Abstract
The toxin-coregulated pilus (TCP) of Vibrio cholerae and the soluble TcpF protein that is secreted via the TCP biogenesis apparatus are essential for intestinal colonization. The TCP biogenesis apparatus is composed of at least nine proteins but is largely uncharacterized. TcpC is an outer membrane lipoprotein required for TCP biogenesis that is a member of the secretin protein superfamily. In the present study, analysis of TcpC in a series of strains deficient in each of the TCP biogenesis proteins revealed that TcpC was absent specifically in a tcpQ mutant. TcpQ is a predicted periplasmic protein required for TCP biogenesis. Fractionation studies revealed that the protein is not localized to the periplasm but is associated predominantly with the outer membrane fraction. An analysis of the amount of TcpQ present in the series of tcp mutants demonstrated the inverse of the TcpC result (absence of TcpQ in a tcpC deletion strain). Complementation of the tcpQ deletion restored TcpC levels and TCP formation, and similarly, complementation of tcpC restored TcpQ. Metal affinity pull-down experiments performed using His-tagged TcpC or TcpQ demonstrated a direct interaction between TcpC and TcpQ. In the presence of TcpQ, TcpC was found to form a high-molecular-weight complex that is stable in 2% sodium dodecyl sulfate and at temperatures below 65°C, a characteristic of secretin complexes. Fractionation studies in which TcpC was overexpressed in the absence of TcpQ showed that TcpQ is also required for proper localization of TcpC to the outer membrane.
Vibrio cholerae is a gram-negative bacterium that causes the gastrointestinal disease cholera, which is spread via the fecal-oral route. Upon passage through the stomach, the bacterium utilizes its single polar flagellum to swim from the lumen through the intestinal mucous layer to reach the epithelial surface in the crypts of the intestine where it colonizes. Toxin-coregulated pilus (TCP) serves as the primary factor involved in the colonization and persistence of the bacteria in the small intestine (18, 32, 39, 40). TCP is classified as a type 4 pilus based on the N-terminal homology of the monomeric (pilin) subunit, TcpA. Type 4 pili are long, filamentous appendages expressed by a number of gram-negative bacteria and are classified by sequence homology within the hydrophobic N-terminal region of the mature pilin and a methylated N-terminal residue (10, 25, 37). There are two subclasses of type 4 pili, namely, 4A and 4B, with TCP representing the latter (35). TCP has been demonstrated to be necessary for the colonization of humans, and the colonization defect of tcp mutants is paralleled in the infant mouse model (12). Upon colonization the bacterium produces cholera toxin, which is an ADP-ribosylating toxin composed of one A subunit and five B subunits encoded by the ctxAB operon. Cholera toxin is secreted by a type II extracellular protein secretion (Eps) system (16, 33). The ctxAB operon is part of the genome of the cholera toxin bacteriophage (CTXφ), a 7-kb lysogenic filamentous bacteriophage which utilizes TCP as its receptor (42).
TCP fibers are homopolymers of TcpA pilin, encoded by the first gene in the tcp biogenesis operon. The operon is located on a large pathogenicity island, the Vibrio pathogenicity island, located on chromosome 1. Biogenesis of type 4 pilus fibers involves polymerization and secretion of the pilin subunit by a mechanism involving a set of proteins that compose an assembly and export apparatus (reviewed in reference 37). Biogenesis of TCP requires the activities of nine TCP-specific proteins in addition to the pilin itself that are encoded by genes located in the tcp operon. The apparatus is not only responsible for the biogenesis of TCP but is also required for the extracellular secretion of TcpF, a soluble colonization factor (17). TcpF secretion is a two-step process, akin to the type II protein secretion systems. The general secretory pathway translocates TcpF to the periplasm, and then outer membrane translocation is achieved via the TCP biogenesis machinery. The functions of only three of the nine TCP-specific gene products have been reported. The tcpJ gene encodes the type 4 prepilin peptidase, required for the processing of TcpA (21). The tcpT gene encodes the putative cytoplasmic ATPase, which is peripherally tethered to the inner membrane TCP apparatus components (14, 23). The tcpC gene encodes an outer membrane lipoprotein required for pilus biogenesis and also contributes to resistance to complement (30).
