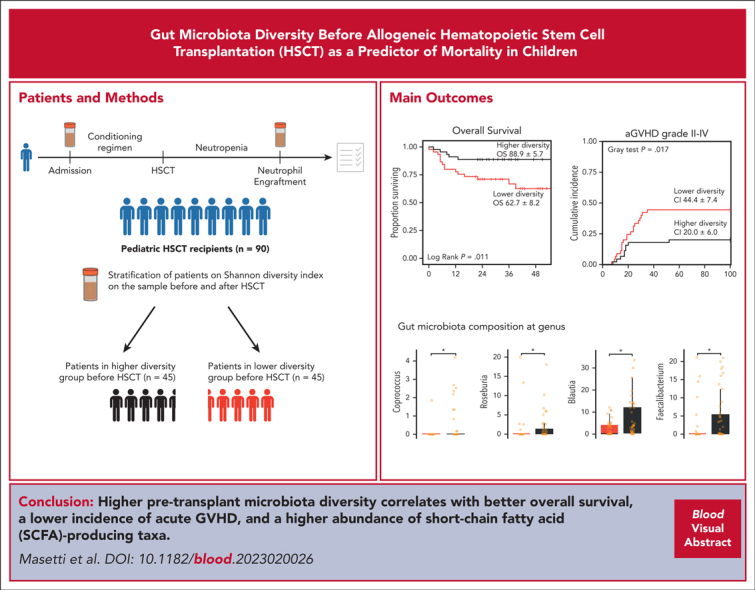
Key Points
-
•
Higher gut microbiota diversity before transplantation correlates with better overall survival and lower acute GVHD incidence.
-
•
In pediatric patients, higher pretransplant diversity is associated with higher abundance of short-chain fatty acid–producing taxa.
Visual Abstract
Abstract
The correlation existing between gut microbiota diversity and survival after allogeneic hematopoietic stem cell transplantation (allo-HSCT) has so far been studied in adults. Pediatric studies question whether this association applies to children as well. Stool samples from a multicenter cohort of 90 pediatric allo-HSCT recipients were analyzed using 16S ribosomal RNA amplicon sequencing to profile the gut microbiota and estimate diversity with the Shannon index. A global-to-local networking approach was used to characterize the ecological structure of the gut microbiota. Patients were stratified into higher- and lower-diversity groups at 2 time points: before transplantation and at neutrophil engraftment. The higher-diversity group before transplantation exhibited a higher probability of overall survival (88.9% ± 5.7% standard error [SE] vs 62.7% ± 8.2% SE; P = .011) and lower incidence of grade 2 to 4 and grade 3 to 4 acute graft-versus-host disease (aGVHD). No significant difference in relapse-free survival was observed between the 2 groups (80.0% ± 6.0% SE vs 55.4% ± 10.8% SE; P = .091). The higher-diversity group was characterized by higher relative abundances of potentially health-related microbial families, such as Ruminococcaceae and Oscillospiraceae. In contrast, the lower-diversity group showed an overabundance of Enterococcaceae and Enterobacteriaceae. Network analysis detected short-chain fatty acid producers, such as Blautia, Faecalibacterium, Roseburia, and Bacteroides, as keystones in the higher-diversity group. Enterococcus, Escherichia-Shigella, and Enterobacter were instead the keystones detected in the lower-diversity group. These results indicate that gut microbiota diversity and composition before transplantation correlate with survival and with the likelihood of developing aGVHD.
Masetti and colleagues report on a multicenter observational study of 90 children that investigated the role of the microbiome in pediatric allogeneic hematopoietic stem cell transplantation (allo-HSCT). The authors’ results indicate that patients with high fecal microbial diversity prior to allo-HSCT have longer overall survival and lower incidence of acute graft-versus-host disease compared with patients with low microbial diversity. Higher pretransplant diversity is associated with higher abundance of short-chain fatty acid–producing organisms, suggesting a potentially causative relationship, as has also been observed in adult allo-HSCT recipients.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is a potentially curative treatment for many pediatric hematologic malignancies and for a variety of nonmalignant diseases.1,2 Nevertheless, transplant-related mortality (TRM) due to associated complications, including acute graft-versus-host disease (aGVHD) and infections, limits therapeutic benefits.3,4 During allo-HSCT, the patients’ gut microbiota (GM) is severely injured because of a combination of factors, including conditioning regimen, antibiotic exposure, and dietary changes.5,6 This alteration, the so-called dysbiosis, is characterized by a dramatic decrease in alfa diversity, a shift in beta diversity, a loss of health-associated commensals, and an expansion of potentially pathogenic bacteria.7 Recently, data from adult multicenter cohorts have linked a lower alfa diversity at the time of neutrophil engraftment with a higher mortality risk.8,9 The relative abundance of specific bacterial taxa was also associated with clinical outcomes, such as Enterococcus expansion, which correlated with higher aGVHD-related mortality,10 whereas increased amounts of Blautia were associated with reduced aGVHD lethality.11 Moreover, the pretransplant GM profile of patients was shown to differ from that of healthy controls because of prior treatments and the disease itself,12 and it was significantly correlated with several clinical outcomes.9,13, 14, 15, 16, 17, 18 Therefore, GM alterations before allo-HSCT may as well be implicated in the genesis of transplant-related complications. In the pediatric allo-HSCT setting, fewer studies have examined the correlation between GM configuration at different time points during transplantation and clinical outcomes,17, 18, 19, 20, 21, 22, 23 and data on the relationship between GM and mortality have not been published yet. The GM of children differs from that of adults, and this accounts for the need for specific pediatric studies on the GM–and–allo-HSCT relationship.24, 25, 26 Although the burden of allo-HSCT complications, such as aGVHD and TRM, is lower than that observed in adults,27, 28, 29, 30, 31 the risk is still significant, with most fatal events being attributable to organ failure and infections.32,33 Furthermore, the long-term impact of complications may be dramatic in children.34 In this scenario, we aimed to evaluate, in a multicenter cohort of pediatric allo-HSCT recipients, the association between GM diversity and clinical outcomes before allo-HSCT and at neutrophil engraftment, particularly focusing on overall survival (OS) rates, incidence of relapse, and aGVHD.
Methods
Patients and specimen collection
Pediatric patients undergoing allo-HSCT for any indication were enrolled in single- and multicenter protocols for stool collection, coordinated by the IRCCS Azienda Ospedaliero-Universitaria di Bologna (Bologna, Italy). The participating centers were the University Hospital of Bologna, Italy; the University Hospital of Pavia, Italy; the Bambino Gesù Children’s Hospital of Rome, Italy; the University Hospital of Verona, Italy; and the University Hospital of Wrocław, Poland. Stool samples were collected following different sampling protocols from 2013 to 2020, as specified in previously published studies,26,35,36 in which 1 pretransplant sample and several postallo-HSCT samples were collected for each patient. In the analysis of this work, only patients with a stool sample available both before allo-HSCT and at neutrophil engraftment were included, for a total of 90 patients and 180 fecal samples. The protocols were approved by the Ethics Committee CE-AVEC Emilia-Romagna, Italy (reference number 19/2013/U/Tess) and by the institutional review board of each participating center. Written informed consent was obtained from all participants/legal guardians by the treating physicians. The study was conducted in accordance with the Declaration of Helsinki and the European data protection regulation.
