Abstract
Radical cations (holes) produced in DNA by ionizing radiation and other oxidants yield DNA–protein cross-links (DPCs). Detailed studies of DPC formation in chromatin via this process are lacking. We describe here a comprehensive examination of DPC formation within nucleosome core particles (NCPs), which are the monomeric component of chromatin. DNA holes are introduced at defined sites within NCPs that are constructed from the bottom-up. DPCs form at DNA holes in yields comparable to those of alkali-labile DNA lesions that result from water trapping. DPC-forming efficiency and site preference within the NCP are dependent on translational and rotational positioning. Mass spectrometry and the use of mutant histones reveal that lysine residues in histone N-terminal tails and amino termini are responsible for the DPC formation. These studies are corroborated by computational simulation at the microsecond time scale, showing a wide range of interactions that can precede DPC formation. Three consecutive dGs, which are pervasive in the human genome, including G-quadruplex-forming sequences, are sufficient to produce DPCs that could impact gene expression.
Graphical Abstract

INTRODUCTION
DNA–protein cross-links (DPCs) block transcription and replication and as such pose a significant biological threat.1,2 The first known DNA-dependent proteases that are postulated to be constituents of a general DPC repair pathway have been discovered in the past decade.3–7 DPC repair deficiency has already been associated with the Ruijs–Aalfs syndrome, which gives rise to premature aging and early onset liver cancer.3,7,8 DPCs are produced by a variety of chemical agents, including aldehydes and chemotherapeutic agents.9–15 Transient DPCs formed between histone proteins and the epigenetic base 5-formylcytosine (5fC) play a role in regulating transcription in cells.16–18 DPCs also result from nucleobase oxidation, including ionization induced by a variety of UV-absorbing photochemical reagents and the direct effect of ionizing radiation, which is used to treat more than 50% of cancer patients.19–26 Ionizing radiation produces DPCs in greater quantities than either DNA–DNA interstrand cross-links or double-strand breaks.27,28 Despite the prevalence and biological significance of radiation-induced DPC formation within chromatin, there is a gap in our knowledge regarding their formation from nucleobase radical cations that are generated via direct ionization. We report our investigation of this chemistry in nucleosome core particles (NCPs), the monomeric component of chromatin.
Ionization produces mobile radical cations (“holes”) that can migrate over hundreds of angstroms and even longer distances via charge transfer in free DNA of appropriate sequences (Scheme 1).29–31 The holes localize on 2′-deoxyguanosine (dG) in sequences of (2 to 3) consecutive dGs on account of their relatively favorable redox potentials and give rise to dG lesions due to water trapping.32–36 Although the role of nucleophilic amino acids within chromatin or even monomeric NCPs on charge transfer is not well understood (Scheme 1), the reaction of one electron-oxidized dG with peptides and proteins is well studied.37–40 Product and computational model studies, respectively, reveal that the ε-amine of lysine and methyl amine preferentially add to the C8-position of dG (e.g., C8-K-dG) in the presence of a one-electron oxidant (Scheme 2).39–43 The initially formed amine adducts are susceptible to further oxidation, resulting in products derived from addition at the C5-position of guanine (e.g., C8-K-dG-Ox). DPC formation under oxidative conditions has also been attributed to reaction between a one-electron-oxidized amino acid and DNA.28 A more complex pathway has been proposed in NCPs in which a hole at guanine is reduced by histone 3 tyrosine 41 (H3-Y41), and the resulting tyrosyl radical forms a DPC by reacting with a proximal pyrimidine.44 Similarly, in addition to direct nucleophilic attack of an amine on the dG-radical cation (dG•+), computational experiments support the possibility that the deprotonated dG-radical cation (dG•) is reduced by a protonated lysine and that the resulting nitrogen radical reacts to form the DPC between dG and lysine.45 An important conclusion from the same study is that the reactivity of even the peptide trilysine with a hole in DNA is nonrandom and will be governed by specific sequence interactions, of which there are many possibilities within NCPs in cellular DNA.
Scheme 1.
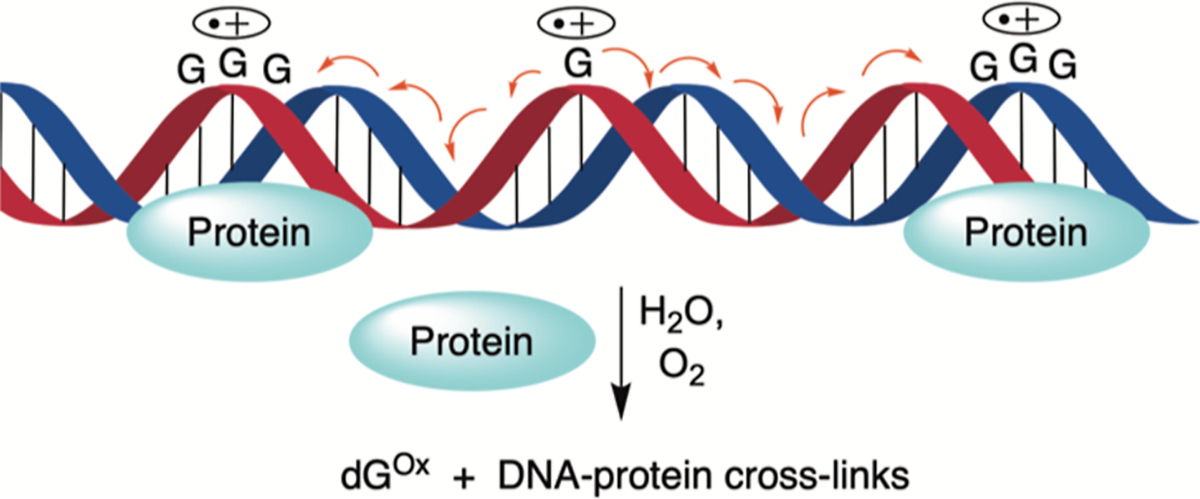
Charge Migration through DNA and the Formation of Hole Trapping Products on dG
Scheme 2.
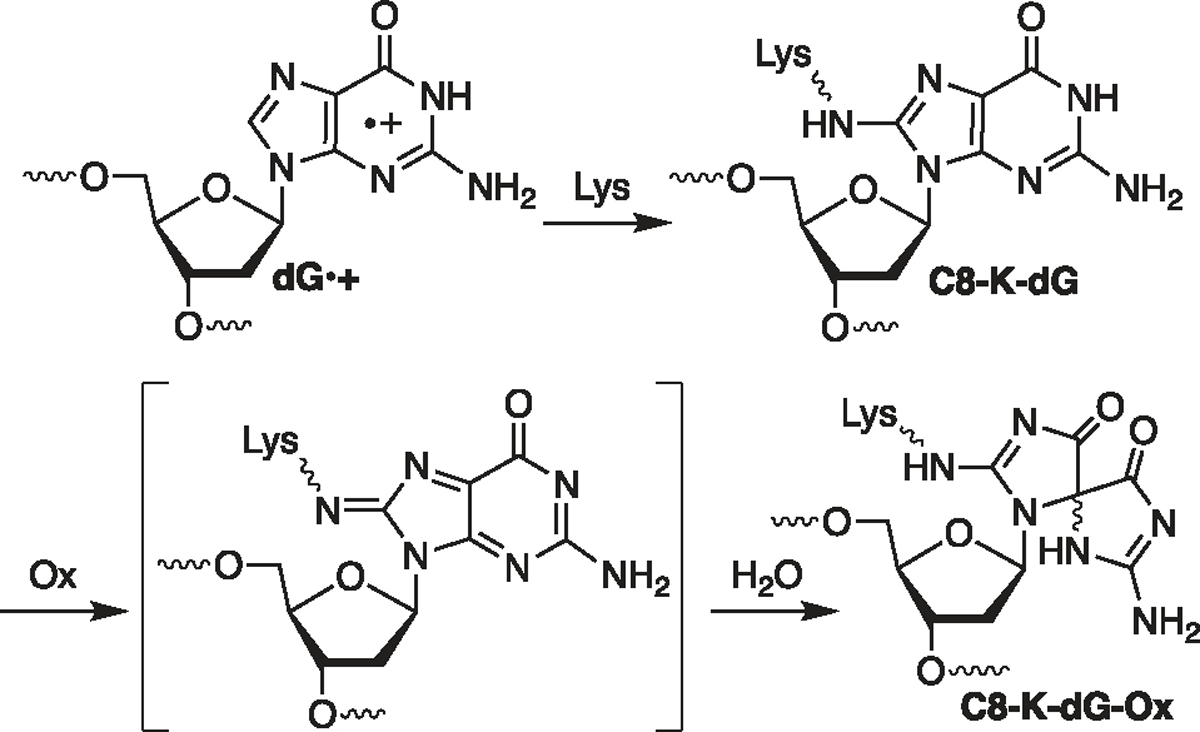
Lysine Trapping and Further Oxidation of dG•+
The product studies referred to above were typically carried out in the presence of a large excess of peptide or protein and often do not account for DNA hole transfer. The DNA–protein complexes are also structurally distinct from that in chromatin where the majority of binding is with the phosphate backbone and transient interactions with lysine-rich histone tails.46,47 Although DPC formation was not the focus, studies have been carried out on nucleosomes (and NCPs) to better understand charge transfer in chromatin.48–51 Disparate conclusions drawn from these investigations were recently adjudicated by examining hole migration in NCPs prepared from the bottom-up.50 Holes were injected into nucleosomal DNA by independently generating dA• at a defined position via UV-photolysis of 1, which is incorporated in chemically synthesized oligonucleotides via a corresponding phosphoramidite (Scheme 3A).52,53 Hole formation is driven by the effect of a stacked dA on the pKa of the nitrogen radical (Scheme 3B).54–57 The efficiency with which holes are transformed into alkali-labile lesions (Gox) at consecutive dGs depended upon the position within the NCP and is modulated by properties that include DNA unwrapping and quenching by H3-Y41.52 DPCs were not detected in this investigation because the experiments were carried out under conditions (pH 5) at which the lysines are completely protonated.31 Protein nucleophiles, such as the ε-amine of lysine side chains, could compete with water to form DPCs.38,40,58 Lysine-rich histone proteins that make up the core of NCPs are proximal to DNA. Their proximity to DNA, particularly those lysines within flexible tails, provides ample opportunities to trap holes and form DPCs. Below, we describe the competition of histones with water to form DNA–protein cross-links at pH 7.5 in NCPs. By introducing holes at defined positions in nucleosomal DNA, this study provides a comprehensive analysis of DPC formation from DNA ionization within NCPs.
Scheme 3.
