Abstract
The light-scattering properties of submicroscopic metal particles ranging from 40 to 120 nm in diameter have recently been investigated. These particles scatter incident white light to generate monochromatic light, which can be seen either by the naked eye or by dark-field microscopy. The nanoparticles are well suited for detection in microchannel-based immunoassays. The goal of the present study was to detect Helicobacter pylori- and Escherichia coli O157:H7-specific antigens with biotinylated polyclonal antibodies. Gold particles (diameter, 80 nm) functionalized with a secondary antibiotin antibody were then used as the readout. A dark-field stereomicroscope was used for particle visualization in poly(dimethylsiloxane) microchannels. A colorimetric quantification scheme was developed for the detection of the visual color changes resulting from immune reactions in the microchannels. The microchannel immunoassays reliably detected H. pylori and E. coli O157:H7 antigens in quantities on the order of 10 ng, which provides a sensitivity of detection comparable to those of conventional dot blot assays. In addition, the nanoparticles within the microchannels can be stored for at least 8 months without a loss of signal intensity. This strategy provides a means for the detection of nanoparticles in microchannels without the use of sophisticated equipment. In addition, the approach has the potential for use for further miniaturization of immunoassays and can be used for long-term archiving of immunoassays.
Immunoassays are based on specific antibody-antigen reactions (26). Quantification of immunoassays is generally achieved by measuring the specific activity of a label, for example, radioactivity, fluorescence, chemiluminescence, bioluminescence, or electrical conductivity (20, 21, 26). However, these labels share a common drawback, which is that they are not suitable for long-term preservation (26). While different isotopes have various half-lives, the use of radioactivity is more difficult because of issues of disposition and potential harmful health effects. Furthermore, fluorescence suffers from the issue of photobleaching (15).
Recently, nanoparticles, especially gold and silver particles, have been successfully applied for labeling because of their easily controlled size distribution, long-term stability, and compatibility with biological macromolecules, including proteins and nucleic acids (4, 11). Multiple nanoparticle detection methods have been developed, including scanning and transmission electron microscopy (12, 25), Raman spectroscopy (6, 9, 27), and the naked eye (16, 24). Detection by the naked eye may be preferable, as other techniques are expensive and require specialized equipment and additional preparations. Both a DNA microarray and a heterogeneous immunoassay have been developed with 10-nm gold particles amplified with silver, which can be detected by the naked eye (16, 24).
Most recently, the light-scattering properties of submicroscopic metal particles, such as gold nanoparticles, have been investigated (28). These particles scatter incident white light to generate monochromatic light and can be seen either by the naked eye or by dark-field microscopy (29). The intensity generated by a nanoparticle is 100,000 times that generated by a fluorescein-labeled molecule (29). The approach has been used with success in DNA hybridization arrays, immunohistochemistry, and immunoassays (29). The resonance light-scattering (RLS) properties of nanoparticles render them well suited for use in microchannel-based immunoassays (11).
The goal of this study was to develop a resonance light-scattering nanoparticle-labeled immunoassay with a microfluidic platform and dark-field zoom stereomicroscopy as a simple and inexpensive strategy for the detection of the gastrointestinal microbial pathogens Helicobacter pylori and Escherichia coli O157:H7.
MATERIALS AND METHODS
Bacterial growth conditions and antigen preparations.
The procedures used for bacterial culture and antigen preparation were described previously (13, 14). Briefly, H. pylori strain ATCC 49503 was cultured on Columbia agar plates containing 5% sheep blood, and the plates were incubated at 37°C under microaerophilic conditions (5% oxygen, 85% nitrogen, and 10% carbon dioxide) for 72 h (13) Bacteria were then inoculated into brucella broth supplemented with 10% fetal bovine serum and were grown overnight with gentle shaking under microaerophilic conditions at 37°C. E. coli O157:H7 strain CL-56 and nonpathogenic E. coli strains K-12, HB101, F18, and C25 were cultured on 5% sheep blood agar plates at 37°C overnight and were then stored at 4°C (14). The bacteria were then cultured in static Penassay broth (Difco Laboratories, Detroit, Mich.) overnight at 37°C.
For use as an additional negative control, Lactobacillus rhamnosus strain R011 (Institut Rosell-Lallemand Inc., Montreal, Quebec, Canada) was cultured on 5% sheep blood agar plates at 37°C overnight, stored at 4°C, and subsequently cultured in static Mann-Ragosa-Sharpe broth (Difco) overnight at 37°C (13).
