Abstract
Marine bacterioplankton transform dimethylsulfoniopropionate (DMSP) into the biogeochemically important and climatically active gas dimethylsulfide. In order to identify specific bacterial taxa mediating DMSP processing in a natural marine ecosystem, we amended water samples from a southeastern U.S. salt marsh with 20 μM DMSP and tracked community shifts with flow cytometry (FCM) coupled to 16S rRNA gene analyses. In two out of four seasons studied, DMSP amendments induced the formation of distinct bacterioplankton populations with elevated nucleic acid (NA) content within 24 h, indicative of cells actively utilizing DMSP. The 16S rRNA genes of the cells with and without elevated NA content were analyzed following cell sorting and PCR amplification with sequencing and terminal restriction fragment length polymorphism approaches. Compared to cells in the control FCM populations, bacteria with elevated NA content in the presence of DMSP were relatively enriched in taxa related to Loktanella, Oceanicola, and Sulfitobacter (Roseobacter lineage, α-Proteobacteria); Caulobacter (α-Proteobacteria); and Brachymonas and Xenophilus (β-Proteobacteria) in the May-02 sample and to Ketogulonicigenium (Roseobacter lineage, α-Proteobacteria) and novel γ-Proteobacteria in the Sept-02 sample. Our study suggests that diverse bacterioplankton participate in the metabolism of DMSP in coastal marine systems and that their relative importance varies temporally.
Dimethylsulfoniopropionate [DMSP; (CH3)2-S-CH2-CH2-COOH] is an osmoprotectant synthesized by marine algae and vascular plants (25, 26, 33). As a consequence of senescence, grazing, and viral activity, intracellular DMSP can be released from algal and plant cells into the water column, where it undergoes microbial transformations. Bacterially mediated DMSP transformations can generate over 90% of oceanic dimethylsulfide (DMS, H3C-S-CH3), and DMS accounts for at least half of the global biogenic sulfur flux to the atmosphere (30, 31). Along with its role as a precursor for DMS, DMSP is a ubiquitous source of organic matter in marine surface waters and can provide up to 15% of the carbon requirement of heterotrophic bacteria and fulfill most of the bacterial cellular sulfur demand (14, 29, 39).
Previous studies have made significant progress in identifying which bacterial taxa mediate DMSP degradation in natural marine communities. Culturing approaches have identified DMSP-utilizing bacteria in the Roseobacter lineage (α-Proteobacteria) and in the genera Alcaligenes (β-Proteobacteria) and Pseudomonas (γ-Proteobacteria) (1, 5, 6, 12, 21, 22, 38). Culture-independent studies of surface waters from estuaries and open ocean sites with high DMSP turnover rates have revealed DMSP-utilizing bacteria in the Bacteroidetes, Roseobacter, β-Proteobacteria, and γ-Proteobacteria lineages (1, 13, 16, 24, 35, 39, 40). Most of these field studies, however, have targeted a broad taxonomic grouping (at the phylum or class level), and few have resolution at the level of genus and below.
Flow cytometry (FCM) discriminates between populations within complex bacterial communities based on fluorescence and size differences among the cells. In conjunction with a sorting unit, defined FCM populations can be physically separated and then subjected to further taxonomic analysis (2, 3, 23, 28, 32, 36). However, the combined methods are challenging to use, primarily because of the low bacterioplankton densities typical of seawater and the large number of sorted cells needed for subsequent DNA-based analysis.
Coastal salt marshes have one of the highest natural DMS emission rates and represent an active DMSP-degrading ecosystem (4). In this study, a culture-independent approach was employed to identify bacteria involved in DMSP degradation at the species level. We amended water from a salt marsh tidal creek with DMSP and identified cells responding to the addition by FCM sorting of cells with elevated nucleic acid (NA) content. Methods were optimized to obtain DNA template from as few as 25,000 FCM sorted preserved bacterial cells. Terminal restriction fragment length polymorphism (T-RFLP) analysis and sequencing of 16S rRNA genes in the sorted populations were used to identify bacterial taxa involved in DMSP degradation.
MATERIALS AND METHODS
Sample collection and processing.
Surface water samples were collected from Dean Creek (a salt marsh tidal creek) on Sapelo Island, Georgia, in acid-washed Nalgene carboys at 4-month intervals from May 2002 to May 2003 (samples May-02, Sept-02, Jan-03, and May-03). Once collected, water samples were stored in the dark at 4°C and processed within 3 days. To isolate a bacterioplankton size fraction and exclude bacteriovores, water samples were sequentially filtered through 47-mm-diameter, 3.0- and 1.0-μm-pore-size polycarbonate filters (Poretics Products, Livermore, Calif.) under pressures lower than 250 mm Hg. The filtrate was then amended with a mixture of inorganic nitrogen and phosphorus (5 μM NH4Cl, 5 μM NaNO3, and 1 μM NaH2PO4, final concentrations) and incubated in the dark at 25°C with 100-rpm shaking for 48 h to establish carbon-limited conditions. Microcosms were established by adding 150 ml of the preincubated water to each of six 250-ml Erlenmeyer flasks. DMSP (20 μM, final concentration) was added to three of the microcosms, and the remaining three microcosms were used as no-addition controls. Flasks were incubated in the dark at 25°C with 100-rpm shaking for 48 h. Five-milliliter subsamples from each microcosm were collected at 0, 1, 3, 6, 12, 24, and 48 h after DMSP amendments; preserved with 1% paraformaldehyde (final concentration) at 4°C for 1 h; and then stored in the dark at −20°C until flow-cytometric analysis. All glassware was ashed at 550°C prior to use.
FCM.
Flow-cytometric analysis was performed with a MoFlo flow cytometer (DakoCytomation, Glostrup, Denmark). Prior to running on the instrument, preserved bacterioplankton were stained with Sybr Green II (1:5,000 dilution of the commercial stock; Molecular Probes Inc.) in the dark at room temperature for 20 min (18) and then mixed with an internal bead standard (1-μm-diameter yellow-green fluorescent beads; Fluoresbrite YG Microspheres; Polysciences, Warrington, Pa.). A sterilized phosphate-buffered saline (PBS) solution (Puraflo; Dakocytomation, Fort Collins, Colo.) served as the sheath fluid. Data acquisition was triggered by green fluorescence (FL1). All signals were collected with logarithmic amplification.
