|---|---|
| Protein kinase C-binding protein NELL2 | NELL2 | Q99435 | 1 | 5.10E–03 | 0.647 | 1.30E–03 |
| 0 | 1.00E–01 | 1.191 | ||||
| Growth/differentiation factor 15 | GDF15 | Q99988 | 1 | 2.30E–03 | 0.629 | 1.80E–03 |
| 0 | 2.80E–01 | 1.125 | ||||
| Matrilin-2 | MATN2 | O00339 | 1 | 6.10E–03 | 0.652 | 1.90E–03 |
| 0 | 1.30E–01 | 1.174 | ||||
| Peptidyl-prolyl cis-trans isomerase C | PPIC | P45877 | 1 | 8.70E–03 | 0.659 | 4.20E–03 |
| 0 | 2.10E–01 | 1.141 | ||||
| EGF-containing fibulin-like extracellular matrix protein 1 | EFEMP1 | Q12805 | 1 | 1.10E–02 | 0.683 | 5.70E–03 |
| 0 | 2.20E–01 | 1.142 | ||||
| DnaJ homolog subfamily B member 9 | DNAJB9 | Q9UBS3 | 1 | 1.70E–02 | 0.68 | 8.30E–03 |
| 0 | 2.40E–01 | 1.131 | ||||
| WAP four-disulfide core domain protein 2 | WFDC2 | Q14508 | 1 | 1.20E–02 | 0.679 | 8.40E–03 |
| 0 | 3.10E–01 | 1.114 | ||||
| Defensin-5 | DEFA5 | Q01523 | 1 | 1.60E–02 | 0.681 | 9.20E–03 |
| 0 | 2.80E–01 | 1.12 | ||||
| CD48 antigen | CD48 | P09326 | 1 | 1.80E–02 | 0.694 | 9.60E–03 |
| 0 | 2.50E–01 | 1.13 | ||||
| R-spondin-1 | RSPO1 | Q2MKA7 | 1 | 1.70E–02 | 0.687 | 9.70E–03 |
| 0 | 2.80E–01 | 1.122 | ||||
| Inter-alpha-trypsin inhibitor heavy chain H3 | ITIH3 | Q06033 | 1 | 1.50E–02 | 0.682 | 9.90E–03 |
| 0 | 3.10E–01 | 1.113 | ||||
| Apolipoprotein F | APOF | Q13790 | 1 | 1.40E–02 | 0.685 | 1.00E–02 |
| 0 | 3.30E–01 | 1.111 | ||||
| Triggering receptor expressed on myeloid cells 2 | TREM2 | Q9NZC2 | 1 | 2.20E–02 | 0.705 | 1.20E–02 |
| 0 | 2.60E–01 | 1.129 | ||||
| Leucine-rich repeat transmembrane protein FLRT2 | FLRT2 | O43155 | 1 | 2.30E–02 | 0.703 | 1.30E–02 |
| 0 | 2.80E–01 | 1.121 | ||||
| Twisted gastrulation protein homolog 1 | TWSG1 | Q9GZX9 | 1 | 1.70E–02 | 0.696 | 1.30E–02 |
| 0 | 3.50E–01 | 1.106 | ||||
| Sushi, von Willebrand factor type A, EGF, and pentraxin domain-containing protein 1 | MASP1 | Q4LDE5 | 1 | 2.20E–02 | 0.695 | 1.30E–02 |
| 0 | 3.00E–01 | 1.115 | ||||
| Tumor necrosis factor receptor superfamily member 11B | TNFRSF11B | O00300 | 1 | 2.00E–02 | 0.707 | 1.50E–02 |
| 0 | 3.50E–01 | 1.108 | ||||
| Kallikrein-11 | KLK11 | Q9UBX7 | 1 | 2.60E–02 | 0.705 | 1.50E–02 |
| 0 | 3.00E–01 | 1.116 | ||||
| Brorin | VWC2 | Q2TAL6 | 1 | 3.00E–02 | 0.696 | 1.60E–02 |
| 0 | 2.70E–01 | 1.121 | ||||
| Thrombospondin-2 | THBS2 | P35442 | 1 | 1.10E–02 | 0.689 | 1.60E–02 |
| 0 | 5.40E–01 | 1.069 | ||||
| SPARC-related modular calcium-binding protein 1 | SMOC1 | Q9H4F8 | 1 | 3.20E–02 | 0.7 | 1.80E–02 |
| 0 | 3.00E–01 | 1.114 | ||||
| Ribonuclease 4 | RNASE4 | P34096 | 1 | 2.90E–02 | 0.697 | 1.80E–02 |
| 0 | 3.30E–01 | 1.107 | ||||
| 0 | 1.90E–01 | 0.87 | ||||
| Neuroblastoma suppressor of tumorigenicity 1 | NBL1 | P41271 | 1 | 2.90E–02 | 0.72 | 2.00E–02 |
| 0 | 3.40E–01 | 1.108 | ||||
| Slit homolog 2 protein | SLIT2 | O94813 | 1 | 3.30E–02 | 0.717 | 2.20E–02 |
| 0 | 3.30E–01 | 1.109 | ||||
| Inhibin beta B chain | INHBA | P09529 | 1 | 3.40E–02 | 0.711 | 2.20E–02 |
| 0 | 3.50E–01 | 1.104 | ||||
| C-X-C motif chemokine 9 | CXCL9 | Q07325 | 1 | 2.40E–02 | 0.717 | 2.60E–02 |
| 0 | 4.80E–01 | 1.08 | ||||
| Interleukin-15 receptor subunit alpha | IL15RA | Q13261 | 1 | 3.80E–02 | 0.731 | 2.70E–02 |
| 0 | 3.70E–01 | 1.101 | ||||
| Macrophage metalloelastase | MMP12 | P39900 | 1 | 4.20E–02 | 0.721 | 3.20E–02 |
| 0 | 4.20E–01 | 1.089 | ||||
| Elafin | PI3 | P19957 | 1 | 4.40E–02 | 0.722 | 3.20E–02 |
| 0 | 4.00E–01 | 1.092 | ||||
| Brain-specific serine protease 4 | PRSS22 | Q9GZN4 | 1 | 3.70E–02 | 0.732 | 3.20E–02 |
| 0 | 4.30E–01 | 1.089 | ||||
| Collagen alpha-3(VI) chain: bovine pancreatic trypsin inhibitor/Kunitz inhibitor domain, isoform 1 | COL6A3 | P12111 | 1 | 4.10E–02 | 0.721 | 3.20E–02 |
| 0 | 4.30E–01 | 1.087 | ||||
| HLA class I histocompatibility antigen, alpha chain G | HLA-G | P17693 | 1 | 3.60E–02 | 0.738 | 3.40E–02 |
| 0 | 4.50E–01 | 1.088 | ||||
| Acylphosphatase-2 | ACYP2 | P14621 | 1 | 4.00E–02 | 0.733 | 3.60E–02 |
| 0 | 4.60E–01 | 1.084 | ||||
| Ganglioside GM2 activator | GM2A | P17900 | 1 | 4.50E–02 | 0.733 | 3.80E–02 |
| 0 | 4.50E–01 | 1.084 | ||||
| Cathepsin Z | CTSZ | Q9UBR2 | 1 | 4.60E–02 | 0.746 | 3.80E–02 |
| 0 | 4.20E–01 | 1.093 | ||||
| Insulin-like growth factor-binding protein 2 | IGFBP2 | P18065 | 1 | 5.50E–02 | 0.737 | 4.00E–02 |
| 0 | 4.10E–01 | 1.091 | ||||
| Interleukin-18-binding protein | IL18BP | O95998 | 1 | 5.20E–02 | 0.745 | 4.00E–02 |
| 0 | 4.20E–01 | 1.09 | ||||
| Trefoil factor 3 | TFF3 | Q07654 | 1 | 3.70E–02 | 0.7312 | 4.20E–02 |
| 0 | 5.60E–01 | 1.066 | ||||
| Tumor necrosis factor receptor superfamily member 1B | TNFRSF1B | P20333 | 1 | 3.90E–02 | 0.729 | 4.30E–02 |
| 0 | 5.70E–01 | 1.064 | ||||
| Desmocollin-2 | DSC2 | Q02487 | 1 | 4.80E–02 | 0.739 | 4.30E–02 |
| 0 | 4.80E–01 | 1.079 | ||||
| Coiled-coil domain-containing protein 80 | CCDC80 | Q76M96 | 1 | 6.00E–02 | 0.748 | 4.30E–02 |
| 0 | 4.00E–01 | 1.093 | ||||
| Alpha-parvin | PARVA | Q9NVD7 | 1 | 5.80E–02 | 0.744 | 4.50E–02 |
| 0 | 4.40E–01 | 1.086 | ||||
| Vascular endothelial growth factor D | VEGFA | O43915 | 1 | 4.70E–02 | 0.755 | 4.60E–02 |
| 0 | 4.80E–01 | 1.083 | ||||
| Sushi, von Willebrand factor type A, EGF, and pentraxin domain containing protein 1 | MASP1 | Q4LDE5 | 1 | 7.30E–02 | 0.747 | 5.00E–02 |
| 0 | 4.00E–01 | 1.091 |
MASP1 is represented twice because two separate aptamers were identified
*Stress: 1 = upper tertile, 0 = lower/middle tertile
**p value for test of HR within stress status
***Hazard ratios were estimated from least square means from Cox proportional hazard model, adjusting for age, BMI, fasting glucose, smoking (pack-years), alcohol consumption (oz/month), and physical activity index
****p value for interaction term of FOXO3 genotype and stress on mortality for comparison of hazard ratios of FOXO3 genotype (FOXO3 rs12212067 G allele carriers, TG/GG vs. common allele homozygote TT) with mortality in stressed (1) and non-stressed (0) subjects
Fig. 1.