Liquid cultures were centrifuged in a Beckman GPR centrifuge at 1,600 × g for 15 min, and the supernatants were removed. The bacterial pellets were washed four times in sterile phosphate-buffered saline (PBS; pH 7.6). The cell pellets were then resuspended in RIPA buffer (10 mM Tris HCl [pH 8.0], 150 mM NaCl, 0.5% Triton X-100) containing a mixture of proteinase inhibitors (Roche Molecular Biochemicals, Mannheim, Germany) for 40 min at 4°C. The lysates were then centrifuged at 10,000 × g for 15 min at 4°C, and the supernatants with whole bacterial proteins were collected. The samples were then assayed for their protein contents and were stored at −70°C. These lysate antigens were then diluted to concentrations of 0.6, 6, 60, and 600 ng/μl in a coating buffer consisting of 0.03 M NaHCO3 and 0.02 M Na2CO3 at pH 9.6. The diluted antigens were immobilized onto solid substrates for subsequent immunoassays (13).
Electron microscopic measurements of gold nanoparticles.
A 5-μl aliquot of the RLS particle suspension (from the One-Color Microarray Toolkit [Invitrogen Life Technologies, Carlsbad, Calif.]) was fixed with 5 μl of a universal fixative containing 4% paraformaldehyde and 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) on a Formvar-coated copper grid for 2 min. The grid was blotted with a piece of filter paper and washed with 10 μl of distilled water. For transmission electron microscopy, the wash was immediately followed by negative staining with uranyl acetate. The grid was examined with a JEM-1230 transmission electron microscope (JEOL Corp., Tokyo, Japan). Digital images were acquired directly with a charge-coupled-device camera system (1,024 by 1,024 pixels; Advanced Microscopy Techniques Corp., Danvers, Mass.). For scanning electron microscopy, the grid was allowed to air dry after the wash with distilled water and was sputtered coated with chromium before examination in a JSM-6700F field emission scanning electron microscope (JEOL Corp.).
Replica molding of PDMS microchannels.
Fused-silica capillaries with an outer diameter of 360 μm (Polymicro Technologies, Phoenix, Ariz.) were cut to a length of 2 cm and placed onto a clean plastic petri dish (Fisher Scientific, Nepean, Ontario, Canada) for use as a mold. The procedures for replica molding of poly(dimethylsiloxane) (PDMS) layers were similar to those published previously (13, 18). Briefly, a 10:1 (wt/wt) mixture of PDMS prepolymer and curing agent (Sylgard 184 silicone elastomer kit; Dow Corning, Midland, Mich.) was mixed thoroughly in a plastic weighing dish. The PDMS microchannel device was then cast by pouring the mixture over the mold in the petri dish, which was placed in a vacuum desiccator and degassed at 20 torr to remove the air bubbles created during mixing and was then cured for 7 h at 75°C. After the device was cured, the PDMS replica of the master device was cut into rectangular slabs containing a pair of straight channels and was then peeled from the petri dish. A hole of 3 mm in diameter was bored through the PDMS slab on one end of each channel for use as a waste reservoir. The capillaries were then removed to expose the microchannels formed in PDMS.
To mount the PDMS, glass slides (Micro Slides; Corning Glass Works, Corning, N.Y.) were soaked in acetone for 5 min and then rinsed in filtered (syringe filter; pore size, 0.2 μm; Pall Corporation, Ann Arbor, Mich.) distilled water. The slides were first placed onto a hot plate to facilitate evaporation, and then the PDMS slab containing microchannels and a glass slide were placed into a plasma sterilizer (model PDC-32G; Harrick Scientific, Ossining, N.Y.) for 1 min to allow surface oxidation. Immediately after removal from the sterilizer, the PDMS slabs were brought into contact with the glass slides and an irreversible seal was formed spontaneously (18). The channels were then allowed to set for an additional 30 min at room temperature to promote surface rearrangements, which resulted in the appearance of new hydrophobic groups on the PDMS surface (18).
Microchannel nanoparticle immunoassay.
The delivery of reagent solutions through the PDMS microchannels was accomplished with a pressure-driven flow, as described previously (13). A 1-ml syringe with a 30-gauge needle (both from Becton Dickinson, Franklin Lakes, N.J.) was inserted directly into the microchannel at the end opposite the waste reservoir, and each solution was injected in turn. A 5 ml-syringe with a 25-gauge needle (Becton Dickinson) was inserted at the waste reservoir to aspirate waste liquid.
Lysate antigens of H. pylori and E. coli O157:H7 (from 0.6 to 600 ng/μl), L. rhamnosus strain R011 (60 ng/μl), and coating buffer alone were injected into each of the microchannels and passively adsorbed overnight at 4°C. Excess lysate antigens were washed off with PBS containing 0.05% Tween 20, and the channels were incubated in blocking solution containing PBS with 1% (wt/vol) bovine serum albumin (pH 7.6) for 45 min at room temperature. For H. pylori antigen detection, a commercially available biotin-conjugated rabbit polyclonal anti-H. pylori antibody (Biodesign International, Saco, Maine) diluted to 1 part in 1,000 parts of blocking solution was used to probe the coated microchannels for 1 h at room temperature. For E. coli O157:H7 antigen detection, a commercially available biotin-conjugated goat polyclonal anti-E. coli O157:H7 antibody (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) diluted to 1 part in 250 parts of blocking solution was incubated in the coated channels for 1 h at room temperature.
