Abstract
The peptidoglycan of Mycobacterium spp. reportedly has some unique features, including the occurrence of N-glycolylmuramic rather than N-acetylmuramic acid. However, very little is known of the actual biosynthesis of mycobacterial peptidoglycan, including the extent and origin of N glycolylation. In the present work, we have isolated and analyzed muramic acid residues located in peptidoglycan and UDP-linked precursors of peptidoglycan from Mycobacterium tuberculosis and Mycobacterium smegmatis. The muramic acid residues isolated from the mature peptidoglycan of both species were shown to be a mixture of the N-acetyl and N-glycolyl derivatives, not solely the N-glycolylated product as generally reported. The isolated UDP-linked N-acylmuramyl-pentapeptide precursor molecules also contain a mixture of N-acetyl and N-glycolyl muramyl residues in apparent contrast to previous observations in which the precursors isolated after treatment with d-cycloserine consisted entirely of N-glycolyl muropeptides. However, nucleotide-linked peptidoglycan precursors isolated from M. tuberculosis treated with d-cycloserine contained only N-glycolylmuramyl-tripeptide precursors, whereas those from similarly treated M. smegmatis consisted of a mixture of N-glycolylated and N-acetylated residues. The full pentapeptide intermediate, isolated following vancomycin treatment of M. smegmatis, consisted of the N-glycolyl derivative only, whereas the corresponding M. tuberculosis intermediate was a mixture of both the N-glycolyl and N-acetyl products. Thus, treatment with vancomycin and d-cylcoserine not only caused an accumulation of nucleotide-linked intermediate compounds but also altered their glycolylation status, possibly by altering the normal equilibrium maintained by de novo biosynthesis and peptidoglycan recycling.
The peptidoglycan of Mycobacterium spp. is classified as A1γ, unmodified peptidoglycan, as is that of Escherichia coli and many other bacterial species (32). However, the peptidoglycan of mycobacteria contains a variety of reported modifications including, invariably, an N-glycolyl (NGlyc) instead of an N-acetyl (NAc) function on the muramic acid (Mur), amidation of the carboxylic acids, and additional glycine or serine residues (18, 20). The peptidoglycan biosynthetic pathway of E. coli has been well studied (34, 35), and the mycobacterial pathway is assumed to be the same, based on limited biochemical analysis of some years ago (25, 28, 33) and more recent comparative genomics (2, 5, 6, 21). Analysis of the various precursors of peptidoglycan biosynthesis in Mycobacterium spp. could provide important information on the biosynthetic origin of these modifications and the notable refractoriness of Mycobacterium tuberculosis to β-lactam antibiotics and help in the search for alternative drug regimens for the treatment of multiple-drug-resistant forms of tuberculosis (10, 16).
UDP-N-acetylmuramyl-l-alanyl-d-glutamyl-meso-diaminopimelyl-d-alanyl-d-alanine (UDP-MurNAc-pentapeptide) is the final cytosolic precursor of peptidoglycan biosynthesis in E. coli. Because of the relatively low abundance of these precursors in untreated cells, drug treatment was used in order to obtain sufficient amounts for chemical characterization in mycobacterial studies. An incomplete precursor, UDP-N-glycolylmuramyl-l-alanyl-d-glutamyl-diaminopimelic acid (UDP-MurNGlyc-tripeptide) was isolated and analyzed from M. tuberculosis and Mycobacterium phlei following accumulation after treatment with d-cycloserine (28, 33). During a comparative study, we observed that the muramic acid residues isolated from mature peptidoglycan of M. tuberculosis and Mycobacterium smegmatis were a mixture of the N-acetyl and N-glycolyl derivatives, not solely the N-glycolylated product as generally reported (4, 19, 20). Since the nucleotide-linked precursors should reflect the nature of mature peptidoglycan, we hypothesized that the d-cycloserine treatment used in earlier studies had altered the composition of these molecules. In the present work, we have been able to isolate and analyze the nucleotide-linked precursors from M. tuberculosis and M. smegmatis with and without drug treatment, thereby allowing direct comparison.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
M. smegmatis mc2155 and M. tuberculosis H37Rv were obtained from the American Type Culture Collection (Manassas, Va.). Nutrient broth was from EM Science (Gibbstown, N.J.). Middlebrook 7H9 and 7H11 media were from Becton Dickinson (Sparks, Md.). UDP-N-acetylglucosamine (UDP-GlcNAc), vancomycin, d-cycloserine, trichloroacetic acid (TCA), 3-[N-morpholino]propanesulfonic acid (MOPS), mutanolysin, diaminopimelic acid (DAP), ninhydrin, and β-N-acetylhexosaminidase from Aspergillus oryzae were purchased from Sigma (St. Louis, Mo.). Sephadex G-25 and the Superdex peptide 10/300 GL column were from Amersham Biosciences (Piscataway, N.J.). 2-Mercaptoethanol was from J. T. Baker Inc. (Phillipsburg, N.J.). High-performance liquid chromatography (HPLC)-grade acetonitrile, water, and methanol were from Burdick and Jackson (Muskegon, Mich.). Trifluoroacetic acid (TFA) and the methanolic-HCl (3 N) kit were purchased from Supelco Inc. (Bellefonte, Pa.). Tri-Sil reagent was obtained from Pierce (Rockford, Ill.). The Hypersil ODS (C18) column was purchased from Phenomenex (Torrance, Calif.). All other reagents were American Chemical Society grade or better. Pronase was purchased from Roche (Nutley, N.J.). Chalaropsis spp. muramidase was purified as described previously (11, 12). UDP-MurNAc, UDP-MurNAc-tripeptide, and UDP-MurNAc-pentapeptide were prepared as described elsewhere, using recombinant E. coli Mur A, B, C, D, E, and F (30, 37).