Forest plots of hazard ratios of contrasting FOXO3 genotypes and mortality in stressed (S) and non-stressed (N) subjects for top 44 proteins that increase with aging. *p value for comparison of hazard ratios of FOXO3 genotype (FOXO3 rs12212067) G allele carriers (TG/GG) vs. major allele homozygote (TT) with mortality in stressed and non-stressed subjects. “Stressed” refers to those subjects with upper tertile of mortality-associated proteins. Note: MASP1 (last entry in Table) is represented twice because two separate aptamers were identified
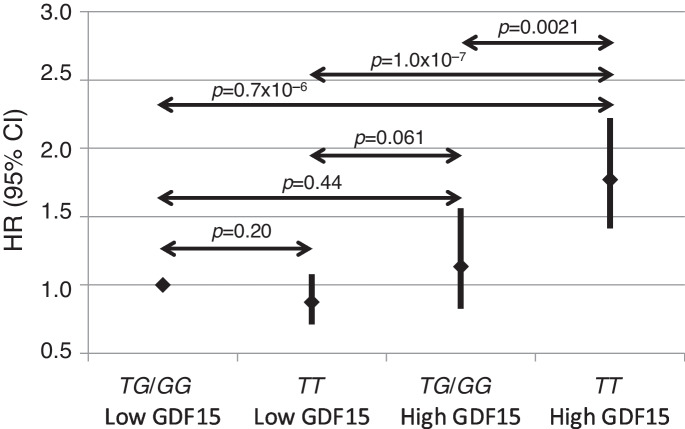
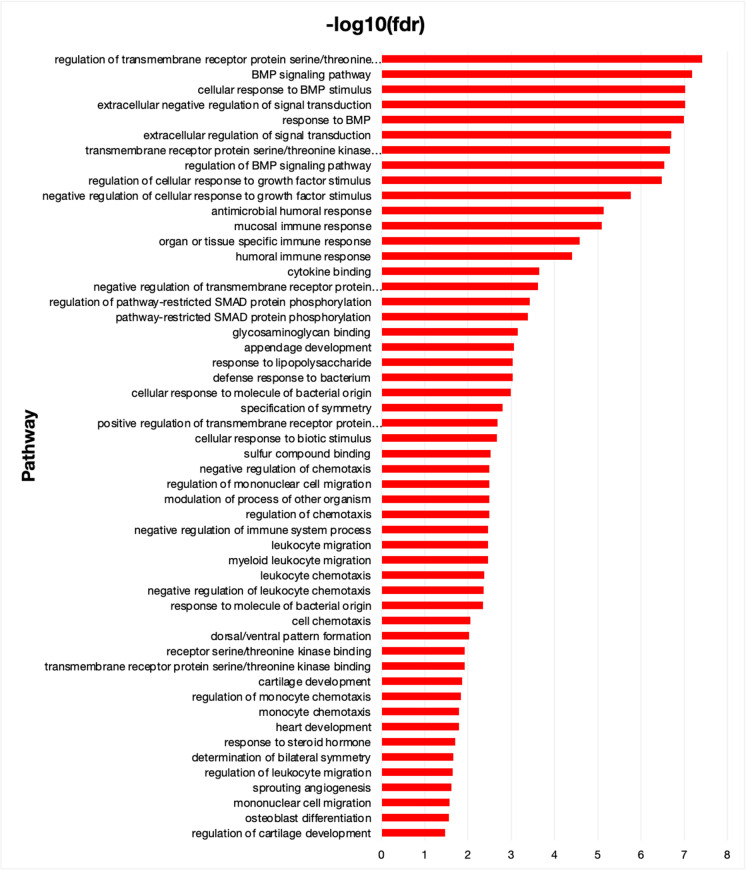
To demonstrate the effect of FOXO3 genotype on mortality in the high-risk group in a more visual manner, we chose to plot survival curves for high and low levels of the well-established aging-related, mortality-associated protein, growth/differentiation factor 15 (GDF15) (Fig. 2). The mortality risk hazard ratios are shown in Fig. 3. The expected high risk of mortality for high GDF was greatly abated in carriers of the minor (G) allele of the longevity genotype of rs12212067. Figure S2 shows survival curves based purely upon the FOXO3 genotype irrespective of circulating GDF15 levels.
Fig. 2.
Survival curves for carriers of the FOXO3 longevity (G) allele of SNP rs12212067 comparing subjects with high vs. low GDF15 protein levels. Survival curves spanning the period from baseline (1991–1993) to December 31, 2019 for subjects with “Low” GDF15 and “High” GDF15 according to whether they were carriers of the FOXO3 longevity-associated (G) allele or were major allele homozygotes (TT) of FOXO3 SNP rs12212067. The survival probabilities were estimated from the Cox proportional hazard model (see “Methods” section): h(t) = h(t0)*exp(β1*Age + β2*BMI + β3*glucose + β4*High_GDF15 + β5*FOXO3_G + β6* (High_GDF15*FOXO3_G)) by fixing age at 75 years, BMI at the mean, 23.8 kg/m2, and glucose at the mean, 111 mg/dL (where β6 is the effect of the interaction of high GDF15 with FOXO3 genotype (G-allele carriers vs. TT genotype) on mortality, giving (β6) = 0.0013). The p values for comparison of survival curves for the group with a Low GDF15 for FOXO3-G carriers vs. FOXO3-TT genotype, and comparison of survival curves for the group with a High GDF15 for FOXO3 G-allele carriers vs. FOXO3 TT genotype, were p=0.20 and p=0.0021, respectively. The p values for comparison of survival curves for FOXO3-TT genotype or for FOXO3 G-allele carriers for those with a Low GDF15 versus those with High GDF15 were p < 1.0 × 10–7 and p = 0.44, respectively
Fig. 3.
Mortality risk expressed as hazard ratio (HR) for subjects with low (lower/middle tertile) and high (upper tertile) GDF15 protein levels and FOXO3 genotype. Subjects were grouped according to GDF15 level and FOXO3 genotypes to compare their risks for mortality. HRs were estimated from Cox models adjusting for age, BMI and glucose level. This showed that the men under stress (with high GDF15) and with FOXO3 longevity genotype (TG/GG) had: (1) similar risk for mortality as those not under stress (with low GDF15) regardless of FOXO3 genotype (p = 0.44, p = 0.061); (2) significantly reduced risk for mortality compared to the men under stress (with high GDF15) and FOXO3 genotype TT (p = 0.0021)
The FOXO3 genotype had a direct effect on the actual protein levels of only one protein, CCDC80 (Table S2). CCDC80 is a coiled-coil domain-containing protein that is predicted to enable glycosaminoglycan binding activity. It may act upstream of or within extracellular matrix organization. It confers positive regulation of cell–substrate adhesion and response to bacteria, and is predicted to be located in the extracellular matrix.
Biological pathways that mediate risk of FOXO3 genotype on mortality
We used gene function prediction algorithms [28] to assess pathways involved in FOXO3-related mortality. Our analyses of FOXO3 gene/protein resilience identified various pathways, some of which had not been specifically ascribed to FOXO3 targets previously. Table 2 and Fig. 4 show that the resilience effect of FOXO3 genetic variance relies ultimately on a combination of
Immune response
Bone morphogenic protein (BMP) signaling
Signal transduction
Leukocyte migration
Growth factor response.
Table 2.