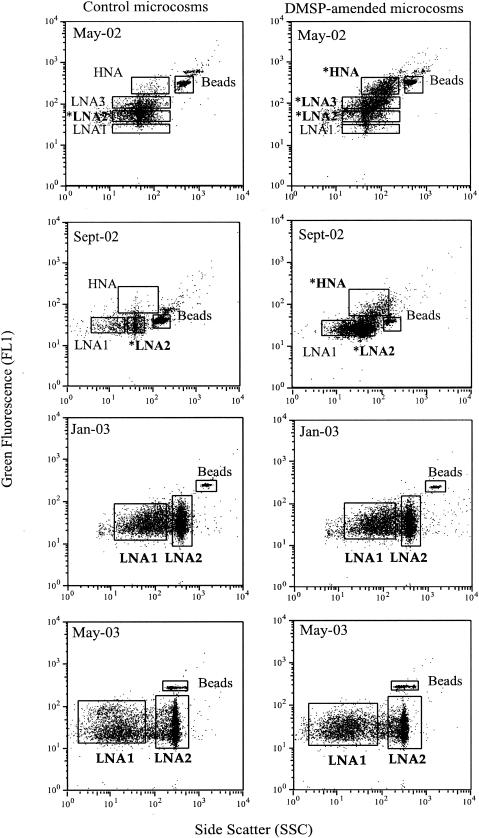
All FCM populations were gated initially on the FCM cytogram of one DMSP replicate after 24 h of incubation, and the gating frames were kept fixed through all the measurements within a single sample date. Gate notation was based on the value of FL1, which is related to the cell NA content. The FCM populations that exhibited high FL1 values were designated the “high-NA-content groups” (HNAs), and the others were designated the “low-NA-content groups” (LNAs). The FCM populations observed for each sample are shown in Fig. 1. Because the FCM population gates chosen were specific to each date, the gate notation refers to different ranges of fluorescence and side scatter for each sample date.
FIG. 1.
Flow-cytometric analysis of control (left panel) and DMSP-amended (right panel) samples collected after 24 h of incubation in May 2002, September 2002, January 2003, and May 2003. Beads and HNA and LNA cells are delimited by gates on each cytogram. Boldface letters are used to label subpopulations that were analyzed with T-RFLP. Asterisks are used to label FCM populations used for clone library construction.
Flow sorting of selected FCM populations was performed on a MoFlo sorter in the “purify 1-2 drop” mode. Two FCM populations were sorted at one time. The sorted cells were collected in sterile 1.5-ml polypropylene test tubes containing 1 ml of PBS solution. Sorting was terminated when the sorted cells were over 500,000 counts for each FCM population or at 30 min.
DNA extraction and PCR amplifications.
Sorted cells from each FCM population were filtered onto a 25-mm-diameter, 0.2-μm-pore-size polycarbonate filter (Poretics Products) and washed three times with 2 ml of PBS followed by 2 ml of deionized water. The filter was air dried, and its edge was trimmed off. One quarter of the trimmed filter was stored at −20°C, while the other three quarters were cut into pieces and transferred into an 0.5-ml tube. Ten microliters of Lyse-N-Go PCR reagent (Pierce Inc., Rockford, Ill.) was added to the tube to lyse the bacterial cells on the filter according to the manufacturer's protocol. A second lysis step was conducted by adding 82 μl of autoclaved water to the same tube and subjecting the tube to three freeze-thaw cycles. The resulting lysate served as the DNA template for PCR amplification.
16S rRNA gene amplification was carried out with Ready-To-Go PCR beads (Amersham Pharmacia, Piscataway, N.J.) with 0.4 μM concentrations of both forward (8F, 5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse (1492R, 5′-TACGGYTACCTTGTTACGACTT-3′) primers. A touchdown PCR program was performed with annealing temperature sequentially decreasing from 62 to 52°C by 1°C per cycle, followed by 15 cycles at 52°C. In each cycle, denaturing (at 95°C), annealing (at 62 to 52°C), and extension (at 72°C) steps were of 1-min duration. An initial 3-min denaturation and final 10-min extension step were also included in the PCR program. PCR amplification was confirmed by electrophoresis on ethidium bromide-stained 1% agarose gels. The amplicons were excised from the gel and cleaned with the QIAquick gel extraction kit (Qiagen, Valencia, Calif.) before subsequent T-RFLP and sequencing analysis.
T-RFLP analysis.
Direct PCR amplification of 16S rRNA genes from sorted cells with a 6-carboxyfluorescein (FAM)-labeled forward primer was inconsistent, probably due to the difficulty in binding the FAM-labeled primer to the low-copy-number templates. Instead, 16S rRNA genes of sorted cells were amplified first with nonlabeled primers 8F and 1492R as described above and then reamplified with 0.4 μM FAM-labeled 8F and nonlabeled 785R (5′-CTACCAGGGTATCTAATCC-3′) primers with two Ready-To-Go PCR beads in 50-μl-volume reaction mixtures with a single annealing temperature of 59°C. PCR amplification was confirmed by electrophoresis on ethidium bromide-stained 1% agarose gels. The reamplified amplicons were excised from the gel, cleaned with the QIAquick gel extraction kit, and digested with CfoI restriction enzyme (Roche, Indianapolis, Ind.) at 37°C for 3 h, after which an ethanol precipitation was performed. The restricted amplicons were resuspended in 12 μl of deionized formamide plus 0.7 μl of DNA fragment length standard (GeneScan 2500 TAMRA; Applied Biosystems, Warrington, United Kingdom). The terminal restriction fragment (T-RF) lengths were determined on an ABI Prism 310 genetic analyzer (Applied Biosystems, Foster City, Calif.) in GeneScan mode.
T-RFLP data were obtained successfully for most sorted bacterial FCM populations. The exceptions were as follows: (i) one set of the May-02 triplicates was used up in the process of optimizing the conditions for DNA extraction and PCR amplification, (ii) some FCM populations were too small to provide enough template for PCR amplification of 16S rRNA genes (Fig. 1), and (iii) one FCM population (Sept-02, LNA1) failed to amplify even though cell numbers were relatively high.
Identification of bacteria associated with responsive T-RFs.
16S rRNA genes directly amplified from sorted cells were used to construct clone libraries of 16S rRNA genes for selected samples (May-02 and Sept-02). Libraries were constructed using a TA cloning kit (Invitrogen Corporation, Carlsbad, Calif.) with the pCR 2.1 vector according to the manufacturer's instructions.
Sequencing was carried out on an ABI PRISM 310 genetic analyzer in sequencing mode, with the BigDye terminator cycle-sequencing kit (PE Biosystems, Foster City, Calif.). 8F served as the sequencing primer. The average length of usable sequence was 400 bp. Bacterial identities were determined using the GenBank blastn search (http://www.ncbi.nlm.nih.gov/BLAST/) and the RDP-II sequence match program (http://rdp.cme.msu.edu/html/analyses.html).
The bacteria associated with T-RFs showing significant differences between control and DMSP-amended microcosms (responsive T-RFs) were putatively assigned using the following process. Initially, 20 clones were randomly selected from each library for sequencing, and the length of the T-RF for each clone was calculated based on sequence data with TRFLPtools, a Visual Basic program for Microsoft Excel (13, 34). If any responsive T-RFs remained unidentified, a clone pool approach was used to rapidly screen the 16S rRNA gene library. Briefly, five random clones were pooled prior to plasmid extraction with the QIAprep Spin Miniprep kit (Qiagen Inc.). T-RFLP analysis was carried out by the method described previously with 500 ng of extracted plasmid DNAs as templates for the FAM-labeled PCR amplification. If any matching T-RFs were found, each of the five clones was analyzed individually to identify the specific clone(s) of interest in the pooled sample. This process continued until the identities of all T-RFs were assigned or more than 200 clones were screened. Then, clones putatively responsible for responsive T-RFs were coinjected with community amplicons in a single T-RFLP analysis. Finally, the inserts of the clones of interest were sequenced to determine taxonomic identity.