GM analysis
Details of the methods used for DNA extraction, 16S ribosomal RNA (rRNA) amplicon sequencing, and data processing were previously reported.35, 36, 37 In brief, V3-V4 hypervariable regions of the 16S rRNA gene were amplified, purified, and sequenced on an Illumina MiSeq platform using the 2 × 250 bp paired-end protocol, following the manufacturer’s instructions (Illumina, San Diego, CA). For this work, raw sequencing data were gathered from public repositories, and sequences were filtered based on the length and assembled using PANDASeq.38 Quality filtering and read binning into amplicon sequence variants (ASVs) were achieved by implementing DADA239 in the QIIME 2 pipeline.40 Taxonomic classification of ASVs was performed using the hybrid method implementing VSEARCH and a q2-classifier against the SILVA 138.1 database.41,42 Raw ASV count tables were processed to apply correction for any bias caused by the fact that the sequences were generated in different sequencing runs over the years (yet all at the same center in Bologna). Accordingly, we implemented the ComBat43 function from the sva R package version 3.46.044 to apply correction for biases relative to the batch covariate of the sequencing run for each group of samples taken from the different studies. All the subsequent analyses made use of the derived bias-adjusted data. Alfa diversity was then estimated with the Shannon index, and beta diversity was determined using the Bray-Curtis distances with the qiime diversity module. Samples were categorized into higher- and lower-diversity groups before allo-HSCT and at neutrophil engraftment separately, based on the median of Shannon diversity values of the overall cohort at the corresponding time point, as recently done in a large-scale multicenter study of adult patients who underwent allo-HSCT.9
Networking analysis
GM networks were first built separately for pre–allo-HSCT and neutrophil engraftment time points. Correlation networks were based on Spearman correlation tests performed with the “rcorr” function from the Hmisc R package at the genus-level relative abundance table filtered for genera with at least 0.3% relative abundance in at least 5% of samples at the time point under investigation.45 Only significant (P < .05) correlations were considered further, and the resulting matrices were used for computing modularity and cohesion values (ie, the ratio of negative to positive cohesions [N:P ratio] and total cohesion [TC]) according to previous studies.46,47 Briefly, modularity quantifies the connection between and within modules, measuring whether the modules are more independent (high modularity) or connected, based on the concept that the impact of an external stressor on a node (ie, genus) is likely to affect only the members of its module. Therefore, the higher the modularity, the lower the spread of the stressor effect in the community. TC represents network complexity, considering both positive and negative cohesions, and therefore reflects the global cohesive density of the community. In contrast, the N:P ratio quantifies the extent to which the ecosystem is allowed to exhibit negative interactions; under stress conditions, such interactions are crushed by environmental pressure, and the ecosystem is forced to rely mainly on cooperative interactions.46, 47, 48 Network adjacency structures from correlation data were obtained with the igraph R package version 1.3.4.49 Network modularity was computed using “igraph,” and the modules were detected by implementing a statistical mechanics spin-glass model and simulated annealing with the following additional parameters: “update.rule = simple,” “implementation = neg,” and “spins = 200.”50 Microbial keystone taxa were detected implementing a brute leave-1-out approach, as previously proposed,48,51 removing each taxon at a time, recomputing the network, and evaluating the ones causing the highest reduction of the TC value. Network plots were obtained from a global network, that is, a network computed considering all 180 samples together, and plotted with Cytoscape 3.9.1.52 Node size was set proportional to the overabundance of that genus in the considered group (higher vs lower diversity, either pre–allo-HSCT or at neutrophil engraftment) and computed as the average relative abundance of the taxon in that group divided by the average relative abundance of the taxon across the entire data set. Additionally, edge width was plotted proportionally to the corresponding computed Spearman ρ.
Outcomes and statistical analysis
The primary end point was OS. Secondary end points were incidence of relapse, relapse-free survival, TRM, cumulative incidence of all grade aGVHD, grade 2 to 4 aGVHD, grade 3 to 4 aGVHD, gut aGVHD, and bloodstream infections (BSIs). TRM was defined as death from causes unrelated to the underlying disease. aGVHD was diagnosed and graded according to Glucksberg criteria.53 Neutrophil engraftment was defined as the first of 3 consecutive days with an absolute neutrophil count ≥ 0.5 × 109/L. For the analysis of clinical primary and secondary outcomes, patients were stratified into higher- and lower-diversity groups before allo-HSCT and at neutrophil engraftment, as described earlier. The probability of OS and relapse-free survival was calculated with the Kaplan-Meier method, and groups were compared using log-rank analysis. Relapse, aGVHD, BSI, and TRM were calculated as cumulative incidence, and groups were compared using the Gray test. The association between GM diversity and clinical outcomes was calculated with Cox proportional hazards univariate and multivariate models (adjusted by age, graft source, donor type, intensity of conditioning regimen, center, and disease type). Spearman correlation was used to investigate the association between GM diversity and patient age. Wilcoxon rank-sum test and Fisher exact test were used to compare continuous and categorical clinical and GM features, respectively. The Shapiro-Wilk test was used to test the normality of the data. Student t test was used to compare the means of normal distributions; otherwise, the Mann-Whitney test was used. Welch analysis of variance test was used to evaluate the difference in means among 3 groups when variances were not homogeneous. Tukey multiple comparison test was used to compare continuous variables for different centers. A compositional analysis with phylogenetic classification at the family and genus level was then performed between the higher- and lower-diversity groups, as well as among those patients who did develop aGVHD and those who did not, using Wilcoxon rank-sum tests to assess significant differences in relative taxon abundances. Permutational analysis of variance with pseudo-F ratio (Adonis Permanova from “vegan” R package54) was used to test data separation54 from Bray-Curtis distance matrices, and the t-distributed stochastic neighbor embedding (t-SNE) algorithm for dimensionality reduction from the “Rtsne” R package55 was used to visualize the distribution of genus-level GM profiles in relation to a given variable. Regarding the analysis of the impact of antibiotics on GM diversity and the relative abundance of the Enterobacteriaceae and Enterococcaceae families, pre–allo-HSCT exposure was considered evaluable if it occurred within 30 days before admission and at least for 3 days at least. Pre-engraftment exposure was considered evaluable if the antibiotic was administered for at least 3 days before the collection of the stool sample at engraftment. All GM statistical analyses were conducted in R56 and graphical representations were produced using the ggplot2 R package,57 whereas survival and cumulative incidence analyses were conducted using SPSS (IBM Corp, 2016. IBM SPSS Statistics for Windows, version 24.0. Armonk, NY: IBM Corp) and NCSS (NCSS 12 Statistical Software [2018]). NCSS, LLC. Kaysville, UT). Analyses on the antibiotic exposure were conducted using GraphPad Prism 9 (GraphPad Prism version 9.5.1 for Mac, GraphPad Software, San Diego, CA). All P values were corrected for multiple comparisons using the Benjamini-Hochberg false discovery rate method; P < .05 was considered statistically significant, whereas P < .1 was considered a trend.
Results
Characteristics of patients
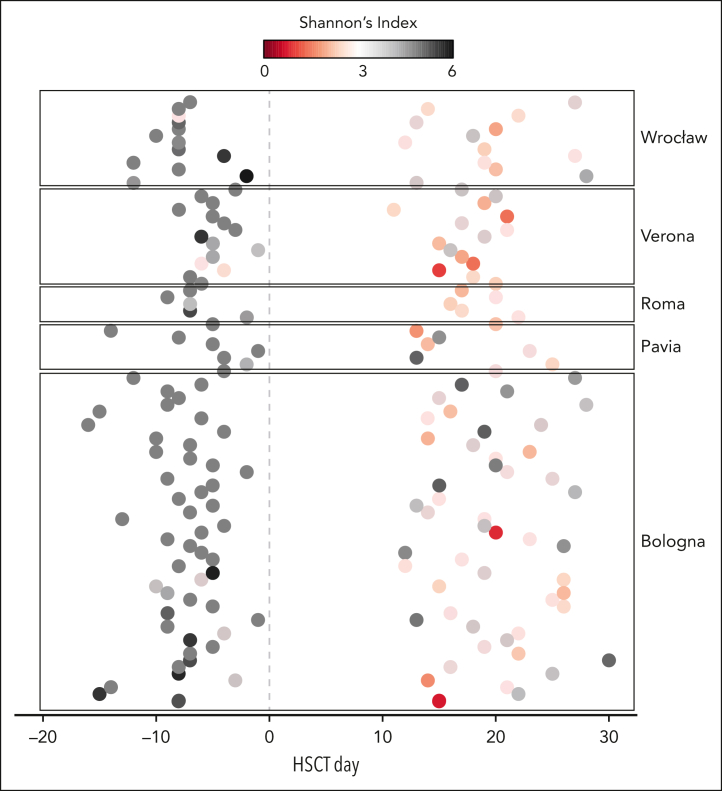
A total of 90 patients from previous studies35, 36, 37 had a stool sample available both before allo-HSCT and at neutrophil engraftment, for a total of 180 stool samples included in this work. A mean of 26 973 (± 5873 standard deviation) high-quality 16S rRNA gene sequences was obtained per sample. The clinical characteristics of the patients included in the study are reported in Table 1. The temporal distribution of the samples for each patient is shown in Figure 1. The median days of sampling were 7.00 ± 0.40 (SE of the mean) before transplant and 19.00 ± 0.92 after HSCT at neutrophil engraftment.
