Abstract
Antibody detection with a recombinant COOH portion of the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid (N) protein, N13 (amino acids 221 to 422), was demonstrated to be more specific and sensitive than that with the full-length N protein, and an N13-based antigen-capturing enzyme-linked immunosorbent assay providing a convenient and specific test for serodiagnosis and epidemiological study of SARS was developed.
Severe acute respiratory syndrome (SARS) is a serious threat to public health and the economy on a global scale. A novel coronavirus, the SARS coronavirus (SARS-CoV), has been identified as the etiological agent for SARS (6). For serodiagnosis of the SARS-CoV infection in clinical microbiology laboratories, the recombinant antigen-based enzyme-linked immunosorbent assay (ELISA) for detecting specific antibodies is well known to offer higher reproducibility and to be more easily standardized and less laborious than either an indirect immunofluorescence assay based on virus-infected cells or an ELISA based on whole-virus lysate and does not require cultivation of the SARS-CoV (12). Since the specific antibodies to nucleocapsid (N) protein are most abundant in the sera from SARS patients (5), several groups have developed some recombinant N protein-based ELISAs. In general, these new assays are more specific and sensitive than the ELISA based on whole-virus lysate, but some false-positive results still occurred with sera from non-SARS patients or healthy people, even with sera collected several years before the SARS outbreak (3, 7, 8, 10-12).
Since the N proteins of the known coronaviruses are relatively conserved, the expressed N protein of the SARS-CoV in Escherichia coli cross-reacts with polyclonal antisera of some known animal coronaviruses in antigenic group I, including transmissible gastroenteritis virus, feline infectious peritonitis virus, and canine coronavirus (9), which raised concerns of potential false positives when the recombinant N protein of the SARS-CoV, whole-virus antigen extracts, or virus-infected cells are used as reagents. However, this concern is very minimal, since the two known human coronaviruses (strains 229E and OC43) do not cause severe clinical diseases and we still do not have data about the prevalence of the antibodies to other coronaviruses or relevant microorganisms in human populations (4, 6, 8). Therefore, it is important to identify the immunoreactive epitope of the N protein or other proteins specific to the SARS-CoV for serodiagnosis of SARS.
In our previous study, the antigenicity of different regions of the SARS-CoV N protein was analyzed by using a protein microarray. Four important regions with possible epitopes were identified, and the full-length N protein and two truncated N proteins, N8 (comprising amino acids [aa] 51 to 422) and N13 (aa 221 to 422), reacted with all 52 sera from SARS patients (1). Among these recombinant proteins, N13 is the shortest one, which decreases the possibility of cross-reaction with antibodies to other microorganisms. The objective of this study was to compare the specificity and sensitivity of N13 with those of N and then to develop an antigen-capturing ELISA for detection of antibodies to the SARS-CoV.
Cloning, expression, and purification of six-His-tagged recombinant proteins N and N13 were reported previously (1). These two proteins expressed in E. coli with pET32a as the expression vector were used as coating antigens, while the N protein expressed in E. coli with pPROEX HTb as the expression vector was used for horseradish peroxidase conjugate preparation.
An N- or N13-based antigen-capturing ELISA was established for the detection of the SARS-CoV antibodies in sera collected from patients with probable SARS in Beijing, People's Republic of China, during the 2003 SARS outbreak; probable SARS cases were determined based on the case definition of the World Health Organization. After 5 ml of blood was collected from each of the patients, the sera were separated by centrifugation and stored at −80°C. Ninety-six well microplates (Nalge Nunc International) were coated with the purified proteins N or N13 at concentrations of 1 μg/ml in 50 mmol of carbonate buffer (pH 9.6) per liter at 4°C overnight and subsequently blocked with blocking buffer (1% bovine serum albumin, 0.1% casein in phosphate-buffered saline [PBS; pH 7.0]) for 2 h at 37°C. Fifty microliters of serum diluted with 50 μl of blocking buffer was added to the well coated with recombinant protein, and the plate was incubated at 37°C for 30 min and then washed five times with PBS containing 0.05% Tween 20. One hundred microliters of horseradish peroxidase-N protein conjugate diluted with 1% bovine serum albumin in PBS (pH 7.0) was added, and the plate was incubated for another 30 min, followed by washing as before. Then, 100 μl of tetramethylbenzidine hydrochloride substrate solution (0.1 mg of tetramethylbenzidine hydrochloride/ml, 0.01% H2O2 in 0.1 mol of acetate buffer/liter [pH 5.8]) was added, the mixture was incubated at 37°C for 20 min, the reaction was terminated by adding 50 μl of a 2-mol/liter concentration of sulfuric acid, and the optical density at 450 nm (OD450) was determined.