T-RFLP analysis of small populations.
We determined whether apparent bacterial community structures were significantly affected by the number of cells used in the T-RFLP analysis, since sorted FCM populations differed in cell number by as much as 25-fold (Table 1). The total number of cells in the May-02 water sample was determined by epifluorescence microscopy counting after 4′,6′-diamidino-2-phenylindole (DAPI) staining. Aliquots containing various numbers of cells (2 × 109, 2 × 107, 2 × 106, 2 × 105, and 2 × 103) were filtered onto 25-mm-diameter, 0.2-μm-pore-size polycarbonate filters. PCR and T-RFLP analysis of the 16S rRNA genes were then performed according to the procedure described above.
TABLE 1.
Cell number and percentage of the total cell count for HNA and LNA subpopulations in four salt marsh bacterioplankton samples after 24 h of incubationa
| Sample | Population | Control (avg ± SD)
|
DMSP (avg ± SD)
|
Significant differencec | ||||
|---|---|---|---|---|---|---|---|---|
| No. of cells (106 ml−1) | % of total cells | No. of cells sorted (103)b | No. of cells (106 ml−1) | % of total cells | No. of cells sorted (103)b | |||
| May-02 | HNA | 0.1 ± 0.1 | 2.1 ± 0.5 | 0.8 ± 0.1 | 5.2 ± 0.6 | 28.2 ± 1.7 | 49.3 ± 1.8 | Yes |
| LNA3 | 1.1 ± 0.3 | 16.8 ± 1.0 | 9.2 ± 0.3 | 6.4 ± 0.4 | 34.6 ± 1.0 | 59.2 ± 1.5 | Yes | |
| LNA2 | 5.0 ± 0.2 | 76.8 ± 0.8 | 27.2 ± 1.7 | 5.4 ± 0.4 | 29.2 ± 0.8 | 24.8 ± 0.9 | Yes | |
| LNA1 | 0.3 ± 0.1 | 4.4 ± 0.5 | 1.6 ± 0.1 | 1.5 ± 0.5 | 8.0 ± 1.8 | 6.6 ± 0.3 | No | |
| Sept-02 | HNA | 0.1 ± 0.1 | 2.1 ± 0.5 | 11.8 ± 1.9 | 5.8 ± 1.5 | 9.7 ± 2.6 | 80.2 ± 25.0 | Yes |
| LNA2 | 0.2 ± 0.1 | 11.2 ± 1.0 | 62.9 ± 0.6 | 40.7 ± 1.5 | 68.6 ± 2.5 | 566.2 ± 79.8 | Yes | |
| LNA1 | 1.5 ± 0.1 | 80.5 ± 1.6 | 455.9 ± 50.0 | 9.1 ± 0.3 | 15.4 ± 0.5 | 126.4 ± 11.6 | Yes | |
| Jan-03 | LNA2 | 2.6 ± 0.2 | 59.2 ± 1.9 | 78.1 ± 2.5 | 3.4 ± 0.2 | 56.1 ± 2.7 | 96.0 ± 5.2 | No |
| LNA1 | 1.8 ± 0.3 | 40.8 ± 5.1 | 53.9 ± 7.1 | 2.7 ± 0.2 | 43.9 ± 3.5 | 75.1 ± 9.8 | No | |
| May-03 | LNA2 | 5.5 ± 0.8 | 64.6 ± 3.9 | 99.5 ± 7.1 | 11.4 ± 0.9 | 63.6 ± 5.0 | 82.0 ± 8.2 | No |
| LNA1 | 3.2 ± 0.2 | 36.7 ± 4.4 | 56.5 ± 5.3 | 6.5 ± 1.0 | 36.1 ± 4.7 | 46.6 ± 7.6 | No | |
Averages and standard deviations were generated based on independent analysis of three replicate incubations.
Actual number of cells available for molecular analysis. Boldface indicates FCM populations that were able to provide DNA templates for subsequent molecular analysis.
“Significant difference” indicates whether the number of cells differed between DMSP and control treatments for a given subpopulation at P < 0.05.
Statistical analysis.
A t test for two samples of unequal variance was performed to compare total bacterial abundance as well as abundance in each of the FCM populations between control and DMSP microcosms. The t test was also performed to compare the relative area of T-RFs between the sorted FCM populations in control and DMSP-amended microcosms to determine responsive T-RFs. The confidence interval was set at 95%, and significant differences were reported when P was <0.05.
A hierarchical cluster analysis was performed using Primer v5 (Primer-E Ltd, Plymouth, United Kingdom) to make quantitative comparisons of T-RFLP profiles among bacterial FCM populations. Before analysis, T-RFLP output data were standardized as described previously (34), except that relative peak area instead of peak height was used as a proxy for the relative abundance of bacterial taxa associated with each T-RF. T-RFs with lengths greater than 600 bp or relative peak areas less than 2% of total area were excluded from the analysis.
Nucleotide sequence accession numbers.
The GenBank accession numbers for 16S rRNA gene sequences determined in this study are AF547399, AF547405 to AF547409, AF547413 to AF547415, AF547417, AF547418, AF547421, AF547422, AF547424, AF547425, AF547427, AY476738 to AY476741, AY476749 to AY476751, AY476763, AY476764, AY476767, AY476769 to AY476771, AY476774 to AY476776, AY476778 to AY476781, AY476784, AY476785, AY476794, AY476795, AY476797, AY476799 to AY476802, AY476804, and AY476805.
RESULTS
FCM analysis of sorted populations.
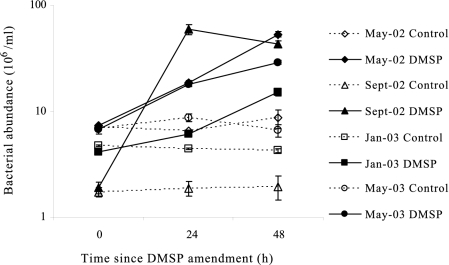
Significant growth of bacterioplankton was observed over the 48-h incubation in microcosms amended with 20 μM DMSP but not in the control microcosms for all four experiments (May-02, Sept-02, Jan-03, and May-03), indicating stimulated bacterial activity following the DMSP amendments. At the 24-h time point, DMSP-amended microcosms had cell numbers 2.9-, 32-, 1.4-, and 2.1-fold higher than did control microcosms in the May-02, Sept-02, Jan-03, and May-03 experiments, respectively (Fig. 2).
FIG. 2.
Total bacterial cell numbers (means ± standard deviations) counted by FCM in control and DMSP treatments over the 48-h incubation for each sample date.