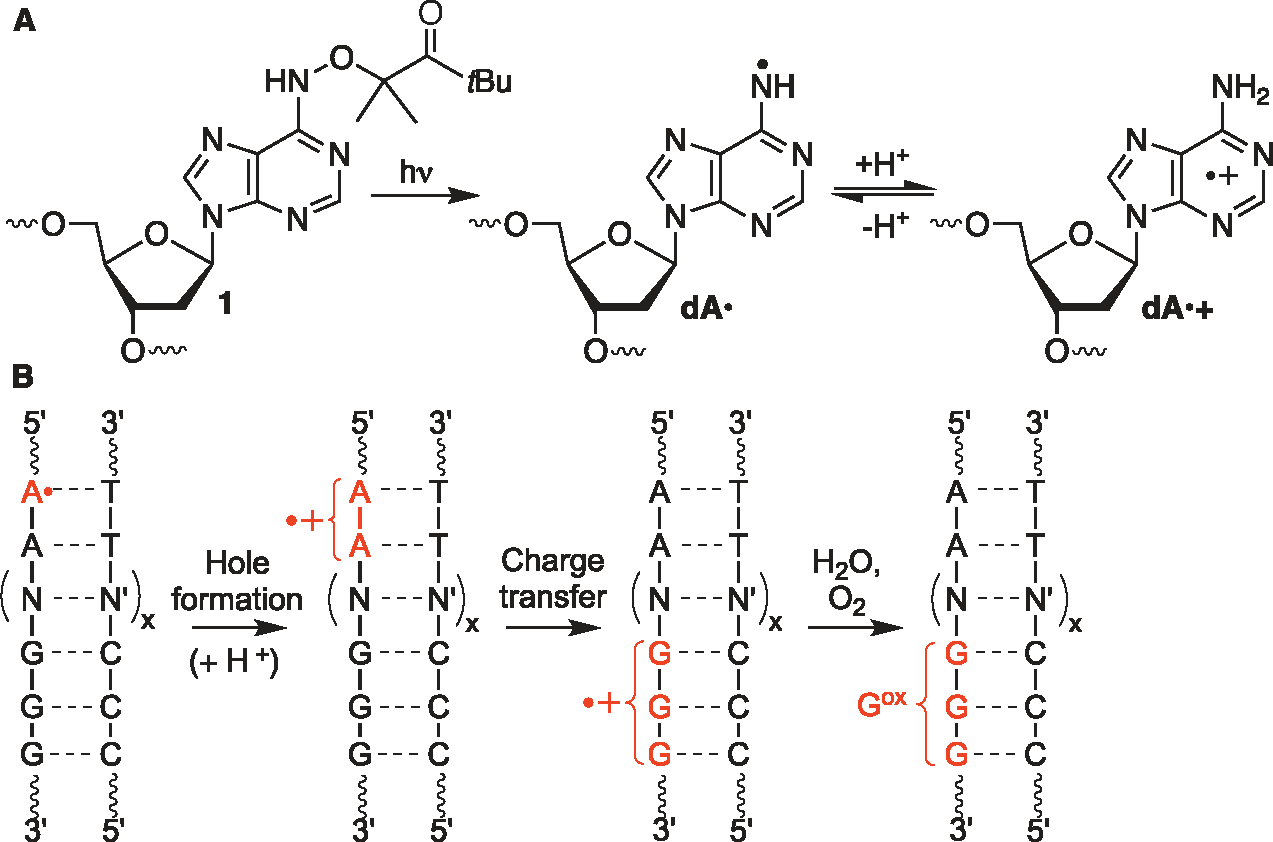
Hole Generation from 1 and Subsequent Charge Transfer in DNA
RESULTS
DNA–Peptide Cross-Link Formation between DNA and a Histone H4 N-Terminal Peptide.
Model studies were carried out using an H4 N-terminal peptide (H4 1–11, 2) and duplex DNA (25 bp) containing either the dA• precursor (1) flanked by a 5′-dA (4) to promote hole formation(dA•+) or dA at the comparable position as 1 as a control (5).54–56 Irradiation (350 nm, 8 h) of 5′-32P-4 (5 μM) in the presence of a large excess of 2 (500 μM) yielded the DNA–peptide crosslink (DpepC) (7.7 ± 0.2%, Figure 1A). DpepCs were detected on denaturing PAGE as two slower moving products that decomposed upon proteinase K treatment. In contrast, photolysis of the comparable duplex containing dA (5′-32P-5) yielded a single DpepC band (3.7 ± 0.3%) that comigrated with the slower of the two products observed from 5′-32P-4. The DpepCs that appeared when 1 is photolyzed exhibited significantly different chemical properties compared to the control. Isolation of the faster moving product band in DpepCs from irradiation of 5′-32P-4 followed by piperidine treatment (1 M, 90 °C, 30 min) yielded 70 ± 1% cleavage (Figures 1B and S1). Cleavage is predominantly at dG10 and dG9, consistent with the hole transfer damage pattern detected in 5′-32P-4 following irradiation in the absence of peptides.54 In contrast, only 11 ± 2% cleavage was observed upon comparable treatment of the DpepCs isolated from photolysis of 5′-32P-5 (Figures 1B and S1).
Figure 1.
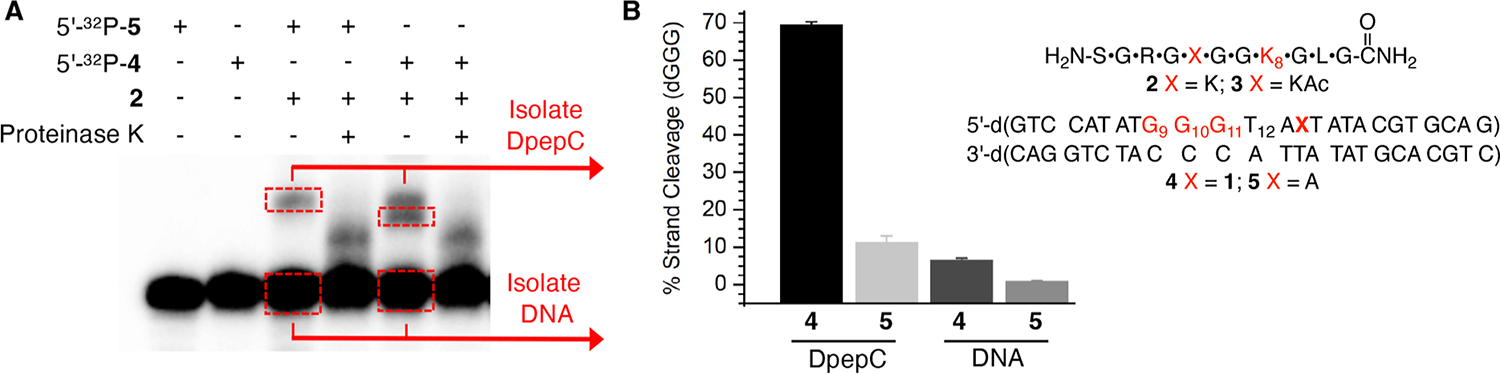
DNA–peptide cross-link formation. (A) Representative 20% denaturing PAGE showing DpepC formation between 5′-32P-4 or 5′-32P-5 and H4 1–11 peptide (2) at pH 7.5 upon photolysis. (B) Percent of piperidine cleavage at the dGGG sequence in isolated DpepC or isolated uncross-linked DNA plotted as the ave. ± std. dev of 3 independent reactions.
In addition to DpepC, water trapping of dG•+ was detected upon piperidine treatment of isolated DNA from photolyzed 5′-32P-4 that did not participate in DpepC formation (Figure S1). After accounting for background alkaline lability in irradiated DNA not containing 1 (5′-32P-5) and DpepC yield, the percent yield of alkali-labile lesions attributable to water trapping of holes resulting from dA• was calculated to be 5.3 ± 0.6% (Figure 1B).59 Together, these data indicate that peptide (2) trapping (back-ground-corrected DpepC yield: 4.0 ± 0.4%) effectively competes with water for hole trapping in photolyzed 5′-32P-4.
LC–MS/MS was used to identify the amino acid(s) involved in the cross-link and the cross-link structure in DpepC (Figures 2, S3). A less polar Lys5 acetylated form of H4 1–11 (H4 1–11-K5Ac, 3) was used for these experiments. The background-corrected yield of DpepCs from the photolyzed 25 bp duplex containing 1 (4) decreased from 4.0 ± 0.4 to 1.5 ± 0.1% upon acetylation of K5 (3) (Figure S2). The faster migrating DpepC band in 5′-32P-4 was isolated, and the DNA was digested to nucleosides prior to LC–MS/MS analysis. Previous studies showed that lysine adds to the C8 (C8-Lys-dG, Scheme 2) in the presence of one electron oxidant such as benzophenone or riboflavin.39–41 The initially formed adducts readily undergo further oxidation and water addition to form C8-Lys-dG-ox under oxidative conditions such as aerobic photolysis (Scheme 2). Identification of the C8-Lys-dG- and C8-Lys-dG-ox-modified peptides was challenging because possible in-source fragmentation to the respective nucleobase forms prior to MS1 measurement.60 Deglycosylation and further adduct decomposition may also occur during higher-energy collisional dissociation for MS2 fragment ion generation.61,62 Consequently, peptides containing such modifications can elude conventional MS analysis engines. To avoid this challenge, we used the NuXL workflow developed by the Urlaub group with search parameters adapted to our experiment.61,63 To account for glycosidic bond fragmentation, the method is designed to search for the exact mass of the precursors (e.g., H4 1–11-K5Ac (3) cross-linked to mononucleotides, nucleosides, or nucleobases) in the MS1 spectra. In addition, the masses of the associated fragment ions are also searched for in the MS2 spectra. Extracted ion chromatograms as well as MS1 and MS2 spectra support modification at the amino terminus and ε-amine of the internal lysine (Figure S3). Tentative assignments were made on the basis of the observation of peptides containing dG modifications C8-K-dG and C8-K-dG-Ox in the base or nucleoside form (Scheme 2).
Figure 2.
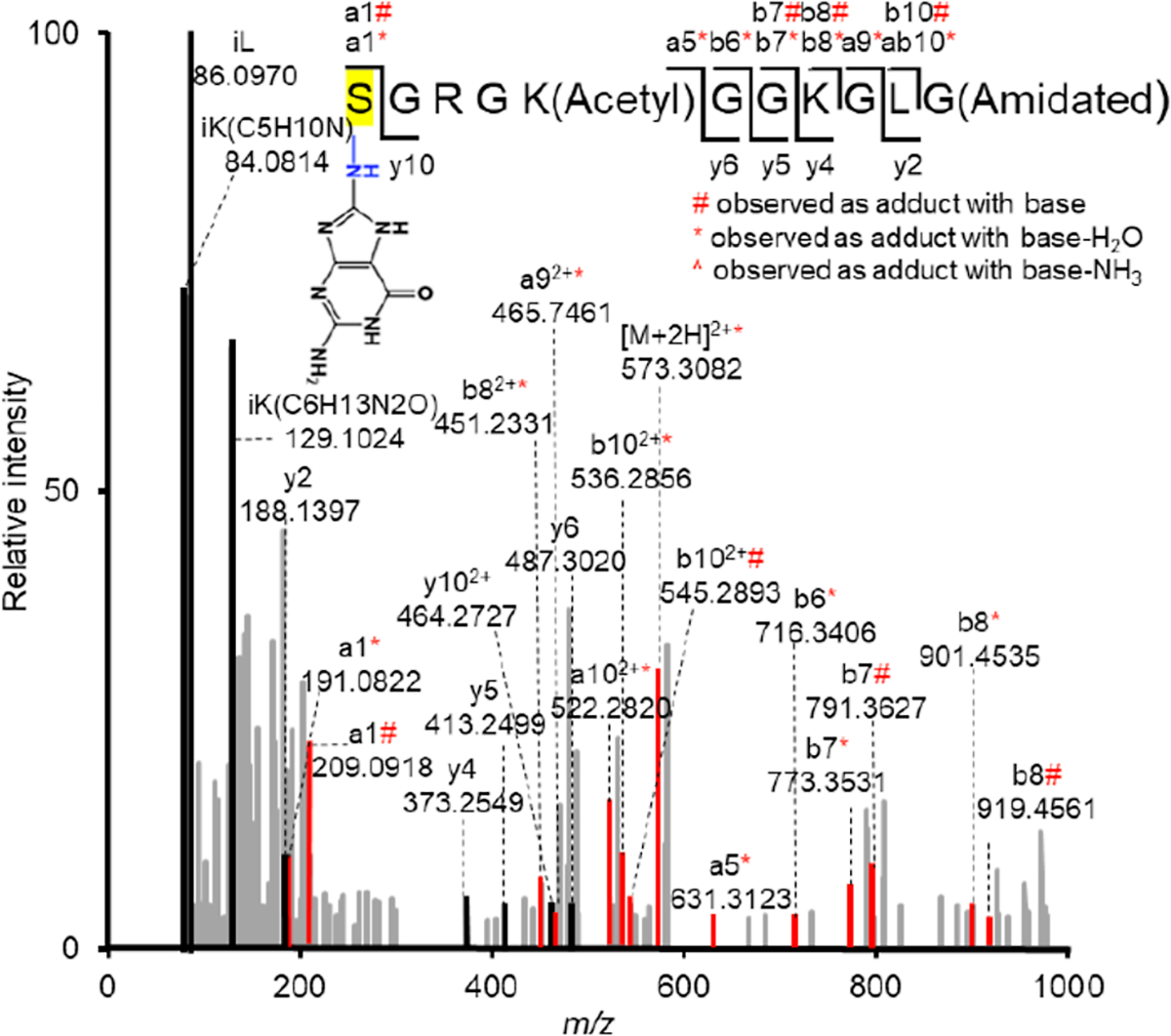
MS/MS spectrum of H4 1–11-K5-Ac (M = 1013.5720) containing C8-Lys-dG (M of base = 149.0338) obtained from photolysis of 3 and 4 detected by LC–MS/MS (exp. m/z = 582.3145, calcd m/z = 582.3107). Color code: Ions containing modification, red; unmodified ion, black; ion from other peptide(s), gray.