The two antibodies without biotin conjugation (i.e., the anti-H. pylori [DAKO, Glostrup, Denmark] and anti-E. coli O157:H7 [Kirkegaard & Perry Laboratories] antibodies) were used as negative controls for the primary antibodies. Additional negative controls included normal rabbit immunoglobulin G (IgG; 1:1,000; Santa Cruz Biotechnology, Santa Cruz, Calif.) and goat serum (1:250; Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.).
After incubation with primary antibodies, the microchannels were washed thoroughly with 0.5% wash solution A supplied with the One-Color Microarray Toolkit. A blocking solution from the kit was incubated in the channel for 20 min, which was followed by a wash step with 0.5% solution A. The RLS gold nanoparticles functionalized with the goat antibiotin antibody supplied in the kit were used as the secondary antibody (2:1 dilution) and were incubated in the microchannels for 1 h at room temperature. After incubation, the channels were washed with 0.5% solution A.
Images of the PDMS microchannels were taken by using a zoom stereomicroscope (SMZ 800; Nikon, Tokyo, Japan) and a color digital camera (Micropublisher, version 3.3; Qimaging, Burnaby, British Columbia, Canada) at ×25 magnification. The proprietary archive solution was subsequently delivered into the channel, and the channel was stored at 4°C in a cold room for long-term preservation. After 7 weeks and 8 months, the microchannels were rehydrated with the archive solution and images were again taken by using dark-field microscopy. All experiments were performed in duplicate on three separate occasions (n = 3), except for experiments demonstrating the linear range of E. coli O157:H7 detection (n = 2) and the specificity of enterohemorrhagic E. coli O157:H7 detection (n = 1 for each of the nonpathogenic E. coli strains). Color changes within the microchannels were then quantified by using novel software developed specifically for this study.
Color quantification.
The red-green-blue values of the pixels of the color digital images of the PDMS microchannels were first transformed to a perceptually uniform color space established by the Commission Internationale de L'Eclairage (CIE) (2, 23). An extended version of the CIE color space, referred as CIE l*a*b* space, was used, as described previously (13). Briefly, background pixel values were determined by using two narrow strips each with a five-pixel width, one on the top and the other one on the bottom of the images. The difference between the value of each pixel within the channel and the background (i.e., the Euclidean distance) was then calculated. The average difference across the whole channel was computed, and the average distance was normalized against the channel length. For the purpose of this study, the intensities of brown color were used for quantification.
Data analyses.
Results of color quantification were expressed as means ± standard error of the mean. Statistical analyses were performed by using InStat software (version 3.01; GraphPad Software Inc., San Diego, Calif.). To test for statistical significance between multiple groups, a one-way analysis of variance (ANOVA) was used, followed by post-hoc comparison by the Tukey-Kramer multiple-comparison test (19). A P value of <0.05 was considered statistically significant.
RESULTS
Electron microscope measurements of gold nanoparticles.
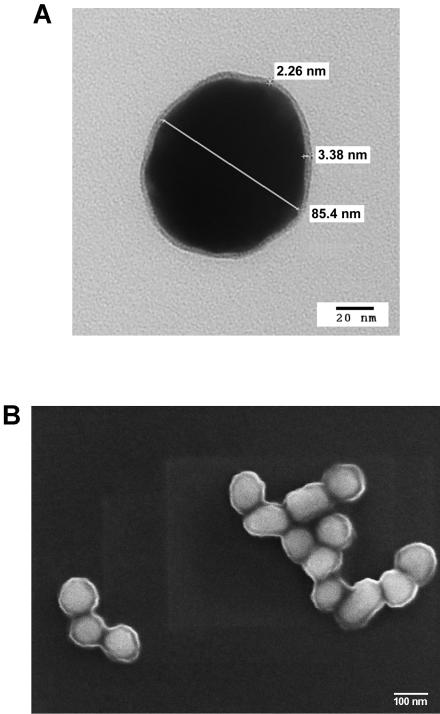
As shown in Fig. 1, the functionalized gold nanoparticles were visualized by electron microscopy. The particles were observed to be surrounded by an antibody halo by both transmission electron microscopy (Fig. 1A) and scanning electron microscopy (Fig. 1B). By transmission electron microscopy, the mean size of the gold nanoparticles was 85.6 ± 2.1 nm (n = 7) and the mean size of the antibody layer 3.5 ± 0.7 nm (n = 3). The shape of the particles was circular, and the particles appeared as both clusters and single particles when they were in suspension.
FIG. 1.
Electron microscopy of the functionalized gold nanoparticles. (A) Transmission electron microcopy of gold particles with measurements (magnification, ×400,000); (B) scanning electron microscopy of gold particles (magnification, ×100,000).