Predicted functions of genes for which FOXO3 genotype reduces risk of mortality (age increased, mortality increased, risk decreased by genotype)
| Function | FDR | Genes in network | Genes in genome |
|---|---|---|---|
| Antimicrobial humoral response | 2.61E–15 | 13 | 97 |
| Humoral immune response | 4.22E–13 | 14 | 197 |
| Defense response to bacterium | 4.16E–12 | 12 | 135 |
| BMP signaling pathway | 2.29E–06 | 7 | 70 |
| Cellular response to BMP stimulus | 2.92E–06 | 8 | 122 |
| Response to BMP | 3.57E–06 | 8 | 128 |
| Regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 4.30E–06 | 9 | 201 |
| Peptidase inhibitor activity | 4.30E–06 | 8 | 135 |
| Endopeptidase regulator activity | 5.14E–06 | 8 | 141 |
| Mucosal immune response | 1.07E–05 | 5 | 26 |
| Regulation of BMP signaling pathway | 1.31E–05 | 6 | 59 |
| Extracellular negative regulation of signal transduction | 1.31E–05 | 4 | 10 |
| Peptidase regulator activity | 1.56E–05 | 8 | 170 |
| Endopeptidase inhibitor activity | 2.19E–05 | 7 | 115 |
| Extracellular regulation of signal transduction | 2.35E–05 | 4 | 12 |
| Organ or tissue specific immune response | 2.37E–05 | 5 | 33 |
| Enzyme inhibitor activity | 3.64E–05 | 9 | 281 |
| Transmembrane receptor protein serine/threonine kinase signaling pathway | 3.64E–05 | 9 | 282 |
| Regulation of cellular response to growth factor stimulus | 8.49E–05 | 8 | 222 |
| Response to molecule of bacterial origin | 5.73E–04 | 7 | 195 |
| Response to lipopolysaccharide | 1.08E–03 | 6 | 135 |
| Cellular response to molecule of bacterial origin | 1.22E–03 | 6 | 139 |
| Negative regulation of cellular response to growth factor stimulus | 1.35E–03 | 5 | 78 |
| Cellular response to biotic stimulus | 2.63E–03 | 6 | 161 |
| Modulation of process of other organism | 4.45E–03 | 5 | 101 |
| Negative regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 4.50E–03 | 5 | 102 |
| Regulation of pathway-restricted SMAD protein phosphorylation | 1.03E–02 | 4 | 58 |
| Pathway-restricted SMAD protein phosphorylation | 1.13E–02 | 4 | 60 |
| Transmembrane receptor protein serine/threonine kinase binding | 1.73E–02 | 3 | 23 |
| Receptor serine/threonine kinase binding | 1.73E–02 | 3 | 23 |
| Appendage development | 2.00E–02 | 4 | 71 |
| Negative regulation of growth | 3.47E–02 | 5 | 163 |
| Specification of symmetry | 3.47E–02 | 4 | 83 |
| Positive regulation of transmembrane receptor protein serine/threonine kinase signaling pathway | 4.24E–02 | 4 | 88 |
| Cytokine binding | 6.77E–02 | 4 | 100 |
| Mesonephros development | 8.54E–02 | 3 | 42 |
| Mesonephric tubule development | 8.54E–02 | 3 | 42 |
| Mesonephric epithelium development | 8.54E–02 | 3 | 42 |
| Mesoderm development | 8.93E–02 | 3 | 43 |
Possible gene association network and gene functions of the top 30 proteins/genes. In silico analyses were performed using the GeneMANIA (gene function prediction using a multiple association network integration algorithm)—an integrated interaction network program that predicts gene functions and possible interaction networks using many large publicly available datasets including protein–protein and genetic interaction networks
FDR, false discovery rate
Fig. 4.
Biological pathways that mediate risk of FOXO3 genotype on mortality. Aging-increased, mortality-increased, and FOXO3 genotype–reduced risk (of mortality) genes (n = 43) were predicted to be involved in signal transduction, BMP signaling pathway, leukocyte migration, immune response, and growth factor response. A full list is available in Table 1 and Table S2
Table S3 lists the top 35 resilience proteins/genes and their functions. Table 3 provides a more general model of FOXO3 resilience. The literature indicates that FOXO3 is a stress-response protein implicated in ameliorating effects of ROS, radiation, endoplasmic reticulum stress, heat, mitochondrial dysfunction, starvation, hypoxia, NAD+/NADH imbalance, and inflammation (see review: [7]).
Table 3.
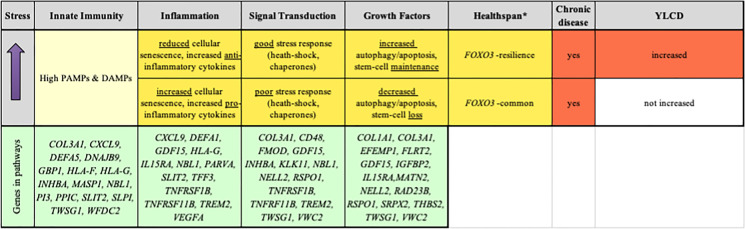
Proposed model for effect of FOXO3 resilience genotype on mortality
Protein levels were assessed for age-related changes and increased mortality rates in order to identify proteins associated with long-term stress. High-stress proteins were identified and examined for effects involving the high-risk persons (upper tertile). Age-increased, genotype-reduced risk (of mortality) genes (n = 44) were predicted to be involved in signal transduction, BMP signaling pathway, leukocyte migration, immune response, and growth factor response. Key proteins/genes are listed below their predicted biological system, but may be included at multiple points. See Table 1 and Table S2 for the full list of proteins
*FOXO3 resilience genotype responds to stressful lifestyle leading to improved stress response and reduced mortality. FOXO3 resilience genotype is a component of the longevity haplotype
PAMPs, pathogen-associated molecular pattern molecules; DAMPs, danger-associated molecular pattern molecules
Discussion
Newly discovered FOXO3 pathways
Based on our findings, we propose a FOXO3 resilience model using previous knowledge as well as the new data obtained here. The key factors involved in reducing mortality risk include the following:
Response to innate immunity: Increasing levels of cellular stress, resulting in moderating levels of danger-associated molecular patterns (DAMPs) and pathology-associated molecular patterns (PAMPs).
Reduced inflammation: The molecules involved serve to initiate formation of inflammasomes and other stress-response structures, resulting in release of inflammatory cytokines and activation of other signaling pathways.
Moderated signal transduction: Signaling pathways are implemented in order to respond to damaged organelles, cells, and extracellular structures and tissues.
Moderated growth factor levels: Damaged proteins are removed by autophagy and damaged cells are removed by apoptosis, thus increasing levels of growth factors necessary for cellular replacement and tissue repair via stem cell differentiation, without leading to stem-cell exhaustion.
Chronic disease: While chronic disease is not delayed, the FOXO3 resilience genotype delays mortality by extending the functionality of those systems shown above.
The model in Table 3 includes those stress proteins that have been implicated in the present study, along with their roles in resilience. Note that a given protein and/or its gene may be present at multiple points in the model, a prime example being GDF15.
FOXO3 longevity versus resilience genotype
It should be noted that we found SNP rs12212067 to be the only FOXO3 variant of the eight longevity variants currently tested that showed a statistically significant association with stress-protein resilience. While this variant was in linkage disequilibrium with other longevity SNPs, the equilibrium is incomplete. We believe that carriers of the rs12212067 variant may represent resilience genotypes that are a component of the longevity haplotype which comprises at least 14 SNPs [4]. The longevity-associated haplotype frequency was much higher (0.24 vs. 0.09 (from dbSNP JPN)) than the “resilience genotype” (Fig. S1). The rs12212067 resilience variant has previously been reported, by genome-wide association studies, to be associated with a milder course of Crohn’s disease and rheumatoid arthritis [29, 30]. The variant is associated with food preference/avoidance in Crohn’s disease patients. A neighboring SNP (rs12196996) has been reported to affect mRNA processing, including the expression of circFOXO3, a circular RNA associated with CHD risk [31].
New pathways identified in the current study include extracellular signal transduction, BMP signaling, leukocyte migration, and growth factor response. Extracellular signal transduction is a broad category of molecules traveling from one cell to another and most often involves protein phosphorylation, but may also include growth factors (e.g., transforming growth factor β, TGF-β), chemokines (e.g., interleukins), hormones (e.g., estrogen), survival factors (e.g., insulin-like growth factor 1, IGF1), and extracellular matrix proteins (e.g., integrin). At the top of the list of proteins modified by FOXO3 genotype is neural epidermal growth factor-like like 2 (NELL2) that causes cell proliferation and inhibition of apoptosis when overexpressed in benign prostate hyperplasia [32]. NELL2 is, moreover, required for neuron survival through the modulation of MAPK pathways.
BMPs are a group of growth factors also known as cytokines and as metabologens that are members of the TGF-β family [33]. These proteins transduce their signals through type I and type II serine-threonine kinase receptors and their intracellular downstream effectors, including small mothers against decapentaplegic (SMAD) proteins [34]. Also, at the top of our list was GDF15, which cooperates with other proteins to mediate the innate immune response to bacterial lipopolysaccharide and to viruses [35]. GDF15 is considered an anti-inflammatory protein at low levels [36]. However, its levels are known to increase in chronic disease [37] and may be an indirect indicator of cellular stress [38]. GDF15 is a mitokine, where mitokines are soluble molecules produced in response to mitochondrial stress, immunosenescence, and inflammaging [39]. Mitokines respond to the increase in stress-related inflammatory cytokines induced by interleukins IL-1β and IL-2, TNF-α, and TGF-β, but may directly lower levels of IL-18 and IL-1β [40]. Studies in transgenic mice have shown that GDF15 lowers expression of pro-inflammatory cytokines IL-18, IL-1β, TNF-α, KC, IL-6, and MCP1 [41]. GDF15 is a secreted ligand of the TGF-β superfamily of proteins. Ligands of this family bind various TGF-β receptors leading to recruitment and activation of SMAD family transcription factors. GDF15 is expressed in a broad range of cell types, acts as a pleiotropic cytokine, and is involved in the stress response program of cells after cellular injury. Increased GDF15 levels are associated with disease states such as tissue hypoxia, inflammation, acute injury, and oxidative stress [39]. Both intracellular GDF15 and the circulating mature GDF15 are implicated in biological processes such as energy homeostasis and body weight regulation [41]. GDF15 is one of a number of biomarkers of frailty onset which together highlight the importance of inflammation and nutrient sensing in this condition [42]. Among GDF15’s related pathways are CREB and ERK signaling.