Determination of the MICs of antibiotics.
The MICs of vancomycin and d-cycloserine were determined by the agar dilution method. In brief, Middlebrook 7H11 agar medium supplemented with oleic acid-albumin-dextrose-catalase was prepared according to the manufacturer's instructions. Drugs were added to the medium at desired concentrations and plated, and the plates were inoculated with 5 μl of M. tuberculosis H37Rv culture with an optical density at 600 nm of 0.3 to 0.5. The minimum concentration of a drug capable of completely suppressing the growth of the organism compared to that in culture without drug was defined as the MIC of that drug. The MICs of vancomycin and d-cycloserine were 12.5 and 20 μg/ml, respectively.
Growth conditions and drug treatment for large-scale culture.
M. tuberculosis H37Rv was grown in Middlebrook 7H9 broth containing 0.05% Tween 80 supplemented with 10% oleic acid-albumin-dextrose-catalase to mid-log phase. For drug treatment, either vancomycin or d-cycloserine (50 and 20 μg/ml, respectively) was added to the mid-log-phase cultures 24 h before harvesting. M. smegmatis mc2155 was grown in nutrient broth to mid-log phase, and either vancomycin (50 μg/ml) or d-cycloserine (20 μg/ml) was added to the mid-log-phase cultures 4 h before harvesting. These concentrations of antibiotics were chosen as they caused maximal accumulation of nucleotide-linked peptidoglycan precursors in both species.
Preparation of cell lysates.
M. smegmatis cells (10 g) were resuspended in 30 ml of 50 mM MOPS buffer (pH 8.0) containing 10 mM MgCl2 and 5 mM 2-mercaptoethanol (buffer A) and broken by ultrasonic probe sonication as previously described (24). The sonicate was centrifuged at 27,000 × g for 30 min at 4°C. The supernatant was transferred to a Teflon centrifuge tube, and TCA was added to achieve a 10% final concentration. The mixture was stirred for 30 min on ice followed by centrifugation at 15,000 × g for 30 min at 4°C. The clear supernatant was transferred to a new tube, and TCA was removed by three extractions with diethyl ether. The remaining aqueous phase was dried on a rotary evaporator and reconstituted in 5 ml of water. In the case of M. tuberculosis H37Rv, 10 g of harvested cells (wet weight) was resuspended in buffer A, disrupted in a mini bead beater with 0.1- to 0.15-mm zirconium beads (Biospec Products, Bartlesville, Okla.) and 25 1-min pulses with 1 min of cooling on ice between the cycles. The lysate was centrifuged twice at 5,000 × g to remove unbroken cells and zirconium beads and processed as described for M. smegmatis.
Isolation of the nucleotide-linked precursors of peptidoglycan biosynthesis from the cell lysate.
A Sephadex G-25 (116- by 2.5-cm) column was equilibrated with 75 mM ammonium acetate (pH 5.0) and calibrated with authentic UDP-MurNAc-pentapeptide and UDP-MurNAc-tripeptide. The reconstituted lysates from M. smegmatis and M. tuberculosis were loaded on the column and eluted with equilibration buffer. Fractions were monitored for absorption at 262 nm, pooled, and lyophilized three times to remove ammonium acetate. The amino acid compositions of the isolated nucleotide precursors were determined at the Protein Structure Core Facility, University of Nebraska Medical Center, Omaha, where the samples were subjected to vapor hydrolysis using 6 N HCl with phenol and sodium sulfite for 20 to 24 h at 110°C under argon. Amino acids were then analyzed with a Beckman 6300 analyzer (Beckman Coulter, Fullerton, Calif.) using postcolumn ninhydrin detection.