Table 1.
Clinical characteristics of patients included in the study
| Characteristics | Overall (N = 90) | Bologna (N = 50) | Wrocław (N = 13) | Verona (N = 15) | Rome (N = 5) | Pavia (N = 7) |
|---|---|---|---|---|---|---|
| Age at allo-HSCT, y (± SD) | 9.0 ± 5.5 | 10.0 ± 5.6 | 7.2 ± 4.1 | 8.0 ± 5.3 | 8.8 ± 7.9 | 8.1 ± 6.3 |
| Male sex, n (%) | 53 (59) | 29 (58) | 11 (85) | 6 (40) | 1 (20) | 6 (86) |
| Disease, n (%) | ||||||
| Acute lymphoblastic leukemia | 33 (37) | 19 (38) | 5 (38) | 6 (40) | 3 (60) | 0 (0) |
| Acute myeloid leukemia | 19 (21) | 14 (28) | 0 (0) | 2 (13) | 1 (20) | 2 (29) |
| Non-Hodgkin lymphoma | 2 (2) | 2 (4) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| MDS or JMML | 10 (11) | 7 (14) | 2 (15) | 1 (7) | 0 (0) | 0 (0) |
| Nonmalignant disease | 26 (29) | 8 (16) | 6 (49) | 6 (40) | 1 (20) | 5 (71) |
| Type of donor (%) | ||||||
| Unrelated | 55 (61) | 30 (60) | 8 (62) | 13 (87) | 1 (20) | 3 (43) |
| HLA-haploidentical relative | 18 (20) | 10 (20) | 2 (15) | 0 (0) | 3 (60) | 3 (43) |
| Identical sibling | 17 (19) | 10 (20) | 3 (23) | 2 (13) | 1 (20) | 1 (14) |
| Stem cell source (%) | ||||||
| Bone marrow | 68 (76) | 47 (94) | 5 (38) | 10 (67) | 2 (49) | 4 (57) |
| PBSC | 21 (23) | 3 (6) | 8 (62) | 4 (27) | 3 (60) | 3 (43) |
| Cord blood | 1 (1) | 0 (0) | 0 (0) | 1 (7) | 0 (0) | 0 (0) |
| Intensity of conditioning (%) | ||||||
| Ablative | 83 (92) | 49 (98) | 10 (77) | 12 (80) | 5 (100) | 7 (100) |
| Reduced intensity | 7 (8) | 1 (2) | 3 (23) | 3 (20) | 0 (0) | 0 (0) |
| Follow-up of survivors, mo | ||||||
| Median | 52.4 | 55.4 | 23 | 39.2 | 78.5 | 79.4 |
| Interquartile range | 26.3-79.0 | 24.0-78.5 | 22.8-24.0 | 24.0-33.0 | 78.0-79.0 | 78.5-80.0 |
JMML, juvenile myelomonocytic leukemia; MDS, myelodysplastic syndrome; PBSC, peripheral blood stem cell; SD, standard deviation.
Figure 1.
Temporal distribution of the samples from the multicenter pediatric allo-HSCT cohort analyzed in this study. Each point in the graph represents 1 of 180 fecal samples for the 90 patients included in this study, sampled before transplantation and at neutrophil engraftment. Samples are plotted based on the time relative to allo-HSCT (day 0) on the horizontal axis. The color code is based on alfa diversity as measured by the Shannon index.
Association between alfa diversity and clinical outcomes
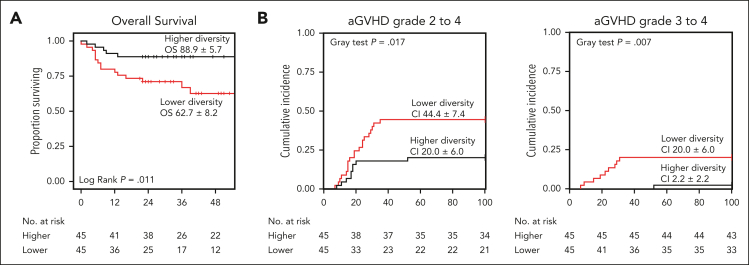
To assess the relationship between GM diversity and clinical outcomes, alfa diversity was analyzed both before transplantation and at the time of neutrophil engraftment. GM diversity significantly decreased from before allo-HSCT to neutrophil engraftment in the whole cohort (P < .0001) as well as in all participating centers (P ≤ .01) (supplemental Figure 1, available on the Blood website). Comparison of diversity among centers revealed no differences either before allo-HSCT or at neutrophil engraftment (P ≥ .14). Additionally, GM diversity correlated significantly with age (Spearman ρ ≥ 0.36; P ≤ .0019; supplemental Figure 2), being lower in patients ≤3 years before and after allo-HSCT (P = .002 and P = .013, respectively) (supplemental Figure 3). The median Shannon diversity value of GM before allo-HSCT and at neutrophil engraftment, used to stratify patients into the 2 groups, was 4.04 and 2.86, respectively. supplemental Tables 1 and 2 show the clinical characteristics of patients in the 2 groups for both time points; no significant differences were found in the variables considered, with the exceptions of the center for groups based on pre–allo-HSCT diversity and patient age for groups based on diversity at neutrophil engraftment (P ≤ .011). Association of clinical variables with OS did not show significant results (P ≥ .09; supplemental Table 3). Focusing on the relationship between GM diversity before allo-HSCT and clinical outcomes, the OS significantly differed between groups (P = .011), with patients with higher diversity showing a higher OS than those with less diversity (hazard ratio, 0.29; 95% confidence interval, 0.11-0.80; Figure 2A). This association was confirmed by a multivariate Cox proportional-hazards analysis adjusted for age, graft source, donor type, intensity of conditioning regimen, center, the and type of disease (hazard ratio, 0.26; 95% confidence interval, 0.09-0.75; supplemental Table 4). The estimated OS at 52 months after allo-HSCT was 88.9% ± 5.7% (standard error [SE]) and 62.7% ± 8.2% (SE) for the higher- and lower-diversity groups, respectively (Figure 2A). There were no statistically significant differences between the higher- and lower-diversity groups in the cumulative incidence of relapse (13.3% ± 5.1% [SE] vs 24.4% ± 6.4% [SE]; P = .1) and relapse-free survival (80.0% ± 6.0% [SE] vs 55.4% ± 10.8% [SE]; P = .091; supplemental Figure 4). The cumulative incidence of grade 2 to 4 aGVHD (20.0% ± 6.0% [SE] vs 44.4% ± 7.4% [SE]; P = .017) as well as the incidence of grade 3 to 4 aGVHD (2.2% ± 2.2% [SE] vs 20.0% ± 6.0% [SE]; P = .007) was significantly lower in the higher-diversity group than in the lower-diversity group, (Figure 2B). Patients with higher diversity showed a trend toward lower gut aGVHD (11.1% ± 4.7% [SE] vs 24.4% ± 6.4% [SE]; P = .098), whereas no differences were found between the groups in terms of BSI (23.8% ± 6.6% [SE] vs 20.9% ± 6.2% [SE]; P = .735) and TRM (8.9% ± 4.2% [SE] vs 15.6% ± 6.2% [SE]; P = .473; supplemental Figure 4). An analysis of aGVHD-related mortality was not performed because of the low number of deaths due to this complication. The results of the univariate and multivariate Cox proportional hazards analysis for all variables considered are shown in supplemental Table 4. As for GM diversity at neutrophil engraftment, no significant differences in outcomes were found between the higher- and lower-diversity groups in terms of OS, relapse-free survival, grade 2 to 4 and 3 to 4 aGVHD, gut aGVHD, BSI, and TRM (supplemental Figure 5; supplemental Table 5).
Figure 2.
Overall survival and aGVHD incidence in respect to pre-allo-HSCT GM diversity. Kaplan-Meier plot for OS (A) and cumulative incidence plots for grade 2 to 4 and 3 to 4 aGVHD (B) for the higher- and lower-diversity groups before allo-HSCT. CI, confidence interval.