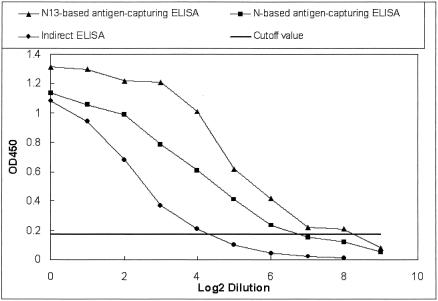
To establish the baseline for the tests, 29 sera taken from healthy people more than 1 year before the 2003 SARS outbreak were analyzed by full-length N- and N13-based antigen-capturing ELISAs. The cutoff OD450 value for a positive diagnosis in both tests was 0.17 (mean + 5 standard deviations). With this cutoff value, no cross-reaction with the sera from healthy people was found by the N13-based ELISA, while 3.4% (1 of 29) of the sera was shown to be positive by the N-based ELISA. Of another 84 sera taken from non-SARS pneumonia patients more than 1 year before the 2003 SARS outbreak, 1.2% (1 of 84) and 6.0% (5 of 84) of them were positive by N13- and N-based antigen-capturing ELISAs, respectively, while 4.8% (4 of 84) were positive by an indirect ELISA based on whole-virus lysate. All the above-described positive reactions were confirmed to be false by immunofluorescence and neutralization assays. To compare the sensitivities of these three tests, one serum sample from a SARS patient was serially diluted twofold with SARS-CoV-negative serum and then analyzed by these assays. As Fig. 1 shows, the positive results were recorded at maximum dilutions of 1:256, 1:64, and 1:16 by the N13-based antigen-capturing ELISA, the N-based ELISA, and the indirect ELISA, respectively, and the OD450 values from the N13-based antigen-capturing ELISA were always higher than those from the other two assays. All the results described above show that the ELISA based on the recombinant COOH portion of the N protein (i.e., N13) was more specific and sensitive for the detection of the SARS-CoV antibodies than either the ELISA based on the full-length N protein or the indirect ELISA based on whole-virus lysate.
FIG. 1.
Detection of antibodies to SARS-CoV by the N13-based antigen-capturing ELISA, the N-based ELISA, and the indirect ELISA based on whole-virus lysate in one twofold serially diluted serum sample from a SARS patient. The cutoff OD450 values for these assays were 0.17, 0.17, and 0.18, respectively.
The N13-based antigen-capturing ELISA and the indirect ELISA based on whole-virus lysate were applied to the detection of antibodies in 280 SARS patient sera. As shown in Table 1, the total positive rate of detection of SARS-CoV antibodies by the antigen-capturing ELISA was higher than that by the indirect ELISA. With the antigen-capturing ELISA, the SARS-CoV antibodies were detected in 14.3, 52.0, 91.7, and 94.8% of the sera from patients with probable SARS collected 12 to 14, 15 to 21, 22 to 28, and 29 to 35 days, respectively, after fever onset. For sera taken more than 35 days after fever onset, the positive rates of detection of SARS-CoV antibodies rose to 100%, which were highly concordant with the clinical diagnoses. The SARS-CoV antibodies were not detected by either assay in either of two sera that were obtained from patients at 43 and 47 days after fever onset. The medical records show that these two patients did not have histories of contact with patients with suspected or probable SARS, suggesting that the diagnoses of SARS for these two patients were wrong.
TABLE 1.
Detection of antibodies to the SARS-CoV in sera from patients with probable SARS by the N13-based antigen-capturing ELISA and the indirect ELISA based on a whole-virus lysate
| Day of sample collection after fever onset | No. of samples | No. (%) of samples positive by:
|
|||
|---|---|---|---|---|---|
| Indirect ELISA
|
Antigen- capturing ELISA | ||||
| IgG | IgM | IgG + IgM | |||
| 12-14 | 7 | 1 (14.3) | 1 (14.3) | 1 (14.3) | 1 (14.3) |
| 15-21 | 25 | 14 (56.0) | 9 (36.0) | 15 (60.0) | 13 (52.0) |
| 22-28 | 60 | 55 (91.7) | 47 (78.3) | 56 (93.3) | 55 (91.7) |
| 29-35 | 96 | 85 (88.5) | 62 (64.6) | 88 (91.7) | 91 (94.8) |
| 36-42 | 51 | 45 (88.2) | 30 (58.8) | 46 (90.2) | 51 (100.0) |
| 43-49 | 26 | 23 (88.5) | 15 (57.7) | 23 (88.5) | 24 (92.3) |
| 50-72 | 15 | 12 (80.0) | 6 (40.0) | 12 (80.0) | 15 (100.0) |
| Total | 280 | 235 (83.9) | 170 (60.7) | 241 (86.1) | 250 (89.3) |
All types of antibodies (immunoglobulin G [IgG], IgM, IgA, etc.) from humans and different animals can be detected simultaneously by an antigen-capturing ELISA (2, 8), which makes it convenient to search for a natural animal reservoir for the SARS-CoV by use of the present N13-based antigen-capturing ELISA.
