Abstract
By using 1,4-dioxane as the sole source of carbon, a 1,4-dioxane-degrading microorganism was isolated from soil. The fungus, termed strain A, was able to utilize 1,4-dioxane and many kinds of cyclic ethers as the sole source of carbon and was identified as Cordyceps sinensis from its 18S rRNA gene sequence. Ethylene glycol was identified as a degradation product of 1,4-dioxane by the use of deuterated 1,4-dioxane-d8 and gas chromatography-mass spectrometry analysis. A degradation pathway involving ethylene glycol, glycolic acid, and oxalic acid was proposed, followed by incorporation of the glycolic acid and/or oxalic acid via glyoxylic acid into the tricarboxylic acid cycle.
The cyclic ether 1,4-dioxane, an organic solvent, is a suspected carcinogen (as determined by the U.S. Environmental Protection Agency) (5) that has been detected in groundwater around landfill sites (13, 26, 27). Its occurrence is thought to be related to its use as a solvent in the chemical synthesis of surfactants and to the disposal of the wastes from these chemical processes as landfill (18). Subsequent leaching of the chemicals from landfills has resulted in contamination of groundwater with 1,4-dioxane and has raised human health issues concerning this harmful chemical.
Previous work has indicated that 1,4-dioxane is resistant to microbial breakdown (3), and there have been very few studies describing its degradation. Parales et al. (15) isolated a 1,4-dioxane-utilizing Actinomycete strain from soil; although they showed that this strain was able to utilize the dioxane as the sole source of carbon, they did not propose a degradation pathway. All other studies on the degradation of 1,4-dioxane have been done with microorganisms that cometabolized tetrahydrofuran and/or supplemented nutrients (1, 19, 20, 28). There has not been, however, any investigation into the use of fungi to degrade 1,4-dioxane, either by itself or in cometabolism with structural analogs or supplemented nutrients.
Cordyceps sinensis is a fungus (division, Eumycota; subdivision, Ascomytina; class, Pyrenomycetes; order, Clavicipitales; family, Clavicipitaceae) (8), and the habitat of the genus Cordyceps is insects. C. sinensis has been identified as mushrooms on host larvae of Hepialus armoricanus from mountainous regions of China. Usually it makes a fruiting body, and it has been consumed as an herbal medicine in Asia. That it contains antioxidant activity has been recognized (14), but it has not yet been used in bioremediation.
In this study, we report the isolation from soil of a strain of C. sinensis (strain A) that is able to utilize 1,4-dioxane as the sole carbon source. We describe the substrate specificity of the fungus for cyclic ethers and identify the degradation products of 1,4-dioxane.
MATERIALS AND METHODS
Isolation and cultivation conditions.
The mineral medium used in this study was basal salt medium (BSM) with 0.0125 × 10−3 M 1, 4-dioxane as the sole source of carbon (15) and without nitrilotriacetic acid. The pH was adjusted to 6.8 with HCl and/or NaOH. For the preparation of a solid medium, 1.5% agar was added to the aforementioned BSM. For liquid cultivation, 3 ml of culture medium was placed in a 15-ml screw-cap vial with a Teflon inner liner and autoclaved for 15 min at 121°C. Filter-sterilized 1,4-dioxane and substrates were added after sterilization. Cultures were grown at 30°C with constant stirring. Growth was estimated from the protein content of the culture medium by the Bradford method (2). Bovine serum albumin (Wako Pure Chemicals, Ltd., Osaka, Japan) was used for the calibration standard.
Identification of the isolated fungus by 18S rRNA gene sequencing.
Under sterile conditions, the fungal cells were disrupted in 500 μl of saline-EDTA buffer (0.1 M NaCl and 0.1 M EDTA [pH 8.0]) with a small amount of aluminum oxide by use of a pestle and a mortar on an ice bath, and the homogenate was centrifuged. Proteinase K (10 μl of a 1-mg/ml solution) (Nippon Gene Co., Ltd., Tokyo, Japan) and 10% sodium dodecyl sulfate (10 μl) were added to a portion (200 μl) of the supernatant, and the mixture was allowed to stand for 60 min at 60°C. The resultant crude extract of genome DNA was purified by phenol-chloroform treatment. To use general primer sets, about 1,600 bp of PCR product was amplified from purified genome DNA, and the PCR product was purified with the MicroSeq 16S rDNA Kit (Applied Biosystems, Foster City, Calif.). The amplified 18S rRNA gene product was sequenced at Hokkaido System Science Co., Ltd. The sequence obtained was identified by using the database of the DNA Data Bank of Japan (DDBJ).