Leukocyte migration (extravasation) involves the passage of cells through the intact vessel wall and the movement of leukocytes out of the circulatory system and toward the site of tissue damage or infection as part of the innate immune response [43]. This process relies heavily on the maintenance of the extracellular matrix. Matrilin-2 (MATN2) [44] and EGF receptor (EGF)-containing fibulin-like extracellular matrix 1 (EFEMP1) are members in this pathway [45].
Model of FOXO3 mortality resilience
Our model of FOXO3 mortality resilience (Table 3) includes four major biological systems in aging in which FoxO3 has been shown to be involved: (1) innate immunity, (2) inflammation (e.g., senescence-associated secretory phenotype (SASP)), (3) signal transduction, and (4) growth factor regulation and response (e.g., stem-cell maintenance, autophagy, and apoptosis). We have attempted to explain how the various stress-related proteins/genes fit into each of these systems. However, many of them play multiple roles and are involved in several pathways. The information on gene function presented below, but not the references, was taken largely from GeneCards [46] and is further detailed in Table S3.
Innate immunity
Innate immunity involves receptors, such as toll-like receptors (TLRs), heat-shock proteins, and high-mobility group proteins, that recognize protein patterns foreign to the cell and thereby present as invading pathogens [47]. These pathogenic (microbial)-associated molecular patterns (PAMPs) are small molecular motifs conserved within a class of microbes [48]. DAMPs are molecules within cells that are a component of the innate immune response released from damaged or dying cells due to trauma or an infection by a pathogen [49, 50]. Once a DAMP is released from the cell, it promotes a noninfectious inflammatory response by binding to a pattern-recognition receptor [50]. Inflammation is a key aspect of the innate immune response because it is used to help mitigate future damage to the organism by removing harmful invaders from the affected area and start the healing process [51]. In various model organisms, FoxO proteins modulate the innate immune system, including in mouse [52], Drosophila [53], Caenorhabditis elegans [54], and Hydra [55].
Foxo1 and 3 (DAF-16)-mediated immunity in adult C. elegans requires SMK-1, a regulatory subunit of the PP4 protein phosphatase complex [54]. The human homolog of SMK-1 is PPP4R3A, a component of the PP4 complex that dephosphorylates H2AX. Phosphorylation of the Ser-139 residue of the histone variant H2AX, forming γH2AX, is an early cellular response to the induction of DNA double-strand breaks [56] and is known to increase with age. dFOXO, the Drosophila equivalent of human FOXO3, promotes tolerance to hypoxia via the innate immunity transcription factor NF-κB/relish [57]. FOXO transcription factors are involved in the cellular responses to bacterial stimuli and act as central regulators of innate immune functions in respiratory epithelial cells [52].
In response to protein misfolding and endoplasmic reticulum (ER) stress, FOXO proteins integrate upstream ER stress and unfolded protein response (UPR) signals with the transcriptional machinery to decrease translation, promote cell survival/termination, and increase the levels of ER-resident chaperones and of ER-associated degradation (ERAD) components to restore ER homeostasis [58].
One of the stress proteins we identified was peptidase inhibitor 3 (PI3), which functions as an antimicrobial protein against Gram-positive and Gram-negative bacteria, and fungal pathogens. CCCL9 is part of a chemokine superfamily that encodes secreted proteins involved in immunoregulatory and inflammatory processes [59]. PI3 is thought to be involved in T-cell trafficking [60]. Defensins such as DEFA5 are a family of antimicrobial and cytotoxic peptides thought to be involved in host defense [61]. They are abundant in the granules of neutrophils [62] and are also found in the epithelia of mucosal surfaces such as those of the intestine, respiratory tract, urinary tract, and vagina. Defensins possess antimicrobial activity against Gram-negative and Gram-positive bacteria [63]. They are thought to kill microbes by permeabilizing their plasma membrane. DNAJB9 is localized within the endoplasmic reticulum, is induced by ER stress, and protects stressed cells from apoptosis [64]. Among its related pathways are metabolism of proteins and the UPR. DNAJB9 is required for survival of B-cell progenitors and for normal antibody production. MASP1 is a serine protease that functions as a component of the lectin pathway of complement activation. The complement pathway plays an essential role in the innate and adaptive immune response. PPIC is a peptidyl-prolyl cis-trans isomerase (PPIase) that catalyzes the cis-trans isomerization of proline imidic peptide bonds in oligopeptides and accelerate that this the folding of proteins.
Inflammation
The NLRP3 inflammasome is a critical component of the innate immune system [65]. It mediates caspase-1 activation and the secretion of proinflammatory cytokines IL-1β and IL-18 in response to microbial infection and cellular damage. However, the aberrant activation of the NLRP3 inflammasome has been linked to several inflammatory disorders, which include cryopyrin-associated periodic syndromes, Alzheimer’s disease, diabetes, and atherosclerosis [65]. FOXO3 restores autophagy flux and attenuates the activation of the NLRP3 inflammasome in Kupffer cells by promoting the transcription of Bim (BCL2L11) [66], suggesting that this could be a potential therapeutic target in non-alcoholic fatty liver disease and other obesity-related diseases. BCL2L11 induces apoptosis, possibly through a caspase-mediated pathway. FOXO3 [67] and FOXO4 [68] are effective inhibitors of NF-κB signaling and can reduce immune responses.
Cellular senescence is a process of permanent cell-cycle arrest during which cells are unable to re-enter the cell cycle despite the presence of growth factors, therefore limiting the lifespan of mammalian cells and preventing unlimited proliferation. Cellular senescence is beneficial during normal embryonic development and tissue damage because of its ability to promote tissue remodeling and renewal [69]. Overexpression of FOXO3 suppressed the senescence process of cerebral microvascular endothelial cells (CMECs) under replicative stress by re-activating the transcription of antioxidant genes and thereby inhibiting ROS generation [70].
CXCL9 is part of a chemokine superfamily that encodes secreted proteins involved in immunoregulatory and inflammatory processes [71]. It is thought to be involved in T-cell trafficking. CXCL9 binds to C-X-C motif chemokine 3 and is a chemoattractant for lymphocytes but not for neutrophils. This cytokine affects the growth, movement, or activation state of cells that participate in immune and inflammatory response. HLA-G is a non-classical major histocompatibility class Ib molecule involved in immune regulatory processes at the maternal–fetal interface [72]. Upon interaction with KIR2DL4 and LILRB1 receptors on decidual NK cells, it triggers NK cell senescence–associated secretory phenotype as a molecular switch to promote vascular remodeling and fetal growth in early pregnancy. TNFRSF1B and TNFRSF11B are members of the TNF-receptor superfamily. Among related pathways are cytokine signaling in immune system and osteoblast signaling. Gene Ontology (GO) annotations include signaling receptor activity and cytokine activity. TREM2 functions in immune response and may be involved in chronic inflammation by triggering the production of constitutive inflammatory cytokines [73]. It regulates microglial proliferation by acting as an upstream regulator of the Wnt/beta-catenin signaling cascade. REM2 also regulates microglial chemotaxis and process outgrowth, and also the microglial response to oxidative stress and lipopolysaccharide. It suppresses PI3K and NF-κB signaling in response to lipopolysaccharide, thus promoting phagocytosis, suppressing pro-inflammatory cytokine and nitric oxide production, inhibiting apoptosis, and increasing expression of IL10 and TGFβ [74]. During oxidative stress, REM2 promotes anti-apoptotic NF-κB signaling and ERK signaling. It also has a role in microglial mTOR activation and metabolism.
Signal transduction, growth factors, and extracellular matrix
Signal transduction is a broad category of inter- and intra-cellular communications of molecular events commonly involving phosphorylation catalyzed by protein kinases. The most consequential pathway involving FOXO is insulin/IGF-1 signaling in which nutrients are sensed in a balance between proliferation versus quiescence. Activation of the PI3K/AKT pathways leads to the inhibition of FOXOs and activation of mTOR, thus regulating the cell cycle (see review: [75]). The PI3K/AKT pathways cause activation of FOXOs, leading to cell-cycle arrest and apoptosis [76]. The MAPK/ERK pathway plays an integral part in cell-cycle entry and proliferation by integrating mitogen and stress signals [77].