Preparation and purification of muropeptides.
The nucleotide-linked precursors purified with Sephadex G-25 were resuspended in 2 M TFA and incubated at 60°C for 1 h. The hydrolysate was cooled and dried under vacuum to remove TFA. The resulting muropeptides were further purified by size exclusion chromatography on a Superdex peptide 10/300 GL column connected to a Waters (Milford, Mass.) model 600 controller, a model 600 pump, and a model 2487 UV detector. The column was equilibrated and eluted with 30% acetonitrile containing 0.1% TFA with a flow rate of 0.5 ml/min. The absorbance of the effluent was monitored at 214 nm. The muropeptide-containing fractions were dried under vacuum and stored at −20°C.
MS analysis of the muropeptides.
HPLC-purified muropeptides from the isolated nucleotides were suspended in HPLC-grade water at a concentration of about 10 μM for liquid chromatography-mass spectrometry (LC-MS) analysis. An aliquot (20 μl) of the suspension was applied to a 2- by 150-mm Hypersil ODS (C18) column connected to an Agilent 1100 (Agilent Technologies, Palo Alto, Calif.) HPLC system. The muropeptides were eluted with a 2-to-30% linear gradient of acetonitrile in 0.5% formic acid at a flow rate of 320 μl/min. The effluent was introduced directly into a Finnigan LCQ Duo electrospray mass spectrometer (Finnigan-Thermoquest, San Jose, Calif.), and the muropeptides were analyzed by MS and tandem MS. The electrospray needle was operated at 4 kV with a sheath gas flow of N2 at 40 lb/in2 and a capillary temperature of 200°C. Tandem MS was performed on the fly of the most dominant ion of the previous MS scan or from a preselected list of [M+H]+ ions. For this purpose, standard MurNAc-pentapeptide was prepared from UDP-MurNAc-pentapeptide by subjecting it to mild acid hydrolysis with 2 M TFA at 60°C for 1 h to remove UDP. The hydrolyzed samples were cooled to room temperature, dried under vacuum to remove TFA, and dissolved in HPLC-grade water prior to LC-MS analysis.
Preparation and analysis of amino sugars from nucleotide-linked peptidoglycan precursors.
The muropeptides obtained after Superdex peptide column chromatography were resuspended in 200 μl of 50 mM 2-[N-morpholino]ethanesulfonate buffer (pH 6.0) containing 1 mM MgCl2, followed by the addition of 10 U of mutanolysin and incubation for 16 h at 37°C to remove the peptide moiety. The reaction was deproteinated by ethanol precipitation, and the supernatant was transferred to a 13- by 100-mm glass tube and dried under vacuum. Scyllo-inositol was added as an internal standard, and the sample was dissolved in 3 N methanolic-HCl, tightly capped, heated at 80°C for 1 h, cooled to room temperature, and dried under a stream of N2. Tri-Sil reagent was added, the sealed tubes were heated at 70°C for 20 min and cooled to room temperature, and the reagent was evaporated under a stream of N2. The derivatized products were dissolved in hexane and analyzed in a trace GC 2000 gas chromatograph (Finnigan-Thermoquest) linked with a Polaris mass detector (Finnigan-Thermoquest). MurNAc and GlcNAc standards were prepared in the same way and analyzed for comparison.
Preparation and analysis of amino sugars from peptidoglycan.
Peptidoglycan was prepared according to procedures previously described (7, 18, 29) with some modifications. In brief, the cell lysate was prepared and centrifuged at 27,000 × g and the pelleted cell wall was repeatedly extracted with 2% sodium dodecyl sulfate (SDS) in phosphate-buffered saline (PBS). The SDS-insoluble peptidoglycan was digested with pronase to remove any residual proteins (13) and extracted again with SDS. The resulting mycolylarabinogalactan-peptidoglycan complex was subjected to mild base (0.5% KOH in methanol) followed by mild acid (0.05 M H2SO4) to remove mycolic acids and arabinogalactan, respectively (7). The purified peptidoglycan was washed with water and lyophilized, and 2 mg was suspended in 0.5 ml of 10 mM sodium acetate (pH 5.0). Purified Chalaropsis spp. muramidase (25 μg) was added, followed by incubation at 37°C for 16 h (36). The soluble muropeptides were recovered after centrifugation at 27,000 × g, and the enzyme was removed by ultrafiltration. The muropeptides were further digested with mutanolysin as described above to remove the peptide side chains. The resulting disaccharide was digested with β-N-acetylhexosaminidase to obtain the amino sugars. The enzyme was removed by alcohol precipitation, and the amino sugars were derivatized with Tri-Sil prior to analysis by gas chromatography and MS (GC-MS), as described above.