Analytical procedure.
The cyclic ethers, including 1,4-dioxane, in the culture medium were determined by high-performance liquid chromatography (21). The chromatograph was equipped with a pump (LC-9A) and a UV detector (SPD-6A; Shimazu, Kyoto, Japan) at 200 nm or a refractive-index detector (Shodex RI SE-51; Showa Denko K. K., Tokyo, Japan); separations were performed on an Inertsil ODS-3 column (GL Science, Tokyo, Japan) with 10% acetonitrile as the mobile phase at 40°C and a flow rate of 1 ml/min.
The degradation products of 1,4-dioxane were derivatized with phenyl boronate (9). One volume of the culture medium was diluted with an equal volume of methanol and centrifuged for 5 min (10,000 × g) at room temperature. The supernatant was removed, and an equal volume of an 80 mM methanol solution of phenyl boronate was added to it. The mixture was shaken for 5 min at room temperature. The sample was then dried by the addition of anhydrous Na2SO4 and analyzed by gas chromatography-mass spectrometry (GC-MS) (GC-MATE [JEOL Ltd., Tokyo, Japan] and HP 6890 [Agilent Technologies, Palo Alto, Calif.]). Separations were carried out on a DB-5MS column (30 m by 0.25 mm [inner diameter]; J&W Scientific, Folsom, Calif.), and the analytes were ionized in electron impact mode. The oven temperature program was as follows: 80°C (hold for 2 min), 150°C (20°C/min), and 300°C (10°C/min). 1,3-Propanediol was not used as an internal standard because of the problem of overlap with the organic acids. Calibration was carried out with authentic standards.
Chemicals.
Deuterium-labeled 1,4-dioxane-d8 (DML-28-5X1) and ethylene glycol-d4 (DLM-1243-5) were purchased from Cambridge Isotope Laboratory, Inc., Andover, Mass. Methyl-substituted 1,4-dioxanes were synthesized by the method of Ratier et al. (17) or Wojtowicz et al. (25). All other chemicals were AR grade.
Nucleotide sequence accession number.
The DDBJ accession number of the sequence determined in this study is AB187268.
RESULTS AND DISCUSSION
Isolation of a 1,4-dioxane-degrading fungus.
A 1,4-dioxane-utilizing fungus was isolated from garden soil from the National Institute for Environmental Studies (NIES). The isolate, named strain A, could be continuously cultivated on BSM with 1,4-dioxane as the sole source of carbon. Strain A had a septum and spores, as observed by light microscopy (Fig. 1), and was identified as the fungus C. sinensis from 18S rRNA gene homology (99.75% of a partial sequence of 1,617 bp).
FIG. 1.
Light micrograph of strain A.
Cultivation conditions for the isolate.
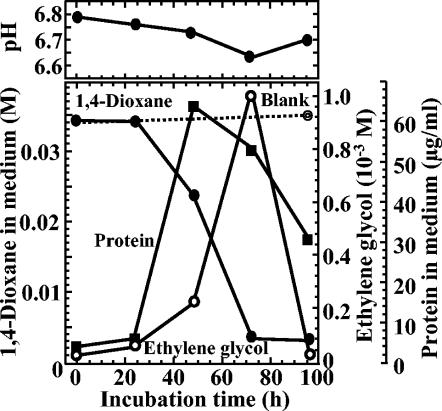
Strain A can grow in medium containing up to 0.09 M 1,4-dioxane (usually microorganisms in wastewater are not affected by 1,4-dioxane, even if its concentration is increased to 0.03 M [12]), but optimal growth was obtained at 0.034 M 1,4-dioxane. When strain A was cultivated in 0.0125 × 10−3 M 1,4-dioxane, a decrease in the dioxane concentration was observed, but traces still remained after 1 week or more (data not shown). This result indicated that the highest concentration of 1,4-dioxane in the actual contaminated wastewater or groundwater can been treated by strain A (1, 19, 20, 28). The time course of the growth of strain A in medium containing 0.034 M 1,4-dioxane is shown in Fig. 2. The decrease of 1,4-dioxane in the medium was essentially complete after 3 days, and there was a concomitant increase in the concentration of the degradation product, ethylene glycol (0.97 × 10−3 M after 3 days). There followed immediately thereafter, however, a rapid decrease in the concentration of ethylene glycol in the medium as it was utilized by the culture, and by the end of the culture period (4 days) the ethylene glycol was almost completely utilized. The resulting ethylene glycol is also toxic and can cause severe poisoning in humans (10), but, as shown above, the concentration of ethylene glycol in the culture medium was relatively low and finally it was completely utilized by strain A.