GM composition across transplant stages
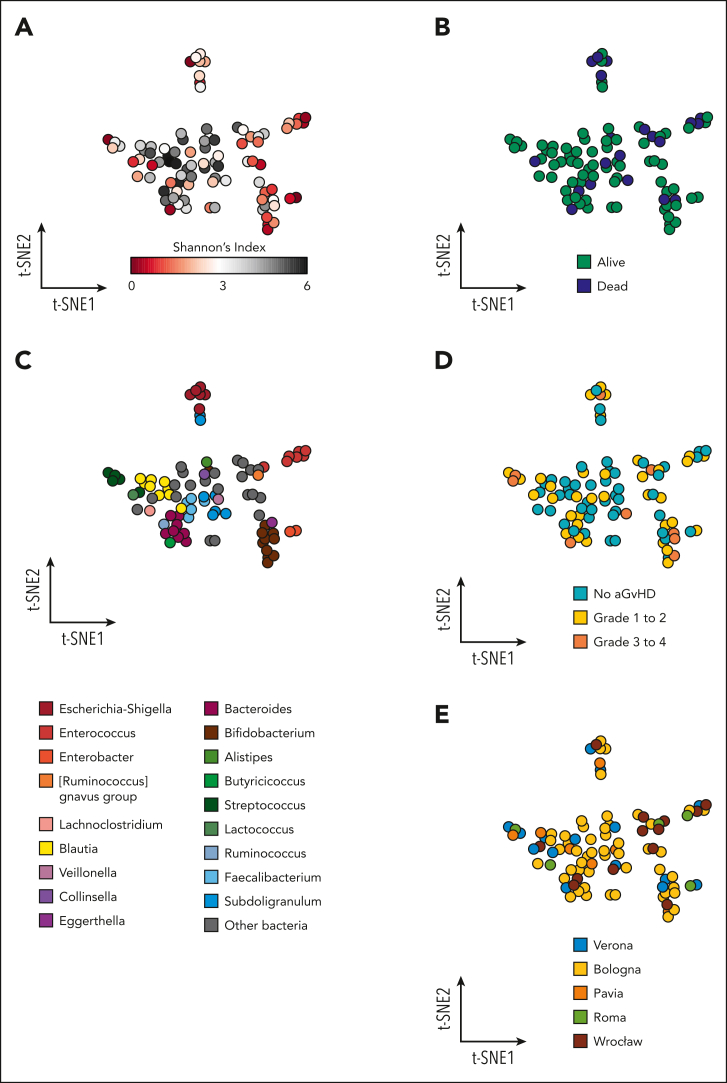
Differences in pre–allo-HSCT GM composition were visualized using t-SNE plots generated from genus-level relative abundance data. Color coding of the t-SNE projections according to the Shannon index of alfa diversity highlighted a large central cluster of high-diversity samples, with an outer rim consisting mostly of lower-diversity projections (Figure 3A). A significant separation between the higher- and lower-diversity groups was, indeed, found using Adonis Permanova on Bray-Curtis distance matrices (P = .001). Color coding based on patient survival outcome showed a colocalization of lower-diversity samples and fatalities (Figure 3B), with GM configurations tending to separate based on outcome (alive vs dead; P = .052). Pre–allo-HSCT alfa diversity was, indeed, significantly higher in patients who had a favorable survival outcome (Figure 4A). In terms of the most abundant genus (Figure 3C), several of the lower-diversity samples were characterized by a higher relative abundance of Escherichia-Shigella, Enterococcus, Enterobacter, and Streptococcus. Furthermore, the GM profiles were segregated based on the occurrence of aGVHD (P = .006), especially with grade 3 to 4 aGVHD tending to colocalize with the lower-diversity samples as well as with the presence of Streptococcus and Bacteroides, but not Blautia, as the most abundant taxa (Figure 3D). In addition, it is noteworthy that the results of the univariate Cox proportional hazards analysis confirm the previously reported link between GM composition and aGVHD onset.7,35 Finally, pre–allo-HSCT samples from all centers were evenly distributed across the different t-SNE clusters, confirming the absence of transplant center-specific effects (P = .592; Figure 3E).
Figure 3.
Pretransplant GM and outcomes in pediatric patients undergoing allo-HSCT. The GM composition at the genus level of the 90 patients before transplantation is represented according to the t-SNE algorithm. Each point represents a stool sample from a single patient, and the axes (t-SNE1 and t-SNE2) have arbitrary units. The more similar the samples are in microbiota composition, the closer they appear on the t-SNE plot. Higher-diversity samples (A) congregate in the center and tend to colocalize with favorable survival outcome (B). The outer ring is mostly composed of lower-diversity samples, which show a higher presence of Escherichia-Shigella, Enterococcus, Enterobacter, and Streptococcus when color-coded for the most abundant taxon detected (C). A similar trend can be observed for the onset of aGVHD (aGVHD) (D), especially grade 3 to 4 aGVHD. Samples from all 5 institutions are well distributed across the t-SNE space (E).
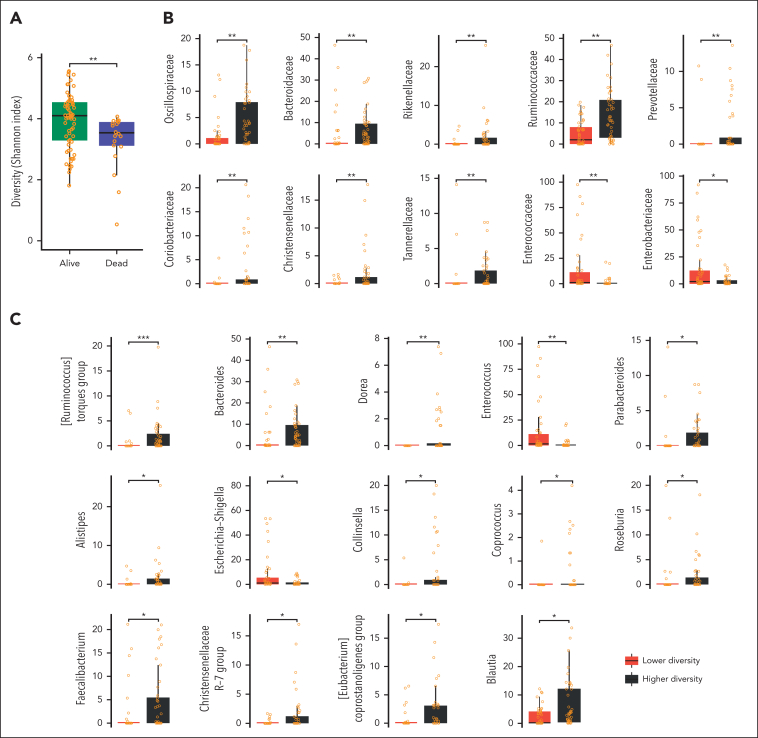
Figure 4.
GM diversity and composition before allo-HSCT. (A) Boxplots showing the distribution of alfa diversity estimated with the Shannon index according to patient outcome (alive vs dead). Significant differences in the GM composition at the family (B) and genus (C) level between the higher- and lower-diversity groups. false discovery rate–corrected Wilcoxon rank-sum tests: ∗∗∗P < .001; ∗∗P < .01; ∗P < .05.
From a compositional point of view, the higher-diversity group before transplantation was characterized by higher relative abundances of the families Oscillospiraceae, Bacteroidaceae, Rikenellaceae, Ruminococcaceae, Prevotellaceae, Coriobacteriaceae, Christensenellaceae, and Tannerellaceae (P < .01; Figure 4B). Conversely, the lower-diversity group showed an overabundance of Enterococcaceae and Enterobacteriaceae (P < .05). Focusing on the genus level, the GM of the higher-diversity group was characterized by higher proportions of Bacteroides, Dorea, Parabacteroides, Alistipes, Collinsella, Coprococcus, Roseburia, Faecalibacterium, Blautia, and members of the Christensenellaceae R-7 group, (Ruminococcus) torques group, and (Eubacterium) coprostanoligenes group (P < .05). In contrast, the lower-diversity group was found to be particularly enriched in Escherichia-Shigella and Enterococcus (P < .05), confirming the t-SNE observations (Figure 4C).
Regarding antibiotic administration before admission, the median antibiotic exposure was significantly longer in the lower-diversity group (P = .028; supplemental Figure 6A). Moreover, in the whole cohort, pre–allo-HSCT antibiotic therapy resulted in a significant reduction in alfa diversity (P = .043; supplemental Figure 6B).