RESULTS
Analysis of the N-acylmuramic acid residues from mature peptidoglycan.
The peptidoglycan-derived N-acylmuramic acids were analyzed by GC-MS as described in Materials and Methods. The retention times of the MurNAc and MurNGlyc residues isolated from M. tuberculosis were 11.37 and 13.25 min, respectively, and the identity of these peaks was confirmed by MS using unique diagnostic ions for the compounds (Fig. 1). MurNGlyc and MurNAc appear to exist in an approximate 7:3 ratio, although the MS data represent a qualitative analysis of the compounds; the actual relative abundance may be somewhat different. Thus, the muramic acid residues from M. tuberculosis peptidoglycan were found to be a mixture of MurNGlyc and MurNAc (Fig. 1), as were those from M. smegmatis peptidoglycan (data not shown).
FIG. 1.
MS analysis of the N-acylmuramic acid residues of mature peptidoglycan isolated from M. tuberculosis. The trimethylsilane derivatives of the N-acylmuramic acids were analyzed by GC-MS as described in Materials and Methods. (A) Total ion chromatogram of the GC run. (B) Mass spectrum of the peak at 11.37 min. The structure of the TMS derivative of N-acetyl muramic acid and the diagnostic fragment (m/z 187.0) used as positive identification are inset. (C) Mass spectrum of the peak at 13.25 min. The structure of the TMS derivative of N-glycolylmuramic acid and the diagnostic fragment with the structure of the diagnostic fragment (m/z 275.1) used as positive identification are inset.
Isolation of nucleotide-linked precursors of peptidoglycan biosynthesis.
The cell lysate obtained from M. smegmatis cells grown without drug treatment showed very little UDP-N-acylmuramyl-pentapeptide based on the absorbance of the Sephadex G-25 column fractions at 262 nm (A262) (Fig. 2A), as did that obtained from M. tuberculosis (data not shown). However, sufficient quantities of the nucleotide precursors were obtained from untreated cells to allow MS analysis. Cell extracts obtained from vancomycin-treated M. smegmatis (Fig. 2C) and M. tuberculosis (data not shown) cells showed an accumulation of a material corresponding to standard UDP-MurNAc-pentapeptide from E. coli. Amino acid analysis of the M. smegmatis-derived material gave an Ala/Glu/DAP ratio of 2.8:1.2:1.0, or an approximate ratio of 3:1:1, as expected. The lysate from d-cycloserine-treated M. smegmatis cells also showed accumulation of a large peak (Fig. 2B), presumably UDP-N-acylmuramyl-tripeptide, since it had an Ala/Glu/DAP ratio of 1.1:1.2:1.0.
FIG. 2.
Elution patterns from Sephadex G-25 columns of the TCA-soluble material from M. smegmatis cell lysates obtained from treated and untreated cells (10 g [wet weight]). The elution was monitored at 262 nm. (A) TCA-soluble material obtained from untreated cells. (B) TCA-soluble material from the cells treated with d-cycloserine. (C) TCA-soluble material from vancomycin-treated cells. The arrows indicate the elution position of nucleotide-linked peptidoglycan precursors.
Analysis of the muropeptides from untreated mycobacteria.