Inhibin subunit β E (INHBA) is an important paralog of the TGF-β gene. INHBA regulates food intake, energy expenditure, and body weight in response to metabolic and toxin-induced stresses. By binding to its receptor, GFRAL, GDF15 activates GFRAL-expressing neurons localized in the area postrema and nucleus tractus solitarius of the brainstem [78]. This then triggers the activation of neurons localized within the parabrachial nucleus and central amygdala, which constitutes part of the “emergency circuit” that shapes feeding responses to stressful conditions. In hepatocytes, GDF15 inhibits growth hormone signaling. Inhibins and activins are involved in regulation of a number of diverse functions such as hypothalamic and pituitary hormone secretion, gonadal hormone secretion, germ cell development and maturation, erythroid differentiation, insulin secretion, nerve cell survival, embryonic axial development, or bone growth, depending on their subunit composition [79].
Neuroblastoma, suppression of tumorigenicity 1 (NBL1) contains a domain resembling the C-terminal cystine knot-like (CTCK) motif found in a number of signaling molecules. These proteins are secreted and act as BMP antagonists by binding to BMPs and preventing them from interacting with their receptors [80].
Neuron-specific EGF-like protein 2 (NELL2) is a glycoprotein containing several von Willebrand factor C domains and EGF-like domains. Studies in mouse suggest that this protein plays a role in neural cell growth and differentiation as well as in oncogenesis and is required for neuron survival through the modulation of MAPK pathways [81].
The extracellular matrix harbors many of the wound-healing cells that include proteins associated with tissue growth and repair [82]. Cluster of differentiation 48 (CD48) is a neutrophil and pancreatic elastase-specific inhibitor. The protein COL6A3 (collagen α-3(VI) chain) contains von Willebrand Factor type A domains that explain its importance in organizing matrix components. EFEMP1 encodes a member of the fibulin family of extracellular matrix glycoproteins that contain tandemly repeated epidermal growth factor-like repeats. Among its related pathways are integrin pathway and ERK signaling. Its binding to EGFR induces EGFR autophosphorylation and the activation of downstream signaling pathways. Fibronectin leucine-rich repeat transmembrane 2 protein (FLRT2) has a role in fibroblast growth factor (FGF)-mediated signaling cascades [83]. It is required for normal organization of the cardiac basement membrane during embryogenesis, and for normal embryonic epicardium and heart morphogenesis. MATN2 is a member of the von Willebrand factor A domain containing protein family. R-spondin-1 (RSPO1) is a secreted activator protein that positively regulates the Wnt signaling pathway. In mice, this protein induces the rapid onset of crypt cell proliferation and increases intestinal epithelial healing, providing a protective effect against chemotherapy-induced adverse effects [84]. RSPO1 is a beta-cell growth factor and insulin secretagogue [85]. Thrombospondin-2 (THBS2) mediates cell-to-cell and cell-to-matrix interactions. This protein has been shown to function as a potent inhibitor of tumor growth and angiogenesis [86]. Twisted gastrulation protein homology 1 (TWSG1) enables TGF-β binding activity. It is involved in several processes, including BMP signaling [87], negative regulation of CD4-positive, α-β T-cell proliferation, positive regulation of pathway-restricted SMAD protein phosphorylation, and TGF-β receptor signaling pathway. THBS2 seems to antagonize BMP signaling by forming ternary complexes with CHRD and BMPs, thereby preventing BMPs from binding to their receptors. In addition to the anti-BMP function, it also has pro-BMP activity, partly mediated by cleavage and degradation of CHRD, which releases BMPs from ternary complexes. THBS2 may be an important modulator of BMP-regulated cartilage development and chondrocyte differentiation. Von Willebrand factor C containing 2 (WC2; also known as brorin) encodes a secreted BMP antagonist. Brorin is possibly involved in neural function and development [88], and may have a role in cell adhesion. BMP antagonist may play a role in neural development [89].
Autophagy
Autophagy controls cellular remodeling and quality control [90]. Dysregulated autophagy has been implicated in obesity, diabetes, cardiovascular disease, neurodegenerative diseases, and cancer. FOXO transcription factors have a multifaceted role in autophagy regulation and dysregulation [6]. Nuclear FOXOs transactivate genes that control the formation of autophagosomes and their fusion with lysosomes. Independently of transactivation, cytosolic FOXO proteins induce autophagy by directly interacting with autophagy proteins. Autophagy is also controlled by FOXOs through epigenetic mechanisms (see review: [3]). FOXO1 is primarily present in the cytosol, while FOXO3 is partitioned equally in both cytoplasm and nucleus.
In muscle, FOXO1 and FOXO3 elevate phagocytosis by increasing the expression of autophagy genes, mainly working as part of the core machinery, and additionally increase protein degradation via the proteasomal pathway [6]. In particular, FOXO3 increases the capacity of the lysosome to degrade incoming cargo, indicating a role for lysosomal function in muscle atrophy [91]. UV excision repair protein RAD23B is a multiubiquitin chain receptor involved in modulation of proteasomal degradation [92]. It may play a role in endoplasmic reticulum–associated degradation (ERAD) of misfolded glycoproteins by association with PNGase and delivering deglycosylated proteins to the proteasome [93, 94].
Apoptosis
Apoptosis is a form of programmed cell death that occurs in multicellular organisms [95]. It can be initiated through one of two pathways. In the intrinsic pathway, the cell kills itself because it senses cell stress, while in the extrinsic pathway the cell kills itself because of signals from other cells. Both pathways induce cell death by activating caspases, which are proteases or other enzymes that degrade proteins. Excessive apoptosis causes atrophy, whereas an insufficient amount results in uncontrolled cell proliferation, such as autoimmune diseases and cancer [96, 97]. Interleukin 15 receptor α subunit (IL15RA) encodes a cytokine receptor that specifically binds interleukin 15 (IL15) with high affinity and that is reported to enhance cell proliferation and expression of apoptosis inhibitor BCL2L1/BCL2-XL and BCL2 [98].
Stem cell maintenance
Stem cells are undifferentiated or partially differentiated cells that can differentiate into various types of cells and proliferate indefinitely to produce more of the same stem cell through a process of self-renewal. FOXO1 and FOXO3 have been reported to regulate germ cells, neural stem cells, hematopoietic stem cells, muscle satellite cells, and cancer stem cells [99].
FOXO3 serves as a core regulator of cellular homeostasis, stress response, and longevity through its ability to modulate a variety of stress responses during nutrient shortage, oxidative stress, hypoxia, heat shock, and DNA damage [100–103]. By reducing oxidative damage responsible for aging, FOXO3-mediated responses to stress are pivotal to health-span and lifespan [104]. Depending on the stress stimulus and subcellular context, once activated, FOXO3 can induce specific sets of nuclear genes, including cell-cycle inhibitors, pro-apoptotic genes, scavengers of ROS, autophagy effectors, and gluconeogenic enzymes [103]. On the other hand, under glucose restriction, FOXO3 translocates to mitochondria to stimulate transcription of oxidative phosphorylation genes, thus restoring cellular ATP levels [103]. FOXO3 target genes and the pathways that their gene products serve are diverse and sometimes antagonistic, meaning FOXO3 is an adaptable player in the dynamic homeostasis of normal and stressed cells [103].
FOXO proteins are critical in response to cellular damage, and help to orchestrate autophagy, the process of marking old proteins/structures, and implementing apoptosis and cellular death, when damage is insurmountable and cannot be corrected [6]. FOXO transcription factors have been shown to regulate metabolic homeostasis, neurogenesis and neuroprotection, cardiac remodeling, skeletal muscle homeostasis, immunity, endocytosis, stem cell homeostasis, and cancer cell growth and invasion [6].
FOXO proteins play pivotal roles in determining whether there are sufficient nutrients for growth or whether cells remain quiescent [105]. It is not clear whether long-lived individuals are born with more stem cells, which may cause them to have a higher prevalence of cancer, or are more adept at maintaining the stem cells they are born with. FOXO3 has been shown to be critical in the maintenance of adult hematopoietic stem cells [99]. It has been found to be the main transcription factor regulating autophagy-related gene expression and pathways in hemopoietic stem cells [106], has been implicated in the maintenance of neural stem cells [107], and is crucial for maintenance of muscle stem cell quiescence [108].
Overlap with other proteomics studies
Other groups have reported serum protein levels that increase with age. However, few are associated with mortality. Findings of three studies [109–111] are shown in Table S4. The proteins of note from these that overlap with our current study, and for which FOXO3 genotype reduces mortality, include coiled-coil domain-containing protein 80 (CCDC80), EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1), GDF15, insulin-like growth factor-binding protein 2 (IGFBP2), macrophage metalloelastase (MMP12), SPARC-related modular calcium-binding protein 1 (SMOC1), thrombospondin-2 (THBS2), and WAP four-disulfide core domain protein 2 (WFDC2).