In this report we identify an outer membrane complex of the TCP biogenesis apparatus, comprised of TcpC and TcpQ. TcpC was originally proposed to function analogously to PapC of the pyelonephritis-associated pilus (pap) of uropathogenic Escherichia coli (29). However, experimental evidence has accumulated suggesting that type 4 pili do not utilize an usher-chaperone system for biogenesis. Rather, the C terminus of TcpC exhibits extensive homology with a well-conserved region of members of the secretin superfamily of proteins, which are utilized by type 4 pilus and type II extracellular secretion systems to translocate pilin subunits and macromolecules across the outer membrane. Secretins are integral outer membrane proteins that form large stable multimeric complexes and usually exhibit a 12-fold symmetry (reviewed in reference 3). Examples of well-characterized secretins include PilQ of Neisseria meningitidis, BfpB of enteropathogenic E. coli (EPEC), XcpQ of Pseudomonas aeruginosa, and PulD of Klebsiella oxytoca (4, 5, 7, 8, 31, 34). The N-terminal domain of secretins is divergent and thought to extend into the periplasm and interact with other components of the secretion or biogenesis machinery (3). These interactions have been shown to have different functions in various systems. Several secretin complexes are dependent on small lipoproteins that play a role in localization and/or multimerization (9, 19, 36), but none of these small lipoproteins have been copurified as part of the secretin complexes. These small lipoproteins do not show a great deal of homology among each other. However, in some systems the small interacting protein is not a lipoprotein. For example, the BfpB secretin is itself a lipoprotein but is dependent on BfpG, a small outer membrane protein, for multimerization (34). Interestingly, the closest BLAST (1) homologs to TcpC, CofD of CFA/III in enterotoxigenic E. coli and CfcD of Citrobacter rodentium, are predicted to be lipoproteins as well (26, 38). The only other secretin that is a lipoprotein, XpsD of Xanthomonas campestris, does not belong to a type 4 pilus biogenesis system (6). The results of the present study are consistent with a role for TcpC as an outer membrane secretin that interacts with a small protein, TcpQ, required for TcpC localization and stability.
MATERIALS AND METHODS
Bacterial strains, plasmids, growth media, and DNA manipulation.
Bacterial strains and plasmids used in this study are listed in Table 1. Derivatives of V. cholerae O395Sm were maintained at −70°C in Luria-Bertani (LB) medium containing 20% (vol/vol) glycerol. V. cholerae strains grown under TCP-expressing conditions were grown in LB broth, pH 6.5, at 30°C for 12 to 15 h. Streptomycin was used at a final concentration of 100 μg/ml, ampicillin was used at 100 μg/ml, kanamycin was used at 45 μg/ml, and arabinose was used at 0.01%. All DNA manipulations were performed by standard molecular and genetic techniques (2, 22).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| Oneshot TOP10 | F−mcrA Δ(mrr-hsdRMS-mrr) recA1 endA1 lon gyrA96 thi-1 supE44 relA1 λ− Δ(lac-proAB) | Invitrogen |
| V. cholerae | ||
| O395Sm | Classical, Ogawa Smr derivative | Laboratory collection |
| RT4031 | O395Sm ΔtcpA11 | 18 |
| RT4368 | O395Sm ΔtcpB1 | 17 |
| RT4369 | O395Sm ΔtcpC1 | 17 |
| RT4370 | O395Sm ΔtcpD1 | 17 |
| RT4371 | O395Sm ΔtcpE1 | 17 |
| RT4372 | O395Sm ΔtcpF1 | 17 |
| RT4373 | O395Sm ΔtcpQ1 | 17 |
| RT4374 | O395Sm ΔtcpR1 | 17 |
| RT4375 | O395Sm ΔtcpS1 | 17 |
| RT4376 | O395Sm ΔtcpT1 | 17 |
| CL101 | O395 CTX-Kmφ | Laboratory collection |
| NB115 | RT4369 pNB10 | This study |
| NB116 | RT4373 pNB10 | This study |
| NB064 | RT4373 pNB11 | This study |
| NB178 | RT4369 pNB11 | This study |
| NB114 | O395Sm pNB10 | This study |
| NB138 | O395Sm pNB11 | This study |
| NB211 | 569B Sm CTX-Kmφ pBADTOPO | This study |
| NB161 | RT4369 pRPA101 | This study |
| Plasmids | ||
| pBADTOPO | pBR322 para-BAD promoter araC Apr | Invitrogen |
| pNB10 | pBADTOPO TcpC-6His Apr | This study |
| pNB11 | pBADTOPO TcpQ-6His Apr | This study |
Autoagglutination assay.
V. cholerae strains were grown under TCP-inducing conditions, removed from the shaker, and allowed to stand on the bench for 30 min. The cultures were inspected without disturbing the tubes. The supernatant clarity and pellet formation were used as a measure of agglutination, with wild type arbitrarily assigned a +++ score.
Preparation and purification of TcpC- and TcpQ-specific antisera.