DMSP amendments resulted in the development of a cluster of cells with high NA content within 24 h in the May-02 and Sept-02 microcosms but not in the Jan-03 or May-03 microcosms (Fig. 1; Table 1). In the May-02 microcosms, HNA and LNA3 groups accounted for 28.2 and 34.6%, respectively, of the total assemblage following DMSP addition; in the control microcosms these populations contained only 2.1 and 16.8% of the total cells, respectively. Likewise for Sept-02, the HNA group accounted for 9.7% of total cells in DMSP-amended microcosms but 2.1% in the controls (Table 1; there was no LNA3 gate defined in the Sept-02 sample). The greatest difference between control and DMSP-amended microcosms in cell NA content (FL1) and to some extent cell size (side scatter) was found after 24 h of incubation for these two experiments. At other time points (0, 3, 6, 12, and 48 h), scatter plots of DMSP-amended microcosms resembled those of control microcosms, even though total cell number was highest at 48 h in some cases (Fig. 2). No NA content or size changes were observed in Jan-03 and May-03 experiments over the course of the incubation, despite the increase in cell numbers. Therefore, flow sorting and subsequent molecular analysis were performed only on subsamples taken 24 h after DMSP amendments.
T-RFLP and sequence analysis of sorted populations.
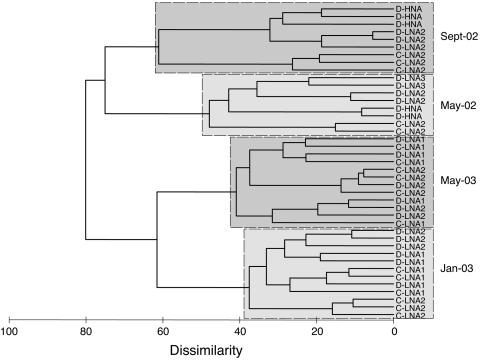
T-RFLP analyses of the replicate subsamples for a given sample date were highly similar as revealed by cluster analysis (Fig. 3). Thus, there was low variability among the replicate incubations within microcosms and consistency in results of molecular analysis. The cluster analysis of T-RFLP data also grouped all samples based on collection date, regardless of the sorting gates used. For the May-02 and Sept-02 samples (those in which a bacterial cell NA-size response to DMSP amendments was evident), FCM populations of control and DMSP-amended samples clustered separately with high dissimilarity values. However, for Jan-03 and May-03 samples (those in which little or no bacterial cell NA-size response to DMSP additions was evident), populations from control and DMSP-amended microcosms were intermingled.
FIG. 3.
Cluster analysis of T-RFLP data after 24 h of incubation. Similarity calculation was based on the Bray-Curtis dissimilarity on square-root-transformed relative peak area. FCM populations from triplicate incubations are shown, except for the May-02 sample, for which only duplicates were available. Sample codes indicate treatment (D, DMSP amendments; C, controls), replicates (1, 2, or 3), and sorted subpopulation (LNA2, LNA3, or HNA).
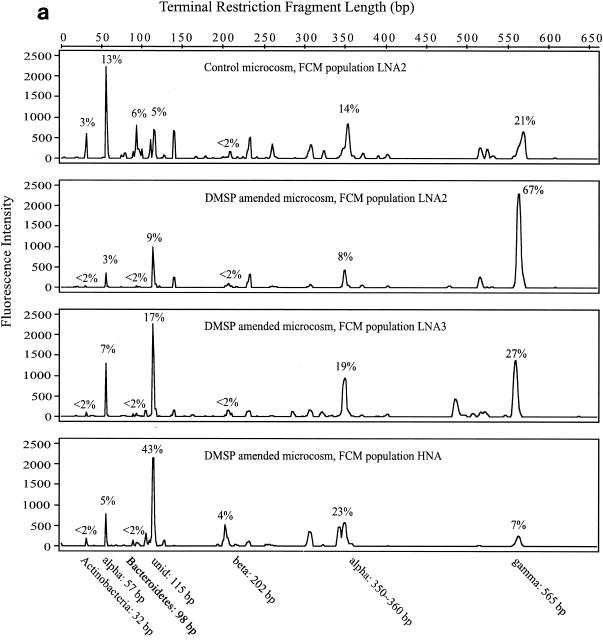
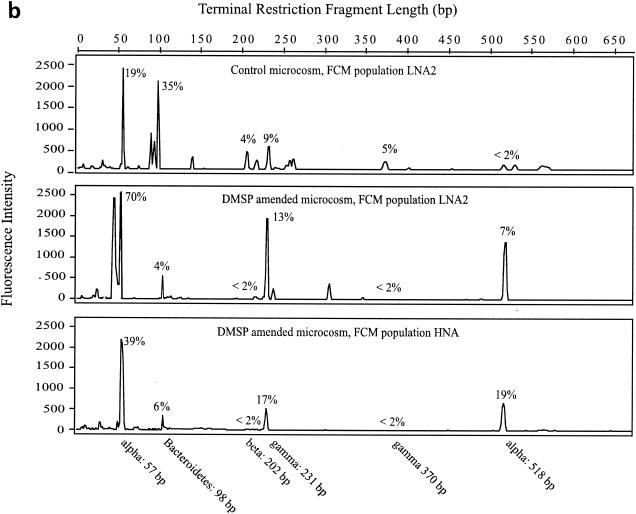
The same major T-RFs were typically found in all FCM populations within a sample, but significant differences in relative abundance of T-RFs in response to DMSP amendments were evident (Fig. 4). In the May-02 samples (Fig. 4a), there were seven responsive T-RFs, six of which were putatively identified from the 16S rRNA gene clone library. They fell into five major phylogenetic groups that are commonly found in coastal marine environments: α-Proteobacteria (mainly Roseobacter lineage), β-Proteobacteria, γ-Proteobacteria, Bacteroidetes, and Actinobacteria (Table 2). The seventh T-RF (length, 115 bp; T-115) was unidentified. Bacteria associated with T-115 (unidentified), T-202 (β-Proteobacteria), and T-350∼360 (α-Proteobacteria) were stimulated by DMSP amendments as evidenced by greater relative abundance in the elevated-NA-larger-size populations. Bacteria associated with T-32 (Actinobacteria), T-57 (α-Proteobacteria, Roseobacter lineage), T-98 (Bacteroidetes), and T-565 (γ-Proteobacteria) decreased in relative abundance in the higher bacterial cell NA-size class after DMSP amendments. In the Sept-02 experiment (Fig. 4b), there were six responsive T-RFs. Bacteria represented by T-231 (γ-Proteobacteria) and T-518 (Roseobacter lineage, α-Proteobacteria) were relatively enriched in FCM populations with elevated NA content after DMSP amendments, while T-57 (Roseobacter lineage, α-Proteobacteria), T-98 (Bacteroidetes), T-202 (β-Proteobacteria), and T-310 (γ-Proteobacteria) reacted in the opposite manner.