Microchannel nanoparticle immunoassay.
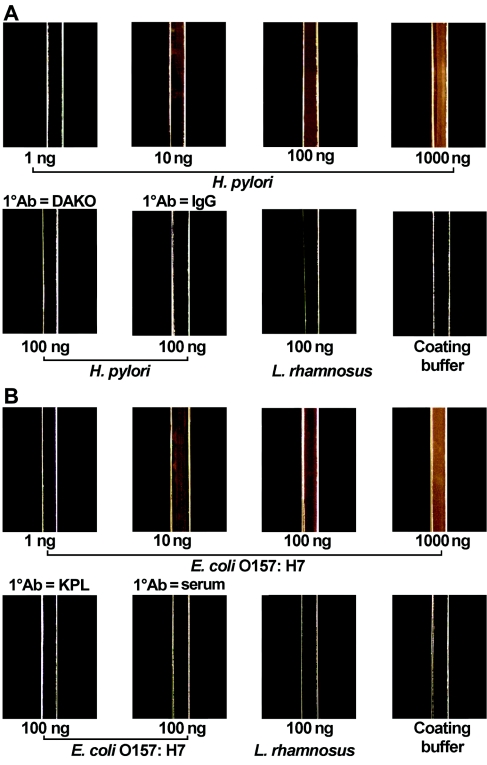
Figure 2 shows that by use of the antibody-nanoparticle conjugate for immune reaction detection, both H. pylori (Fig. 2A) and E. coli O157:H7 (Fig. 2B) antigens were detected at concentrations between 6 and 600 ng/μl, as evidenced by the golden brown color in the microchannels visualized under a dark-field microscope. The volume of the microchannel was approximately 1.5 μl. Hence, the detection limits for H. pylori and E. coli O157:H7 antigen were in the range of 10 to 1,000 ng, which are similar to those of conventional dot enzyme-linked immunosorbent assays (ELISAs) (13, 14).
FIG. 2.
H. pylori and E. coli O157:H7 antigen detection in PDMS microchannels with the RLS nanoparticles as the readout for the heterogeneous immunoassay. Bacterial protein lysates were passively adsorbed onto PDMS microchannels. Polyclonal biotinylated H. pylori and E. coli O157:H7 antibodies were used as primary antibodies (1° Abs), and a goat polyclonal antibiotin antibody-functionalized RLS nanoparticle was used as the secondary antibody. Dark-field microscopy was used for image acquisition. (A) H. pylori lysates were used as positive control antigens, while L. rhamnosus strain R011 and coating buffer alone were used as negative controls. The biotinylated anti-H. pylori antibody was used as a positive control primary antibody, while a nonbiotinylated anti-H. pylori antibody (DAKO) and normal rabbit IgG were used as negative controls. (B) E. coli O157:H7 lysates were used as positive control antigens, whereas L. rhamnosus R011 and coating buffer alone were used as negative controls. Biotinylated anti-E. coli O157:H7 antibody was used as a positive control primary antibody, while a nonbiotinylated anti-E. coli O157:H7 antibody (Kirkegaard & Perry Laboratories [KPL]) and goat serum were used as negative controls.
Validation of the colorimetric quantification software.
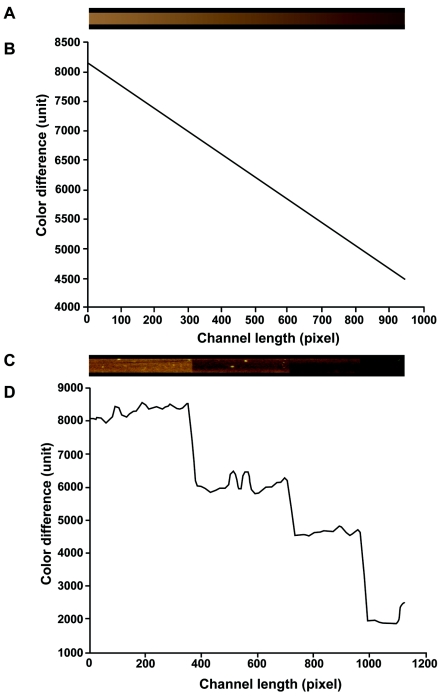
A computer-generated brown strip with decreasing intensity and a black background strip was produced (Fig. 3A) to simulate a real microchannel with a positive reaction (brown). The Euclidean distance was calculated for each brown pixel from the background, and the results were plotted against the locations of the pixels (Fig. 3B). The color difference between the brown and the background strips decreases toward the right side, which correlates with human visual perceptions. Figure 3C represents a composite of four experimental channels with decreasing brown intensities along their lengths, corresponding to experiments with immobilized antigens at concentrations from 600 to 0.6 ng/μl. Figure 3D provides a plot of the Euclidean distance versus the pixel location of the channel shown in Fig. 3C and demonstrates that the Euclidean distance between the pixels within the microchannels and the background decreases in parallel with human visual perceptions of the images.