With regard to the analysis performed on samples collected at the time of neutrophil engraftment, significant separation between the higher- and lower-diversity groups was detected using Adonis Permanova on Bray-Curtis distance matrices (P = .013), but neither genus-level, composition-derived t-SNE representations, nor pairwise Adonis testing, nor Wilcoxon tests of raw alfa diversity values revealed any strong association between GM diversity at this time point and OS (supplemental Figures 7 and 8A-B). Nevertheless, several compositional differences at both the family and genus levels were found at neutrophil engraftment, stratifying based on higher vs lower diversity (supplemental Figure 7B-C). In particular, the higher-diversity group showed higher levels of Lachnospiraceae, Ruminococcaceae, Eggerthellaceae, Erysipelotrichaceae, Tannerellaceae, Christensenellaceae, and Bacteroidaceae (P < .05) and a trend toward lower levels of Enterococcaceae (P = .05). At the genus level, engraftment samples with higher diversity were enriched in Lachnoclostridium, Subdoligranulum, Parabacteroides, and Bacteroides as well as members of the Christensenellaceae R-7 group (P < .05). On the contrary, relative abundances of Enterococcus tended to be higher in the lower-diversity group (P = .05).
Regarding aGVHD, it was not found to be associated with alfa diversity either before transplantation or at neutrophil engraftment (supplemental Figures 8D and 9). However, higher pre–allo-HSCT relative abundances of Blautia and Ruminococcus (P ≤ .05) appeared to be protective against the subsequent aGVHD development, in line with the available literature.35 Such taxonomic signatures were more pronounced, specifically, when looking at grade 3 to 4 aGVHD, the occurrence of which was also associated with higher pre–allo-HSCT proportions of Streptococcus, Actinomyces, Lacticaseibacillus, Rothia, and members of the (Ruminococcus) gnavus group (P ≤ .05). No GM taxonomic signatures related to the onset of grade 1 to 2 or 3 to 4 GVHD were detected at neutrophil engraftment.
Regarding antibiotic administration at the pre-engraftment period of time (ie, from HSCT to engraftment), it was found that patients receiving glycopeptides were enriched with Enterococcaceae (P = .004) but not Enterobacteriaceae (P = .391) in the stool samples collected at engraftment (supplemental Figure 6C). In contrast, the relative abundance of Enterococcaceae was higher in patients receiving piperacillin-tazobactam or meropenem compared with those receiving cefepime or ceftazidime (P = .024; supplemental Figure 6D).
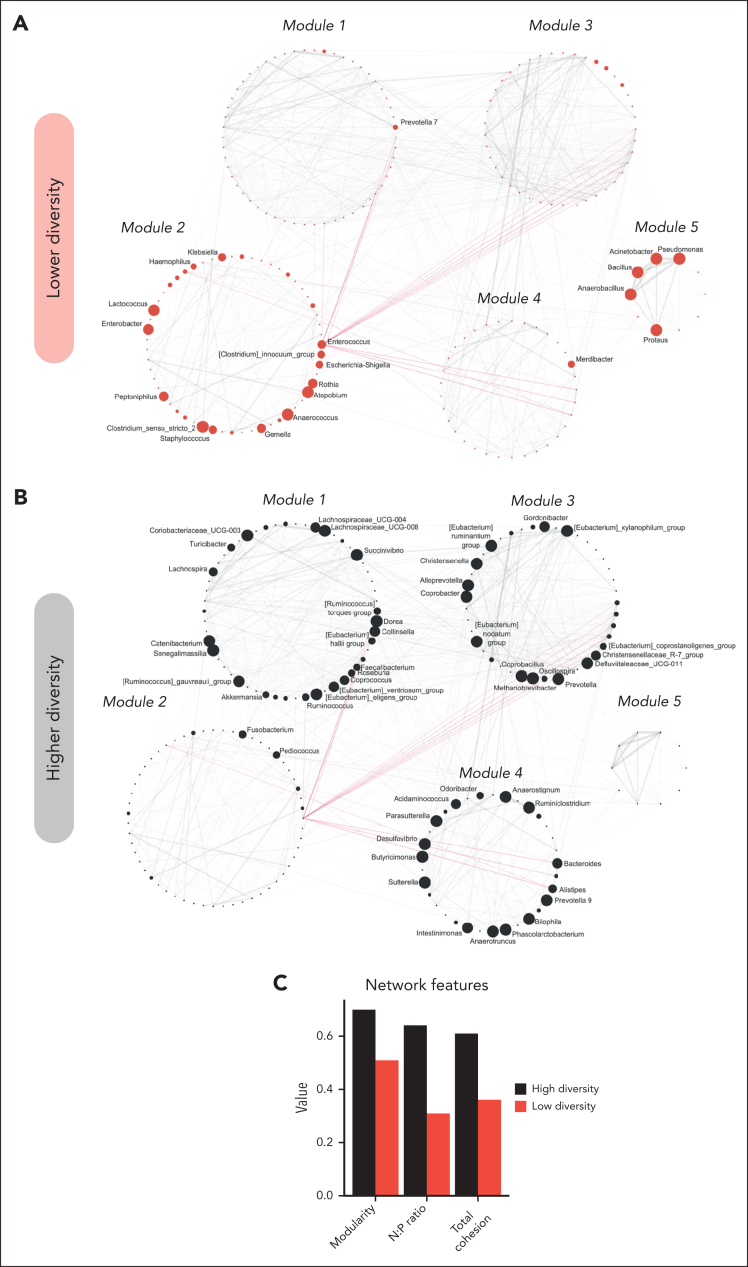
Network analysis
Finally, we explored the GM networks of the higher- and lower-diversity groups before allo-HSCT and at neutrophil engraftment, using a correlation networking approach, as previously reported.48 Before allo-HSCT, the GM network of patients belonging to the lower-diversity group showed Enterococcus, Escherichia-Shigella, Enterobacter, and the (Ruminococcus) gnavus group as keystone taxa (see “Methods”), whereas Bacteroides, Blautia, Faecalibacterium, and Roseburia were detected for the higher-diversity group. Overall, we detected 5 modules, which were clearly differently populated in the 2 diversity groups (Figure 5A-B). The lower-diversity group showed enrichment in network module 2, particularly represented by Enterococcus, Escherichia-Shigella, Rothia, Enterobacter, Anaerococcus, and Klebsiella, and module 5, with Pseudomonas, Anaerobacillus, Bacillus, Proteus, and Acinetobacter. In contrast, the network layout of the higher-diversity group hardly populated these modules, with the overall configuration particularly upheld by modules 1, 3, and 4 containing several short-chain fatty acid (SCFA) producers, as previously outlined (ie, Bacteroides, Coprococcus, Roseburia, Oscillospira, Faecalibacterium, Ruminococcus, and Eubacterium spp), as well as many other gut commensals, with their abundances evenly distributed across modules and nodes. As for network modularity and cohesion before transplantation (ie, N:P ratio and TC), a considerable reduction in all parameters was found in the lower-diversity group (Figure 5C). At the time of neutrophil engraftment, several keystones were maintained, namely Bacteroides in the GM network of patients with higher diversity, and Enterococcus and Escherichia-Shigella in that of patients with lower diversity (supplemental Figure 10). Again, the modularity and N:P ratio were notably reduced in the lower-diversity group, whereas TC showed similar values between groups.
Figure 5.
GM networks of the higher- and lower-diversity groups before allo-HSCT. Correlation networks of interactions reconstructed from genus-level compositional data in the lower- (A) and higher- (B) diversity groups before allo-HSCT. Red lines represent negative interactions, whereas solid gray lines stand for positive ones. Node size is proportional to the overabundance value of the corresponding genus in the GM configuration of the diversity group considered. Only nodes corresponding to genera showing overabundance ≥1.4 were displayed. Labels were displayed only for genera showing an overabundance of at least 1.5. The 5 different modules detected according to a statistical mechanics spin-glass model and simulated annealing are noted with labels. (C) Values of computed network features (ie, modularity, N:P ratio, and TC).
Discussion
To the best of our knowledge, we present the first evidence of an association between pretransplantation lower GM diversity and a poorer outcome in children undergoing allo-HSCT. Moreover, this analysis also involved the largest pediatric cohort studied for GM composition during the allo-HSCT period published to date.