FOXO3 resilience genotype
The FOXO3 longevity genotype is actually a haplotype that includes at least 14 SNPs that have putative functional differences (i.e., transcription factor binding site changes) compared with the more common non-long-lived haplotypes [4]. These SNPs are in a high degree of linkage disequilibrium (LD) (i.e., are closely correlated and co-inherited) in the Japanese population that our subjects belong to. This is because SNP rs12212067, while in LD with the others, has a lower minor allele frequency (0.09 vs 0.26) and may reflect a resilience haplotype that overlaps with the entire FOXO3 longevity haplotype [4]. We reported previously that FOXO3 SNP rs12212067 is located close to a promoter for a transcription variant of FOXO3 that is associated with increased levels of full-length FOXO3 mRNA isoform in peripheral blood and a decrease in truncated FOXO3 full-length mRNA isoforms in skeletal muscle RNA [112]. This may suggest that the current resilience genotype is actually distinct, but part of a longevity haplotype. (Please note that herein “isoform” refers to the various mRNA transcripts that can be generated from a gene.)
In support of the significance of this SNP, Lee and colleagues observed a striking genotype-specific difference in the production of cytokines, with PBMC from rs12212067 minor (G) allele homozygotes secreting less TNF-α (which is proinflammatory) than is secreted by major (T) allele homozygous individuals [29]. PBMCs from minor allele homozygotes also secreted relatively more IL-10 (anti-inflammatory) in response to higher concentrations of lipopolysaccharide binding protein in PBMCs from healthy donors. They showed that FOXO3 increases inflammatory cytokine levels due to several FOXO3 binding sites in the TGF-β1 promoter. The G-allele of rs12212067 was associated with an increased risk of malaria [113]. Others confirmed that the G-allele was associated with an increased inflammatory response to Plasmodium falciparum malaria [113]. The finding of increased risk of Crohn’s disease [29, 30] was independently confirmed by Alonso and colleagues [114].
Limitations
Although 88% of participants were born in Hawaii, there was a theoretical possibility of confounding of FOXO3 genotypes due to geographic origin.
Conclusion
The study found 44 “stress” proteins that influence the association of FOXO3 genotype with reduced mortality. Biological pathways of these proteins suggest that the FOXO3 resilience genotype functions by reducing mortality by mechanisms relating to innate immunity, BMP signaling, leukocyte migration, and growth factor response.
Supplementary information
(DOCX 439 kb)
Author contribution
Conceptualization: T.A.D. Methodology: T.A.D, R.C., E.L., E.K.M., K.F., N.S. Formal analysis and investigation: T.A.D., R.C., E.L., E.K.M., K.F., N.S. Writing—original draft preparation: T.A.D. Writing—review and editing: T.A.D., B.J.M., E.K.M. Funding acquisition: T.A.D., B.J.W. Resources: T.A.D., K.H.M., E.K.M. Supervision: T.A.D., B.J.M., B.J.W.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by the Kuakini Medical Center, the US National Institutes of Health (contract N01-AG-4-2149, Grants 5 U01 AG019349-05, 5R01AG027060 [Kuakini Hawaii Lifespan Study], 5R01AG038707 [Kuakini Hawaii Healthspan Study], 1P20GM125526-01A1 [Kuakini Center of Biomedical Research Excellence for Clinical and Translational Research on Aging]), and contract N01-HC-05102 from the National Heart, Lung, and Blood Institute.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Declarations
Conflict of interest
B.J.W. and T.A.D. currently hold US patent 20130295566 entitled “Method of using FOXO3A polymorphisms and haplotypes to predict and promote healthy aging and longevity.” E.K.M., K.F., and N.S. are employees of BioAge Labs.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Schachter F, Fauredelanef L, Guenot F, et al. Genetic associations with human longevity at the APOE and ACE loci. Nature Genet. 1994;6(1):29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- 2.Willcox BJ, Donlon TA, He Q, et al. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci USA. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris BJ, Willcox BJ, Donlon TA. Genetic and epigenetic regulation of human aging and longevity. Biochim Biophys Acta Mol Basis Dis. 2019;1865(7):1718–1744. doi: 10.1016/j.bbadis.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donlon TA, Morris BJ, Chen R, et al. FOXO3 longevity interactome on chromosome 6. Aging Cell. 2017;16(5):1016-25. [DOI] [PMC free article] [PubMed]
- 5.Chen R, Morris BJ, Donlon TA, et al. Foxo3 longevity genotype mitigates the increased mortality risk in men with a cardiometabolic disease. Aging. 2020;12(23):23509–23524. doi: 10.18632/aging.202175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng Z. The FoxO-autophagy axis in health and disease. Trends Endocrinol Metab. 2019;30(9):658–671. doi: 10.1016/j.tem.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Morris BJ, Willcox DC, Donlon TA, Willcox BJ. FOXO3: a major gene for human longevity - a mini-review. Gerontology. 2015;61(6):515-25. [DOI] [PMC free article] [PubMed]
- 8.Xin Z, Ma Z, Jiang S, et al. FOXOs in the impaired heart: new therapeutic targets for cardiac diseases. Biochim Biophys Acta Mol Basis Dis. 2017;1863(2):486–498. doi: 10.1016/j.bbadis.2016.11.023. [DOI] [PubMed] [Google Scholar]
- 9.Hariharan N, Ikeda Y, Hong C, et al. Autophagy plays an essential role in mediating regression of hypertrophy during unloading of the heart. PLoS One. 2013;8(1):e51632. doi: 10.1371/journal.pone.0051632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao DJ, Jiang N, Blagg A, et al. Mechanical unloading activates FoxO3 to trigger Bnip3-dependent cardiomyocyte atrophy. J Am Heart Assoc. 2013;2(2):e000016. doi: 10.1161/JAHA.113.000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei X, Wu B, Zhao J, et al. Myocardial hypertrophic preconditioning attenuates cardiomyocyte hypertrophy and slows progression to heart failure through upregulation of S100A8/A9. Circulation. 2015;131(17):1506–17. doi: 10.1161/CIRCULATIONAHA.114.013789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119(9):2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pajvani UB, Accili D. The new biology of diabetes. Diabetologia. 2015;58(11):2459–2468. doi: 10.1007/s00125-015-3722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renault VM, Rafalski VA, Morgan AA, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vidal RL, Figueroa A, Court FA, et al. Targeting the UPR transcription factor XBP1 protects against Huntington's disease through the regulation of FoxO1 and autophagy. Hum Mol Genet. 2012;21(10):2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derisbourg MJ, Hartman MD, Denzel MS. Perspective: Modulating the integrated stress response to slow aging and ameliorate age-related pathology. Nat Aging. 2021;1(9):760–768. doi: 10.1038/s43587-021-00112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerville F, De Souto BP, Ader I, et al. Revisiting the hallmarks of aging to identify markers of biological age. J Prev Alzheimers Dis. 2020;7(1):56–64. doi: 10.14283/jpad.2019.50. [DOI] [PubMed] [Google Scholar]
- 18.Worth RM, Kagan A. Ascertainment of men of Japanese ancestry in Hawaii through World War II Selective Service registration. J Chronic Dis. 1970;23:389–397. doi: 10.1016/0021-9681(70)90022-6. [DOI] [PubMed] [Google Scholar]
- 19.Kagan A, Harris BR, Winkelstein W, Jr, et al. Epidemiologic studies of coronary heart disease and stroke in Japanese men living in Japan, Hawaii and California: demographic, physical, dietary and biochemical characteristics. J Chronic Dis. 1974;27:345–364. doi: 10.1016/0021-9681(74)90014-9. [DOI] [PubMed] [Google Scholar]
- 20.Yano K, Reed DM, McGee DL. Ten-year incidence of coronary heart disease in the Honolulu Heart Program. Relationship to biologic and lifestyle characteristics. Am J Epidemiol. 1984;119:653–666. doi: 10.1093/oxfordjournals.aje.a113787. [DOI] [PubMed] [Google Scholar]
- 21.Kagan A, editor. The Honolulu Heart Program: an epidemiological study of coronary heart disease and stroke. Amsterdam, The Netherlands: Harwood Academic Publishers; 1996. [Google Scholar]
- 22.Donlon TA, Curb JD, He Q, et al. FOXO3 gene variants and human aging: coding variants may not be key players. J Gerontol A Biol Sci Med Sci. 2012;67:1132–1139. doi: 10.1093/gerona/gls067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donlon TA, Willcox BJ, Morris BJ. FOXO3 cell resilience gene neighborhood. Aging. 2017; 9(12):2467-8. [DOI] [PMC free article] [PubMed]
- 24.Gold L, Ayers D, Bertino J, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 26.Statistical Analysis System (SAS) version 9.4. SAS Institute, Cary, NC, USA. https://libguides.library.kent.edu/statconsulting/SAS. Accessed 22 Apr 2022
- 27.Stata 16. https://blog.stata.com/2019/06/26/stata-16-released/
- 28.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008; 9(Suppl 1):S4. [DOI] [PMC free article] [PubMed]