The Jameson-Wolf algorithm to predict antigenic index and the Kyte-Doolittle algorithm to predict hydrophobicity were used to select regions in the protein sequence of TcpC and TcpQ that might be antigenic (15, 20). On the basis of this analysis, peptides corresponding to amino acids 352 to 370 in TcpC (SSIETTKDTNTDEETRTVK) and amino acids 52 to 70 in TcpQ (ETFIESEEFEQEESPSKDP) were synthesized and conjugated to keyhole limpet hemagglutinin, and the conjugates were used for immunizing rabbits for the production of polyclonal antisera (Biosynthesis, Inc., Lewisville, Tex.). TcpQ antibody was affinity purified using Sulfolink gel (Pierce) according to the specifications of the manufacturer.
Immunoblot analysis.
Proteins from whole-cell lysates or various cellular fractions were separated on a sodium dodecyl sulfate (SDS)-12.5% polyacrylamide gel. For immunodetection, proteins were electroblotted onto a nitrocellulose membrane at 4°C in transfer buffer (25 mM Tris, 192 mM glycine, 20% methanol, pH 8.3) with a wet transfer apparatus (Bio-Rad Laboratories) (41). TcpC polyclonal antibody was used at a dilution of 1:1,000, and the affinity-purified TcpQ antibody was used at a dilution of 1:100. The horseradish peroxidase-conjugated secondary antibody (Cappel; ICN) was used at a dilution of 1:100,000.
Cellular fractionation.
Cells grown under TCP-expressing conditions were harvested by centrifugation and treated with polymyxin B sulfate (10 mg/ml) in phosphate-buffered saline for 10 min on ice to release periplasmic contents. Spheroplasts were separated from the soluble periplasmic fraction by centrifugation at 10,000 × g for 10 min. The spheroplast fraction was resuspended in 1× SDS-polyacrylamide gel electrophoresis (PAGE) loading buffer, and the supernatant containing the periplasmic contents was diluted with 2× SDS-PAGE loading buffer. In order to obtain total membranes, the spheroplast fraction was resuspended in 10 mM Tris-Cl (pH 8.0) and passed through a French pressure cell at 8,000 lb/in2 to break open the cells. After a low-speed centrifugation at 10,000 × g at room temperature (RT) for 10 min to remove any unbroken cells, the lysate was centrifuged at 100,000 × g at 4°C for 30 min to pellet the membrane fraction. The inner and outer membranes were differentially solubilized as follows. The membrane pellet was resuspended in 10 mM Tris-Cl-100 mM NaCl containing 2.5% Sarkosyl, held at RT for 30 min, and centrifuged at 200,000 × g at RT for 1 h. The supernatant was retained as the inner membrane fraction, and the pellet containing the outer membrane was resuspended in 200 μl of 10 mM Tris-Cl (pH 8.0). Protein estimation was carried out using the BCA protein assay kit (Pierce) with bovine serum albumin as standard.
Metal affinity chromatography.
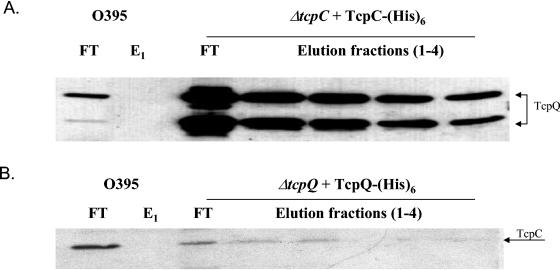
Cells grown under TCP-expressing conditions and induced with arabinose to express His-tagged TcpC or TcpQ were harvested and resuspended in extraction buffer (50 mM sodium phosphate, 300 mM sodium chloride, pH 7.0) containing 1% (vol/vol) Triton X-100. The cells were broken by two passages through a French pressure cell at 8,000 lb/in2. The lysate was loaded onto a Talon metal affinity resin spin column (Clontech), and the protein purification protocol specified by the manufacturer was followed. The initial elution fractions (E1 and E2 [see Fig. 5]) were obtained using an extraction buffer that contained 150 mM imidazole, and the final fractions (E3 and E4) were obtained using a buffer containing 100 mM EDTA. All steps were carried out on ice or at 4°C.
FIG. 5.
TcpC and TcpQ interact directly as demonstrated by metal affinity chromatography. (A) Metal affinity chromatography performed with His-tagged TcpC. Fractions were separated by SDS-PAGE, and TcpQ was detected by immunoblot analysis. Elution fractions 1 and 2 (E1 and E2) were obtained using buffer containing 150 mM imidazole. Fractions 3 and 4 (E3 and E4) were obtained using buffer containing 100 mM EDTA in order to completely strip the beads. (B) The reciprocal experiment with metal affinity chromatography with His-tagged TcpQ and probing of the immunoblot with anti-TcpC antibody. FT, flowthrough.
Cholera toxin phage transduction.