FIG. 3.
Validation of a colorimetric quantification algorithm. (A) A synthetic brown channel of decreasing intensity along its length; (B) a plot of the Euclidean distance versus the location of the pixels along the synthetic channel; (C) a composite of four experimental channels with decreasing brown intensities along their lengths; (D) a plot of the Euclidean distance versus the location of the pixels along the composite channel.
Quantification of microchannel nanoparticle immunoassay.
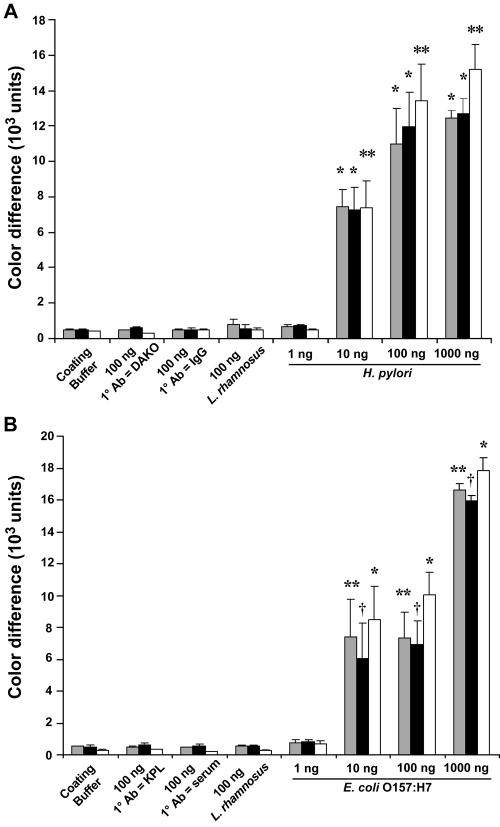
Color differences between the intensities in the microchannels produced by the nanoparticles for H. pylori (Fig. 2A) and E. coli O157:H7 (Fig. 2B) detection and the background were quantified as shown in Fig. 4A and B, respectively. By a one-way ANOVA, there was a statistically significant difference between the immobilized H. pylori antigens at amounts ranging from 10 to 1,000 ng (P < 0.001) and each of the four negative controls (i.e., H. pylori at 100 ng with anti-H. pylori antibody but without biotin conjugation, H. pylori at 100 ng with normal rabbit IgG, L. rhamnosus at 100 ng, and coating buffer alone) taken during the initial experiments (Fig. 4A). Similarly, there was a statistically significant difference between the H. pylori antigens at amounts ranging from 10 to 1,000 ng and each of the negative controls taken at 7 weeks after the initial experiments (P < 0.001) and after 8 months of storage (P < 0.01) (Fig. 4A). There was a significant difference between immobilized E. coli O157:H7 antigens at amounts ranging from 10 to 1,000 ng (P < 0.01) and the negative controls (E. coli at 100 ng with anti-E. coli O157:H7 antibody but without biotin conjugation, E. coli O157:H7 at 100 ng with goat serum, L. rhamnosus at 100 ng, and coating buffer alone) taken during the initial experiments (Fig. 4B). In addition, there was a statistically significant difference between E. coli O157:H7 antigens at amounts ranging from 10 to 1,000 ng and each of the negative controls taken at 7 weeks after the initial experiments (P < 0.05) and after 8 months of storage (P < 0.001) (Fig. 4B). The limit of detection of H. pylori and E. coli O157:H7 antigens by this assay was approximately 10 ng, which is comparable to that of the conventional dot blot ELISA (13, 14). There was no statistically significant difference between the quantification results obtained at 7 weeks and 8 months after the initial experiments and the results obtained at the time of the initial experiment, indicating that the immune reactions detected within the PDMS microchannels are stable at 4°C for at least 8 months. In addition, the linear range of the immune reaction lies between 1 and 50 ng of immobilized E. coli O157:H7 protein antigens (Fig. 5). Beyond this point, the signal intensity becomes saturated.
FIG. 4.
Quantification of RLS gold nanoparticle readout for the detection of H. pylori and E. coli O157:H7 antigens in PDMS microchannels. (A) Quantification of microchannel immunoassay for detection of H. pylori (Fig. 2A) by using the quantification software package; (B) quantification of microchannel immunoassay for detection of E. coli O157:H7 (Fig. 2B). Results are expressed as mean color differences ± standard error of the mean (n = 3). Gray bars, the results of quantification at the time of the initial experiments; black bars, quantification performed at 7 weeks after the initial experiments; open bars, quantification performed at 8 months of storage. There was a significant difference between H. pylori at a concentration of 10 to 1,000 ng and each of the negative controls (*, P < 0.001 at the time of the initial experiments and 7 weeks after the initial experiments; **, P < 0.01 at 8 months of storage [ANOVA]). Similarly, there was a statistically significant difference between E. coli O157:H7 at concentrations of 10 to 1,000 ng and each of the negative controls used (**, P < 0.01 at the time of the initial experiments; †, P < 0.05 at 7 weeks after the initial experiments; *, P < 0.001 at 8 months of storage [ANOVA]). 1° Ab, primary antibody; KPL, Kirkegaard & Perry Laboratories.