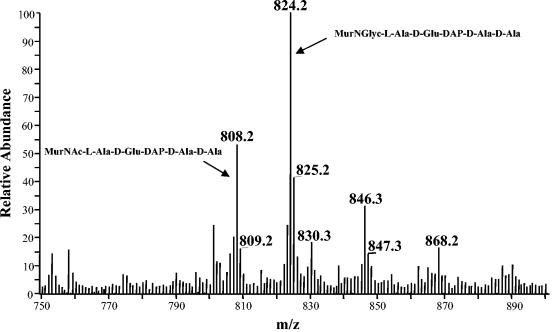
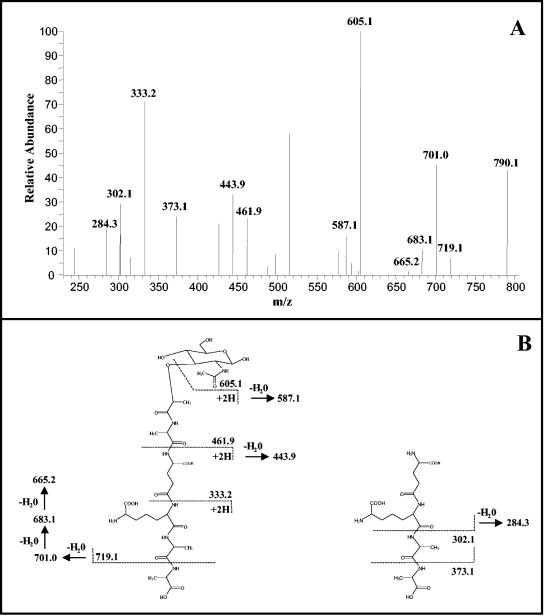
The nucleotide precursors obtained from the Sephadex G-25 column were hydrolyzed with TFA to remove UDP, and the resulting muropeptides were further purified on the Superdex peptide column. This step helped to eliminate several impurities as well as salts, yielding highly purified muropeptides. Those obtained from both drug-treated and untreated cells of M. tuberculosis and M. smegmatis were analyzed by LC-MS. The positive ion mass spectrum of the muropeptides obtained from untreated M. tuberculosis cells were dominated by two molecular ions, m/z 808.2 and m/z 824.2 (Fig. 3). The corresponding monosodium adducts (m/z 830.3 and m/z 846.3) were also observed. The calculated monoisotopic mass of MurNAc-pentapeptide is 807.3. When m/z 808.2 was subjected to tandem MS, it generated a series of fragments (Fig. 4), including m/z 605.1, two y-type (15, 31) peptide fragments (m/z 461.9 and m/z 333.2), a b-type (15, 31) peptide fragment (m/z 719.1), and three internal fragments (m/z 443.9, m/z 373.1, and m/z 302.1), positively identifying MurNAc-pentapeptide. The daughter ions generated from the molecular ion with an m/z of 824.2 were identical to the daughter ions from MurNAc-pentapeptide, except that the b-ion was 16 atomic mass units (amu) larger. Therefore, the additional 16 amu are due to modification of the muramic acid residue (which is only present in this fragment), indicating that the acetyl residue had been replaced with a glycolyl residue. Muropeptides isolated from untreated M. smegmatis produced identical mass spectra.
FIG. 3.
Positive ion mass spectrum of the UDP-N-acylmuramyl-l-Ala-d-Glu-DAP-d-Ala-d-Ala isolated from untreated M. tuberculosis. The m/z 808.2 ion represents MurNAc-l-Ala-d-Glu-DAP-d-Ala-d-Ala, and m/z 830.3 was the corresponding monosodium adduct. m/z 824.2 was identified as MurNGlyc-l-Ala-d-Glu-DAP-d-Ala-d-Ala. Ions corresponding to the mono- and disodium adducts (m/z 846.3 and m/z 868.2, respectively) were also observed.
FIG. 4.
Tandem mass spectrum of the molecular ion m/z 808.2. The structures of daughter ions labeled with m/z values in panel A are indicated in panel B, which shows the deduced fragmentation pattern of MurNAc-l-Ala-d-Glu-DAP-d-Ala-d-Ala. The ion with an m/z value of 790.1 represents the molecular ion after the neutral loss of H2O. The structures of unlabeled ions were not solved.
Analysis of the muropeptides from drug-treated M. tuberculosis and M. smegmatis.
The muropeptides obtained from vancomycin-treated M. tuberculosis consisted of a mixture of ions of m/z 824.2 and m/z 808.2 in a ratio comparable to that in the nontreated cells, which was typically 7:3 but showed some variability from experiment to experiment. GC-MS analysis of this product after mutanolysin treatment and acid hydrolysis yielded both MurNGlyc and MurNAc in a similar ratio. However, muropeptides isolated from M. smegmatis after vancomycin treatment were almost exclusively MurNGlyc-pentapeptide (m/z 824.2) (Table 1).
TABLE 1.
N substitution of the muramic acid residues of mature peptidoglycan or nucleotide-linked peptidoglycan precursors isolated from M. tuberculosis or M. smegmatis
| Drug treatment |
M. tuberculosis
|
M. smegmatis
|
||||||
|---|---|---|---|---|---|---|---|---|
| Peptidoglycan
|
Nucleotide
|
Peptidoglycan
|
Nucleotide
|
|||||
| MurNGlyc | MurNAc | MurNGlyc | MurNAc | MurNGlyc | MurNAc | MurNGlyc | MurNAc | |
| None | + | + | + | + | + | + | + | + |
| d-Cycloserine (20 μg/ml) | NDa | ND | + | − | ND | ND | + | + |
| Vancomycin (50 μg/ml) | ND | ND | + | + | ND | ND | + | − |
ND, not determined.