In conclusion, the expressed truncated N protein N13 (aa 221 to 422) from E. coli appears to be a more sensitive antigen than the full-length N protein for SARS-CoV antibody detection in serially diluted serum and a more specific one than the full-length N protein for SARS-CoV antibody detection, and the N13-based antigen-capturing ELISA provides a convenient and specific test for serodiagnosis of SARS and epidemiological research, although its sensitivity for detecting the SARS-CoV antibody in clinical samples is still to be confirmed by screening a large quantity of samples.
Acknowledgments
We acknowledge Bastin David from Tianjin Biochip Center for carefully reading and revising the manuscript.
This work was supported by grants 2003AA208211 from the National High Technology Research and Development Program of China (863 Program) and 2004035057 from the China Postdoctoral Science Foundation.
REFERENCES
- 1.Chen, Z., D. Pei, L. Jiang, Y. Song, J. Wang, H. Wang, D. Zhou, J. Zhai, Z. Du, B. Li, M. Qiu, Y. Han, Z. Guo, and R. Yang. 2004. Antigenicity analysis of different regions of the severe acute respiratory syndrome coronavirus nucleocapsid protein. Clin. Chem. 50:988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncan, R. J. S. 1988. The use of ELISA for rapid viral diagnosis: antibody detection, p. 309-310. In D. M. Kemeny and S. J. Challacombe (ed.), ELISA and other solid phase immunoassays. Theoretical and practical aspects. John Wiley & Sons, New York, N.Y.
- 3.Guan, M., H. Y. Chen, S. Y. Foo, Y.-J. Tan, P.-Y. Goh, and S. H. Wee. 2004. Recombinant protein-based enzyme-linked immunosorbent assay and immunochromatographic tests for detection of immunoglobulin G antibodies to severe acute respiratory syndrome (SARS) coronavirus in SARS patients. Clin. Diagn. Lab. Immunol. 11:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 5.Krokhin, O., Y. Li, A. Andonov, H. Feldmann, R. Flick, S. Jones, U. Stroeher, N. Bastien, K. V. Dasuri, K. Cheng, J. N. Simonsen, H. Perreault, J. Wilkins, W. Ens, F. Plummer, and K. G. Standing. 2003. Mass spectrometric characterization of proteins from the SARS virus: a preliminary report. Mol. Cell. Proteomics 2:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 7.Liu, X., Y. Shi, P. Li, L. Li, Y. Yi, Q. Ma, and C. Cao. 2004. Profile of antibodies to the nucleocapsid protein of the severe acute respiratory syndrome (SARS)-associated coronavirus in probable SARS patients. Clin. Diagn. Lab. Immunol. 11:227-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi, Y., Y. Yi, P. Li, T. Kuang, L. Li, M. Dong, Q. Ma, and C. Cao. 2003. Diagnosis of severe acute respiratory syndrome (SARS) by detection of SARS coronavirus nucleocapsid antibodies in an antigen-capturing enzyme-linked immunosorbent assay. J. Clin. Microbiol. 41:5781-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun, Z. F., and X. J. Meng. 2004. Antigenic cross-reactivity between the nucleocapsid protein of severe acute respiratory syndrome (SARS) coronavirus and polyclonal antisera of antigenic group I animal coronaviruses: implication for SARS diagnosis. J. Clin. Microbiol. 42:2351-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timani, K. A., L. Ye, Y. Zhu, Z. Wu, and Z. Gong. 2004. Cloning, sequencing, expression, and purification of SARS-associated coronavirus nucleocapsid protein for serodiagnosis of SARS. J. Clin. Virol. 30:309-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woo, P. C., S. K. Lau, H. W. Tsoi, K. H. Chan, B. H. Wong, X. Y. Che, V. K. Tam, S. C. Tam, V. C. Cheng, I. F. Hung, S. S. Wong, B. J. Zheng, Y. Guan, and K. Y. Yuen. 2004. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet 363:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo, P. C. Y., S. K. P. Lau, B. H. L. Wong, H.-W. Tsoi, A. M. Y. Fung, K.-H. Chan, V. K. P. Tam, J. S. M. Peiris, and K.-Y. Yuen. 2004. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 42:2306-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]