Strains were grown according to the procedure described for the autoagglutination assay, and the transduction assay was carried out as previously described (42). Briefly, equal volumes of CTX-Kmφ supernatant and bacterial cultures were mixed and incubated at RT for 30 min. Appropriate dilutions were plated on LB agar containing kanamycin. Transduction efficiency was reported as ratio of test strain transductants to the number of input CFU divided by the ratio of wild-type transductants to the number of input CFU. The coculture assay was performed by growing the CTX-Kmφ-containing strain along with the test strain overnight in LB medium (pH 6.5) at 30°C. Appropriate dilutions were plated on LB agar containing kanamycin, streptomycin, and ampicillin.
RESULTS
TcpC is detectable in all tcp deletion strains except for a tcpQ mutant.
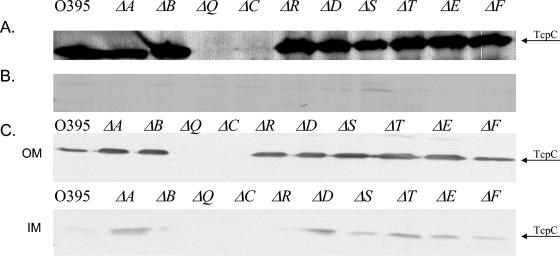
By using a set of nonpolar, in-frame deletions in the tcp genes, it has been previously shown that TcpC is required for TCP biogenesis, and TcpC has been shown to be an outer membrane lipoprotein (17, 30). To investigate possible interactions between TcpC and other components of the biogenesis apparatus, we began by assessing the localization and stability of TcpC in each of the tcp deletion mutants. Western immunoblot analysis of whole-cell lysates revealed that TcpC was present at a level comparable to wild type in all the tcp deletions, except a tcpQ deletion strain (Fig. 1A). TcpC was not mislocalized to the periplasm in any of the biogenesis mutants (Fig. 1B). Membrane fractionation experiments revealed that TcpC localizes to the outer membrane in all the deletion mutants in which it is expressed (Fig. 1C). The tcpC deletion was complemented with a plasmid expressing a TcpC-(His)6 construct, as determined by restoration of autoagglutination and CTXφ transduction (Table 2). The absence of TcpC in a tcpQ deletion strain indicated a possible interaction between the two proteins, and this prompted us to analyze the levels of TcpQ in a tcpC mutant.
FIG. 1.
TcpC stability in tcp deletion strains. (A) Western immunoblot analysis of whole-cell lysates of tcp deletion mutants with TcpC antibody. (B) Periplasmic fractions of the tcp deletion strains. (C) Outer (OM) and inner (IM) membrane fractionations of the tcp deletion strains.
TABLE 2.
TCP-related phenotypes in wild-type, tcp mutant, and complemented strains
| Phenotype | Result for strain type:
|
||||
|---|---|---|---|---|---|
| WTa | ΔtcpC | ΔtcpQ | ΔtcpC pTcpC-His6 | ΔtcpQ pTcpQ-His6 | |
| Agglutinationb | +++ | − | − | + | + |
| CTXφ transduction frequency | 1.6 × 10−7 | 1 × 10−10 | 1 × 10−10 | 2 × 10−7 | 2.8 × 10−7 |
WT, wild type.
+++, wild-type level of autoagglutination; +, moderate autoagglutination.
TcpQ is absent in a tcpC deletion mutant and localizes to the membrane fraction in a wild-type background.
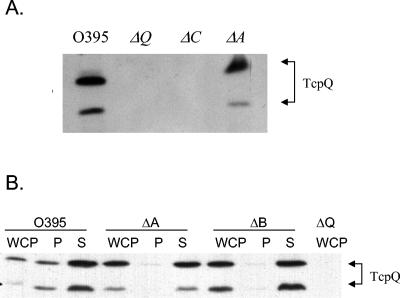
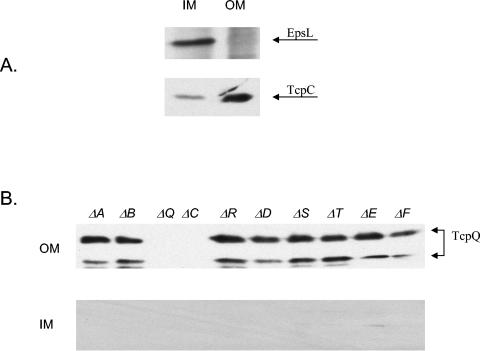
TcpQ is a 15-kDa protein that is predicted to be localized to the periplasm due to the presence of an N-terminal signal sequence along with an overall hydrophilic nature of the mature protein (SignalP) (27). Western immunoblot analysis of whole-cell lysates from wild-type and ΔtcpQ and ΔtcpC strains revealed that TcpQ was completely absent in a tcpC deletion mutant (Fig. 2A). This reciprocal result with respect to the previous experiment that demonstrated an absence of TcpC in a tcpQ mutant further supports the potential interaction between TcpC and TcpQ. Such a protein-protein interaction within an outer membrane complex might contribute to the stability of both the proteins in the complex. Periplasm and spheroplast fractionation experiments showed that the majority of TcpQ fractionates to the spheroplast