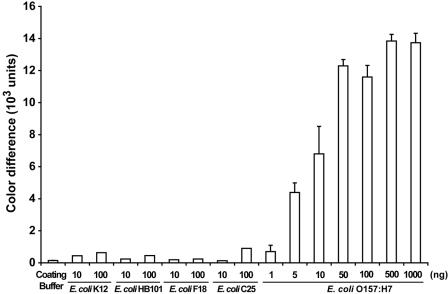
FIG. 5.
Specificity of nanoparticle immunoassay for detection of E. coli O157:H7. Whole-protein lysates of nonpathogenic E. coli strains K-12, HB101, F18, and C25 were used to test the specificity of the nanoparticle immunoassay. Additional dilutions of E. coli O157:H7 antigens were used to establish both the linear range and the saturability of the immunoassay.
The specificity of the E. coli O157:H7 nanoparticle immunoassay was demonstrated (Fig. 5) by using nonpathogenic E. coli strains (strains K-12, HB101, F18, and C25) as negative controls. Whole-protein lysates from each of the nonpathogenic bacterial strains showed minimal reaction with the polyclonal primary antibody used in the nanoparticle immunoassay.
DISCUSSION
A novel heterogeneous PDMS-based microfluidic immunoassay with antibody-functionalized RLS nanoparticles as labels for the detection of gram-negative gastrointestinal pathogens is described in this study. This strategy has the same detection limit as a conventional ELISA system, while it allows long-term preservation. This assay has the potential for further miniaturization and automation without the need for expensive detection equipment.
Miniaturized proof-of-concept PDMS-based microfluidic immunoassay strategies have been demonstrated previously in the literature (1, 5, 7). Recently, accurate detection of H. pylori in biological samples was achieved by using PDMS-based heterogeneous microchannel ELISA (13). However, the enzyme-based label used is not suitable for long-term preservation.
Nanoparticles, which can be synthesized with dimensions ranging from 0.8 to 250 nm, have been used extensively as a biomolecular label in both light and electron microscopy (10). Long-term stability and biocompatibility with a variety of macromolecules, such as DNA, RNA, and proteins, indicate that the metal nanoparticles can be used successfully as a label for a variety of diagnostic approaches. For instance, much work has been done in nucleic acid hybridization assays in which the modified gold nanoparticles are aggregated and then hybridize with target DNA and can be detected by a colorimetric method (3). More recently, surface-enhanced Raman scattering with colloidal silver and gold nanoparticles as enhancing substrates has been developed as a sensitive readout for a heterogeneous immunoassay (6, 9, 27). Although this technique is attractive for the future development of a multiplex microarray platform by using multilabel techniques (27), the detection strategy is costly and complex, as it requires special surface coating and the use of a laser (9). Hence, it is only suitable for use as a research tool.
The light-scattering properties of metal nanoparticles were recently examined by Yguerabide et al. (28, 29) by using previously developed theories (17). When a particle is illuminated by a beam of monochromatic light, it extracts some of the incident energy and scatters the energy in all directions at the frequency of the incident beam (17). However, with white light illumination particles of different sizes and compositions display a distinct scattered light color which corresponds to the peak wavelength of the light-scattering spectral band (28, 29). For example, with incident white light, 40-nm silver particles scatter blue color, while gold particles of the same size scatter green color. However, larger gold particles (78 nm) scatter bright yellow color with white light illumination (28). When the size of the particle is comparable to or larger than the wavelength of the incident light, an increase in the particle diameter results in a rapid increase in scattered light intensity. For example, it has been estimated that the relative light-scattering power of an 80-nm gold particle is 500 times that of a 20-nm particle (28). In addition, it was calculated that the light-scattering power of a 60-nm gold particle is equivalent to about 5 × 105 fluorescein molecules (28). The RLS gold particles used in this study were of 80 nm in diameter, as measured by transmission electron microscopy. The strong signal generated by the relatively large nanoparticles made them suitable for use in miniaturized immunoassays.
Recently, these RLS particles were used in microarrays for gene expression profiling (8, 29) and a sandwiched immunoassay in a microarray format (22). The RLS signals were at least 50 times more intense than confocal laser-based fluorescence signals (8) in DNA microarrays. In protein microarrays, RLS particles provided very sensitive labels for sandwich immunoassays (22).