The positive ion mass spectrum of the M. tuberculosis muropeptides isolated after d-cycloserine treatment was dominated by an ion with an m/z value of 682.2 and the corresponding sodium adducts (Fig. 5). Tandem MS positively identified this molecule as MurNGlyc-tripeptide. On the other hand, mass spectra of samples obtained from d-cycloserine-treated M. smegmatis were dominated by ions of m/z 666.2 and m/z 682.2 (data not shown), indicating the presence of a mixture of MurNAc and MurNGlyc residues. Tandem MS of these molecular ions indicated that the difference of 16 amu was due to the existence of a MurNGlyc residue instead of MurNAc. GC-MS analysis of the amino sugars after removal of the peptide and acid hydrolysis confirmed the LC-MS results.
FIG. 5.
Positive ion mass spectrum of the nucleotide-linked intermediates isolated from M. tuberculosis H37Rv treated with d-cycloserine. The mass spectrum is dominated by the ion with an m/z value of 682.2, which represents MurNGlyc-l-Ala-d-Glu-DAP, and the corresponding monosodium adduct (m/z of 704.2).
DISCUSSION
The pool level of UDP-MurNAc-pentapeptide has been calculated to be about 105 molecules per E. coli cell (22), a figure likely to be similar in other organisms. This low abundance poses major obstacles to the isolation and analysis of these crucial precursors from any organism. Previously, this problem had been addressed by treating bacterial cells with peptidoglycan biosynthesis inhibitors to allow accumulation of the precursors, thus enabling isolation of tangible quantities for analysis (3, 17). Studies of the biosynthetic precursor UDP-N-acylmuramyl-tripeptide from M. tuberculosis and M. phlei (28, 33) determined that all the muramic acid residues were, apparently, completely NGlyc substituted. Since UDP-MurNGlyc-tripeptide is the immediate precursor of the UDP-MurNGlyc-pentapeptide, all of the UDP-N-acylmuramyl-pentapeptides should be NGlyc substituted, as has been the supposition (5, 7, 18). In turn, this scenario points to peptidoglycan containing MurNGlyc exclusively, as has been reported elsewhere (7, 18, 19).
However, our analysis found that the muramic acid residues in mature peptidoglycan of mycobacteria consist of a mixture of the NGlyc and NAc derivatives. We expected that detailed analysis of the nucleotide-linked precursor of peptidoglycan would reflect the nature of mature peptidoglycan. Therefore, we attempted to isolate the final nucleotide-linked precursors from both M. tuberculosis and M. smegmatis, avoiding prior treatment with peptidoglycan biosynthetic inhibitors. This goal was successfully met through the application of a modified version of the protocol of Mengin-Lecreulx et al. (22). On analysis, muropeptides isolated from M. tuberculosis and M. smegmatis were also shown to contain both MurNGlyc and MurNAc residues.
Our analysis of mature peptidoglycan from mycobacterial species is consistent with the mass spectra presented in the original identification of NGlyc residues in peptidoglycan isolated from M. smegmatis (1, 19); however, the observation that M. smegmatis peptidoglycan could contain both MurNGlyc and MurNAc appears to have received little attention after it was reported that mycobacterial nucleotide-linked precursors were exclusively NGlyc substituted (19, 28, 33). In these studies d-cycloserine, an inhibitor of the d-Ala-d-Ala ligase, was used to allow accumulation of these intermediates and facilitate isolation and characterization. We hypothesized that the discrepancy between our results and the earlier ones was due to d-cycloserine treatment, and we tested this idea by isolating the nucleotide-linked intermediates from M. tuberculosis and M. smegmatis cells following d-cycloserine and vancomycin treatment. The results indicated that this is indeed the case (Table 1). Interestingly, vancomycin and d-cycloserine appear to have different effects on M. tuberculosis and M. smegmatis. Muropeptides isolated from M. smegmatis treated with vancomycin were exclusively the NGlyc derivative, while those from M. tuberculosis treated similarly were a mixture (Table 1). However, treatment of M. tuberculosis with d-cycloserine resulted in muropeptides containing exclusively NGlyc-substituted muramic acid as previously reported (28, 33).