FIG. 4.
T-RFLP chromatogram of May-02 (a) and Sept-02 (b) samples. Empirically determined taxonomic identities of T-RFs and their lengths are indicated beneath the chromatogram. The average contribution of each identified T-RF is indicated by percentages. Alpha, α-Proteobacteria; beta, β-Proteobacteria; gamma, γ-Proteobacteria; unid, unidentified. Sample codes are as in Fig. 3.
TABLE 2.
Taxonomic identification of bacterial groups associated with peaks in T-RFLP profiles based on 16S rRNA gene clone library analysisa
| Sample | TRFL (bp) | Clone (accession no.) | Group | Closest relative (accession no.) | Similarity (%) | Closest described relative (accession no.) | Similarity (%) |
|---|---|---|---|---|---|---|---|
| May-02 | 32 | SIMO-1620 (AY476751) | Actinobacteria | Uncultured Actinobacterium MB11A03 (AY033296) | 96 | Streptomyces africanus (AY208912) | 80 |
| 57 | SIMO-277 (AF547399) | Roseobacter | Uncultured α-proteobacterium (AY580445) | 99 | “Citricella thiooxidans” (AY639887) | 95 | |
| SIMO-293 (AF547406) | Roseobacter | Uncultured α-proteobacterium (AY038571) | 97 | Roseovarius nubinhibens (AF098495) | 96 | ||
| SIMO-312 (AF547424) | Sulfitobacter | Arctic sea ice bacterium ARK9994 (AF468380) | 97 | Sulfitobacter pontiacus (Y13155) | 97 | ||
| SIMO-313 (AF547425) | Sulfitobacter | Arctic sea ice bacterium ARK9994 (AF468380) | 97 | Sulfitobacter pontiacus (AY159887) | 97 | ||
| SIMO-1603 (AY476739) | Roseobacter | Uncultured α-proteobacterium (AY580445) | 100 | Ruegeria atlantica (D88527) | 95 | ||
| SIMO-1604 (AY476740) | Sulfitobacter | Sulfitobacter pontiacus (AY159887) | 99 | Sulfitobacter pontiacus (AY159887) | 99 | ||
| 98 | SIMO-295 (AF547408) | Bacteroidetes | Uncultured Bacteroidetes (AY580726) | 92 | Flexibacter canadensis (AB078046) | 76 | |
| SIMO-296 (AF547409) | Bacteroidetes | Uncultured Bacteroidetes (AY580664) | 94 | Flavobacterium columnare (AB015480) | 82 | ||
| SIMO-309 (AF547422) | Bacteroidetes | Uncultured Bacteroidetes (AY580666) | 96 | Flavobacterium limicola (AJ585236) | 82 | ||
| 115 | Unidentified | ||||||
| 202 | SIMO-308 (AF547421) | β-Proteobacteria | Uncultured β-proteobacterium (AB113609) | 93 | Xenophilus azovorans (AF285414) | 92 | |
| SIMO-1616 (AY476750) | β-Proteobacteria | Uncultured bacterium (AF546929) | 95 | Brachymonas petroleovorans (AY275432) | 93 | ||
| 350-360 | SIMO-292 (AF547405) | Roseobacter | Roseovarius sp. strain 2S5-2 (AB114422) | 97 | Sulfitobacter dubius (AY180102) | 96 | |
| SIMO-294 (AF547407) | Roseobacter | Bacterium K2-11 (AY345438) | 98 | Loktanella vestfoldensis (AJ582226) | 95 | ||
| SIMO-300 (AF547413) | Oceanicola | Oceanicola batsensis (AY424898) | 97 | Oceanicola batsensis (AY424898) | 97 | ||
| SIMO-301 (AF547414) | α-Proteobacteria | Uncultured bacterium (AJ459874) | 95 | Caulobacter crescentus (AE006011) | 95 | ||
| SIMO-302 (AF547415) | Roseobacter | Bacterium K2-11 (AY345438) | 99 | Loktanella vestfoldensis (AJ582227) | 95 | ||
| SIMO-1605 (AY476741) | Roseobacter | Bacterium K2-11 (AY345438) | 98 | Loktanella vestfoldensis (AJ582226) | 96 | ||
| 565 | SIMO-1599 (AY476738) | γ-Proteobacteria | Neptunomonas naphthovorans (AF053734) | 93 | Neptunomonas naphthovorans (AF053734) | 93 | |
| Sept-02 | 57 | SIMO-335 (AY476763) | Roseobacter | Unidentified bacterium (Z88582) | 99 | Ruegeria atlantica (D88527) | 95 |
| SIMO-341 (AY476769) | Roseobacter | Unidentified bacterium (Z88582) | 99 | Ruegeria atlantica (D88527) | 95 | ||
| SIMO-342 (AY476770) | Roseobacter | Unidentified α-proteobacterium (AY580445) | 99 | Ruegeria atlantica (D88527) | 94 | ||
| SIMO-343 (AY476771) | Roseobacter | Unidentified α-proteobacterium (AY580445) | 99 | Ruegeria atlantica (D88527) | 96 | ||
| SIMO-346 (AY476774) | Roseobacter | Unidentified bacterium (Z88582) | 98 | “Citricella thiooxidans” (AY639887) | 95 | ||
| SIMO-1606 (AY476802) | Roseobacter | Uncultured α-proteobacterium (AY580466) | 99 | Roseovarius nubinhibens (AF098495) | 96 | ||
| 98 | SIMO-347 (AY476775) | Bacteroidetes | Uncultured Bacteroidetes (AY580722) | 99 | Cytophaga fermentans (M58766) | 84 | |
| SIMO-351 (AY476779) | Bacteroidetes | Uncultured Bacteroidetes (AY580722) | 99 | Cytophaga fermentans (M58766) | 84 | ||
| SIMO-1600 (AY476799) | Bacteroidetes | Uncultured Bacteroidetes (AY580722) | 99 | Cytophaga fermentans (M58766) | 88 | ||
| SIMO-1601 (AY476800) | Bacteroidetes | Uncultured prokaryote (AF477832) | 99 | Cytophaga fermentans (M58766) | 82 | ||
| 202 | SIMO-1617 (AY476804) | Rhodoferax | Rhodoferax antarcticus (AY609198) | 97 | Rhodoferax antarcticus (AY609198) | 97 | |
| SIMO-1618 (AY476805) | β-Proteobacteria | Uncultured bacterium (AF546929) | 96 | Brachymonas petroleovorans (AY275432) | 94 | ||
| 231 | SIMO-1602 (AY476801) | γ-Proteobacteria | Uncultured γ-proteobacterium (AY580764) | 93 | Microbulbifer elongatus (AF500006) | 86 | |
| 370 | SIMO-339 (AY476767) | γ-Proteobacteria | Pseudoalteromonas sp. strain Md 213 (AY461670) | 99 | “Pseudoalteromonas porphyrae” (AF475096) | 95 | |
| SIMO-348 (AY476776) | γ-Proteobacteria | Pseudoalteromonas sp. strain Md 213 (AY461670) | 99 | Pseudoalteromonas prydzensis (U85855) | 95 | ||
| SIMO-350 (AY476778) | γ-Proteobacteria | Uncultured γ-proteobacterium (AY580367) | 95 | Colwellia demingiae (U85844) | 95 | ||
| SIMO-352 (AY476780) | γ-Proteobacteria | Pseudoalteromonas sp. strain Md 213 (AY461670) | 99 | Pseudoalteromonas mariniglutinosa (AJ507251) | 95 | ||
| SIMO-353 (AY476781) | γ-Proteobacteria | Pseudoalteromonas sp. strain Md 213 (AY461670) | 98 | Pseudoalteromonas prydzensis (U85855) | 94 | ||
| SIMO-356 (AY476784) | γ-Proteobacteria | Pseudoalteromonas sp. strain A28 (AF227238) | 92 | “Pseudoalteromonas porphyrae” (AF475096) | 88 | ||
| SIMO-357 (AY476785) | γ-Proteobacteria | Pseudoalteromonas sp. strain Md 213 (AY461670) | 99 | “Pseudoalteromonas porphyrae” (AF475096) | 94 | ||
| SIMO-367 (AY476795) | γ-Proteobacteria | Pseudoalteromonas sp. strain FE1-03 (AJ784130) | 94 | “Pseudoalteromonas porphyrae” (AF475096) | 93 | ||
| 518 | SIMO-366 (AY476794) | Roseobacter | Marine α-proteobacterium AS-19 (AJ391181) | 97 | Ketogulonicigenium vulgare (AF136846) | 93 |
Group affiliations were determined with the RDP-II Sequence Match program (http://rdp.cme.msu.edu/html/analyses.html). The closest relative of each clone was determined with the blastn program of GenBank (http://www.ncbi.nlm.nih.gov/BLAST/). Only one representative is shown if more than one clone had an identical sequence in the same sample.