fraction, in contrast to what would be expected for a periplasmic protein (Fig. 2B). Fractionation experiments were performed on the wild type, a tcpA deletion strain (pilin mutant), and a tcpB deletion strain (random biogenesis mutant). Further membrane fractionation of the spheroplast fraction was performed on the wild type and the biogenesis mutants in order to determine whether TcpQ localized to the inner or the outer membrane. EpsL and TcpC were used as inner and outer membrane controls for the Sarkosyl-based membrane fractionation protocol (Fig. 3A). TcpQ localizes predominantly to the outer membrane fraction in the wild type and all of the biogenesis mutants, which would be consistent with an interaction with TcpC in the outer membrane (Fig. 3B). The Western blot analysis also indicated that TcpQ migrates as two bands. The two bands likely correspond to two forms of TcpQ, since both were detected by an antibody generated against an internal TcpQ peptide sequence and both the protein species are absent in a tcpQ deletion. The smaller band is likely the result of proteolytic degradation, although this not been investigated. Complementation of the tcpQ deletion with a plasmid expressing TcpQ-(His)6 restores expression of TCP (Table 2), and both protein species could be detected using an anti-His antibody (data not shown), suggesting that, if proteolytic degradation occurs, it is likely to be an N-terminal event. Concurrent with restoration of TCP expression, TcpC is partially restored in the tcpQ complementation strain (data not shown).
FIG. 2.
TcpQ stability and localization in wild-type and pilin and biogenesis mutant strains. (A) Whole-cell pellet lysates from wild-type, ΔtcpQ, ΔtcpC, and ΔtcpA strains were electroblotted and probed with TcpQ antibody. (B) Fractionation profile of wild-type, ΔtcpA, and ΔtcpB strains. WCP, whole-cell pellet lysates; P, periplasm; S, spheroplast. Arrows indicate the two forms of TcpQ.
FIG. 3.
TcpQ localization in wild-type and biogenesis mutant strains. (A) Inner and outer membrane fractions from wild-type cells probed with EpsL and TcpC (membrane controls) (B) Membrane fractionation profile of wild-type and biogenesis mutant strains. OM, outer membrane; IM, inner membrane. Arrows indicate the two forms of TcpQ.
Altered protein localization in tcpC and tcpQ mutants.
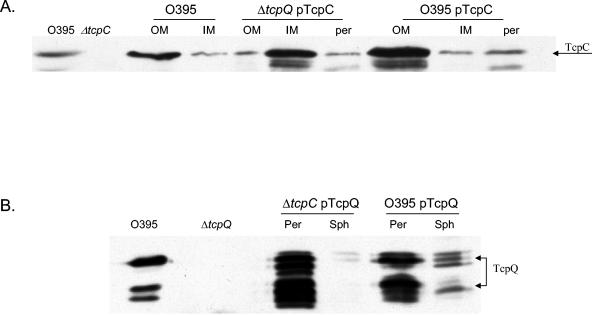
In order to understand the localization pattern of TcpC and TcpQ, we overexpressed TcpC in a tcpQ deletion strain and TcpQ in a tcpC deletion strain. The cross-complementation strains did not make TCP, which was tested by phage transduction and coculture experiments. The TcpC localization pattern in the tcpQ deletion strain was analyzed by cellular fractionation. Western blot analysis indicated that TcpC was mislocalized to the inner membrane fraction, which was different from a wild-type strain overexpressing TcpC, where the localization pattern was consistent with the known pattern for TcpC (Fig. 4A). To confirm TcpQ localization to the periplasm in the absence of TcpC, we overexpressed TcpQ-(His)6 in a tcpC deletion strain. Periplasm and spheroplast fractionation demonstrated that TcpQ localized predominantly to the periplasmic fraction as expected (Fig. 4B).
FIG. 4.
TcpC and TcpQ localization patterns in tcpQ and tcpC deletion strains. (A) Immunoblot analysis of membrane and periplasmic fractions from wild-type (O395), ΔtcpQ (pNB10), and O395 (pNB10) strains with anti-TcpC antibody. (B) Fractionation profile of TcpQ in ΔtcpC (pNB11) and O395 (pNB11) strains with anti-TcpQ antibody. The first two lanes in both panels are wild-type and deletion strain controls. Per or per, periplasm; Sph, spheroplast; OM, outer membrane; IM, inner membrane.
TcpC and TcpQ directly interact.