The effects of the drugs are perhaps due to perturbation of the biosynthesis through a direct effect on the putative NAc/NGlyc hydroxylase (8, 9). Alternatively, the drugs could affect the equilibrium of peptidoglycan recycling (26, 27). The presence of peptidoglycan recycling pathways in Mycobacterium is yet to be established, but recycling similar to that first identified in E. coli has been demonstrated in other organisms (14, 23, 26, 27). Although the mechanism by which d-cycloserine and vancomycin exert their effects on glycolylation of nucleotide-linked peptidoglycan precursors is unclear, we have unequivocally shown that the muramyl peptides of both the mature peptidoglycan and the UDP-muramyl peptides of untreated mycobacteria contain a mixture of MurNGlyc and MurNAc.
Acknowledgments
This work was supported by grants (AI18357 and AI49151) from the National Institute of Allergy and Infectious Disease, National Institutes of Health.
REFERENCES
- 1.Adam, A., J. F. Petit, J. Wietzerbin-Falszpan, P. Sinay, D. W. Thomas, and E. Lederer. 1969. L'acide N-glycolyl-muramique, contituant des parois de Mycobacterium smegmatis: identification par spectrometrie de masse. FEBS Lett. 4:87-92. [DOI] [PubMed] [Google Scholar]
- 2.Belanger, A. E., J. C. Porter, and G. F. Hatfull. 2000. Genetic analysis of peptidoglycan biosynthesis in mycobacteria: characterization of a ddlA mutant of Mycobacterium smegmatis. J. Bacteriol. 182:6854-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billot-Klein, D., D. Shlaes, D. Bryant, D. Bell, R. Legrand, L. Gutmann, and J. van Heijenoort. 1997. Presence of UDP-N-acetylmuramyl-hexapeptides and -heptapeptides in enterococci and staphylococci after treatment with ramoplanin, tunicamycin, or vancomycin. J. Bacteriol. 179:4684-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 64:29-63. [DOI] [PubMed] [Google Scholar]
- 5.Crick, D. C., S. Mahapatra, and P. J. Brennan. 2001. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology 11:107R-118R. [DOI] [PubMed] [Google Scholar]
- 6.De Smet, K. A. L., K. E. Kempsell, A. Gallagher, K. Duncan, and D. B. Young. 1999. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis. Microbiology 145:3177-3184. [DOI] [PubMed] [Google Scholar]
- 7.Draper, P., O. Kandler, and A. Darbre. 1987. Peptidoglycan and arabinogalactan of Mycobacterium leprae. J. Gen. Microbiol. 133:1187-1194. [DOI] [PubMed] [Google Scholar]
- 8.Essers, L., and H. J. Schoop. 1978. Evidence for the incorporation of molecular oxygen, a pathway in biosynthesis of N-glycolylmuramic acid in Mycobacterium phlei. Biochim. Biophys. Acta 544:180-184. [DOI] [PubMed] [Google Scholar]
- 9.Gateau, O., C. Bordet, and G. Michel. 1976. Study of formation of N-glycolylmuramic acid from Nocardia asteroides. Biochim. Biophys. Acta 421:395-405. [DOI] [PubMed] [Google Scholar]
- 10.Green, D. W. 2002. The bacterial cell wall as a source of antibacterial targets. Expert Opin. Ther. Targets 6:1-19. [DOI] [PubMed] [Google Scholar]
- 11.Hash, J. H. 1963. Purification and properties of staphylolytic enzymes from Chalaropsis sp. Arch. Biochem. Biophys. 102:379-388. [DOI] [PubMed] [Google Scholar]
- 12.Hash, J. H., and M. V. Rothlauf. 1967. The N,O-diacetylmuramidase of Chalaropsis species. I. Purification and crystallization. J. Biol. Chem. 242:5586-5590. [PubMed] [Google Scholar]
- 13.Hirschfield, G. R., M. McNeil, and P. J. Brennan. 1990. Peptidoglycan-associated polypeptides of Mycobacterium tuberculosis. J. Bacteriol. 172:1005-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. van Heijenoort, J. T. Park, S. Normark, and J. M. Frere. 1995. AmpD, essential for both beta-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, R. S., S. A. Martin, K. Biemann, J. T. Stults, and J. T. Watson. 1987. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 59:2621-2625. [DOI] [PubMed] [Google Scholar]
- 16.Khasnobis, S., V. E. Escuyer, and D. Chatterjee. 2002. Emerging therapeutic targets in tuberculosis: post-genomic era. Expert Opin. Ther. Targets 6:21-40. [DOI] [PubMed] [Google Scholar]