FIG. 2.
Time course of growth of a strain A culture with 1,4-dioxane in the medium.
Identification of 1,4-dioxane degradation products.
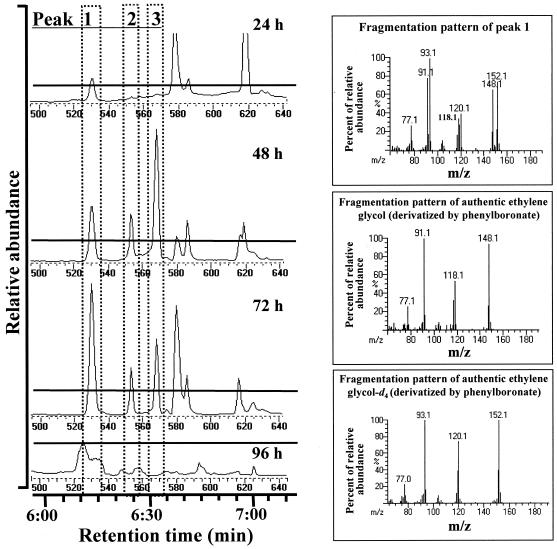
Identification of the 1,4-dioxane degradation products produced by strain A was performed with a phenyl boronate derivatization procedure and the use of 0.031 M 1,4-dioxane-d8. A preliminary identification of ethylene glycol was made by GC-MS following derivatization by N,O-bis(trimethylsilyl)trifluoroacetamide (data not shown). The peak, however, was small, and the presence of many other unidentified derivatives and a high background precluded positive identification of the suspected analyte. To overcome these problems, a more specific method to detect the degradation products was sought. Phenyl boronate can form ether bonds with dihydroxy compounds in water-methanol (9), and these derivatives are amenable to GC-MS analysis. As shown in Fig. 3, peak 1 has the same retention time as nonlabeled ethylene glycol but has an m/z that is 4 mass units greater. Deuterium exchange had occurred (almost half was completely exchanged), and the main derivative was clearly identified as ethylene glycol (-d4) and therefore was derived from 1,4-dioxane-d8. Peak 1 did not occur at time zero or in control experiments using 1,4-dioxane-d8 without inoculation with strain A or without 1,4-dioxane-d8 but with strain A (96 h of incubation at 30°C). Peaks 2 and 3 were assigned to nonlabeled glycolic acid and oxalic acid, respectively (Fig. 4). Exchange of the deuterons on glycolic acid was also observed. Even after just the first step of 1,4-dioxane-d8 degradation, a peak for nonlabeled ethylene glycol (molecular weight, 148) was detected coeluting with ethylene glycol-d4 (molecular weight, 152).
FIG. 3.
Chromatograph of derivatized diols from 1,4-dioxane-d8. The solid lines indicate equal height.
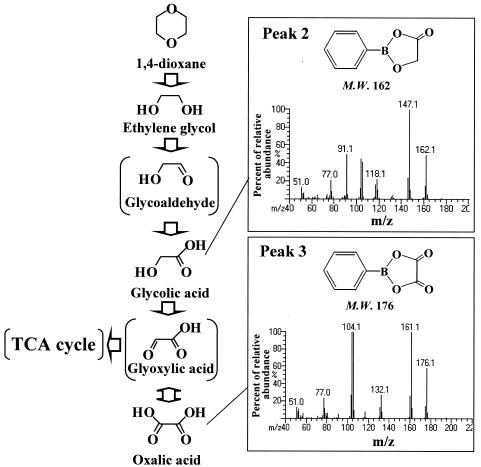
FIG. 4.
Proposed degradation pathway of 1,4-dioxane. Glycolaldehyde, glyoxalate, and the TCA cycle are shown as references for the proposed pathway. M.W., molecular weight.
Proposed 1,4-dioxane degradation pathway.