Based on observed increases in cell counts (Fig. 2), bacteria stimulated by DMSP amendments were likely present in the Jan-03 or May-03 samples. However, no significant change in either cell NA content or size was observed between FCM populations in control and DMSP-amended microcosms. Thus, DMSP-utilizing bacteria could not be sorted from the bulk population for these two samples.
T-RFLP analysis of small populations.
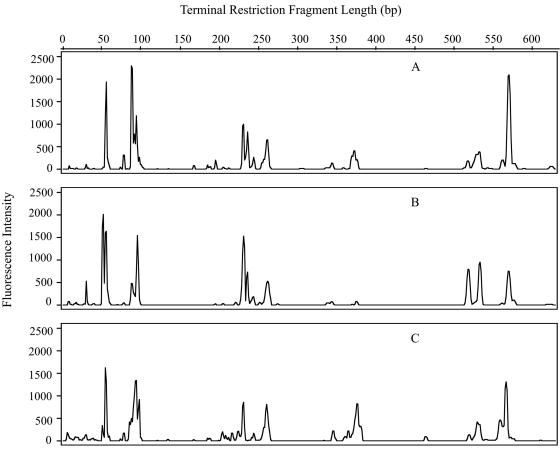
Following optimization of the DNA extraction method, PCR amplification and T-RFLP analysis were carried out on bacterial population sizes as low as 2,000 cells from unpreserved samples and 20,000 cells from preserved samples. Bacterial populations containing 2 × 109, 2 × 106, and 2 × 103 unpreserved cells from the May-02 water sample were analyzed by T-RFLP. Regardless of the initial cell number, T-RFLP chromatograms typically contained the same major peaks in approximately the same relative abundances (Fig. 5). Based on this finding, FCM populations differing in size by as much as 500,000 cells were compared in the T-RFLP analyses.
FIG. 5.
Community structures as revealed by T-RFLP chromatograms from different-sized bacterioplankton populations of 2 × 109 (A), 2 × 106 (B), and 2 × 103 (C) cells collected in May 2002.
DISCUSSION
Determining the identity of bacterial taxa carrying out specific ecological activities, such as turnover of dissolved organic carbon in marine systems, is a central task of microbial ecology. In this study, we successfully employed a culture-independent method involving FCM and cell sorting to identify bacteria stimulated by DMSP amendments. Bacterial cell NA content is widely used as a proxy for bacterial cell activity. With few disagreements (19, 41), cells with higher NA content are considered to be more active than cells with lower NA content and responsible for a significant fraction of bulk activities (8, 9, 19, 20, 27). Based on this consensus, we assume that bacteria increasing their cell NA content above that in no-addition controls were actively responding to DMSP additions.
The taxa identified as responsive to DMSP amendments probably include those assimilating DMSP directly as well as those utilizing products of DMSP degradation. Taxonomic groups assimilating DMSP or its degradation products but maintaining the same NA content and size would be overlooked with our approach (as was the case for cells in the Jan-03 and May-03 samples). The high concentrations of DMSP in amended microcosms (20 μM) relative to natural concentrations (20 to 100 nM) (17) and the preincubation used to establish carbon-limited conditions might have introduced bias. Nonetheless, this approach allowed an initial identification of bacteria that respond to DMSP and that may be important under natural conditions.
T-RFLP analysis was used in this study because it provides an efficient way to view the community composition of many sorted FCM populations. However, like fluorescent in situ hybridization and denaturing gradient gel electrophoresis, this quick examination of community-level 16S rRNA heterogeneity includes a tradeoff with taxonomic resolution (7). To partially overcome this limitation, we also constructed 16S rRNA gene clone libraries for selected samples to access the within-T-RF variability (Table 3). The sequence similarity of clones within dominant T-RFs was highly variable and ranged from as low as 74.1% (for T-98, representing the Bacteroidetes group in May-02) to as high as 99.3% (for T-57, representing the Roseobacter lineage in Sept-02). High variability within Bacteroidetes T-RFs generated with commonly used restriction enzymes has been found previously (7).
TABLE 3.