- 17.Kohlrausch, U., and J. V. Holtje. 1991. One-step purification procedure for UDP-N-acetylmuramyl-peptide murein precursors from Bacillus cereus. FEMS Microbiol. Lett. 62:253-257. [DOI] [PubMed] [Google Scholar]
- 18.Kotani, S., I. Yanagida, K. Kato, and T. Matsuda. 1970. Studies on peptides, glycopeptides and antigenic polysaccharide-glycopeptide complexes isolated from an L-11 enzyme lysate of cell walls of Mycobacterium tuberculosis strain H37Rv. Biken. J. 13:249-275. [PubMed] [Google Scholar]
- 19.Lederer, E. 1971. The mycobacterial cell wall. Pure Appl. Chem. 25:135-165. [DOI] [PubMed] [Google Scholar]
- 20.Lederer, E., A. Adam, R. Ciorbaru, J. F. Petit, and J. Wietzerbin. 1975. Cell walls of mycobacteria and related organisms; chemistry and immunostimulant properties. Mol. Cell Biochem. 7:87-104. [DOI] [PubMed] [Google Scholar]
- 21.Lepage, S., P. Dubois, T. K. Ghosh, B. Joris, S. Mahapatra, M. Kundu, J. Basu, P. Chakrabarti, S. T. Cole, M. Nguyen-Disteche, and J. M. Ghuysen. 1997. Dual multimodular class A penicillin-binding proteins in Mycobacterium leprae. J. Bacteriol. 179:4627-4630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mengin-Lecreulx, D., B. Flouret, and J. van Heijenoort. 1982. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 151:1109-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mengin-Lecreulx, D., J. van Heijenoort, and J. T. Park. 1996. Identification of the mpl gene encoding UDP-N-acetylmuramate:l-alanyl-γ-d-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 178:5347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271:7820-7828. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee, T., D. Basu, S. Mahapatra, C. Goffin, J. vanBeeumen, and J. Basu. 1996. Biochemical characterization of the 49 kDa penicillin-binding protein of Mycobacterium smegmatis. Biochem. J. 320:197-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Normark, S. 1995. β-Lactamase induction in gram-negative bacteria is intimately linked to peptidoglycan recycling. Microb. Drug Resist. 1:111-114. [DOI] [PubMed] [Google Scholar]
- 27.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petit, J. F., A. Adam, and J. Wietzerbin-Falszpan. 1970. Isolation of UDP-N-glycolylmuramyl-(Ala,Glu,Dap) from Mycobacterium phlei. FEBS Lett. 6:55-57. [DOI] [PubMed] [Google Scholar]
- 29.Petit, J. F., A. Adam, J. Wietzerbin-Falszpan, E. Lederer, and J. M. Ghuysen. 1969. Chemical structure of the cell wall of Mycobacterium smegmatis. I. Isolation and partial characterization of the peptidoglycan. Biochem. Biophys. Res. Commun. 35:478-485. [DOI] [PubMed] [Google Scholar]
- 30.Reddy, S. G., S. T. Waddell, D. W. Kuo, K. K. Wong, and D. L. Pompliano. 1999. Preparative enzymatic synthesis and characterization of the cytoplasmic intermediates of murein biosynthesis. J. Am. Chem. Soc. 121:1175-1178. [Google Scholar]
- 31.Roepstorff, P., and J. Fohlman. 1984. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11:601. [DOI] [PubMed] [Google Scholar]
- 32.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takayama, K., H. L. David, L. Wang, and D. S. Goldman. 1970. Isolation and characterization of uridine diphosphate-N-glycolylmuramyl-l-alanyl-gamma-d-glutamyl-meso-alpha,alpha′-diaminopimelic acid from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 39:7-12. [DOI] [PubMed] [Google Scholar]
- 34.van Heijenoort, J. 1994. Biosynthesis of bacterial peptidoglycan unit, p. 39-54. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Biomedical Press, Amsterdam, The Netherlands.
- 35.van Heijenoort, J. 2001. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Product Rep. 18:503-519. [DOI] [PubMed] [Google Scholar]
- 36.Wietzerbin, J., B. C. Das, J. F. Petit, E. Lederer, M. Leyh-Bouille, and J. M. Ghuysen. 1974. Occurrence of d-alanyl-(d)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of mycobacteria. Biochemistry 13:3471-3476. [DOI] [PubMed] [Google Scholar]
- 37.Yagi, T., S. Mahapatra, K. Mikuova, D. C. Crick, and P. J. Brennan. 2003. Polymerization of mycobacterial arabinogalactan and ligation to peptidoglycan. J. Biol. Chem. 278:26497-26504. [DOI] [PubMed] [Google Scholar]