DPC Formation via Charge Transfer in NCPs.
NCPs in which the DNA sequence is based upon the Widom 601 strong positioning sequence were prepared as previously described (Figures S4, S6 and S7).52 Holes were introduced in 3 regions of the core particle by substituting an identical sequence of DNA (“cassette”) for an equal length of DNA in the Widom sequence (Figure 3). Each cassette contains two dG5 runs separated by a dCGC sequence that serve as sinks for the holes and a dA• radical precursor (1) separated from a dG5 run by a single dA. In two regions, 1 was placed separately in the dG-rich strand near the 5′- or 3′-terminus of each cassette to probe hole migration from different directions. The cassette in NCP-26 is near the terminus where DNA unwrapping is relatively facile.64 Hole migration is more efficient in the nucleosomal DNA region encompassed by NCP-35 and NCP-51 than in NCP-55 and NCP-71 where the dG-rich sequence is proximal to H3-Y41, which quenches hole migration in NCPs.52 Control sequences where 1 was replaced by dA (NCP-C26, NCP-C35,51, and NCP-C55,71) were generated to account for any background DPC and/or other DNA damage under the photolysis conditions employed to achieve hole injection. In addition, yields were normalized based on the extent of hole injection, which was determined by exploiting the formation of restriction sites (EcoR1 and Mse1) upon conversion of 1 to dA (Figure S8).
Figure 3.
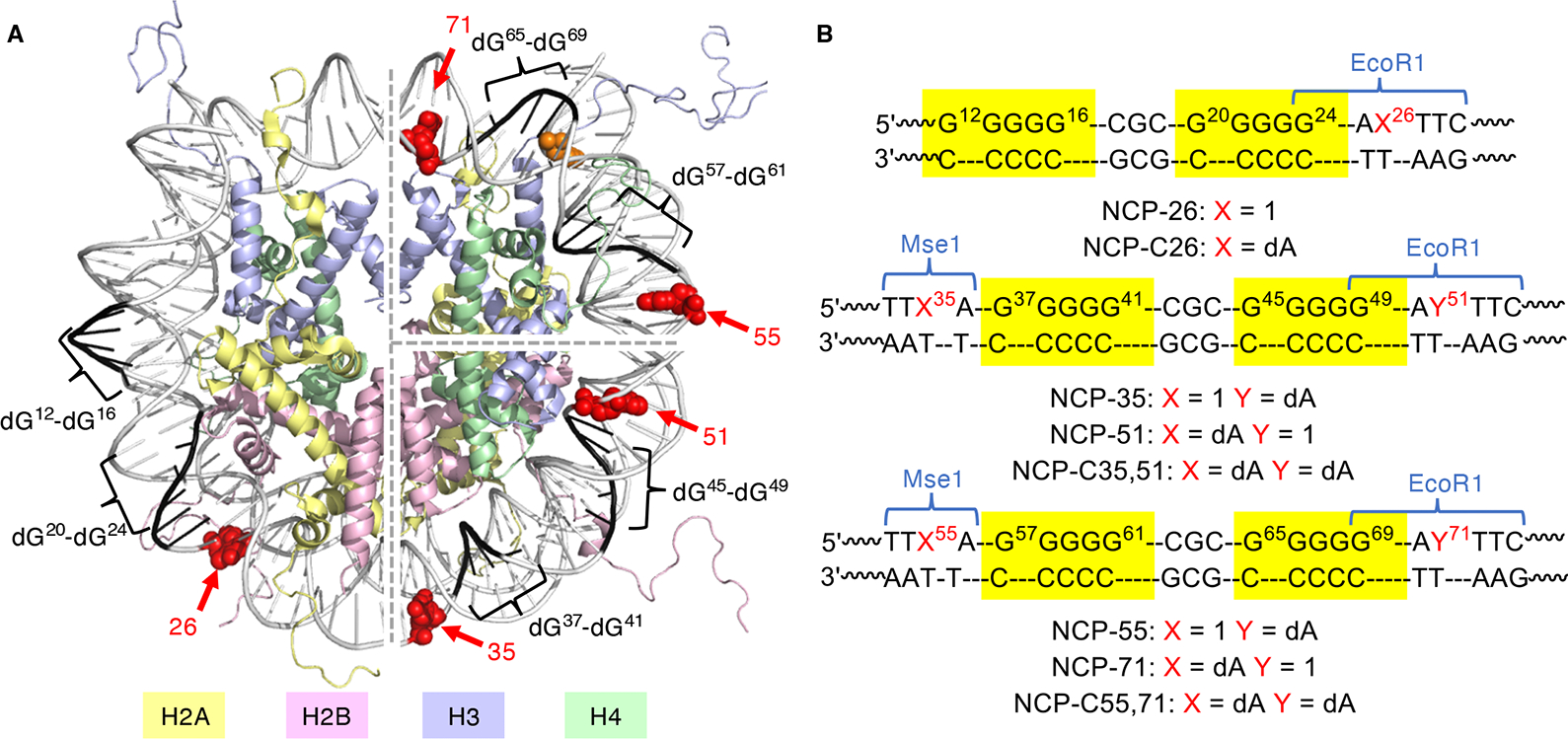
Nucleosome core particles. (A) Core particle structure highlighting dG5 tracts, positions at which 1 is incorporated and H3-Y41. The structure was generated by superimposing two NCP structures (pdb: 1kx5 and 3lz0). (B) Nucleosomal DNA sequences in the region of dG5 tracts. See Figure S4A for the entire nucleosomal DNA sequences.
When dA• was produced at position 35 (NCP-35) at pH 7.5 (Figure 3B), the DPC yield was 9.0 ± 0.7% (Table 1, Figure 4A). The NCP structure is essential for DPC formation as these products were not observed following irradiation when free 5′-32P-DNA was merely mixed with an octamer in the same ratios and concentrations as present in NCP samples (Figure 4A). Photolysis of NCP-51, which introduces the hole on the opposite side of the cassette, generated DPC yields that were within the experimental error of those from NCP-35 (Table 1 and Figure S9A). In contrast to the DNA–peptide model study, the DPCs formed in the control (NCP-C35,51) and those from the NCPs containing the dA• precursor (1) are inseparable by SDS gel electrophoresis (Figure S9A). Consequently, piperidine-induced cleavage of the isolated DPCs from NCP-35 or NCP-51 reflects the reactivity of the DPCs formed via the background reaction and those from holes injected via 1. Denaturing PAGE analysis of piperidine-treated DPCs from NCP-C35,51 resulted in 21 ± 2% strand scission (Figure S10A). Piperidine-induced cleavage of DPCs from NCP-35 and NCP-51 within the region of the two dG5 runs was considerably higher (59 ± 2 and 61 ± 4%, respectively) (Figure 4B). The overall piperidine-induced cleavage correlates well with the DPC yields from these NCPs (NCP-35, NCP-51, and NCP-C35,51) and the different piperidine labilities of the two types of DPCs observed in the model study. Moreover, cleavage was observed at the same set of dGs in NCP-35 and NCP-51 DPCs (dG45-dG47 and dG37-dG38, Figure 4B), indicating that cross-linking sites on DNA are independent of the direction of hole migration. Examination of nucleosome crystal structures and molecular dynamics (MD) simulations indicates that major groove accessibility within the region(s) of hole localization to histone tails contributes significantly to DPC formation (Figure 5). The major cross-linking sites (dG45-dG47) in NCP-35 and NCP-51 are in a rotational orientation in which the major groove is oriented outward from the octamer core and in close proximity to the histone H2B and H4 N-terminal tails (Figure 5E). Previous MDs also support the interaction between H2B and H4 with DNA in this region.47 When considering only the histone core and the DNA as solute (Figure 5A,C), the simulation described here reveals that dG39, dG45, dG46 and dG48 have the largest solvent-accessible surface area (SASA). In addition, these guanines and dG47 experience a larger SASA decrease (8–13 Å2) when the histone tails are included in the solute. These calculations corroborate the greater accessibility of the major cross-linking sites dG45-dG47 to the histone tails.
Table 1.
Yields of Alkali-Labile Lesions and DPCs in Photolyzed NCPs
| substrate | DPC (%)a | H2O trapping (%)a |
|---|---|---|
| NCP-26 | 3.7 ± 0.6 | 8.8 ± 1.4 |
| NCP-35 | 9.0 ± 0.7 | 6.4 ± 0.7 |
| NCP-51 | 10.4 ± 1.0 | 7.1 ± 1.4 |
| NCP-55 | 2.7 ± 0.6 | 0.6 ± 1.4 |
| NCP-71 | N.D. | N.D. |
| H3-Y41F NCP-55 | 7.7 ± 0.9 | 6.0 ± 0.7 |
| H3-Y41F NCP-71 | 6.5 ± 1.2 | 3.8 ± 1.3 |
Yields are background-corrected, normalized for extent hole injection, and expressed as ave. ± std. dev. of 3 samples. N.D. = not detectable.
Figure 4.

DPC formation in NCPs. (A) DPC formation in NCP-35. Denaturing PAGE analysis of piperidine-treated isolated DPCs from (B) NCP-35 and NCP-51, (C) NCP-55, NCP-55, H3-Y41F NCP-71 and H3-Y41F NCP-71, and (D) NCP-26.
Figure 5.

Solvent accessibilities and histone tail interactions at dG5 tracts in NCP-C35,51 and NCP-C55,71. (A,B) SASA of dG5 tracts in (A) NCP-C35,51 and (B) NCP-C55,71. The guanines are colored according to their SASA from white (25 Å) to bright blue (50 Å). (C,D) SASA of guanines within dG5 tracts of (C) NCP-C35,51 and (D) NCP-C55,71 with (blue bar) or without (blue and striped bar) histone tails. (E) dG5 tracts in NCP-C35,51 with 40 conformations of H4 (light green) and H2B (pink) tails from MD simulations. (F) dG tracts in NCP-C51,71 with 40 conformations of H3 (light blue) and H4 (light green) tails from MD simulations. dG5 tracts are highlighted in black. Major cross-linked dG sites within those tracts are highlighted in red. The structures were generated from MD simulations that utilized pdb: 3lz0 as a starting point.