A sensitive heterogeneous immunoassay was developed by using the RLS gold nanoparticles. The immunoassay proved to be capable of detecting H. pylori and E. coli O157:H7 antigens at quantities as low as 10 ng. This detection limit is comparable to that of the conventional dot blot ELISA (13, 14). In addition, detection was achieved by using a simple strategy consisting of dark-field microscopy. The gold particles were delivered by pressure-driven flow into microchannels, demonstrating their suitability for further miniaturization. Furthermore, upon rehydration by using an arching solution, this microchannel immunoassay could be preserved for up to 8 months without losing sensitivity, indicating its suitability for the purposes of achieving a long-term archive of the test results. The commercially available polyclonal anti-E. coli O157:H7 antibody used in the development of this nanoparticle immunoassay was specific for the detection of E. coli O157:H7. A monoclonal antibody raised against a specific bacterial antigenic epitope could be used in the future to potentially further improve the sensitivity of detection.
The image analysis software package developed for quantification of the intensity of the nanoparticles was robust. The algorithm was previously validated and used to quantify the results of a microchannel ELISA with a blue precipitating substrate (13). As shown in the present study, the software was easily modified to quantify the brown colors produced by gold nanoparticles exposed to white light.
The cost of the nanoparticle immunoassay is estimated to be approximately $6.00 per channel, whereas the cost of a conventional microtiter plate format (Premier Platinum HpSA test; Meridian Diagnostics, Cincinnati, Ohio) can be up to $35.00 per well. Therefore, with miniaturization the cost of reagents can be greatly reduced.
In summary, a PDMS microchannel immunoassay has been developed with antibody-functionalized RLS gold nanoparticles visualized by using dark-field microscopy. The assay is able to accurately detect both H. pylori- and E. coli O157:H7-specific antigens at levels similar to those detected by a conventional ELISA. In addition, the immunoassay can be preserved for the long term without losing sensitivity. Furthermore, software has been developed to objectively quantitate the intensities of the nanoparticles in the microchannels. These advances should prove useful in the future development of an automated, miniaturized immunoassay system for use in microbial diagnostics.
Acknowledgments
This study was supported by a Canadian Institutes of Health Research (CIHR)-University Industry (Canadian Association of Gastroenterology/AstraZeneca Canada) Research Initiative Award (to F.Y.H.L.), a CIHR fellowship (to F.Y.H.L.), and operating grants from the Canadian Institutes of Health Research (to P.M.S.) and Natural Sciences and Engineering Research Council (NSERC) of Canada Collaborative Health Research Project (to D.L. and P.M.S.). P.M.S. is the recipient of a Canada Research Chair in Gastrointestinal Disease.
REFERENCES
- 1.Bernard, A., B. Michel, and E. Delamarche. 2001. Micromosaic immunoassays. Anal. Chem. 73:8-12. [DOI] [PubMed] [Google Scholar]
- 2.Connolly, C., and T. Fliess. 1997. A study of efficiency and accuracy in the transformation from RGB to CIELAB color space. IEEE Trans. Image Processing 6:1046-1048. [DOI] [PubMed] [Google Scholar]
- 3.Csáki, A., R. Möller, and W. Fritzsche. 2002. Gold nanoparticles as novel label for DNA diagnostics. Expert Rev. Mol. Diagn. 2:187-193. [DOI] [PubMed] [Google Scholar]
- 4.Daniel, M.-C., and D. Astruc. 2004. Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem. Rev. 104:293-346. [DOI] [PubMed] [Google Scholar]
- 5.Dodge, A., K. Fluri, E. Verpoorte, and N. F. de Rooij. 2001. Electrokinetically driven microfluidic chips with surface-modified chambers for heterogeneous immunoassays. Anal. Chem. 73:3400-3409. [DOI] [PubMed] [Google Scholar]
- 6.Dou, X., T. Takama, Y. Yamaguchi, H. Yamamoto, and Y. Ozaki. 1997. Enzyme immunoassay utilizing surface-enhanced Raman scattering of the enzyme reaction product. Anal. Chem. 69:1492-1495. [Google Scholar]
- 7.Eteshola, E., and D. Leckband. 2001. Development and characterization of an ELISA assay in PDMS microfluidic channels. Sensors Actuators B 72:129-133. [Google Scholar]