There are several papers describing the degradation pathway for ethylene glycol by humans, yeasts, and bacteria (4, 6, 7, 11, 24). However, to our knowledge, no pathway has been proposed for filamentous fungi. A general degradation pathway can be proposed as the sequential production of ethylene glycol, glycolaldehyde, glycolic acid, and oxalic acid, followed by incorporation of the glycolic acid and/or oxalic acid into the tricarboxylic acid (TCA) cycle. As shown in Fig. 3, the degradation products of 1,4-dioxane-d8 (initially at a concentration of 0.035 M) were identified as ethylene glycol-d4 (1.21 × 10−3 M), glycolic acid (0.35 × 10−3 M), oxalic acid (0.57 × 10−3 M), and protein (44 μg/ml) after 3 days. When strain A was cultivated with 0.01 M ethylene glycol or glycolic acid as the sole source of carbon, 0.23 × 10−3 M glycolic acid, 0.54 × 10−3 M oxalic acid, and 24 and 39 μg/ml of protein, respectively, were produced after 3 days. The reverse of the metabolic pathway involving the identified metabolites was not observed. From these results, the order of breakdown was assigned as ethylene glycol to glycolic acid to oxalic acid. At the end of the cultivation period, peaks 2 and 3 (Fig. 3) decreased, presumably because these compounds were metabolized to CO2 and/or components of the cell via the TCA cycle without accumulation of oxalic acid. A degradation pathway for strain A was proposed, as shown in Fig. 4.
Effect of metabolites on degradation of 1,4-dioxane.
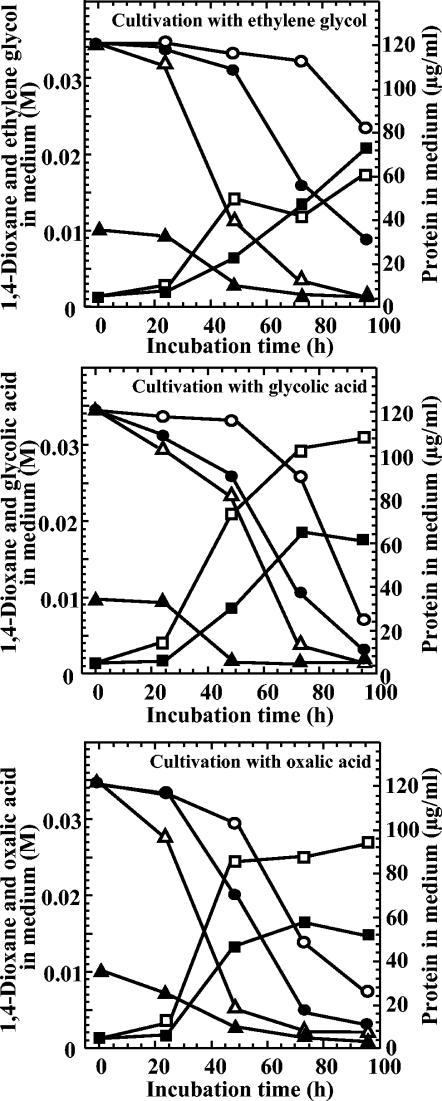
The effect of metabolites on the degradation of 1,4-dioxane was examined (Fig. 5). Ethylene glycol, glycolic acid, or oxalic acid (0.034 or 0.01 M for each compound) was cultivated with 0.034 M 1,4-dioxane for 4 days. Utilization of 1,4-dioxane by strain A occurred after consumption of the added organic acids. These results indicated that there is a catabolite repression for this pathway, so metabolites of 1,4-dioxane as the sole source of carbon (Fig. 2) may be limited to about 5% or less of the added substrate (on a molar basis). The 1,4-dioxane-degrading enzyme and other enzymes probably are encoded on the same operon.
FIG. 5.
Effect of ethylene glycol or organic acids on utilization of 1,4-dioxane. Open symbols, cultivation on 0.034 M 1,4-dioxane with 0.034 M ethylene glycol or organic acids. Closed symbols, cultivation on 0.034 M 1,4-dioxane with 0.01 M ethylene glycol or organic acids. Circles, 1,4-dioxane; triangles, ethylene glycol or organic acids; squares, protein.
Effect of substrate on growth of strain A.