Compared to NCP-35 and NCP-51, the DPC yields from NCP-55 and NCP-71 are much lower, with the latter within an experimental error of zero (Table 1 and Figure S9B). This is consistent with efficient hole reduction in this region by H3-Y41.52 Substituting phenylalanine (H3-Y41F) for tyrosine 41 to create H3-Y41F NCP-55 and H3-Y41F NCP-71 increased DPC yields to 7.7 ± 0.9 and 6.5 ± 1.2%, respectively (Table 1 and Figure S9B). Piperidine cleavage of isolated DPCs showed that the increased cleavage was concentrated within the dG5 tracts (Figure 4C). We attribute the increased level of DPC formation in H3-Y41F NCP-55 and H3-Y41F NCP-71 to charge transfer following the generation of dA•+. In both NCPs, the vast majority of cross-links formed at positions dG57-dG59 and dG65-dG67 (Figure 4C). The greater quantity of cross-links at dG57-dG59 are likely due to extensive interactions with the H4 tail (Figure 5F). In terms of solvent accessibility, the two dG5 tracts are equivalent (Figure 5B,D), with slightly larger accessibility for dG57. Overall, the impact of the interaction with the histone tails is less important than that for NCP-C35,51 (Figure 5C,D). This is also indicated by fewer observed contacts between the tails and guanines in positions 55 to 71 (see the Supporting Information: Movies,seq_35,51 and seq_55,71). The difference in the extent of histone tail interactions is reflected in the DPC yields within the corresponding NCPs (Table 1).
Piperidine treatment of the DPCs from NCP-55 and H3-Y41F NCP-55 also yields minor strand breaks at T53, T54, and the nucleotide at which dA• is generated (Figure 4C). The DPCs formed within the 5′-TT1 local sequence are independent of the charge-transfer process and explain the low yield of DPCs observed in NCP-55. The DPCs within 5′-TT1 are proposed to result in part from formation of 5-formyl-2′-deoxyuridine (5fU) at position 54 via hydrogen atom abstraction from the thymine methyl group by dA• (Scheme 4A), which is a facile process.57 The peroxyl radical derived from O2 trapping of T• (Tp•) yields 5fU. Based upon the reaction of Tp• and other pyrimidine nucleobase peroxyl radicals, we speculate that this peroxyl radical can react with the 5′-adjacent thymidine (T53) to produce a variety of damaged thymidines, including 5fU.57,65–68 Like 5fC, 5fU forms Schiff bases with histone lysines that are stabilized via hydride reduction and further characterized by LC–MS/MS (Scheme 4B and Figure S11).16,17,69 Two peptides consistent with the tentative assignment of stabilized DPC (Scheme 4B) were detected in the extracted ion chromatogram and MS1 spectra. While we were only able to obtain an MS2 spectrum for one of these peptides, its fragmentation pattern was also consistent with the tentative assignment.
Scheme 4.
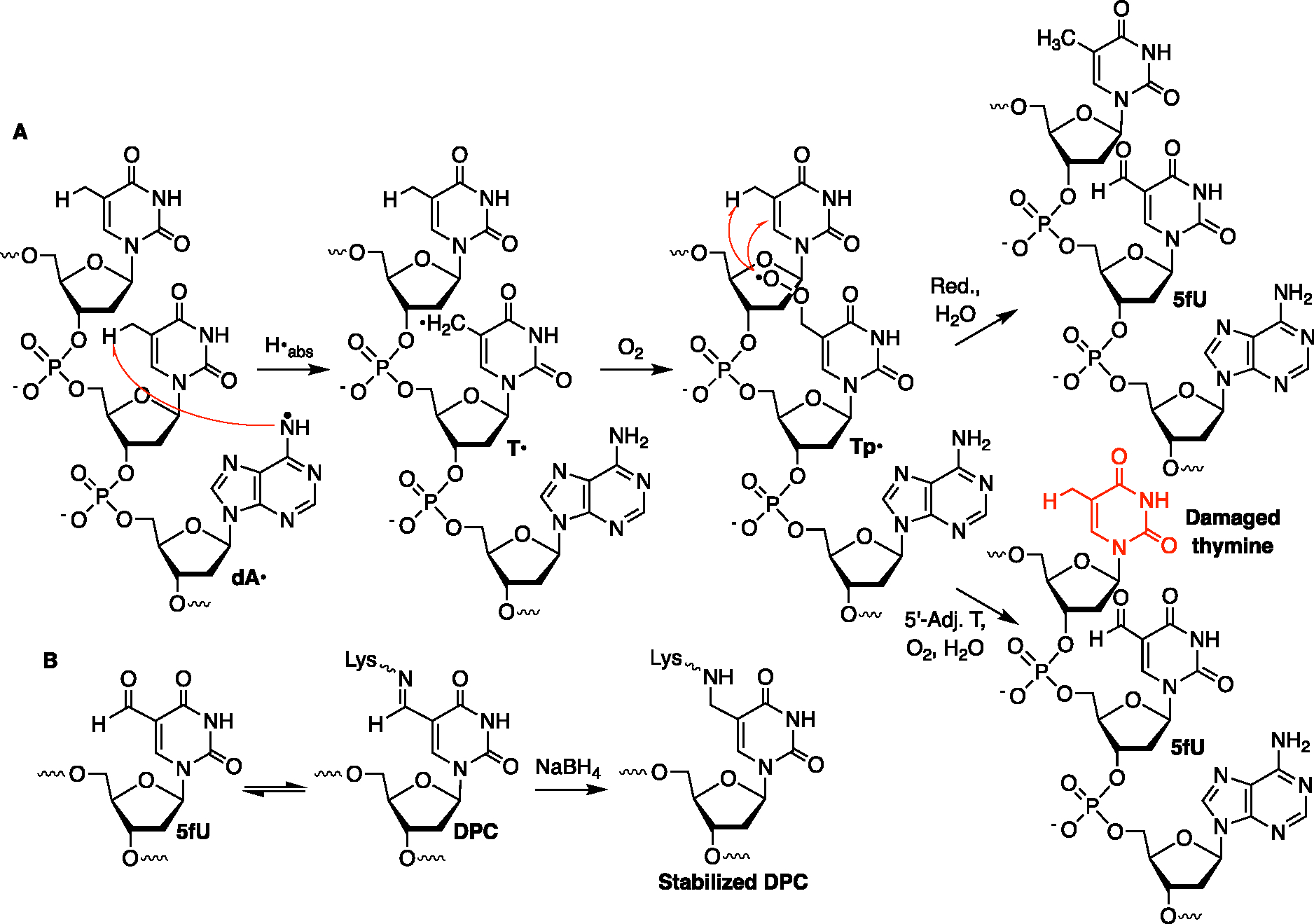
DPC and Tandem Lesion Formation dA• Generation in 5′-dTT1 Sequences
DPCs were formed in low yield (3.7 ± 0.6%) upon photolysis of NCP-26, which contains dG tracts at the entry/exit where DNA is dynamic and unwraps from the histone octamer (Table 1 and Figure S9C).64 In contrast to the other two regions (35/51, 55/71), piperidine-induced cleavage of DPCs from NCP-26 indicated that cross-linking was distributed more evenly within the two dG5 tracts of the cassette (Figure 4D). This relatively diffuse cross-linking pattern is consistent with access to a broader distribution of dGs due to the dynamics of DNA unwrapping. Interestingly, cross-link sites extended to a more distal hole sink sequence (dG36-dG38) that is 10 bp away from the hole transfer initiation site (126). This demonstrates the ability to form DPCs via longer-range hole migration in NCPs.
DPC Formation Results Directly from Hole Trapping in NCPs.
Charge-transfer-induced DPCs were anticipated to result from nucleophilic addition to C8 of dG•+. Due to the prolonged photolysis in NCPs (14 h), we also considered the possibility that DPCs were a secondary product, resulting from photooxidation of 8-oxodGuo that is produced from water trapping of dG•+.39,40 Consequently, the DPC yield and the extent of hole injection in NCP-51 were measured as a function of irradiation time (Figure S12AB). The latter was determined by exploiting formation of an EcoR1 restriction site upon conversion of 151 to dA. If DPCs are a secondary oxidation product, one would expect their formation to lag hole injection. However, the DPC yield and conversion of 1 to EcoR1 susceptible material increase linearly with respect to time. Furthermore, the ratio of DPC formed: photoconverted 1 remains constant over the course of the photolysis (Figure S12C), consistent with cross-links resulting directly from hole trapping in the nucleosomal DNA.
Water Trapping of DNA Holes in NCPs.
In addition to DPCs, the water trapping of holes was detected in the form of piperidine labile lesions within the DNA of isolated NCPs that did not participate in cross-link formation. These products were detected in photolyzed NCP-26, NCP-35, NCP-51, H3-Y41F NCP-55 and H3-Y41F NCP-71 (Figure S10A,C) but not NCP-55 and NCP-71 where H3-Y41 quenches hole transfer (Figure S9B).52 In contrast to the oligonucleotide–peptide model system, alkali-labile lesions are formed in lower yield than DPCs in NCP-35/51, H3-Y41F NCP-55/71 (Table 1). The more efficient DPC formation in these NCPs may be ascribed to the abundance of nucleophilic amino acids on histone proteins and their proximity to DNA holes. In NCP-26, DNA unwrapping decreases interactions with the histone core but does not adversely affect the access of water to dG•+. In contrast to the positional preferences for DPC formation, water trapping is unselective toward specific dGs within the two dG5 tracts, regardless of the direction of hole migration in all of the NCPs (Figure S10A–C). Together, these data indicate that protein trapping of holes in NCPs is largely dictated by the accessibility of histone proteins rather than the preferred sites of hole localization.
Analysis of Histone and Amino Acid Participation in DPCs.
The participation of individual histone proteins in DPC formation within NCP-51 was quantified using fluorescent staining following DNA digestion and protein separation by Tricine-SDS PAGE (Figure S13). Absolute quantification of individual histones was carried out by establishing standard curves using the corresponding purified proteins. After accounting for the contributions of individual histone proteins due to background DPCs using NCP-C35,51, H2B and H4 were determined to be the most significant contributors to DPCs resulting from hole transfer in NCP-51 (Figure 6). These two proteins contributed roughly equally and together accounted for almost 70% of the DPCs. The predominant involvement of H2B and H4 is supported by MDs analyses, which suggest that the N-terminal tail lysines of H2B and H4 bind at dG45-dG47 where the majority of DPCs form in NCP-51 (Figure 7). In MD simulations, lysines interact with the guanines in the major groove and to a lesser extent with the 2′-deoxyguanosine phosphate groups (Figures 7 and S15). For dG37-dG41 and the H2B tail, we count contacts mostly between the N-terminal alanine and lysines closest to the amino terminus (lysines located between residues 2and 12 of H2B). dG45-dG49 interact more with the lysines located from position 17 to 25 of H2B. This region of the cassette is also in contact with the amino terminus and lysines 5, 8, and 12 of H4. Interactions at dG45-dG49 involve a larger assortment of lysines and are generally more frequent and shorter lived than the interactions between nucleophilic residues and dG37-dG41 (Figure S14A). Furthermore, some lysines show similar interaction profiles with adjacent guanine pairs (e.g., H2B-K17 with dG47 and dG48; Figures 7A, S14A and S15A). Examining the trajectories indicates that the lysine ammonium group is positioned between the two guanines, interacting simultaneously with O6 or N7 from each nucleobase. Close lysine amino group contacts with O6 and N7 (less than 3 Å) are common, representing 5.9 and 4.3% of the total amount of counted guanine contacts, respectively, in the region between positions 35 and 51 (5.7 and 2.1% in the region between positions 55 and 71). Such contacts require only a small shift in lysine position to trigger DPC formation at C8 after DNA oxidation. The proximity of C8 to the phosphate group where interactions occur may also promote DPC formation (Figure S15B). A comparable interaction has been detected in MD simulations describing binding between RNA containing 8-oxo-7,8-dihydroguanosine, and protein.70
Figure 6.