- 8.Francois, P., M. Bento, P. Vaudaux, and J. Schrenzel. 2003. Comparison of fluorescence and resonance light scattering for highly sensitive microarray detection of bacterial pathogens. J. Microbiol. Methods 55:755-762. [DOI] [PubMed] [Google Scholar]
- 9.Grubisha, D. S., R. J. Lipert, H.-Y. Park, J. Driskell, and M. D. Porter. 2003. Femtomolar detection of prostate-specific antigen: an immunoassay based on surface-enhanced Raman scattering and immunogold labels. Anal. Chem. 75:5936-5943. [DOI] [PubMed] [Google Scholar]
- 10.Horisberger, M. 1981. Colloidal gold: a cytochemical marker for light and fluorescent microscopy and for transmission and scanning light microscopy. Scanning Electron Microsc. 2:9-31. [PubMed] [Google Scholar]
- 11.Jain, K. K. 2003. Nanodiagnostics: application of nanotechnology in molecular diagnostics. Expert Rev. Mol. Diagn. 3:153-161. [DOI] [PubMed] [Google Scholar]
- 12.Levit-Binnun, N., A. B. Lindner, O. Zik, Z. Eshhar, and E. Moses. 2003. Quantitative detection of protein arrays. Anal. Chem. 75:1436-1441. [DOI] [PubMed] [Google Scholar]
- 13.Lin, F. Y. H., M. Sabri, D. Erickson, J. Alirezaie, D. Li, and P. M. Sherman. 2004. Development of a novel microfluidic immunoassay for the detection of Helicobacter pylori infection. Analyst 129:823-828. [DOI] [PubMed] [Google Scholar]
- 14.Lin, F. Y. H., P. M. Sherman, and D. Li. 2004. Development of a novel hand-held immunoassay for the detection of enterohemorrhagic Escherichia coli O157:H7. Biomed. Microdevices 6:125-130. [DOI] [PubMed] [Google Scholar]
- 15.Longin, A., C. Souchier, M. French, and P.-A. Bryon. 1993. Comparison of anti-fading agents used in fluorescence microscopy: image analysis and laser confocal microscopy study. J. Histochem. Cytochem. 41:1833-1840. [DOI] [PubMed] [Google Scholar]
- 16.Ma, Z., and S.-F. Sui. 2002. Naked-eye sensitive detection of immunoglobulin G by enlargement of Au nanoparticles in vitro. Angew. Chem. Int. Ed. 41:2176-2179. [PubMed] [Google Scholar]
- 17.Mishchenko, M. I., L. D. Travis, and A. A. Lacis. 2002. Scattering, absorption, and emission of light by small particles. Cambridge University Press, Cambridge, United Kingdom.
- 18.Ng, J. M. K., I. Gitlin, A. D. Stroock, and G. M. Whitesides. 2002. Components for integrated poly(dimethylsiloxane) microfluidic systems. Electrophoresis 23:3461-3473. [DOI] [PubMed] [Google Scholar]
- 19.Norman, G. R., and D. L. Streiner. 2000. Biostatistics: the bare essentials, 2nd ed., p. 71-72. B. C. Decker, Hamilton, Ontario, Canada.
- 20.Rossier, J. S., and H. H. Girault. 2001. Enzyme linked immunosorbent assay on a microchip with electrochemical detection. Lab. Chip 1:153-157. [DOI] [PubMed] [Google Scholar]
- 21.Santandrew, M., S. Alegret, and E. Fàbregas. 1999. Determination of b-HCG using amperometric immunosensors based on a conducting immunocomposite. Anal. Chim. Acta 396:181-188. [Google Scholar]
- 22.Saviranta, P., R. Okon, A. Brinker, M. Warashina, J. Eppinger, and B. H. Geierstanger. 2004. Evaluating sandwich immunoassays in microarray format in terms of the ambient analyte regime. Clin. Chem. 50:1907-1920. [DOI] [PubMed] [Google Scholar]
- 23.Sharma, G., and J. Trussell. 1997. Digital color imaging. IEEE Trans. Image Processing 6:901-932. [DOI] [PubMed] [Google Scholar]
- 24.Taton, T. A., C. A. Mirkin, and R. L. Letsinger. 2000. Scanometric DNA array detection with nanoparticle probes. Science 289:1757-1760. [DOI] [PubMed] [Google Scholar]
- 25.Thanh, N. T. K., and Z. Rosenzweig. 2002. Development of an aggregation-based immunoassay for anti-protein A using gold nanoparticles. Anal. Chem. 74:1624-1628. [DOI] [PubMed] [Google Scholar]
- 26.Wild, D. (ed.) 2001. The immunoassay handbook, 2nd ed. Nature Publishing Group, New York, N.Y.
- 27.Xu, S., X. Ji, W. Xu, X. Li, L. Wang, Y. Bai, B. Zhao, and Y. Ozaki. 2004. Immunoassay using probe-labelling immunogold nanoparticles with silver staining enhancement via surface-enhanced Raman scattering. Analyst 129:63-68. [DOI] [PubMed] [Google Scholar]
- 28.Yguerabide, J., and E. E. Yguerabide. 1998. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications. I. Theory. Anal. Biochem. 262:137-156. [DOI] [PubMed] [Google Scholar]
- 29.Yguerabide, J., and E. E. Yguerabide. 1998. Light-scattering submicroscopic particles as highly fluorescent analogs and their use as tracer labels in clinical and biological applications. II. Experimental characterization. Anal. Biochem. 262:157-176. [DOI] [PubMed] [Google Scholar]