- 29.Lee JC, Espéli M, Anderson CA, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155:57–69. doi: 10.1016/j.cell.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marlow G, Han DY, Triggs CM, Ferguson LR. Food intolerance: associations with the rs12212067 polymorphism of FOXO3 in Crohn's disease patients in New Zealand. J Nutrigenet Nutrigenomics. 2015;8(2):70–80. doi: 10.1159/000435783. [DOI] [PubMed] [Google Scholar]
- 31.Zhou YL, Wu WP, Cheng J, et al. CircFOXO3 rs12196996, a polymorphism at the gene flanking intron, is associated with circFOXO3 levels and the risk of coronary artery disease. Aging. 2020;12(13):13076-89. [DOI] [PMC free article] [PubMed]
- 32.Liu J, Liu D, Zhang X, et al. NELL2 modulates cell proliferation and apoptosis via ERK pathway in the development of benign prostatic hyperplasia. Clin Sci (Lond). 2021;135(13):1591–1608. doi: 10.1042/CS20210476. [DOI] [PubMed] [Google Scholar]
- 33.Anusuya GS, Kandasamy M, Jacob Raja SA, Sabarinathan S, Ravishankar P, Kandhasamy B. Bone morphogenetic proteins: signaling periodontal bone regeneration and repair. J Pharm Bioallied Sci. 2016;8(Suppl 1):S39–s41. doi: 10.4103/0975-7406.191964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyazono K, Kamiya Y, Morikawa M. Bone morphogenetic protein receptors and signal transduction. J Biochem. 2010;147(1):35–51. doi: 10.1093/jb/mvp148. [DOI] [PubMed] [Google Scholar]
- 35.Rochette L, Zeller M, Cottin Y, Vergely C. GDF15: an emerging modulator of immunity and a strategy in COVID-19 in association with iron metabolism. Trends Endocrinol Metab. 2021;32(11):875–889. doi: 10.1016/j.tem.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luan HH, Wang A, Hilliard BK, et al. GDF15 is an inflammation-induced central mediator of tissue tolerance. Cell. 2019;178(5):1231–44.e11. doi: 10.1016/j.cell.2019.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D, Day EA, Townsend LK, Djordjevic D, Jørgensen SB, Steinberg GR. GDF15: emerging biology and therapeutic applications for obesity and cardiometabolic disease. Nat Rev Endocrinol. 2021;17(10):592–607. doi: 10.1038/s41574-021-00529-7. [DOI] [PubMed] [Google Scholar]
- 38.Patel S, Alvarez-Guaita A, Melvin A, et al. GDF15 provides an endocrine signal of nutritional stress in mice and humans. Cell Metab. 2019;29(3):707–18.e8. doi: 10.1016/j.cmet.2018.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim KH, Lee MS. GDF15 as a central mediator for integrated stress response and a promising therapeutic molecule for metabolic disorders and NASH. Biochim Biophys Acta Gen Subj. 2021;1865(3):129834. doi: 10.1016/j.bbagen.2020.129834. [DOI] [PubMed] [Google Scholar]
- 40.Conte M, Martucci M, Chiariello A, Franceschi C, Salvioli S. Mitochondria, immunosenescence and inflammaging: a role for mitokines? Semin Immunopathol. 2020;42(5):607–617. doi: 10.1007/s00281-020-00813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baek SJ, Eling T. Growth differentiation factor 15 (GDF15): a survival protein with therapeutic potential in metabolic diseases. Pharmacol Ther. 2019;198:46–58. doi: 10.1016/j.pharmthera.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonçalves R, Maciel ÁCC, Rolland Y, Vellas B, de Souto BP. Frailty biomarkers under the perspective of geroscience: a narrative review. Ageing Res Rev. 2022;81:101737. doi: 10.1016/j.arr.2022.101737. [DOI] [PubMed] [Google Scholar]
- 43.Nourshargh S, Alon R. Leukocyte migration into inflamed tissues. Immunity. 2014;41(5):694–707. doi: 10.1016/j.immuni.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Korpos É, Deák F, Kiss I. Matrilin-2, an extracellular adaptor protein, is needed for the regeneration of muscle, nerve and other tissues. Neural Regen Res. 2015;10(6):866–869. doi: 10.4103/1673-5374.158332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou YH, Hu Y, Yu L, et al. Weaponizing human EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1) for 21(st) century cancer therapeutics. Oncoscience. 2016;3(7-8):208–219. doi: 10.18632/oncoscience.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.GenCards: the human gene database. Version 5.10. 2022 https://www.genecards.org
- 47.Imler JL, Hoffmann JA. Toll receptors in innate immunity. Trends Cell Biol. 2001;11(7):304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- 48.Taghavi M, Khosravi A, Mortaz E, Nikaein D, Athari SS. Role of pathogen-associated molecular patterns (PAMPS) in immune responses to fungal infections. Eur J Pharmacol. 2017;808:8–13. doi: 10.1016/j.ejphar.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 49.Tang D, Kang R, Coyne CB, Zeh HJ, Lotze MT. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev. 2012;249(1):158–175. doi: 10.1111/j.1600-065X.2012.01146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahla RS, Reddy MC, Prasad DV, Kumar H. Sweeten PAMPs: role of sugar complexed PAMPs in innate immunity and vaccine biology. Front Immunol. 2013;4:248. doi: 10.3389/fimmu.2013.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, Liu S, Goraya MU, Maarouf M, Huang S, Chen JL. Host immune response to influenza A virus infection. Front Immunol. 2018;9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seiler F, Hellberg J, Lepper PM, et al. FOXO transcription factors regulate innate immune mechanisms in respiratory epithelial cells. J Immunol (Baltimore). 2013;190(4):1603–1613. doi: 10.4049/jimmunol.1200596. [DOI] [PubMed] [Google Scholar]
- 53.Becker T, Loch G, Beyer M, et al. FOXO-dependent regulation of innate immune homeostasis. Nature. 2010;463:369–373. doi: 10.1038/nature08698. [DOI] [PubMed] [Google Scholar]
- 54.McHugh DR, Koumis E, Jacob P, et al. DAF-16 and SMK-1 contribute to innate immunity during adulthood in Caenorhabditis elegans. G3 (Bethesda) 2020;10(5):1521–1539. doi: 10.1534/g3.120.401166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boehm AM, Khalturin K, Erxleben FA, et al. FoxO is a critical regulator of stem cell maintenance in immortal Hydra: Proc Natl Acad Sci USA. 2013;109:19697-19602, 2012. [DOI] [PMC free article] [PubMed]
- 56.Mah LJ, El-Osta A, Karagiannis TC. GammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24(4):679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 57.Barretto EC, Polan DM, Beevor-Potts AN, Lee B, Grewal SS. Tolerance to hypoxia is promoted by FOXO regulation of the innate immunity transcription factor NF-κB/relish in Drosophila. Genetics. 2020;215(4):1013–1025. doi: 10.1534/genetics.120.303219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alasiri G, Fan LY, Zona S, et al. ER stress and cancer: the FOXO forkhead transcription factor link. Mol Cell Endocrinol. 2018;462(Pt B):67–81. doi: 10.1016/j.mce.2017.05.027. [DOI] [PubMed] [Google Scholar]
- 59.Hughes CE, Nibbs RJB. A guide to chemokines and their receptors. FEBS J. 2018;285(16):2944–2971. doi: 10.1111/febs.14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johansen KH, Golec DP, Thomsen JH, Schwartzberg PL, Okkenhaug K. PI3K in T cell adhesion and trafficking. Front Immunol. 2021;12:708908. doi: 10.3389/fimmu.2021.708908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kagan BL, Ganz T, Lehrer RI. Defensins: a family of antimicrobial and cytotoxic peptides. Toxicology. 1994;87(1-3):131–149. doi: 10.1016/0300-483x(94)90158-9. [DOI] [PubMed] [Google Scholar]
- 62.Rice WG, Ganz T, Kinkade JM, Jr, Selsted ME, Lehrer RI, Parmley RT. Defensin-rich dense granules of human neutrophils. Blood. 1987;70(3):757–765. [PubMed] [Google Scholar]
- 63.Lehrer RI, Lichtenstein AK, Ganz T. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Annu Rev Immunol. 1993;11:105–128. doi: 10.1146/annurev.iy.11.040193.000541. [DOI] [PubMed] [Google Scholar]
- 64.Lee HJ, Kim JM, Kim KH, Heo JI, Kwak SJ, Han JA. Genotoxic stress/p53-induced DNAJB9 inhibits the pro-apoptotic function of p53. Cell Death Differ. 2015;22(1):86–95. doi: 10.1038/cdd.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int J Mol Sci. 2019;20(13):3328. doi: 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, Zhang W, Wu X, Gong J. Foxo3a-dependent Bim transcription protects mice from a high fat diet via inhibition of activation of the NLRP3 inflammasome by facilitating autophagy flux in Kupffer cells. Oncotarget. 2017;8(21):34258–34267. doi: 10.18632/oncotarget.15946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin L, Hron JD, Peng SL. Regulation of NF-κB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 68.Zhou W, Cao Q, Peng Y, et al. FoxO4 inhibits NF-κB and protects mice against colonic injury and inflammation. Gastroenterology. 2009;137(4):1403–14. [DOI] [PMC free article] [PubMed]