Higher GM diversity before transplantation, but not at the time of neutrophil engraftment, correlated with a better probability of OS and a lower cumulative incidence of grade 2 to 4 and grade 3 to 4 aGVHD. Our data partly differ from those reported in adults, whose diversity both before and after transplantation (ie, at neutrophil engraftment) was significantly related to transplant outcomes.9 In particular, Peled et al observed that lower GM diversity at the time of neutrophil engraftment predicted poor OS and correlated with higher risks of TRM and death attributable to aGVHD in 2 independent cohorts.9 Higher diversity in the pre–allo-HSCT period was associated with increased OS and reduced TRM in only 1 cohort. Therefore, the finding that pretransplant GM features may predict allo-HSCT outcomes is consistent between pediatric and adult cohorts. However, our data differ from those of adults on the predictive nature of posttransplant GM composition. This finding may be due to age-related variation in the GM and possibly to the different abilities of the GM to withstand external stressors between adults and children. Children, in general, have significantly lower GM diversity, higher relative abundances of Bacteroides and Bifidobacterium spp, and lower relative abundances of Blautia than adults.24,58 Functional properties of the GM also vary based on age, with a predominance of catabolic over biosynthetic pathways in the pediatric period.58 In addition, children’s GM is more plastic and malleable to modification through environmental factors, undergoing larger shifts when exposed to external stressor.24 Considering these different ecological properties compared with those in adults, we hypothesize that allo-HSCT–induced dysbiosis in the pediatric setting may imply loss of age-related GM signatures, including alfa diversity, with high interpatient variability. We have previously reported that pediatric patients are particularly prone to lose age-specific GM signatures during allo-HSCT, probably in an individual manner,7 in line with the so-called Anna Karenina principle, stating that each dysbiotic GM is dysbiotic in its own way.59
Interestingly, the higher-diversity group before allo-HSCT exhibited favorable GM compositional features, namely higher relative abundances of SCFA producers, such as Blautia, Roseburia, Faecalibacterium, Bacteroides, Coprococcus, and Parabacteroides.60, 61, 62, 63, 64, 65, 66, 67 SCFAs have a pivotal trophic effect on the intestinal epithelial barrier, being an energy source for enterocytes and increasing the gene expression of tight-junction proteins and mucins,68,69 and play a crucial role in the cross talk between GM and host immune cells.70,71 Higher proportions of butyrate-producing species and increased SCFA production were linked, in several cohorts, to a lower incidence and severity of transplant-related complications, mainly aGVHD and viral infections.18,72, 73, 74, 75, 76, 77 The higher-diversity group also showed increased relative abundances of members of the (Eubacterium) coprostanoligenes group, generally known for its metabolic asset capable of bio-transforming cholesterol into coprostanol, influencing host fat metabolism, which may be a focus in future metagenomic studies.78,79 In contrast, the lower-diversity group was characterized by the families Enterobacteriaceae, which includes many facultative aerobic pathobionts, and Enterococcus. Both these taxa were associated with a higher risk of bacteremia and viremia as well as increased aGVHD-related mortality10 in pediatric allo-HSCT recipients.18,80
The reconstruction of GM networks allowed us to detect significant differences between the higher- and lower-diversity groups in terms of network topology and network properties, which can be linked to potential ecological interactions within GM communities. In particular, the GM of the lower-diversity group before transplantation lacked modularity and stability and showed less competitive interactions (indicated by a lower N:P ratio), thus being more fragile, less resilient, and likely unable to maintain eubiotic cross talk with the host during and after allo-HSCT. By contrast, the ecosystem found in the higher-diversity GM group was more modular and robust. Moreover, this ecosystem was held up by the presence of SCFA producers as keystone taxa (ie, Blautia, Bacteroides, Faecalibacterium, and Roseburia), all of which are well-known components of a healthy child GM.81,82
In conclusion, our findings underscore the importance of pretransplant GM diversity and compositional structure in influencing allo-HSCT–related clinical outcomes in the pediatric setting. In particular, our data emphasize the importance of an overall healthy network, rather than the abundance of specific families or genera, in preventing complications and unfavorable outcomes.
Our study has some limitations. First, the different clinical practices of the participating centers resulted in a heterogeneous cohort encompassing several diseases, conditioning regimens, and transplant practices. Second, although it is the largest pediatric cohort to date, the sample size is still small and not comparable with what is available for adults. Moreover, the age range is wide, including both infants and older children, whose GM likely follows different ecological patterns. Large-scale multicenter cohort studies are needed to fully uncover the clinical significance of the age-related GM configuration in determining patient outcomes. Furthermore, more frequent and longer sampling may enable the implementation of additional approaches, such as that proposed by Raman et al,83 to efficiently detect a so-called ecogroup, that is, an ensemble of taxa that act as a core microbiota during the whole process, whose presence and functionality might provide further insights to the dynamics occurring during transplantation. This ecogroup would most likely include SCFA producers, which could act not only as markers of a healthy ecosystem but also as drivers of favorable associations. Unlike in the cohort of Peled et al,9 in our cohort, we were unable to define a significant relationship with a specific cause of death, probably because of the relatively low number of events.
Although our study provides novel and important information regarding GM in pediatric allo-HSCT recipients, many questions remain unanswered. For instance, our study, like most of the published ones, characterizes the GM only from a compositional point of view. Functional modifications with metagenomics sequencing and metabolomics should be addressed along with the clinical implications. Moreover, this information should be integrated with other GM components, namely fungi and viruses, whose roles are not fully understood yet. Our study also defines opportunities for GM-targeted therapeutics in children, such as nutritional modulation or even fecal microbiota transplantation.84 In particular, our results provide information regarding the best timeframe to act in order to improve the GM configuration and, in cascade, transplant outcomes, which was found to be the one before allo-HSCT, unlike the adult counterpart. Importantly, this increases the possibility of preventing GM injury starting from the diagnosis of the oncological or hematologic disease. Early personalized precision interventions could therefore be proposed to patients based on their GM composition. However, their safety and effectiveness should be specifically addressed in interventional studies in pediatric allo-HSCT recipients. Furthermore, the lower GM diversity and the higher relative abundance of Enterococcaceae observed in pre–allo-HSCT samples of patients receiving antibiotics (especially glycopeptides) suggest that antibiotic-sparing strategies in this key timeframe could contribute to maintaining eubiosis.
Conclusions
To our knowledge, this is the first study demonstrating an association between GM diversity before transplantation and OS in children given allo-HSCT. In addition, we found significant associations between higher pretransplant diversity and the presence of SCFA-producing taxa, as well as a network structure with higher modularity and plasticity. GM modulation during this critical time window related to allo-HSCT could, therefore, possibly influence patients’ prognosis.66,67
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
This work was supported by grants from Fondazione Cassa di Risparmio in Bologna (19431-2021.0082) (R.M.), Federide ONLUS association (R.M.), Fondazione Regionale per la Ricerca Biomedica (Plagencell N° CP2_10/2018) (M.Z.), and Italian Ministry of Health (Ricerca Corrente 08069119 and 08045818) (M.Z.).
Authorship
Contribution: R.M., D.Z., and P.B. conceptualized the study; D.L., E.M., and M.F. performed formal analysis, visualized the data, and wrote the original draft; F.B., F.G., T.B., M.U., J.F., S.C., M.Z., P.M., and F.L. provided the resources; R.M., D.Z., F.D., M.C., A. Pession, F.L., A. Prete, and S.T. reviewed and edited the manuscript; R.M., A. Prete, and P.B. supervised the study; and R.M., M.Z., A. Prete, and P.B. acquired the funding.
Footnotes
∗R.M. and D.L. contributed equally as joint first authors.
Quality-filtered sequences used in this study are available in the European Nucleotide Archive (project code PRJEB23820), MG-RAST (accession number 11624), and National Center for Biotechnology Information Sequence Read Archive (project code PRJNA592853).