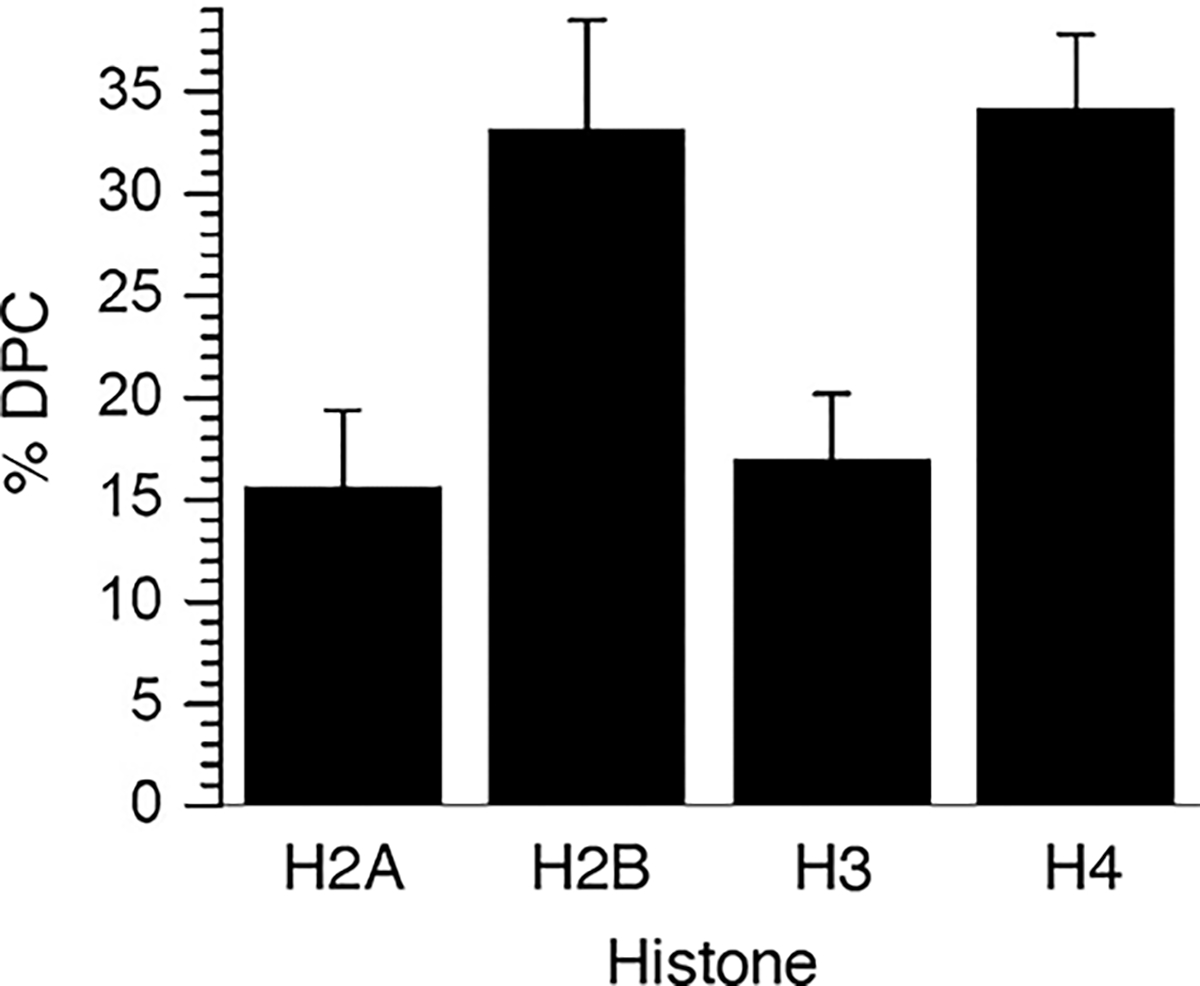
Individual histone protein contribution to DPCs formed by hole trapping upon the photolysis of WT NCP-51.
Figure 7.
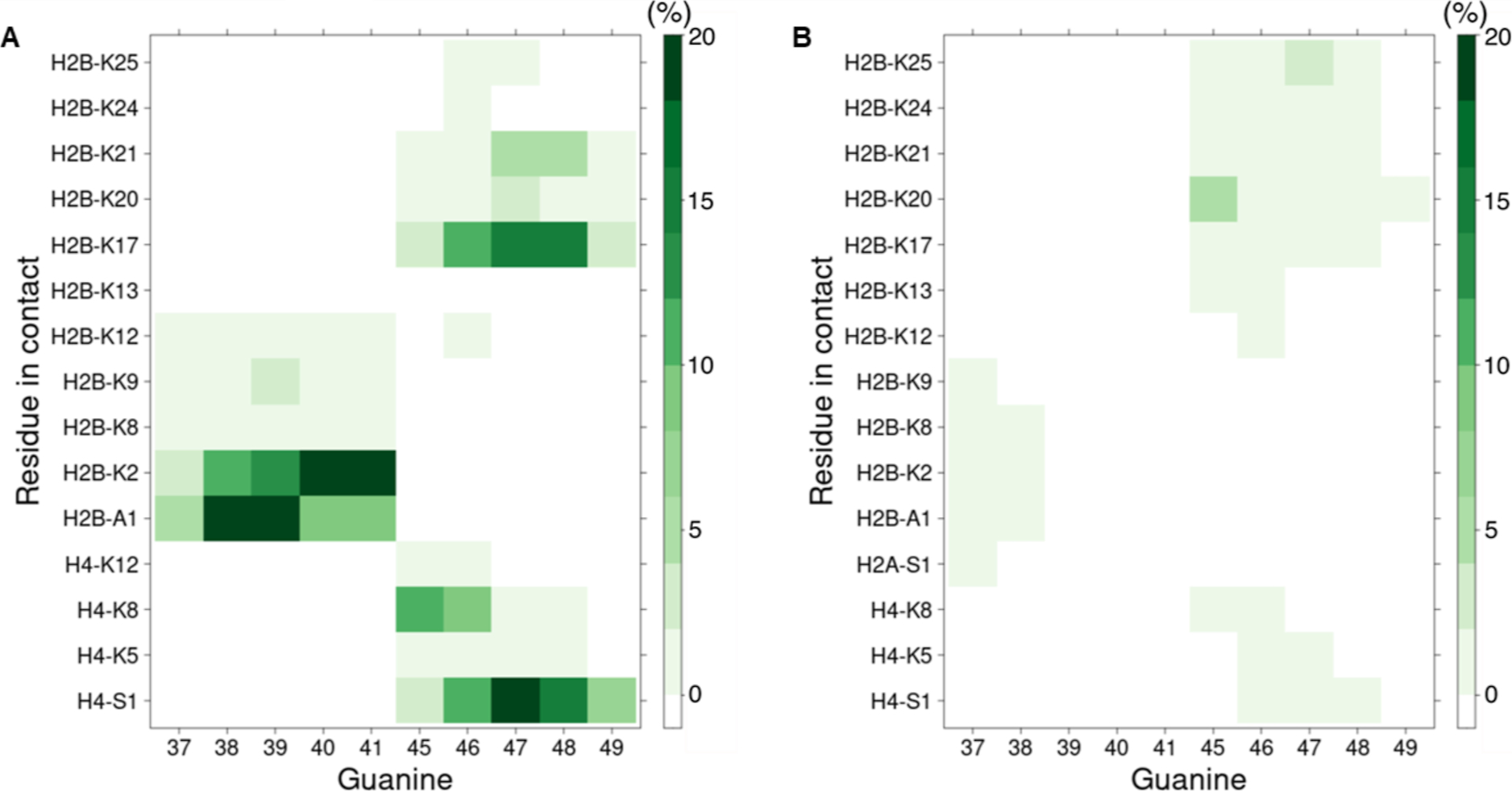
Contact map between dG5 tracts and histone tails in NCP-C35,51. (A) Contact map between ammonium group of lysines or of N-terminal residues and major groove exposed heavy atoms of guanines. (B) Contact map between the ammonium group of lysines or N-terminal residues and phosphate groups. The heat map extends from white (no contact) to dark green (at least one contact is observed for more than 20% of the trajectory frames).
The remaining 30% of DPCs from NCP-51 were composed of approximately equal amounts of H2A and H3 (Figure 6). Previous simulations indicate that H3 compensates for its less frequent interactions with dG45-dG47 by exhibiting the longest residence time of the N-terminal histone tails and that the minor cross-link site in NCP-51, dG37-dG39, interacts with H2A and H2B.47 However, no interactions between the lysines within the N-terminal tails of H2A and H3 are detected in the current simulations, except for a few contacts between H2A-S1 and dG37 (Figure 7B). We cannot rule out the possibility that the minor DPCs detected involving H2A and H3 result from incomplete background subtraction and/or are due to sliding of the modified sequence of the 601 DNA within the NCP.
Inferential support for the significant contribution of N-terminal histone tails to DPC formation was gleaned by comparing product formation in NCP-51 in 100 mM NaCl (Table 1) to that in 400 mM NaCl (DPC, 4.2 ± 1.0%; H2O trapping, 5.0 ± 0.4%; Figure S14). Higher ionic strength weakens interactions between histone tails and DNA.71 DPC and water trapping yields declined in NCP-51 at higher salt concentrations. Decreased H2O trapping in the NCP is qualitatively consistent with a previous report on hole transfer in free DNA and a nucleosome.49 Importantly, the almost 3-fold decrease in DPC yield was considerably greater than that of H2O trapping, even after accounting for ~20% of the NCP decomposition at 400 mM NaCl.
To identify which amino acids are responsible for cross-linking in NCP-51 following charge transfer, the isolated DPCs from 4 independently prepared samples were subjected to trypsin digestion, DNA digestion, followed by LC–MS/MS analysis. Two samples were subjected to in-gel propionylation prior to treatment with trypsin. Background DPCs resulting from photolysis were accounted for and subtracted during the MS/MS analysis. Specifically, MS2 spectra from DPC (NCP-51) were removed if the same precursor ion appears in DPC (NCP-C31,51) with comparable retention time and intensity.61 C8-K-dG and C8-K-dG-ox (Scheme 2) were tentatively identified from all 4 histones (Figure 8), consistent with the histone quantification experiment. Modifications were tentatively assigned at 13 lysines based on as many as 6 MS2 spectra for a single site.59 All of the modifications were on lysine residues within the corresponding N-terminal tail. Modifications were detected throughout the N-terminal tails of H2B and H4. In contrast, modifications were clustered closer to the respective N-termini within the H2A and H3 tails. This is consistent with tail flexibility enabling interactions with DNA at positions that are farther from where the histone tails exit the core. In MD simulations, H2B adopts conformations, which can be characterized by the folding of the tail and the position(s) of DNA interaction(s) (Figure S17A). When contacts are observed with the dG37-dG41 tract, a flexible loop is formed by the tail when it exits the core with a turn involving residues 14 to 18, and only the N-terminal part of the tail can explore the major groove. In another typical conformation, the region closest to the N-terminus is stacked on the minor groove at position 51–55, and the middle portion of the tail (residues 12 to 25) interacts with the dG45-dG49 major groove. Contacts with H4 are only present when the tail is extended and are limited to residues 1 to 13 (Figure S17B).