Substrate specificity for the growth of strain A was estimated by using commercial and synthesized cyclic ethers (Table 1). Of the commercial cyclic ethers, all compounds were utilized by strain A, but 18-crown-6 was not a good substrate for enzymatic degradation, although it was a good substrate for growth of strain A. The best substrate was tetrahydrofuran, and it was utilized to the extent of 96% after 3 days. Synthesized cyclic ethers were also tested. 2-Hydroxymethyl-1,4-dioxane was not utilized by strain A within 3 days, and other synthesized cyclic ethers, although utilized, produced poor growth rates. Growth was apparently related to the extent of methylation of the 1,4-dioxane carbons, with the highest growth rates being associated with tetramethyl-1,4-dioxane. It is possible that monomethylated cyclic ethers are not suitable substrates for strain A or do not serve as suitable substrates for the relevant enzymes. These results indicated that strain A can degrade secondary ethers, and our future studies will investigate these processes by using purified enzymes.
TABLE 1.
Substrate specificity for growth of strain A
| Sole source of carbona | % Utilization after 3 days | Protein content in medium (μg/ml) |
|---|---|---|
| 1,4-Dioxane | 90 | 53 |
| 1,3-Dioxane | 91 | 42 |
| Tetrahydrofuran | 96 | 78 |
| Tetrahydropyran | 89 | 39 |
| s-Trioxane | 90 | 25 |
| 18-Crown-6 | 40 | 61 |
| 2-Methyl-1,4-dioxane | 25 | 52 |
| 2,3-Dimethyl-1,4-dioxane | 32 | 56 |
| 2,3,5-Trimethyl-1,4-dioxane | 32 | 20 |
| 2,3,5,6-Tetramethyl-1,4-dioxane | 54 | 33 |
| 2-Hydroxymethyl-1,4-dioxane | 17 | 5 |
A 0.034 M concentration of each chemical was added to the medium.
At present, such enzymes have not been identified, but etherases and oxidases are likely candidates for the degradation of 1,4-dioxane by strain A (16, 22, 23). From the study of cultivation with 2,3,5,6-tetramethyl-1,4-dioxane, it has been estimated that strain A has an enzyme which is not affected by methyl residues. Also, from the observation of consistent deuterium exchange (all or nothing) of the degradation product, ethylene glycol, there is a possibility that ethylene glycol formation occurs via two hemiacetals at one side of the 1,4-dioxane molecule by etherase-type reactions (22).
Acknowledgments
This study was supported by the National Institute for Environmental Studies.
REFERENCES
- 1.Bernhardt, D., and H. Diekmann. 1991. Degradation of dioxane tetrahydrofuran and other cyclic ethers by an environmental Rhodococcus strain. Appl. Microbiol. Biotechnol. 36:120-123. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Burback, B. L., and J. J. Perry. 1993. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. Appl. Environ. Microbiol. 59:1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clay, K. L., and R. C. Murphy. 1977. On the metabolic acidosis of ethylene glycol intoxication. Toxicol. Appl. Pharmacol. 39:39-49. [DOI] [PubMed] [Google Scholar]
- 5.Derosa, C. T., S. Wilbur, J. Holler, P. Richter, and Y. W. Stevens. 1996. Health evaluation of 1,4-dioxane. Toxicol. Ind. Health. 12:1-43. [DOI] [PubMed] [Google Scholar]
- 6.Gabow, P. A., K. Clay, J. B. Sullivan, and R. Lepoff. 1986. Organic acids in ethylene glycol intoxication. Ann. Intern. Med. 105:16-20. [DOI] [PubMed] [Google Scholar]
- 7.Harada, T., and T. Hirabayashi. 1968. Utilization of alcohols by Hansenula miso. Agric. Biol. Chem. 32:1175-1180. [Google Scholar]
- 8.Hawksworth, D. L., P. M. Kirk, B. C. Sutton, and D. N. Pegler. 1995. Ainsworth & Bisby's dictionary of fungi, 8th ed., p. 109. CAB International, Wallingford, United Kingdom.