Data are available on request from the corresponding author, Marco Fabbrini (m.fabbrini@unibo.it).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Supplementary Material
References
- 1.Copelan EA, Chojecki A, Lazarus HM, Avalos BR. Allogeneic hematopoietic cell transplantation; the current renaissance. Blood Rev. 2019;34:34–44. doi: 10.1016/j.blre.2018.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Masetti R, Muratore E, Gori D, Prete A, Locatelli F. Allogeneic hematopoietic stem cell transplantation for pediatric acute myeloid leukemia in first complete remission: a meta-analysis. Ann Hematol. 2022;101(11):2497–2506. doi: 10.1007/s00277-022-04965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Algeri M, Merli P, Locatelli F, Pagliara D. The role of allogeneic hematopoietic stem cell transplantation in pediatric leukemia. J Clin Med. 2021;10(17) doi: 10.3390/jcm10173790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrara JLM, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zama D, Biagi E, Masetti R, et al. Gut microbiota and hematopoietic stem cell transplantation: where do we stand? Bone Marrow Transplant. 2017;52(1):7–14. doi: 10.1038/bmt.2016.173. [DOI] [PubMed] [Google Scholar]
- 6.Zama D, Gori D, Muratore E, et al. Enteral versus parenteral nutrition as nutritional support after allogeneic hematopoietic stem cell transplantation: a systematic review and meta-analysis. Transplant Cell Ther. 2021;27(2):180.e1–180.e8. doi: 10.1016/j.jtct.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Masetti R, Zama D, Leardini D, et al. The gut microbiome in pediatric patients undergoing allogeneic hematopoietic stem cell transplantation. Pediatr Blood Cancer. 2020;67(12) doi: 10.1002/pbc.28711. [DOI] [PubMed] [Google Scholar]
- 8.Taur Y, Jenq RR, Perales M-A, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peled JU, Gomes ALC, Devlin SM, et al. Microbiota as predictor of mortality in allogeneic hematopoietic-cell transplantation. N Engl J Med. 2020;382(9):822–834. doi: 10.1056/NEJMoa1900623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stein-Thoeringer CK, Nichols KB, Lazrak A, et al. Lactose drives Enterococcus expansion to promote graft-versus-host disease. Science. 2019;366(6469):1143–1149. doi: 10.1126/science.aax3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenq RR, Taur Y, Devlin SM, et al. Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant. 2015;21(8):1373–1383. doi: 10.1016/j.bbmt.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masetti R, Muratore E, Leardini D, et al. Gut microbiome in pediatric acute leukemia: from predisposition to cure. Blood Adv. 2021;5(22):4619–4629. doi: 10.1182/bloodadvances.2021005129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montassier E, Al-Ghalith GA, Ward T, et al. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016;8(1) doi: 10.1186/s13073-016-0301-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rashidi A, Kaiser T, Holtan SG, Weisdorf DJ, Khoruts A, Staley C. Pre-transplant recovery of microbiome diversity without recovery of the original microbiome. Bone Marrow Transplant. 2019;54(7):1115–1117. doi: 10.1038/s41409-018-0414-z. [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Frank DN, Horch M, et al. Associations between acute gastrointestinal GvHD and the baseline gut microbiota of allogeneic hematopoietic stem cell transplant recipients and donors. Bone Marrow Transplant. 2017;52(12):1643–1650. doi: 10.1038/bmt.2017.200. [DOI] [PubMed] [Google Scholar]
- 16.Doki N, Suyama M, Sasajima S, et al. Clinical impact of pre-transplant gut microbial diversity on outcomes of allogeneic hematopoietic stem cell transplantation. Ann Hematol. 2017;96(9):1517–1523. doi: 10.1007/s00277-017-3069-8. [DOI] [PubMed] [Google Scholar]
- 17.Masetti R, Biagi E, Zama D, et al. Early modifications of the gut microbiome in children with hepatic sinusoidal obstruction syndrome after hematopoietic stem cell transplantation. Sci Rep. 2021;11 doi: 10.1038/s41598-021-93571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaitkute G, Panic G, Alber DG, et al. Linking gastrointestinal microbiota and metabolome dynamics to clinical outcomes in paediatric haematopoietic stem cell transplantation. Microbiome. 2022;10(1) doi: 10.1186/s40168-022-01270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingham AC, Kielsen K, Mordhorst H, et al. Microbiota long-term dynamics and prediction of acute graft-versus-host disease in pediatric allogeneic stem cell transplantation. Microbiome. 2021;9(1):148. doi: 10.1186/s40168-021-01100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingham AC, Kielsen K, Cilieborg MS, et al. Specific gut microbiome members are associated with distinct immune markers in pediatric allogeneic hematopoietic stem cell transplantation. Microbiome. 2019;7(1):131. doi: 10.1186/s40168-019-0745-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Amico F, Soverini M, Zama D, et al. Gut resistome plasticity in pediatric patients undergoing hematopoietic stem cell transplantation. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-42222-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simms-Waldrip TR, Sunkersett G, Coughlin LA, et al. Antibiotic-induced depletion of anti-inflammatory Clostridia is associated with the development of graft-versus-host disease in pediatric stem cell transplantation patients. Biol Blood Marrow Transplant. 2017;23(5):820–829. doi: 10.1016/j.bbmt.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Morkis IVC, Vicente BdM, Habigzang M, et al. Should we evaluate intestinal microbiota of pediatric patients undergoing hematopoietic stem cell transplantation? Bone Marrow Transplant. 2019;55(7):1506–1508. doi: 10.1038/s41409-019-0727-6. [DOI] [PubMed] [Google Scholar]
- 24.Derrien M, Alvarez AS, de Vos WM. The gut microbiota in the first decade of life. Trends Microbiol. 2019;27(12):997–1010. doi: 10.1016/j.tim.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Zama D, Bossù G, Leardini D, et al. Insights into the role of intestinal microbiota in hematopoietic stem-cell transplantation. Ther Adv Hematol. 2020;11 doi: 10.1177/2040620719896961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masetti R, D’Amico F, Zama D, et al. Febrile neutropenia duration is associated with the severity of gut microbiota dysbiosis in pediatric allogeneic hematopoietic stem cell transplantation recipients. Cancers (Basel) 2022;14(8) doi: 10.3390/cancers14081932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger M, Lanino E, Cesaro S, et al. Feasibility and outcome of haploidentical hematopoietic stem cell transplantation with post-transplant high-dose cyclophosphamide for children and adolescents with hematologic malignancies: an AIEOP-GITMO retrospective multicenter study. Biol Blood Marrow Transplant. 2016;22(5):902–909. doi: 10.1016/j.bbmt.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Zeiser R, Blazar BR. Acute graft-versus-host disease - biologic process, prevention, and therapy. N Engl J Med. 2017;377(22):2167–2179. doi: 10.1056/NEJMra1609337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eapen M, Horowitz MM, Klein JP, et al. Higher mortality after allogeneic peripheral-blood transplantation compared with bone marrow in children and adolescents: the Histocompatibility and Alternate Stem Cell Source Working Committee of the International Bone Marrow Transplant Registry. J Clin Oncol. 2004;22(24):4872–4880. doi: 10.1200/JCO.2004.02.189. [DOI] [PubMed] [Google Scholar]
- 30.Davies SM, Wang D, Wang T, et al. Recent decrease in acute graft-versus-host disease in children with leukemia receiving unrelated donor bone marrow transplants. Biol Blood Marrow Transplant. 2009;15(3):360–366. doi: 10.1016/j.bbmt.2008.12.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dini G, Zecca M, Balduzzi A, et al. No difference in outcome between children and adolescents transplanted for acute lymphoblastic leukemia in second remission. Blood. 2011;118(25):6683–6690. doi: 10.1182/blood-2011-05-354233. [DOI] [PubMed] [Google Scholar]
- 32.Kong SG, Jeong S, Lee S, Jeong JY, Kim DJ, Lee HS. Early transplantation-related mortality after allogeneic hematopoietic cell transplantation in patients with acute leukemia. BMC Cancer. 2021;21(1) doi: 10.1186/s12885-021-07897-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mateos MK, O’Brien TA, Oswald C, et al. Transplant-related mortality following allogeneic hematopoeitic stem cell transplantation for pediatric acute lymphoblastic leukemia: 25-year retrospective review. Pediatr Blood Cancer. 2013;60(9):1520–1527. doi: 10.1002/pbc.24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobsohn DA. Acute graft-versus-host disease in children. Bone Marrow Transplant. 2008;41(2):215–221. doi: 10.1038/sj.bmt.1705885. [DOI] [PubMed] [Google Scholar]
- 35.Biagi E, Zama D, Rampelli S, et al. Early gut microbiota signature of aGvHD in children given allogeneic hematopoietic cell transplantation for hematological disorders. BMC Med Genom. 2019;12(1):49. doi: 10.1186/s12920-019-0494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biagi E, Zama D, Nastasi C, et al. Gut microbiota trajectory in pediatric patients undergoing hematopoietic SCT. Bone Marrow Transplant. 2015;50(7):992–998. doi: 10.1038/bmt.2015.16. [DOI] [PubMed] [Google Scholar]