Contribution of major bacterial taxa to the total bacterioplankton assemblage in control microcosms after 24 h of incubation based on percentage of total area under the T-RFLP chromatogram
| Group | T-RF(s) | Contribution to each sample (%)
|
|||
|---|---|---|---|---|---|
| May-02 | Sept-02 | Jan-03 | May-03 | ||
| α-Proteobacteria | T-57, T-350∼360, T-518 | 32 | 19 | 37 | 34 |
| β-Proteobacteria | T-202 | 0 | 4 | 25 | 30 |
| γ-Proteobacteria | T-231, T-370, T-565 | 21 | 14 | 0 | 0 |
| Bacteroidetes | T-98 | 6 | 35 | 0 | 0 |
| Actinobacteria | T-32 | 3 | 0 | 0 | 0 |
| Unidentified bacteria | 38 | 28 | 38 | 36 | |
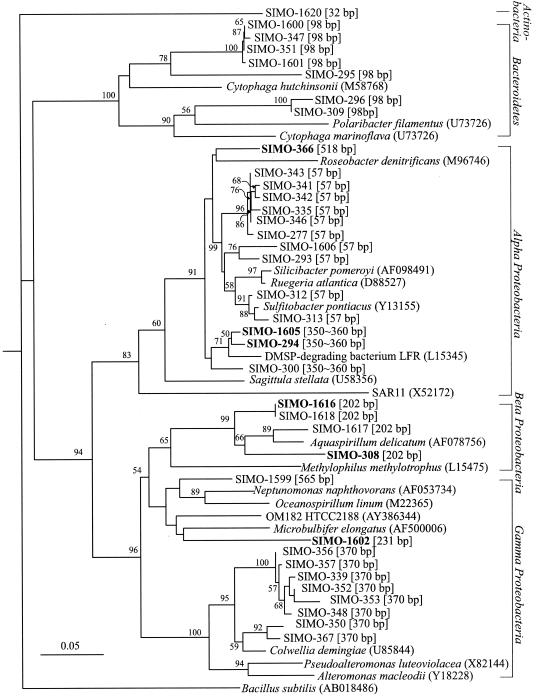
The Roseobacter lineage (α-Proteobacteria) is ubiquitous in marine environments and is a major taxonomic group in many marine bacterioplankton communities (10). Recently, evidence from both lab and field studies has suggested that members of the Roseobacter lineage carry out DMSP transformations in marine systems (12, 22, 37). Roseobacter group isolates can degrade DMSP by two distinct pathways (cleavage and demethylation-demethiolation [1, 12, 15]) and may be important in regulating DMS formation in the surface ocean (31). During blooms of DMSP-producing algae in the open ocean, members of the Roseobacter lineage have been found in high abundance (13, 39). More recently, they have been directly linked to DMSP utilization by microautoradiography-fluorescent in situ hybridization methodology (24, 35). Several members of Roseobacter lineages identified in the present study as responding positively to DMSP additions had high 16S rRNA gene sequence similarity to previously identified DMSP-utilizing bacteria, including Ruegeria atlantica and Sulfitobacter pontiacus (1, 11-13) (Table 1). However, the response of other members of the Roseobacter lineage indicated that they either were uninvolved with DMSP metabolism or were outcompeted by other taxa (Table 2; Fig. 6). This intragroup difference demonstrates the challenge of assigning biogeochemical functions to large and diverse marine bacterioplankton groups and clearly would be missed by the use of analysis methods with lower taxonomic resolution.
FIG. 6.
Phylogenetic tree based on partial sequences of 16S rRNA genes showing the relationship among clones from FCM populations. Clones from the HNA populations formed after DMSP addition are labeled with boldface. Clones from LNA populations in the DSMP and control treatments are shown in lightface. The tree was constructed using Jukes-Cantor distance and neighbor-joining methods excluding positions with <50% sequence conservation. Bacillus subtilis was used as the outgroup. Bootstrap values higher than 50% are indicated at the branch nodes. The scale bar indicates the amount of genetic change in terms of the number of nucleotide substitutions per site. The GenBank accession numbers of the reference sequences are shown in parentheses. Empirically determined T-RFLP fragment lengths are shown in brackets. Only one representative is shown if more than one clone had identical sequences in one sample.
DMSP is not utilized exclusively by bacteria of the Roseobacter lineage (24, 25). In previous studies, DMSP-degrading β- and γ-Proteobacteria were readily isolated from estuarine water and surface sediment in a coastal marsh (1). Also, γ-Proteobacteria have been found to be abundant in the bacterioplankton community associated with algal blooms in the ocean (13, 40) and can assimilate DMSP in natural bacterial assemblages (35). In this study, bacteria related to Brachymonas and Xenophilus (β-Proteobacteria) in the May-02 sample and a novel γ-proteobacterium in the Sept-02 sample were enriched in the FCM populations with elevated NA content after DMSP addition, indicating active linkage to DMSP utilization. In contrast, bacteria related to Actinobacteria, Flexibacter and Flavobacterium (Bacteroidetes), and Neptunomonas (γ-Proteobacteria) in the May-02 sample and Cytophaga (Bacteroidetes), Brachymonas, Rhodoferax (β-Proteobacteria), and Pseudoalteromonas and Colwellia (γ-Proteobacteria) in the Sept-02 sample were concentrated in the FCM populations with unchanged or low NA content (LNA1 and LNA2) (Table 2; Fig. 4b). However, because not all DMSP-utilizing bacteria responded with an increase in bacterial cell NA content or size (Fig. 2), we cannot assume that these taxa play no role in DMSP degradation.
Cluster analysis grouped bacterial FCM populations into four clusters coincident with sampling dates, indicating that temporal factors had a major influence on bacterial community composition. The bacterial assemblages in the control microcosms for the two samples in which an NA-size response to DMSP additions was evident (May-02 and Sept-02) were similar, with relatively low contributions from β-Proteobacteria (Table 3). The bacterial assemblages in the control microcosms of the other two samples (with no NA-size response to DMSP; Jan-03 and May-03) were likewise similar to one another but distinct from the May-02 and Sept-02 samples, with a higher abundance of β-Proteobacteria and few γ-Proteobacteria or Bacteroidetes (Table 3). These differences in bacterial community composition may underlie the disparity in cell parameter response to DMSP amendments observed among samples, and they underscore the influence of temporal variability in the composition and physiology of the bacterioplankton that transform DMSP in coastal environments.
Acknowledgments
We thank Julie Nelson for assistance with FCM and cell sorting; Brian Binder for advice on data analysis; Wendy Ye, Edward Sheppard, and Erin Biers for assistance with sampling at Sapelo Island; and Nasreen Bano for advice on phylogenetic tree construction.
This study was supported by NSF grant 0084164 to the Sapelo Island Microbial Observatory (SIMO).
REFERENCES
- 1.Ansede, J. H., R. Friedman, and D. C. Yoch. 2001. Phylogenetic analysis of culturable dimethyl sulfide-producing bacteria from a Spartina-dominated salt marsh and estuarine water. Appl. Environ. Microbiol. 67:1210-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bihari, N., R. Batel, and R. K. Zahn. 1999. Flow cytometry in marine environmental research. Period. Biol. 101:151-155. [Google Scholar]
- 3.Collier, J. L., and L. Campbell. 1999. Flow cytometry in molecular aquatic ecology. Hydrobiologia 401:33-53. [Google Scholar]
- 4.Dacey, J. W. H., G. M. King, and S. G. Wakeham. 1987. Factors controlling emission of dimethylsulfide from salt marshes. Nature 330:643-645. [Google Scholar]
- 5.De Souza, M. P., and D. C. Yoch. 1995. Comparative physiology of dimethyl sulfide production by dimethylsulfoniopropionate lyase in Pseudomonas doudoroffii and Alcaligenes sp. strain M3a. Appl. Environ. Microbiol. 61:3986-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Souza, M. P., and D. C. Yoch. 1995. Purification and characterization of dimethylsulfoniopropionate lyase from an Alcaligenes-like dimethyl sulfide-producing marine isolate. Appl. Environ. Microbiol. 61:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eiler, A., and S. Bertilsson. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228-1243. [DOI] [PubMed] [Google Scholar]
- 8.Gasol, J. M., and P. A. Del Giorgio. 2000. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64:197-224. [Google Scholar]
- 9.Gasol, J. M., U. L. Zweifel, F. Peters, J. A. Fuhrman, and Å. Hagström. 1999. Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl. Environ. Microbiol. 65:4475-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovannoni, S. J., and M. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84 In D. L. Kirchman (ed.), Microbial ecology of the oceans. John Wiley & Sons, New York, N.Y.