Figure 8.

Correspondence between modified residues detected by LC–MS/MS of NCP-51 and residues interacting with dG5 tracts in MD simulations of NCP-C35,51. (A) Two perspectives of the NCP. Modified residues detected only by LC–MS/MS (red), interacting residues detected only computationally (cyan), or residues detected both computationally and by LC–MS/MS (dark violet). Asterisks (*) indicate that MS2 spectra do not contain enough fragment ions to distinguish modification at this lysine from that at the adjacent lysine. The structures were generated from an MD simulation snapshot based upon pdb: 3lz0. (B) Histone N-terminal tail sequences [same color assignment as in (A)].
Results from H3-Y41F NCP-71 show a similar pattern, where almost all lysines on H3 and H4 tails contribute to cross-linking by reacting with proximal, major groove-accessible holes at dG57-dG61 and dG65-dG69 (Figure S18). For the more distal H2B tail, modification is detected only at the N-terminal amine and a few neighboring lysines (K8, K12, K13). Fewer lysine—DNA contacts are identified in the MD simulations of NCP-C55,71 than of NCP-C35,51 (Figure S19). Again, this is in qualitative agreement with the lower DPC yield measured in the former NCP (Table 1). H4 tail interactions with this region are detected, mostly with dG57. However, most of the time, the H4 tail is solvent-exposed, extending toward nucleobases 45–49 or interacting within the minor groove of dG57-dG61 and dG65-dG69 (see the Supporting Information: Movies,seq_35,51 and seq_55,71 and Figure S17B). The H4 lysine interaction profile (Figures S14B and S19C,D) is in agreement with the fuzzy interaction previously described.72 Furthermore, dG65-dG69 have contact only with H3 lysines, with H4 being too short to reach these positions. Finally, after considering the contacts between lysines and phosphate groups, the overall interaction profile for NCP-C55,71 from MD simulations is in good agreement with the LC–MS/MS data (Figure S18).
MS identification of modified lysines was complemented by experiments in which the effects of histone mutations on DPC yields in NCPs were determined. As described above, LC–MS/MS determined that the N-amino terminus of the H4 tail peptide contributes to DpepC formation. However, uncovering cross-linking to DNA by N-amino termini in NCPs is challenging due to proximal Arg residues that give rise to short peptides upon trypsin digestion that elude LC–MS/MS detection. Consequently, inferential evidence for hole trapping by the N-amino termini was sought by constructing N-terminal capped histone proteins that were prepared through N-terminal serine mutation and subsequent thiazolidone formation (Figure S16).23 Capping histone H2B, and then additionally H4 and H2A, gradually reduced the DPC yield in NCP 51 from 10.4 ± 1.0 to 7.5 ± 0.5% (Table 2). Although DNA cross-linking was not detected by LC–MS/MS to any of these N-terminal amines, these experiments, as well as the contact profile observed in MD simulations (Figure 7), suggest that they contribute to DPC formation.
Table 2.
Effect of Histone Mutations on the DPC Yields in NCP-51
| NCP-51 | DPC (%)a |
|---|---|
| WT | 10.4 ± 1.0 |
| H2B N-cap | 9.0 ± 0.6 |
| H2B N-cap, H2A N-cap, H4 N-cap | 7.5 ± 0.5 |
| H2B Del1–22, K24,25,28,31A, N-cap, H2A N-cap, H4 N-cap | 5.1 ± 0.5 |
Yields are background-subtracted and expressed as ave. ± std. dev. of 3 samples.
Employing a truncated (Del 1–22), N-terminal capped form of histone H2B in which the lysines within the histone binding region were also mutated (K24,25,28,31A) reduced the DPC yield to 5.1 ± 0.5%, consistent with the involvement of lysines on the H2B tail detected in the LC–MS/MS experiments. While DPC yields in NCPs reconstituted with mutant histones correlate with the presence of nucleophiles in N-terminal histone tails, we cannot draw quantitative conclusions concerning contributions of individual amines due to the ability of histone tails to compensate for changes in the binding of other mutated proteins.73
GGG Is Sufficient For DPC Formation in NCPs upon Charge Transfer.
The 13-nucleotide sequence of the dG-rich cassettes used above is only expected to be found ~100 times in the human genome. To explore the generality of DPC formation in NCPs, we prepared a series of NCPs (Figure 9) containing a dG3 tract [5′-d(GGGA1)]. (The NCPs are named according to the position of 1.) dG3 is the most readily oxidized native trinucleotide sequence and is statistically expected to be present once every 32 base pairs in DNA, almost 30 million times within the human genome.32 We initially examined hole formation from this sequence when the dG3 tract was positioned at dG45-dG47, which was the favored site for DPC formation in NCP-35,51. DPC was observed at these sites and was favored at dG45-dG46 over dG47 (Figure S10E). The selectivity is perhaps fortuitously consistent with the generally preferred hole localization at the 5′- and middle-nucleotides within a dG3 sequence.33 The DPC and H2O trapping product yields (Table 3) were on par with those observed from NCP-35,51 (Table 1), suggesting that a dG3 tract is sufficient for generating a considerable amount of DPCs in NCPs. Placing a dG3 tract opposite dG45-dG47 (NCP-248) gave rise to a comparable DPC yield, corroborating the hypothesis that DPC formation was strongly dependent on major groove accessibility to N-terminal histone tails. NCP-191 contains dG3 at dG187-dG189, a site that is symmetric with respect to dG45-dG47 in the NCP and interacts with the N-terminal tails of the other copy of H2B and H4.47 The DPC yield in NCP-191 is similar to those in NCP-49 and NCP-248. This is consistent with the convergence on tail–DNA binding sites observed from the symmetrical nucleosome structure. The yield decreases in NCP-183 when dG3 is located at dG179-dG181, suggesting that the additional contact with the H2A tail, which contains fewer lysines, does not compensate for decreased interaction with H2B and H4 tails. The DPC yield from NCP-223, in which the dG3 tract major groove is positioned outward at the dyad axis, was considerably lower than those at other locations. The lower DPC yield in NCP-223 is consistent with the calculation that histone tails make relatively infrequent contacts at the dyad axis.47 Overall, these data suggest that pervasive dG3 tracts are sufficient for DPC formation and that the absolute yield is dependent on their rotational and translation position.
Figure 9.

Nucleosome core particles containing dG3 tracts. The structure was generated by superimposing two NCP structures (pdb: 1kx5 and 3lz0). The local DNA sequences can be found in the Supporting Information.
Table 3.
DPC Formation and H2O Trapping in NCPs Containing dG3 Tracts
| NCP | DPC (%)a | H2O trapping (%)a |
|---|---|---|
| NCP-223 | 3.0 ± 0.6 | 3.3 ± 0.5 |
| NCP-49 | 8.3 ± 1.3 | 9.4 ± 0.7 |
| NCP-248 | 9.6 ± 1.0 | 10.3 ± 0.8 |
| NCP-191 | 9.7 ± 1.1 | 7.5 ± 0.8 |
| NCP-183 | 6.7 ± 0.4 | 6.7 ± 0.3 |
Yields are background-subtracted and expressed as ave. ± std. dev. of 3 samples.
SUMMARY
DNA–protein cross-links are a biologically important family of DNA lesions produced as a result of ionizing radiation and other modes for damaging DNA via single-electron oxidation. The physiological significance of DPCs in human cells has been bolstered by the discovery and investigation of proteases, such as Spartan. Spartan is implicated in replication-dependent and -independent DPC repair, and deficiency of this protein can result in premature aging and early onset liver cancer.5,6,8 Although thorough studies of DPC formation between peptides and individual proteins have been reported, histone protein–DNA interactions in chromatin are distinct. This is partly due to the lack of direct interactions between nucleobases and the histone proteins that enable the octameric core to bind DNA without sequence specificity. However, only a small number of studies concerning DPC formation have been carried out on NCPs.44,49 To help elucidate DPC formation induced by direct ionization of DNA in chromatin, we synthesized nucleosome core particles from the bottom-up containing a photolabile nucleotide that yields a radical cation (“hole”) at a defined site within the nucleosomal DNA. The NCPs were constructed so as to contain dG-rich sequences to favor hole transfer and localization within specific regions of nucleosomal DNA.
DPC formation strongly depended on translational and rotational positions within the NCP. For instance, the DPC yield in the entry/exit region was significantly lower than in regions in which DNA was more tightly bound to the octameric histone core. DPC formation was also compromised when holes were introduced near the NCP dyad axis, which does not make frequent contacts with histone tails.47 Within dG-tracts, DPC formation was favored at positions where the major groove faced away from the octameric core and is more accessible to lysine-rich histone tails. In MD simulations, lysines in histone tails can transiently form hydrogen bonds with the O6 and N7 atoms of one or two guanines. The histone tail lysines also interact within the minor groove or with the DNA backbone, albeit less frequently (Figure S19C,D). The site preference for DPC formation agrees with computational studies that indicated that dG•+ reaction with nucleophiles is kinetically favored at the C8-position, which lies in the major groove than at either the C5- or C4-positions. The latter is accessible from the minor groove.41,42 dG•+ reaction with the lysine-rich histone tails is modulated by the octameric core in a similar way as hydroxyl radical reactivity, albeit with the added requirement of histone tail proximity.74 Hydroxyl radical-mediated strand scission results predominantly from reaction in the minor groove. The periodicity of nucleosomal DNA cleavage arises from the minor groove accessibility to the freely diffusible hydroxyl radical in solution. The solvent exposed minor groove is, in turn, determined by DNA wrapping around the octameric histone core.