The absence of TcpC in a tcpQ deletion strain and the absence of TcpQ in a tcpC deletion strain suggested a possible interaction between the two proteins. The complementation data indicated that the potential interaction was required for TCP biogenesis. To confirm the interaction, we performed a pull-down experiment using the His-tagged version of TcpC as well as the reverse using a His-tagged version of TcpQ. Whole-cell extracts were prepared from the ΔtcpC pTcpC-(His)6 strain, membrane proteins were solubilized using a detergent, and the resulting lysates were incubated with Talon metal affinity resin. The eluates from the metal affinity column were subjected to Western blot analysis. It was found that TcpC-(His)6 was able to pull down both forms of TcpQ (Fig. 5A) and TcpQ-(His)6 was able to pull down TcpC (Fig. 5B). These data suggest that TcpC and TcpQ are directly associated with one another.
TcpC forms a multimeric complex in the outer membrane.
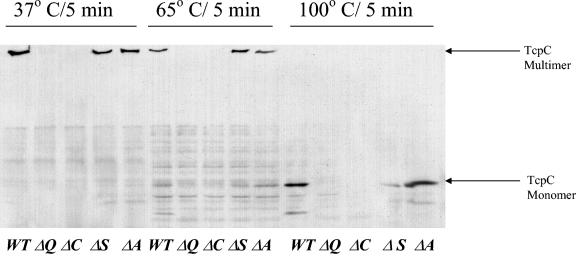
A general property of secretins such as BfpB, the bundle-forming pilus (BFP) biogenesis secretin of EPEC, as well as others, is that they form a stable multimer that does not dissociate unless heated to temperatures of 65°C or higher in protein gel sample buffer containing 4% SDS (34). To determine if TcpC is part of a stable multimeric complex, whole-cell lysates from O395, ΔtcpA, ΔtcpQ, ΔtcpC, and ΔtcpS strains were incubated in protein gel sample buffer containing 4% SDS at 37, 65, and 100°C and examined for multimer formation by Western analysis. A ΔtcpS strain was included in the analysis because, like TcpQ, TcpS is another predicted periplasmic protein in the biogenesis apparatus that might potentially influence TcpC. In the wild-type background TcpC was found to migrate as a multimer that did not enter the resolving gel unless subjected to 100°C. The high-molecular-weight complex was still present in the ΔtcpA and ΔtcpS strain lysates, indicating that the complex formation is independent of the presence of the TcpA pilin and that TcpS, the other predicted periplasmic protein, does not play a role in complex formation (Fig. 6). As expected the complex was absent in the tcpQ strain lysate.
FIG. 6.
TcpC forms a high-molecular-weight complex similar to that seen for other secretins. The whole-cell lysates from wild-type (WT), ΔtcpC, ΔtcpQ, ΔtcpA, and ΔtcpS strains were resuspended in SDS-PAGE loading buffer and treated at 37 or 65°C or boiled for 5 min. Following SDS-PAGE, Western blot analysis with TcpC antibody was performed.
DISCUSSION
Type 4 pili are critical virulence factors for many pathogenic bacteria and play a variety of roles in pathogenesis such as host adhesion, twitching motility, and microcolony formation. Similar to type II secretion systems of gram-negative bacteria, type 4 pili require a multicomponent assembly apparatus for pilus biogenesis. However, our understanding of the functional characteristics of type 4 pili greatly surpasses our understanding of their biogenesis. Characterization of the type 4 pilus biogenesis apparatus has a number of important implications in understanding molecular mechanisms of pathogenesis and will help us better understand the strikingly similar type II secretion machinery present in gram-negative bacteria.
In the present study, we sought to characterize the outer membrane lipoprotein, TcpC, within the context of the biogenesis apparatus. Using a set of in-frame deletions, we found that TcpC levels are comparable to wild type in all the mutants except a tcpQ deletion strain. TcpQ is a predicted periplasmic protein with a molecular mass of 15 kDa and some similarity to pilus operon genes located in the same position, like CfcC, CofC, and BfpG of C. rodentium, enterotoxigenic E. coli, and EPEC, respectively. Interestingly, localization studies found TcpQ to be present predominantly in the outer membrane rather than the periplasm except in a tcpC mutant. In such a mutant TcpQ was completely absent, the reciprocal of the result with respect to TcpC in a tcpQ mutant, suggesting the existence of a TcpC-TcpQ protein complex. Metal affinity chromatography performed using His-tagged TcpC and TcpQ confirmed the same.