- 9.Houze, P., J. Chaussard, P. Harry, and M. Pays. 1993. Simultaneous determination of ethylene glycol, propylene glycol, 1,3-butylene glycol and 2,3-butylene glycol in human serum and urine by wide-bore column gas chromatography. J. Chromatogr. 619:251-257. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsen, D., T. P. Hewlett, R. Webb, S. T. Brown, A. T. Ordinario, and K. E. McMartin. 1988. Ethylene glycol intoxication: evaluation of kinetics and crystalluria. Am. J. Med. 84:145-152. [DOI] [PubMed] [Google Scholar]
- 11.Kataoka, M., M. Sasaki, A. R., Hidalgo, M. Nakano, and S. Shimizu. 2001. Glycolic acid production using ethylene glycol-oxidizing microorganisms. Biosci. Biotechnol. Biochem. 65:2265-2270. [DOI] [PubMed] [Google Scholar]
- 12.Klečka, G. M., and S. J. Gonsior. 1986. Removal of 1,4-dioxane from wastewater. J. Hazardous Materials 13:161-168. [Google Scholar]
- 13.Lesage, S., R. E. Jackson, M. W., Priddle, and P. G. Riemann. 1990. Occurrence and fate of organic solvent residues in anoxic groundwater at the Gloucester Landfill, Canada. Environ. Sci. Technol. 24:559-566. [Google Scholar]
- 14.Li. S. P., P. Li, T. T. Dong, and K. W. Tsim. 2001. Anti-oxidation activity of different types of natural Cordyceps sinensis and cultured Cordyceps mycelia. Phytomedicine 8:207-212. [DOI] [PubMed] [Google Scholar]
- 15.Parales, R. E., J. E. Adamus, N. White, and H. D. May. 1994. Degradation of 1,4-dioxane by an Actinomycete in pure culture. Appl. Environ. Microbiol. 60:4527-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce, B. A., and M. T. Heydeman. 1980. Metabolism of di(ethylene glycol) [2-(2′-hydrocyethoxy)ethanol] and other short poly(ethylene glycol)s by gram-negative bacteria. J. Gen. Microbiol. 118:21-27. [Google Scholar]
- 17.Ratier, M., B. Delmond, and J. C. Pommier. 1972. Synthesis of dioxygen heterocycles via organotin compounds. dioxanes and dioxolanes. Bull. Soc. Chim. Fr. 1972:1593-1594. [Google Scholar]
- 18.Robinson, J. J., and E. W. Ciurczak. 1980. Direct gas chromatographic determination of 1,4-dioxane in ethoxylated surfactants. J. Soc. Cosmet. Chem. 31:329-337. [Google Scholar]
- 19.Roy, D., G. Anagnostu, and P. Chaphalkar,. 1994. Biodegradation of dioxane and diglyme in industrial waste. Environ. Sci. Health A 29:129-147. [Google Scholar]
- 20.Roy, D., G. Anagnostu, and P. Chaphalkar. 1995. Analysis of respirometric data to obtain kinetic coefficients for biodegradation of 1,4-dioxane. J. Environ. Sci. Health A 30:1775-1790. [Google Scholar]
- 21.Scalia, S., M. Guarneri, and E. Menegatti. 1990. Determination of 1,4-dioxane in cosmetic products by high-performance liquid chromatography. Analyst 115:929-931. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt, S., R. M. Wittich, P. Fortnagel, D. Erdmann, and W. Francke. 1992. Metabolism of 3-methyldiphenyl ether by Sphingomonas sp. SS31. FEMS Microbiol. Lett. 96:253-258. [DOI] [PubMed] [Google Scholar]
- 23.White, G. F., N. J. Russell, and E. C. Tidswell. 1996. Bacterial scission of ether bonds. Microbiol. Rev. 60:216-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willetts, A. 1981. Bacterial metabolism of ethylene glycol. Biochim. Biophys. Acta 677:194-199. [DOI] [PubMed] [Google Scholar]
- 25.Wojtowicz, J. A., R. J. Polak, and J. A. Zaslowsky. 1971. Synthesis of 3-alkoxyoxetanes. J. Org. Chem. 36:2232-2237. [Google Scholar]
- 26.Yasuhara, A., H. Shiraishi, M. Nishikawa, T. Yamamoto, O. Nakasugi, T. Okumura, K. Kenmotsu, H. Fukui, M. Nagase, and Y. Kawagoshi. 1999. Organic components in leachates from hazardous waste disposal sites. Waste Manag. Res. 17:186-197. [Google Scholar]
- 27.Yasuhara, A., H. Shiraishi, M. Nishikawa, T. Yamamoto, T. Uehiro, O. Nakasugi, T. Okumura, K. Kenmotsu, H. Fukui, M. Nagase, Y. Ono, Y. Kawagoshi, K. Baba, and Y. Noma. 1997. Determination of organic components in leachates from hazardous waste disposal sites in Japan by gas chromatography-mass spectrometry. J. Chromatogr. A 774:321-332. [Google Scholar]
- 28.Zenker, M. J., R. C. Borden, and M. A. Barlaz. 2000. Mineralization of 1,4-dioxane in the presence of a structural analog. Biodegradation 11:239-246. [DOI] [PubMed] [Google Scholar]