- 11.González, J. M., J. S. Covert, W. B. Whitman, J. R. Henriksen, F. Mayer, B. Scharf, R. Schmitt, A. Buchan, J. A. Fuhrman, R. P. Kiene, and M. A. Moran. 2003. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., dimethylsulfoniopropionate-demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 53:1261-1269. [DOI] [PubMed] [Google Scholar]
- 12.González, J. M., R. P. Kiene, and M. A. Moran. 1999. Transformation of sulfur compounds by an abundant lineage of marine bacteria in the alpha-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 65:3810-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González, J. M., R. Simó, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedrós-Alió, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiene, R. P., and L. J. Linn. 2000. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 45:849-861. [Google Scholar]
- 15.Kiene, R. P., L. J. Linn, and J. A. Bruton. 2000. New and important roles for DMSP in marine microbial communities. J. Sea Res. 43:209-224. [Google Scholar]
- 16.Kiene, R. P., L. J. Linn, J. González, M. A. Moran, and J. A. Bruton. 1999. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 65:4549-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiene, R. P., and S. K. Service. 1991. Decomposition of dissolved DMSP and DMS in estuarine waters—dependence on temperature and substrate concentration. Mar. Ecol. Prog. Ser. 76:1-11. [Google Scholar]
- 18.Lebaron, P., N. Parthuisot, and P. Catala. 1998. Comparison of blue nucleic acid dyes for flow cytometric enumeration of bacteria in aquatic systems. Appl. Environ. Microbiol. 64:1725-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebaron, P., P. Servais, H. Agogué, C. Courties, and F. Joux. 2001. Does the high nucleic acid content of individual bacterial cells allow us to discriminate between active cells and inactive cells in aquatic systems? Appl. Environ. Microbiol. 67:1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebaron, P., P. Servais, A. C. Baudoux, M. Bourrain, C. Courties, and N. Parthuisot. 2002. Variations of bacterial-specific activity with cell size and nucleic acid content assessed by flow cytometry. Aquat. Microb. Ecol. 28:131-140. [Google Scholar]
- 21.Ledyard, K. M., and J. W. H. Dacey. 1994. Dimethylsulfide production from dimethylsulfoniopropionate by a marine bacterium. Mar. Ecol. Prog. Ser. 110:95-103. [Google Scholar]
- 22.Ledyard, K. M., E. F. Delong, and J. W. H. Dacey. 1993. Characterization of a DMSP-degrading bacterial isolate from the Sargasso Sea. Arch. Microbiol. 160:312-318. [Google Scholar]
- 23.Legendre, L., C. Courties, and M. Troussellier. 2001. Flow cytometry in oceanography 1989-1999: environmental challenges and research trends. Cytometry 44:164-172. [DOI] [PubMed] [Google Scholar]
- 24.Malmstrom, R. R., R. P. Kiene, and D. L. Kirchman. 2004. Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49:597-606. [Google Scholar]
- 25.Mulholland, M. M., and M. L. Otte. 2001. The effects of nitrogen supply and salinity on DMSP, glycine betaine and proline concentrations in leaves of Spartina anglica. Aquat. Bot. 71:63-70. [Google Scholar]
- 26.Pakulski, J. D., and R. P. Kiene. 1992. Foliar release of dimethylsulfonioproprionate from Spartina alterniflora. Mar. Ecol. Prog. Ser. 81:277-287. [Google Scholar]
- 27.Servais, P., E. O. Casamayor, C. Courties, P. Catala, N. Parthuisot, and P. Lebaron. 2003. Activity and diversity of bacterial cells with high and low nucleic acid content. Aquat. Microb. Ecol. 33:41-51. [Google Scholar]
- 28.Shapiro, H. M. 2000. Microbial analysis at the single-cell level: tasks and techniques. J. Microbiol. Methods 42:3-16. [DOI] [PubMed] [Google Scholar]
- 29.Simó, R., S. D. Archer, C. Pedrós-Alió, L. Gilpin, and C. E. Stelfox-Widdicombe. 2002. Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 47:53-61. [Google Scholar]
- 30.Simó, R., C. Pedrós-Alió, G. Malin, and J. O. Grimalt. 2000. Biological turnover of DMS, DMSP and DMSO in contrasting open-sea waters. Mar. Ecol. Prog. Ser. 203:1-11. [Google Scholar]
- 31.Simó, R. 2001. Production of atmospheric sulfur by oceanic plankton: biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 16:287-294. [DOI] [PubMed] [Google Scholar]
- 32.Steen, H. B. 2000. Flow cytometry of bacteria: glimpses from the past with a view to the future. J. Microbiol. Methods 42:65-74. [DOI] [PubMed] [Google Scholar]
- 33.Stefels, J. 2000. Physiological aspects of the production and conversion of DMSP in marine algae and higher plants. J. Sea Res. 43:183-197. [Google Scholar]
- 34.Stepanauskas, R., M. A. Moran, B. A. Bergamaschi, and J. T. Hollibaugh. 2003. Covariance of bacterioplankton composition and environmental variables in a temperate delta system. Aquat. Microb. Ecol. 31:85-98. [Google Scholar]
- 35.Vila, M., R. Simó, R. P. Kiene, J. Pinhassi, J. M. González, M. A. Moran, and C. Pedrós-Alió. 2004. Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl. Environ. Microbiol. 70:4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallner, G., B. Fuchs, S. Spring, W. Beisker, and R. Amann. 1997. Flow sorting of microorganisms for molecular analysis. Appl. Environ. Microbiol. 63:4223-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoch, D. C. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68:5804-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoch, D. C., J. H. Ansede, and K. S. Rabinowitz. 1997. Evidence for intracellular and extracellular dimethylsulfoniopropionate (DMSP) lyases and DMSP uptake sites in two species of marine bacteria. Appl. Environ. Microbiol. 63:3182-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]
- 40.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2002. Rapid turnover of dissolved DMS and DMSP by defined bacterioplankton communities in the stratified euphotic zone of the North Sea. Deep-Sea Res. Part II Top. Stud. Oceanogr. 49:3017-3038. [Google Scholar]
- 41.Zubkov, M. V., B. M. Fuchs, P. H. Burkill, and R. Amann. 2001. Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol. 67:5210-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]