TcpC was found to form a stable high-molecular-weight complex like all other characterized secretins, and the complex is formed in the absence of the pilin and other biogenesis components, except TcpQ. All these results point to features of TcpC that are consistent with its forming a secretin complex in the outer membrane. Most of the biochemically characterized secretins require the presence of a small outer membrane (lipo)protein which plays a role in stabilizing the secretin and/or outer membrane localization (9, 11, 19, 36). TcpQ was found to be required for TcpC stability, and biochemical fractionation demonstrated a role in outer membrane localization as well, suggesting a possible role as a chaperone and/or a pilot protein. TcpQ could serve as a protein plug, gating the secretin pore, but experimental evidence does not support this hypothesis. Antibiotic sensitivity studies performed in EPEC suggested that BfpB was an incompletely gated pore, and E. coli expressing the f1 phage protein pIV had a moderate increase in maltodextrin diffusion into the periplasm (24, 34). However, overexpression of TcpC in the absence of TcpQ does not result in increased outer membrane permeability (data not shown). This could be attributed to the mislocalization of TcpC. The possibility that a portion of TcpC flaps back into the pore and serves as a plug, as for K. oxytoca, where the N-terminal portion of the secretin, PulD, is thought to perform the function of a plug (28), does exist. It is possible that TcpQ could play a role in the opening and closing of the secretin pores by possibly utilizing mechanically transferred energy from ATP hydrolysis, which occurs at the cytoplasmic face of the inner membrane. Recent biochemical evidence in the BFP system, obtained using cross-linking and pull-down studies performed using a His-tagged version of BfpB, suggests the possible existence of a complex, which spans the periplasmic space (13). Such a complex could play a structural or functional role in type 4 pilus biogenesis systems.
Although TcpC belongs to the secretin superfamily, there are several features that distinguish it from the typical members of the secretin family. While retaining the overall C-terminal homology to the secretin family members, TcpC lacks the highly conserved secretin signature sequence, (V,I)PXL(S,G)XIPXXGXLF, present in the C-terminal region. BfpB lacks this motif as well, and it has been previously suggested by Schmidt et al. (34) that BfpB might define a distinctive subclass of outer membrane proteins that intersect with the secretin superfamily. Most characterized secretins, with the exception of BfpB, require a small outer membrane lipoprotein for the stability and/or outer membrane localization of the secretin. TcpC falls into the category of putative secretins that are themselves lipoproteins, which includes BfpB, CofD, and CfcD. However, TcpC requires the small periplasmic protein TcpQ to stabilize it. TcpQ does not possess a lipoprotein consensus motif. Finally, BfpB monomers are stable in the absence of BfpG (34), whereas TcpC is completely degraded in the absence of TcpQ. These differences indicate that TcpC and BfpB might belong to a subclass in the secretin superfamily, but the secretin complex biogenesis pathways might be different. It also suggests that TcpQ might play a role slightly different in secretin biogenesis from that of the set of characterized small proteins. These differences between TcpC and BfpB are further reiterated by the failure of BfpB to complement a tcpC deletion (data not shown).
It was previously known that TcpC is required for TCP biogenesis. The results of the present study suggest that it is the secretin in the biogenesis apparatus. A recent study demonstrated that TcpF, a secreted colonization factor, utilizes TCP to reach the extracellular environment. TcpF was found to accumulate in the periplasm in a TcpC deletion strain, but it is unclear whether the requirement is a direct one or whether the lack of formation of TCP leads to the accumulation of TcpF in the periplasm. Further studies to characterize the TCP biogenesis apparatus might lead to a better understanding of the type 4 pilus biogenesis apparatus as well as an understanding of the possible convergence of the type 4 pilus and type II secretion systems in V. cholerae.
Acknowledgments
We thank Anu Daniel and M. S. Donnenberg for the BFP complementation studies and Maria Sandkvist for antibody directed against EpsL.
This work was supported by grant AI R01 25096 from the NIH to R.K.T.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. Greene Publishing Associates and John Wiley & Sons, New York, N.Y.