- 37.D’Amico F, Biagi E, Rampelli S, et al. Enteral nutrition in pediatric patients undergoing hematopoietic SCT promotes the recovery of gut microbiome homeostasis. Nutrients. 2019;11(12):2958. doi: 10.3390/nu11122958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinf. 2012;13(4):31. doi: 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4(10) doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bokulich NA, Kaehler BD, Rideout JR, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leek JT, Johnson WE, Parker HS, et al. sva: Surrogate variable analysis. R package. Version 3.48.0 Bioconductor. 2022. [DOI]
- 45.Harrell Jr FE. Hmisc: Harrell miscellaneous. Version 4.7-2. 2022. https://CRAN.R-project.org/package=Hmisc
- 46.Hernandez DJ, David AS, Menges ES, Searcy CA, Afkhami ME. Environmental stress destabilizes microbial networks. ISME J. 2021;15(6):1722–1734. doi: 10.1038/s41396-020-00882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herren CM, McMahon KD. Cohesion: a method for quantifying the connectivity of microbial communities. ISME J. 2017;11(11):2426–2438. doi: 10.1038/ismej.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fabbrini M, D’Amico F, Leardini D, et al. Levofloxacin prophylaxis and parenteral nutrition have a detrimental effect on intestinal microbial networks in pediatric patients undergoing HSCT. Commun Biol. 2023;6(1):36. doi: 10.1038/s42003-023-04436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Csárdi G, Nepusz T, Müller K, et al. igraph: Network Analysis and Visualization in R. Version 1.5.0. 2023. [DOI]
- 50.Traag VA, Bruggeman J. Community detection in networks with positive and negative links. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80(3 pt 2):036115. doi: 10.1103/PhysRevE.80.036115. [DOI] [PubMed] [Google Scholar]
- 51.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glucksberg H, Storb R, Fefer A, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18(4):295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 54.Oksanen J, Simpson G, Blanchet F, et al. vegan: community ecology package. Version 2.6-4. 2022. https://CRAN.R-project.org/package=vegan
- 55.Krijthe J. Rtsne: T-distributed stochastic neighbor embedding using a Barnes-Hut implementation. Version 0.16. 2015. https://github.com/jkrijthe/Rtsne
- 56.R Core Team R: A language and environment for statistical computing. Version 4.3.0. 2020. https://www.R-project.org/
- 57.Wickham H. Springer-Verlag; 2016. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 58.Radjabzadeh D, Boer CG, Beth SA, et al. Diversity, compositional and functional differences between gut microbiota of children and adults. Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-57734-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zaneveld JR, McMinds R, Vega Thurber R. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol. 2017;2(9) doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 60.Lei Y, Tang L, Liu S, et al. Parabacteroides produces acetate to alleviate heparanase-exacerbated acute pancreatitis through reducing neutrophil infiltration. Microbiome. 2021;9(1) doi: 10.1186/s40168-021-01065-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reichardt N, Duncan SH, Young P, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8(6):1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bornet E, Westermann AJ. The ambivalent role of Bacteroides in enteric infections. Trends Microbiol. 2022;30(2):104–108. doi: 10.1016/j.tim.2021.11.009. [DOI] [PubMed] [Google Scholar]
- 63.Yang J, Li Y, Wen Z, Liu W, Meng L, Huang H. Oscillospira - a candidate for the next-generation probiotics. Gut Microbes. 2021;13(1):1987783. doi: 10.1080/19490976.2021.1987783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peterson CT, Perez Santiago J, Iablokov SN, Chopra D, Rodionov DA, Peterson SN. Short-chain fatty acids modulate healthy gut microbiota composition and functional potential. Curr Microbiol. 2022;79(5) doi: 10.1007/s00284-022-02825-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang M, Zhou L, Wang Y, et al. Faecalibacterium prausnitzii produces butyrate to decrease c-Myc-related metabolism and Th17 differentiation by inhibiting histone deacetylase 3. Int Immunol. 2019;31(8):499–514. doi: 10.1093/intimm/dxz022. [DOI] [PubMed] [Google Scholar]
- 66.Nie K, Ma K, Luo W, et al. Roseburia intestinalis: a beneficial gut organism from the discoveries in genus and species. Front Cell Infect Microbiol. 2021;11:757718. doi: 10.3389/fcimb.2021.757718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu X, Mao B, Gu J, et al. Blautia—a new functional genus with potential probiotic properties? Gut Microbes. 2021;13(1):1–21. doi: 10.1080/19490976.2021.1875796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Masetti R, Zama D, Leardini D, et al. Microbiome-derived metabolites in allogeneic hematopoietic stem cell transplantation. Int J Mol Sci. 2021;22(3):1197. doi: 10.3390/ijms22031197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romick-Rosendale LE, Haslam DB, Lane A, et al. Antibiotic exposure and reduced short chain fatty acid production after hematopoietic stem cell transplant. Biol Blood Marrow Transplant. 2018;24(12):2418–2424. doi: 10.1016/j.bbmt.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 70.Chang P v, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111(6):2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nastasi C, Candela M, Bonefeld CM, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci Rep. 2015;5(1) doi: 10.1038/srep16148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Docampo MD, da Silva MB, Lazrak A, et al. Alloreactive T cells deficient of the short-chain fatty acid receptor GPR109A induce less graft-versus-host disease. Blood. 2022;139(15):2392–2405. doi: 10.1182/blood.2021010719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meedt E, Hiergeist A, Gessner A, et al. Prolonged suppression of butyrate-producing bacteria is associated with acute gastrointestinal graft-vs-host disease and transplantation-related mortality after allogeneic stem cell transplantation. Clin Infect Dis. 2022;74(4):614–621. doi: 10.1093/cid/ciab500. [DOI] [PubMed] [Google Scholar]
- 74.Burgos da Silva M, Ponce DM, Dai A, et al. Preservation of the fecal microbiome is associated with reduced severity of graft-versus-host disease. Blood. 2022;140(22):2385–2397. doi: 10.1182/blood.2021015352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mathewson ND, Jenq R, Mathew A V, et al. Gut microbiome–derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat Immunol. 2016;17(5):505–513. doi: 10.1038/ni.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Markey KA, Schluter J, Gomes ALC, et al. The microbe-derived short-chain fatty acids butyrate and propionate are associated with protection from chronic GVHD. Blood. 2020;136(1):130–136. doi: 10.1182/blood.2019003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Haak BW, Littmann ER, Chaubard JL, et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood. 2018;131(26):2978–2986. doi: 10.1182/blood-2018-01-828996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freier TA, Beitz DC, Li L, Hartman PA. Characterization of Eubacterium coprostanoligenes sp. nov., a cholesterol-reducing anaerobe. Int J Syst Bacteriol. 1994;44(1):137–142. doi: 10.1099/00207713-44-1-137. [DOI] [PubMed] [Google Scholar]
- 79.Juste C, Gérard P. Cholesterol-to-coprostanol conversion by the gut microbiota: what we know, suspect, and ignore. Microorganisms. 2021;9(9) doi: 10.3390/microorganisms9091881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perez P, Patiño J, Estacio M, Pino J, Manzi E, Medina D. Bacteremia in pediatric patients with hematopoietic stem cell transplantation. Hematol Transfus Cell Ther. 2020;42(1):5–11. doi: 10.1016/j.htct.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roswall J, Olsson LM, Kovatcheva-Datchary P, et al. Developmental trajectory of the healthy human gut microbiota during the first 5 years of life. Cell Host Microbe. 2021;29(5):765–776.e3. doi: 10.1016/j.chom.2021.02.021. [DOI] [PubMed] [Google Scholar]
- 82.Deering KE, Devine A, O’sullivan TA, Lo J, Boyce MC, Christophersen CT. Characterizing the composition of the pediatric gut microbiome: a systematic review. Nutrients. 2020;12(1):16. doi: 10.3390/nu12010016. Page 16. 2019;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Raman AS, Gehrig JL, Venkatesh S, et al. A sparse covarying unit that describes healthy and impaired human gut microbiota development. Science (1979) 2019;365(6449) doi: 10.1126/science.aau4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muratore E, Leardini D, Baccelli F, Venturelli F, Prete A, Masetti R. Nutritional modulation of the gut microbiome in allogeneic hematopoietic stem cell transplantation recipients. Front Nutr. 2022;9 doi: 10.3389/fnut.2022.993668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.