- 69.Tominaga K. The emerging role of senescent cells in tissue homeostasis and pathophysiology. Pathobiol Aging Age Relat Dis. 2015;5:27743. doi: 10.3402/pba.v5.27743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qi XF, Chen ZY, Xia JB, et al. FoxO3a suppresses the senescence of cardiac microvascular endothelial cells by regulating the ROS-mediated cell cycle. J Mol Cell Cardiol. 2015;81:114–126. doi: 10.1016/j.yjmcc.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 71.Tokunaga R, Zhang W, Naseem M, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation – a target for novel cancer therapy. Cancer Treat Rev. 2018;63:40–47. doi: 10.1016/j.ctrv.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Menier C, Rouas-Freiss N, Favier B, LeMaoult J, Moreau P, Carosella ED. Recent advances on the non-classical major histocompatibility complex class I HLA-G molecule. Tissue Antigens. 2010;75(3):201–206. doi: 10.1111/j.1399-0039.2009.01438.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhong L, Chen XF, Wang T, et al. Soluble TREM2 induces inflammatory responses and enhances microglial survival. J Exp Med. 2017;214(3):597–607. doi: 10.1084/jem.20160844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu Z, Zhang X, Dong W, et al. TREM2 suppresses the proinflammatory response to facilitate PRRSV infection via PI3K/NF-κB signaling. PLoS Pathog. 2020;16(5):e1008543. doi: 10.1371/journal.ppat.1008543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hay N. Interplay between FOXO, TOR, and Akt. Biochim Biophys Acta. 2011;1813(11):1965–1970. doi: 10.1016/j.bbamcr.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roy SK, Srivastava RK, Shankar S. Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of FOXO transcription factor, leading to cell cycle arrest and apoptosis in pancreatic cancer. J Mol Signal. 2010;5:10. doi: 10.1186/1750-2187-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35(6):600–604. doi: 10.3109/10799893.2015.1030412. [DOI] [PubMed] [Google Scholar]
- 78.Breit SN, Brown DA, Tsai VW. The GDF15-GFRAL pathway in health and metabolic disease: friend or foe? Annu Rev Physiol. 2021;83:127–151. doi: 10.1146/annurev-physiol-022020-045449. [DOI] [PubMed] [Google Scholar]
- 79.Namwanje M, Brown CW. Activins and inhibins: roles in development, physiology, and disease. Cold Spring Harb Perspect Biol. 2016;8(7). [DOI] [PMC free article] [PubMed]
- 80.Hung WT, Wu FJ, Wang CJ, Luo CW. DAN (NBL1) specifically antagonizes BMP2 and BMP4 and modulates the actions of GDF9, BMP2, and BMP4 in the rat ovary. Biol Reprod. 2012;86(5):1–9. doi: 10.1095/biolreprod.111.096172. [DOI] [PubMed] [Google Scholar]
- 81.Yang X, Cao N, Chen L, Liu L, Zhang M, Cao Y. Suppression of cell tumorigenicity by non-neural pro-differentiation factors via inhibition of neural property in tumorigenic cells. Front Cell Dev Biol. 2021;9:714383. doi: 10.3389/fcell.2021.714383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maquart FX, Monboisse JC. Extracellular matrix and wound healing. Pathol Biol (Paris). 2014;62(2):91–95. doi: 10.1016/j.patbio.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 83.Haines BP, Wheldon LM, Summerbell D, Heath JK, Rigby PW. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev Biol. 2006;297(1):14–25. doi: 10.1016/j.ydbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 84.Kim KA, Kakitani M, Zhao J, et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309(5738):1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- 85.Wong VS, Yeung A, Schultz W, Brubaker PL. R-spondin-1 is a novel beta-cell growth factor and insulin secretagogue. J Biol Chem. 2010;285(28):21292–21302. doi: 10.1074/jbc.M110.129874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hoshino A, Kim HS, Bojmar L, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182(4):1044–61.e18. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brazil DP, Church RH, Surae S, Godson C, Martin F. BMP signalling: agony and antagony in the family. Trends Cell Biol. 2015;25(5):249–264. doi: 10.1016/j.tcb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 88.Koike N, Kassai Y, Kouta Y, Miwa H, Konishi M, Itoh N. Brorin, a novel secreted bone morphogenetic protein antagonist, promotes neurogenesis in mouse neural precursor cells. J Biol Chem. 2007;282(21):15843–15850. doi: 10.1074/jbc.M701570200. [DOI] [PubMed] [Google Scholar]
- 89.Sun M, Forsman C, Sergi C, Gopalakrishnan R, O'Connor MB, Petryk A. The expression of twisted gastrulation in postnatal mouse brain and functional implications. Neuroscience. 2010;169(2):920–931. doi: 10.1016/j.neuroscience.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Milan G, Romanello V, Pescatore F, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nature Commun. 2015;6:6670. [DOI] [PMC free article] [PubMed]
- 92.Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2(9):601–608. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 93.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7(8):766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 94.Can ND, Basturk E, Kizilboga T, et al. Interactome analysis of Bag-1 isoforms reveals novel interaction partners in endoplasmic reticulum-associated degradation. PLoS One. 2021;16(8):e0256640. doi: 10.1371/journal.pone.0256640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.D'Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43(6):582–592. doi: 10.1002/cbin.11137. [DOI] [PubMed] [Google Scholar]
- 96.Grodzicky T, Elkon KB. Apoptosis: a case where too much or too little can lead to autoimmunity. Mt Sinai J Med. 2002;69(4):208–219. [PubMed] [Google Scholar]
- 97.Mortezaee K, Salehi E, Mirtavoos-Mahyari H, et al. Mechanisms of apoptosis modulation by curcumin: implications for cancer therapy. J Cell Physiol. 2019;234(8):12537–12550. doi: 10.1002/jcp.28122. [DOI] [PubMed] [Google Scholar]
- 98.Kurowska M, Rudnicka W, Kontny E, et al. Fibroblast-like synoviocytes from rheumatoid arthritis patients express functional IL-15 receptor complex: endogenous IL-15 in autocrine fashion enhances cell proliferation and expression of Bcl-x(L) and Bcl-2. J Immunol (Baltimore). 2002;169(4):1760–1767. doi: 10.4049/jimmunol.169.4.1760. [DOI] [PubMed] [Google Scholar]
- 99.Liang R, Ghaffari S. Stem cells seen through the FOXO lens: an evolving paradigm. Curr Top Dev Biol. 2018;127:23–47. doi: 10.1016/bs.ctdb.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 100.Kops GJ, Dansen TB, Polderman PE, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 101.Sengupta A, Molkentin JD, Paik JH, DePinho RA, Yutzey KE. FoxO transcription factors promote cardiomyocyte survival upon induction of oxidative stress. J Biol Chem. 2011;286:7468–7478. doi: 10.1074/jbc.M110.179242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014;39:159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fasano C, Disciglio V, Bertora S, Lepore Signorile M, Simone C. FOXO3a from the nucleus to the mitochondria: a round trip in cellular stress response. Cells. 2019;8(9):1110. doi: 10.3390/cells8091110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gurkar AU, Robinson AR, Cui Y, et al. Dysregulation of DAF-16/FOXO3A-mediated stress responses accelerates oxidative DNA damage induced aging. Redox Biol. 2018;18:191–199. doi: 10.1016/j.redox.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andrade J, Shi C, Costa ASH, et al. Control of endothelial quiescence by FOXO-regulated metabolites. Nat Cell Biol. 2021;23(4):413–423. doi: 10.1038/s41556-021-00637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Warr MR, Binnewies M, Flach J, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santo EE, Paik J. FOXO in neural cells and diseases of the nervous system. Curr Top Dev Biol. 2018;127:105–118. doi: 10.1016/bs.ctdb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gopinath SD, Webb AE, Brunet A, Rando TA. FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Reports. 2014;2(4):414–426. doi: 10.1016/j.stemcr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sathyan S, Ayers E, Gao T, et al. Plasma proteomic profile of age, health span, and all-cause mortality in older adults. Aging Cell. 2020;19(11):e13250. doi: 10.1111/acel.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanaka T, Basisty N, Fantoni G, et al. Plasma proteomic biomarker signature of age predicts health and life span. eLife. 2020:9. [DOI] [PMC free article] [PubMed]
- 111.Eiriksdottir T, Ardal S, Jonsson BA, et al. Predicting the probability of death using proteomics. Commun Biol. 2021;4(1):758. doi: 10.1038/s42003-021-02289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frankum R, Jameson TSO, Knight BA, et al. Extreme longevity variants at the FOXO3 locus may moderate FOXO3 isoform levels. GeroScience. 2021. [DOI] [PMC free article] [PubMed]
- 113.Nguetse CN, Kremsner PG, Velavan TP. FOXO3A regulatory polymorphism and susceptibility to severe malaria in Gabonese children. Immunogenetics. 2015;67(2):67–71. doi: 10.1007/s00251-014-0816-z. [DOI] [PubMed] [Google Scholar]
- 114.Alonso A, Domènech E, Julià A, et al. Identification of risk loci for Crohn's disease phenotypes using a genome-wide association study. Gastroenterology. 2015;148(4):794–805. doi: 10.1053/j.gastro.2014.12.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 439 kb)
Data Availability Statement
The datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.