- 3.Bitter, W. 2003. Secretins of Pseudomonas aeruginosa: large holes in the outer membrane. Arch. Microbiol. 179:307-314. [DOI] [PubMed] [Google Scholar]
- 4.Bitter, W., M. Koster, M. Latijnhouwers, H. de Cock, and J. Tommassen. 1998. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol. 27:209-219. [DOI] [PubMed] [Google Scholar]
- 5.Brok, R., P. Van Gelder, M. Winterhalter, U. Ziese, A. J. Koster, H. de Cock, M. Koster, J. Tommassen, and W. Bitter. 1999. The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol. 294:1169-1179. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. Y., D. Y. Chen, J. Miaw, and N. T. Hu. 1996. XpsD, an outer membrane protein required for protein secretion by Xanthomonas campestris pv. campestris, forms a multimer. J. Biol. Chem. 271:2703-2708. [DOI] [PubMed] [Google Scholar]
- 7.Collins, R. F., L. Davidsen, J. P. Derrick, R. C. Ford, and T. Tonjum. 2001. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 183:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, R. F., R. C. Ford, A. Kitmitto, R. O. Olsen, T. Tonjum, and J. P. Derrick. 2003. Three-dimensional structure of the Neisseria meningitidis secretin PilQ determined from negative-stain transmission electron microscopy. J. Bacteriol. 185:2611-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake, S. L., S. A. Sandstedt, and M. Koomey. 1997. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol. Microbiol. 23:657-668. [DOI] [PubMed] [Google Scholar]
- 10.Giron, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 11.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 12.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang, J., D. Bieber, S. W. Ramer, C. Y. Wu, and G. K. Schoolnik. 2003. Structural and topographical studies of the type IV bundle-forming pilus assembly complex of enteropathogenic Escherichia coli. J. Bacteriol. 185:6695-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iredell, J. R., and P. A. Manning. 1997. Translocation failure in a type-4 pilin operon: rfb and tcpT mutants in Vibrio cholerae. Gene 192:71-77. [DOI] [PubMed] [Google Scholar]
- 15.Jameson, B. A., and H. Wolf. 1988. The antigenic index: a novel algorithm for predicting antigenic determinants. Comput. Appl. Biosci. 4:181-186. [DOI] [PubMed] [Google Scholar]
- 16.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirn, T. J., N. Bose, and R. K. Taylor. 2003. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol. Microbiol. 49:81-92. [DOI] [PubMed] [Google Scholar]
- 18.Kirn, T. J., M. J. Lafferty, C. M. Sandoe, and R. K. Taylor. 2000. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35:896-910. [DOI] [PubMed] [Google Scholar]
- 19.Koster, M., W. Bitter, H. de Cock, A. Allaoui, G. R. Cornelis, and J. Tommassen. 1997. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol. Microbiol. 26:789-797. [DOI] [PubMed] [Google Scholar]
- 20.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 21.LaPointe, C. F., and R. K. Taylor. 2000. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. 275:1502-1510. [DOI] [PubMed] [Google Scholar]
- 22.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Manning, P. A. 1997. The tcp gene cluster of Vibrio cholerae. Gene 192:63-70. [DOI] [PubMed] [Google Scholar]
- 24.Marciano, D. K., M. Russel, and S. M. Simon. 1999. An aqueous channel for filamentous phage export. Science 284:1516-1519. [DOI] [PubMed] [Google Scholar]
- 25.Martin, P. R., A. A. Watson, T. F. McCaul, and J. S. Mattick. 1995. Characterization of a five-gene cluster required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol. Microbiol. 16:497-508. [DOI] [PubMed] [Google Scholar]
- 26.Mundy, R., D. Pickard, R. K. Wilson, C. P. Simmons, G. Dougan, and G. Frankel. 2003. Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol. Microbiol. 48:795-809. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 28.Nouwen, N., H. Stahlberg, A. P. Pugsley, and A. Engel. 2000. Domain structure of secretin PulD revealed by limited proteolysis and electron microscopy. EMBO J. 19:2229-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogierman, M. A., and P. A. Manning. 1992. TCP pilus biosynthesis in Vibrio cholerae O1: gene sequence of tcpC encoding an outer membrane lipoprotein. FEMS Microbiol. Lett. 76:179-184. [DOI] [PubMed] [Google Scholar]
- 30.Parsot, C., E. Taxman, and J. J. Mekalanos. 1991. ToxR regulates the production of lipoproteins and the expression of serum resistance in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:1641-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramer, S. W., D. Bieber, and G. K. Schoolnik. 1996. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J. Bacteriol. 178:6555-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhine, J. A., and R. K. Taylor. 1994. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol. Microbiol. 13:1013-1020. [DOI] [PubMed] [Google Scholar]
- 33.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt, S. A., D. Bieber, S. W. Ramer, J. Hwang, C. Y. Wu, and G. Schoolnik. 2001. Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:4848-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, C. E., and R. K. Taylor. 1990. Vibrio cholerae O395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilins. Infect. Immun. 58:3042-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shevchik, V. E., and G. Condemine. 1998. Functional characterization of the Erwinia chrysanthemi OutS protein, an element of a type II secretion system. Microbiology 144:3219-3228. [DOI] [PubMed] [Google Scholar]
- 37.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 38.Taniguchi, T., Y. Akeda, A. Haba, Y. Yasuda, K. Yamamoto, T. Honda, and K. Tochikubo. 2001. Gene cluster for assembly of pilus colonization factor antigen III of enterotoxigenic Escherichia coli. Infect. Immun. 69:5864-5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor, R., C. Shaw, K. Peterson, P. Spears, and J. Mekalanos. 1988. Safe, live Vibrio cholerae vaccines? Vaccine 6:151-154. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regu-lated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]