Quantitative analysis in one nucleosome region confirmed that the majority of DPCs resulted from reaction with histone proteins whose N-terminal tails are calculated to most frequently contact the DNA. A variety of direct and inferential methods were used to identify the portions and specific amino acids of the histone proteins that react with the DNA. An ~3-fold decrease in DPC yield when the NaCl concentration increased from 100 to 400 mM is attributed to reduced histone tail affinity for DNA at higher salt concentrations. Cross-linking by N-terminal tail lysine residues was directly detected via LC–MS/MS analysis of trypsin-digested DPCs and was generally corroborated by MD simulations that identified lysine–DNA interactions. This agreement between the experimental and computational results validates our combined approach and supports the molecular insight given by MD simulation analysis. No modified amino acids were detected within the globular domain of the histone octameric core. In addition, cross-links with the side chains of other amino acids, including tyrosine, were not detected. This is in contrast to a previous report in which DPCs with histone H3 tyrosine 41 were detected when holes were generated within the appropriate region of nucleosomal DNA.44 Furthermore, DPC formation was quenched upon hole-generation in proximity to this residue and is likely due to tyrosine reduction of the hole.54 Utilization of N-terminal capped histones provided indirect support for their trapping of DNA holes, which is consistent with other reports on DPC formation in oxidatively damaged NCPs.23
DPC yield was generally competitive with, and in some instances exceeded, that of alkali-labile lesions attributable to water trapping. Although some dG-rich tracts extended for more than a helical turn, shorter dG3 hole localization sequences were sufficient for generating DPCs, again in comparable yields to water trapping products. Although the calculated barriers for amine addition to dG•+ are significantly lower than for reaction with water, the relative reaction rates are also dependent on concentrations.41,42 It is possible that water competes with histone lysines due to low free amine concentration at pH 7.5, where the reactions are carried out. Our data indicate that DPC formation should be considered when considering DNA damage resulting from hole generation, such as from the direct effect of ionizing radiation. One specific situation in which DPC formation could be biologically significant is in dG-rich regions comprising dG3-tracts that are critical components of potential G-quadruplex sequences (PQSs). Several hundred thousand PQSs have been identified in the human genome.75 PQSs are disproportionately located in gene promoter and 5′-untranslated regions of genes, and the propensity of the guanines in these sequences to undergo oxidation affects transcription.76 Reversible histone–DNA cross-links regulate transcription in cells.18 This investigation provides the impetus to examine whether irreversible histone–DNA cross-links resulting from oxidative stress affect transcription and other cellular events.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for support from the National Institute of General Medical Sciences (GM-131736) to M.M.G. and the Agence Nationale de la Recherche (NucleoMAP Project, ANR-20-CE29–0002-01) to N.G. and M.K. We also gratefully acknowledge support from the PSMN (Pôle Scientifique de Modélisation Numérique) of the ENS de Lyon and GENCIIDRIS (Grand Challenge Jean Zay 2021–101465 and project 2022-A0130800609) to N.G., M.K., and E.D. for the computing resources. We thank Henning Urlaub and Aleksandar Chernev for providing and assisting with the NuXL workflow.
Footnotes
Contributor Information
Tingyu Wen, Department of Chemistry, Johns Hopkins University, Baltimore, Maryland 21218, United States.
Maxime Kermarrec, Université Claude Bernard Lyon 1, Laboratoire de Chimie UMR 5182, ENS de Lyon, CNRS, F-69342 Lyon, France.
Elise Dumont, Institut de Chimie de Nice UMR 7272, Université Côte d’Azur, CNRS, 06108 Nice, France; Institut Universitaire de France, 75005 Paris, France.
Natacha Gillet, Université Claude Bernard Lyon 1, Laboratoire de Chimie UMR 5182, ENS de Lyon, CNRS, F-69342 Lyon, France.
Marc M. Greenberg, Department of Chemistry, Johns Hopkins University, Baltimore, Maryland 21218, United States
REFERENCES
- (1).Wei X; Peng Y; Bryan C; Yang K Mechanisms of DNA-Protein Cross-Link Formation and Repair. Biochim. Biophys. Acta 2021, 1869, 140669. [DOI] [PubMed] [Google Scholar]
- (2).Nakano T; Mitsusada Y; Salem AMH; Shoulkamy MI; Sugimoto T; Hirayama R; Uzawa A; Furusawa Y; Ide H Induction of DNA-Protein Cross-Links by Ionizing Radiation and Their Elimination from the Genome. Mut. Res. 2015, 771, 45–50. [DOI] [PubMed] [Google Scholar]
- (3).Vaz B; Popovic M; Newman JA; Fielden J; Aitkenhead H; Halder S; Singh AN; Vendrell I; Fischer R; Torrecilla I; Drobnitzky N; Freire R; Amor DJ; Lockhart PJ; Kessler BM; McKenna GW; Gileadi O; Ramadan K Metalloprotease Sprtn/Dvc1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair. Mol. Cell 2016, 64, 704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Stingele J; Bellelli R; Alte F; Hewitt G; Sarek G; Maslen SL; Tsutakawa SE; Borg A; Kjær S; Tainer JA; Skehel JM; Groll M; Boulton SJ Mechanism and Regulation of DNA-Protein Crosslink Repair by the DNA-Dependent Metalloprotease Sprtn. Mol. Cell 2016, 64, 688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Weickert P; Li H-Y; Götz MJ; Dürauer S.; Yaneva D.; Zhao S.; Cordes J.; Acampora AC.; Forne I.; Imhof A.; Stingele J. Sprtn Patient Variants Cause Global-Genome DNA-Protein Crosslink Repair Defects. Nat. Commun. 2023, 14, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Weickert P; Stingele J DNA-Protein Crosslinks and Their Resolution. Annu. Rev. Biochem. 2022, 91, 157–181. [DOI] [PubMed] [Google Scholar]
- (7).Lopez-Mosqueda J; Maddi K; Prgomet S; Kalayil S; Marinovic-Terzic I; Terzic J; Dikic I Sprtn Is a Mammalian DNA-Binding Metalloprotease That Resolves DNA-Protein Cross-links. eLife 2016, 5, No. e21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Maskey RS; Flatten KS; Sieben CJ; Peterson KL; Baker DJ; Nam H-J; Kim MS; Smyrk TC; Kojima Y; Machida Y; Santiago A; van Deursen JM; Kaufmann SH; Machida YJ Spartan Deficiency Causes Accumulation of Topoisomerase 1 Cleavage Complexes and Tumorigenesis. Nucleic Acids Res. 2017, 45, 4564–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Tretyakova NY; Groehler A; Ji S DNA-Protein Cross-Links: Formation, Structural Identities, and Biological Outcomes. Acc. Chem. Res. 2015, 48, 1631–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Shang M; Ren M; Zhou C Nitrogen Mustard Induces Formation of DNA-Histone Cross-Links in Nucleosome Core Particles. Chem. Res. Toxicol. 2019, 32, 2517–2525. [DOI] [PubMed] [Google Scholar]
- (11).Ming X; Groehler A; Michaelson-Richie ED; Villalta PW; Campbell C; Tretyakova NY Mass Spectrometry Based Proteomics Study of Cisplatin-Induced DNA-Protein Cross-Linking in Human Fibrosarcoma (Ht1080) Cells. Chem. Res. Toxicol. 2017, 30, 980–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yang K; Park D; Tretyakova NY; Greenberg MM Histone Tails Decrease N7-Methyl-2′-Deoxyguanosine Depurination and Yield DNA-Protein Cross-Links in Nucleosome Core Particles and Cells. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, E11212–E11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yang K; Greenberg MM DNA-Protein Cross-Link Formation in Nucleosome Core Particles Treated with Methyl Methanesulfonate. Chem. Res. Toxicol. 2019, 32, 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Yang K; Sun H; Lowder L; Varadarajan S; Greenberg MM Reactivity of N3-Methyl-2′-Deoxyadenosine in Nucleosome Core Particles. Chem. Res. Toxicol. 2019, 32, 2118–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ren M; Greenberg MM; Zhou C Participation of Histones in DNA Damage and Repair within Nucleosome Core Particles: Mechanism and Applications. Acc. Chem. Res. 2022, 55, 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Ji S; Shao H; Han Q; Seiler CL; Tretyakova NY Reversible DNA-Protein Cross-Linking at Epigenetic DNA Marks. Angew. Chem., Int. Ed. 2017, 56, 14130–14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Li F; Zhang Y; Bai J; Greenberg MM; Xi Z; Zhou C 5-Formylcytosine Yields DNA-Protein Crosslinks in Nucleosome Core Particles. J. Am. Chem. Soc. 2017, 139, 10617–10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Raiber E-A; Portella G; Martínez Cuesta S; Hardisty R; Murat P; Li Z; Iurlaro M; Dean W; Spindel J; Beraldi D; Liu Z; Dawson MA; Reik W; Balasubramanian S 5-Formylcytosine Organizes Nucleosomes and Forms Schiff Base Interactions with Histones in Mouse Embryonic Stem Cells. Nat. Chem. 2018, 10, 1258–1266. [DOI] [PubMed] [Google Scholar]
- (19).Barker S; Weinfeld M; Zheng J; Li L; Murray D Identification of Mammalian Proteins Cross-Linked to DNA by Ionizing Radiation. J. Biol. Chem. 2005, 280, 33826–33838. [DOI] [PubMed] [Google Scholar]
- (20).von Sonntag C Free-Radical-Induced DNA Damage and its Repair; Springer-Verlag: Berlin, 2006. [Google Scholar]
- (21).Hickerson RP; Chepanoske CL; Williams SD; David SS; Burrows CJ Mechanism-Based DNA-Protein Cross-Linking of Muty Via Oxidation of 8-Oxoguanosine. J. Am. Chem. Soc. 1999, 121, 9901–9902. [Google Scholar]
- (22).Johansen ME; Muller JG; Xu X; Burrows CJ Oxidatively Induced DNA-Protein Cross-Linking between Single-Stranded Binding Protein and Oligodeoxynucleotides Containing 8-Oxo-7,8-Dihydro-2’-Deoxyguanosine. Biochemistry 2005, 44, 5660–5671. [DOI] [PubMed] [Google Scholar]
- (23).Bai J; Zhang Y; Xi Z; Greenberg MM; Zhou C Oxidation of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine Leads to Substantial DNA-Histone Cross-Links within Nucleosome Core Particles. Chem. Res. Toxicol. 2018, 31, 1364–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Genereux JC; Barton JK Mechanisms for DNA Charge Transport. Chem. Rev. 2010, 110, 1642–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kanvah S; Joseph J; Schuster GB; Barnett RN; Cleveland CL; Landman U Oxidation of DNA: Damage to Nucleobases. Acc. Chem. Res. 2010, 43, 280–287. [DOI] [PubMed] [Google Scholar]
- (26).Kawai K; Majima T Photoinduced Phenomena in Nucleic Acids Ii: DNA Fragments and Phenomenological Aspects; Barbatti M., Borin AC., Ullrich S., Eds.; Springer International Publishing: Cham, 2015; pp 165–182. [Google Scholar]
- (27).Barker S; Weinfeld M; Murray D DNA Protein Crosslinks: Their Induction, Repair, and Biological Consequences. Mutat. Res. 2005, 589, 111–135. [DOI] [PubMed] [Google Scholar]
- (28).Nakano T; Xu X; Salem AMH; Shoulkamy MI; Ide H Radiation-Induced DNA-Protein Cross-Links: Mechanisms and Biological Significance. Free Radical Biol. Med. 2017, 107, 136–145. [DOI] [PubMed] [Google Scholar]
- (29).Tse ECM; Zwang TJ; Bedoya S; Barton JK Effective Distance for DNA-Mediated Charge Transport between Repair Proteins. ACS Cent. Sci. 2019, 5, 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Slinker JD; Muren NB; Renfrew SE; Barton JK DNA Charge Transport over 34 Nm. Nat. Chem. 2011, 3, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Meggers E; Michel-Beyerle ME; Giese B Sequence Dependent Long Range Hole Transport in DNA. J. Am. Chem. Soc. 1998, 120, 12950–12955. [Google Scholar]
- (32).Saito I; Nakamura T; Nakatani K; Yoshioka Y; Yamaguchi K; Sugiyama H Mapping of the Hot Spots for DNA Damage by One-Electron Oxidation: Efficacy of GG Doublets and GGG Triplets as a Trap in Long-Range Hole Migration. J. Am. Chem. Soc. 1998, 120, 12686–12687. [Google Scholar]
- (33).Yoshioka Y; Kitagawa Y; Takano Y; Yamaguchi K; Nakamura T; Saito I Experimental and Theoretical Studies on the Selectivity of GGG Triplets toward One-Electron Oxidation in B-Form DNA. J. Am. Chem. Soc. 1999, 121, 8712–8719. [Google Scholar]
- (34).Cadet J; Douki T; Ravanat J-L Oxidatively Generated Damage to the Guanine Moiety of DNA: Mechanistic Aspects and Formation in Cells. Acc. Chem. Res. 2008, 41, 1075–1083. [DOI] [PubMed] [Google Scholar]
- (35).Cadet J; Douki T; Gasparutto D; Ravanat J-L; Wagner JR Wiley Series on Reactive Intermediates in Chemistry and Biology; John Wiley & Sons, Inc., 2009; Vol. 2, pp 69–97. [Google Scholar]
- (36).Rokhlenko Y; Geacintov NE; Shafirovich V Lifetimes and Reaction Pathways of Guanine Radical Cations and Neutral Guanine Radicals in an Oligonucleotide in Aqueous Solutions. J. Am. Chem. Soc. 2012, 134, 4955–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Copeland KD; Lueras AMK; Stemp EDA; Barton JK DNA Cross-Linking with Metallointercalator–Peptide Conjugates. Biochemistry 2002, 41, 12785–12797. [DOI] [PubMed] [Google Scholar]
- (38).Kurbanyan K; Nguyen KL; To P; Rivas EV; Lueras AMK; Kosinski C; Steryo M; Gonzalez A; Mah DA; Stemp EDA DNA-Protein Cross-Linking Via Guanine Oxidation: Dependence Upon Protein and Photosensitizer. Biochemistry 2003, 42, 10269–10281. [DOI] [PubMed] [Google Scholar]
- (39).Xu X; Muller JG; Ye Y; Burrows CJ DNA-Protein Cross-Links between Guanine and Lysine Depend on the Mechanism of Oxidation for Formation of C5 Vs. C8 Guanosine Adducts. J. Am. Chem. Soc. 2008, 130, 703–709. [DOI] [PubMed] [Google Scholar]
- (40).Perrier S; Hau J; Gasparutto D; Cadet J; Favier A; Ravanat J-L Characterization of Lysine-Guanine Cross-Links Upon One-Electron Oxidation of a Guanine-Containing Oligonucleotide in the Presence of a Trilysine Peptide. J. Am. Chem. Soc. 2006, 128, 5703–5710. [DOI] [PubMed] [Google Scholar]
- (41).Thapa B; Munk BH; Burrows CJ; Schlegel HB Computational Study of the Radical Mediated Mechanism of the Formation of C8, C5, and C4 Guanine:Lysine Adducts in the Presence of the Benzophenone Photosensitizer. Chem. Res. Toxicol. 2016, 29, 1396–1409. [DOI] [PubMed] [Google Scholar]
- (42).Thapa B; Hebert SP; Munk BH; Burrows CJ; Schlegel HB Computational Study of the Formation of C8, C5, and C4 Guanine:Lysine Adducts Via Oxidation of Guanine by Sulfate Radical Anion. J. Phys. Chem. A 2019, 123, 5150–5163. [DOI] [PubMed] [Google Scholar]
- (43).Bignon E; Chan C-H; Morell C; Monari A; Ravanat J-L; Dumont E Molecular Dynamics Insights into Polyamine-DNA Binding Modes: Implications for Cross-Link Selectivity. Chem.—Eur. J. 2017, 23, 12845–12852. [DOI] [PubMed] [Google Scholar]
- (44).Bjorklund CC; Davis WB Stable DNA-Protein Cross-Links Are Products of DNA Charge Transport in a Nucleosome Core Particle. Biochemistry 2007, 46, 10745–10755. [DOI] [PubMed] [Google Scholar]
- (45).Chan CH; Monari A; Ravanat JL; Dumont E Probing Interaction of a Trilysine Peptide with DNA Underlying Formation of Guanine-Lysine Cross-Links: Insights from Molecular Dynamics. Phys. Chem. Chem. Phys. 2019, 21, 23418–23424. [DOI] [PubMed] [Google Scholar]
- (46).Luger K; Mader AW; Richmond RK; Sargent DF; Richmond TJ Crystal Structure of the Nucleosome Core Particle at 2.8 Å Resolution. Nature 1997, 389, 251–260. [DOI] [PubMed] [Google Scholar]
- (47).Peng Y; Li S; Onufriev A; Landsman D; Panchenko AR Binding of Regulatory Proteins to Nucleosomes Is Modulated by Dynamic Histone Tails. Nat. Commun. 2021, 12, 5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Núñez ME; Noyes KT; Barton JK Oxidative Charge Transport through DNA in Nucleosome Core Particles. Chem. Biol. 2002, 9, 403–415. [DOI] [PubMed] [Google Scholar]
- (49).Davis WB; Bjorklund CC; Deline M Probing the Effects of DNA-Protein Interactions on DNA Hole Transport: The N-Terminal Histone Tails Modulate the Distribution of Oxidative Damage and Chemical Lesions in the Nucleosome Core Particle. Biochemistry 2012, 51, 3129–3142. [DOI] [PubMed] [Google Scholar]
- (50).Bjorklund CC; Davis WB Attenuation of DNA Charge Transport by Compaction into a Nucleosome Core Particle. Nucleic Acids Res. 2006, 34, 1836–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Liu Y; Liu Z; Geacintov NE; Shafirovich V Proton-Coupled Hole Hopping in Nucleosomal and Free DNA Initiated by Site-Specific Hole Injection. Phys. Chem. Chem. Phys. 2012, 14, 7400–7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Sun H; Zheng L; Yang K; Greenberg MM Positional Dependence of DNA Hole Transfer Efficiency in Nucleosome Core Particles. J. Am. Chem. Soc. 2019, 141, 10154–10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Zheng L; Griesser M; Pratt DA; Greenberg MM Aminyl Radical Generation Via Tandem Norrish Type I Photocleavage, Β-Fragmentation: Independent Generation and Reactivity of the 2’-Deoxyadenosin- N6-yl Radical. J. Org. Chem. 2017, 82, 3571–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Zheng L; Greenberg MM DNA Damage Emanating from a Neutral Purine Radical Reveals the Sequence Dependent Convergence of the Direct and Indirect Effects of Γ-Radiolysis. J. Am. Chem. Soc. 2017, 139, 17751–17754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Sun H; Zheng L; Greenberg MM Independent Generation of Reactive Intermediates Leads to an Alternative Mechanism for Strand Damage Induced by Hole Transfer in Poly(dA–T) Sequences. J. Am. Chem. Soc. 2018, 140, 11308–11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Adhikary A; Kumar A; Khanduri D; Sevilla MD Effect of Base Stacking on the Acid-Base Properties of the Adenine Cation Radical [A•+] in Solution: ESR and DFT Studies. J. Am. Chem. Soc. 2008, 130, 10282–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zheng L; Greenberg MM Traceless Tandem Lesion Formation in DNA from a Nitrogen-Centered Purine Radical. J. Am. Chem. Soc. 2018, 140, 6400–6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Madison AL; Perez ZA; To P; Maisonet T; Rios EV; Trejo Y; Ochoa-Paniagua C; Reno A; Stemp EDA Dependence of DNA-Protein Cross-Linking Via Guanine Oxidation Upon Local DNA Sequence as Studied by Restriction Endonuclease Inhibition.Biochemistry 2012, 51, 362–369. [DOI] [PubMed] [Google Scholar]
- (59).See Supporting Information.
- (60).Bae JW; Kim S; Kim VN; Kim J-S Photoactivatable Ribonucleosides Mark Base-Specific RNA-Binding Sites. Nat. Commun. 2021, 12, 6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Stützer A.; Welp LM.; Raabe M.; Sachsenberg T.; Kappert C.; Wulf A.; Lau AM.; David S-S.; Chernev A.; Kramer K.; Politis A.; Kohlbacher O.; Fischle W.; Urlaub H. Analysis of Protein-DNA Interactions in Chromatin by UV Induced Cross-Linking and Mass Spectrometry. Nat. Commun. 2020, 11, 5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Shuck SC; Rose KL; Marnett LJ Mass Spectrometric Methods for the Analysis of Nucleoside-Protein Cross-Links: Application to Oxopropenyl-Deoxyadenosine. Chem. Res. Toxicol. 2014, 27, 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Kramer K; Sachsenberg T; Beckmann BM; Qamar S; Boon K-L; Hentze MW; Kohlbacher O; Urlaub H Photo-Cross-Linking and High-Resolution Mass Spectrometry for Assignment of Rna-Binding Sites in RNA-Binding Proteins. Nat. Methods 2014, 11, 1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Tims HS; Gurunathan K; Levitus M; Widom J Dynamics of Nucleosome Invasion by DNA Binding Proteins. J. Mol. Biol. 2011, 411, 430–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Greenberg MM DNA Damage, DNA Repair and Disease; Dizdaroglu M., Lloyd RS., Eds.; Royal Society of Chemistry: United Kingdom, 2021; Vol. 1, pp 27–60. [Google Scholar]
- (66).Hong IS; Carter KN; Sato K; Greenberg MM Characterization and Mechanism of Formation of Tandem Lesions in DNA by a Nucleobase Peroxyl Radical. J. Am. Chem. Soc. 2007, 129, 4089–4098. [DOI] [PubMed] [Google Scholar]
- (67).Carter KN; Greenberg MM Tandem Lesions Are the Major Products Resulting from a Pyrimidine Nucleobase Radical. J. Am. Chem. Soc. 2003, 125, 13376–13378. [DOI] [PubMed] [Google Scholar]
- (68).Robert G; Wagner JR Tandem Lesions Arising from 5-(Uracilyl)Methyl Peroxyl Radical Addition to Guanine: Product Analysis and Mechanistic Studies. Chem. Res. Toxicol. 2020, 33, 565–575. [DOI] [PubMed] [Google Scholar]
- (69).Runtsch LS; Stadlmeier M; Schon A; Müller M.; Carell T. Comparative Nucleosomal Reactivity of 5-Formyl-Uridine and 5Formyl-Cytidine. Chem.—Eur. J. 2021, 27, 12747–12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Gillet N; Dumont E Dynamics and Energetics of PCBP1 Binding to Severely Oxidized Rna. Front. Mol. Biosci. 2022, 9, 994915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Ohtomo H; Kurita J.-i.; Sakuraba S.; Li Z.; Arimura Y.; Wakamori M.; Tsunaka Y.; Umehara T.; Kurumizaka H.; Kono H.; Nishimura Y. The N-Terminal Tails of Histones H2A and H2B Adopt Two Distinct Conformations in the Nucleosome with Contact and Reduced Contact to DNA. J. Mol. Biol. 2021, 433, 167110. [DOI] [PubMed] [Google Scholar]
- (72).Rabdano SO; Shannon MD; Izmailov SA; Gonzalez Salguero N; Zandian M; Purusottam RN; Poirier MG; Skrynnikov NR; Jaroniec CP Histone H4 Tails in Nucleosomes: A Fuzzy Interaction with DNA. Angew. Chem., Int. Ed. 2021, 60, 6480–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Furukawa A; Wakamori M; Arimura Y; Ohtomo H; Tsunaka Y; Kurumizaka H; Umehara T; Nishimura Y Acetylated Histone H4 Tail Enhances Histone H3 Tail Acetylation by Altering Their Mutual Dynamics in the Nucleosome. Proc. Natl. Acad. Sci. U.S.A. 2020, 117, 19661–19663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Hayes JJ; Tullius TD; Wolffe AP The Structure of DNA in a Nucleosome. Proc. Natl. Acad. Sci. U.S.A. 1990, 87, 7405–7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Fleming AM; Burrows CJ Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J. Am. Chem. Soc. 2020, 142, 1115–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Fleming AM; Ding Y; Burrows CJ Oxidative DNA Damage Is Epigenetic by Regulating Gene Transcription Via Base Excision Repair. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


