Abstract
Staphylococcus epidermidis biofilm formation is associated with the production of the polysaccharide intercellular adhesin (PIA)--poly-N-acetylglucosamine polysaccharide (PNAG) by the products of the icaADBC operon. Recent evidence indicates that SarA, a central regulatory element that controls the production of Staphylococcus aureus virulence factors, is essential for the synthesis of PIA/PNAG and the ensuing biofilm development in this species. Based on the presence of a sarA homolog, we hypothesized that SarA could also be involved in the regulation of the biofilm formation process in S. epidermidis. To investigate this, we constructed nonpolar sarA deletions in two genetically unrelated S. epidermidis clinical strains, O-47 and CH845. The SarA mutants were completely defective in biofilm formation, both in the steady-state conditions of a microtiter dish assay and in the flow conditions of microfermentors. Reverse transcription-PCR experiments showed that the mutation in the sarA gene resulted in downregulation of the icaADBC operon transcription in an IcaR-independent manner. Purified SarA protein showed high-affinity binding to the icaA promoter region by electrophoretic mobility shift assays. Consequently, mutation in sarA provoked a significant decrease in the amount of PIA/PNAG on the cell surface. Furthermore, heterologous complementation of S. aureus sarA mutants with the sarA gene of S. epidermidis completely restored biofilm formation. In summary, SarA appeared to be a positive regulator of transcription of the ica locus, and in its absence, PIA/PNAG production and biofilm formation were diminished. Additionally, we present experimental evidence showing that SarA may be an important regulatory element that controls S. epidermidis virulence factors other than biofilm formation.
Chronic nosocomial infections by biofilm-forming Staphylococcus epidermidis have become more prevalent in recent years with the increased use of prosthetic medical implants. Biofilm formation by S. epidermidis frequently compromises the effectiveness of implanted medical devices by giving rise to persistent and relapsing infections, which are more resistant to the host immune response and antimicrobial chemotherapy (for a review, see reference 18). The formation of S. epidermidis biofilms is proposed to occur in a two-step manner, in which a cellular accumulation process to form the mature biofilm follows rapid initial attachment to an inert synthetic surface (22). Critical to S. epidermidis biofilm formation is the production of a poly-N-acetylglucosamine polysaccharide (PNAG)-polysaccharide intercellular adhesin (PIA) (33, 34). The intercellular adhesin (icaADBC) locus, originally described in S. epidermidis (22, 23) and later found in Staphylococcus aureus (9), contains the genes involved in PIA/PNAG production. The significance of PIA/PNAG as a virulence factor was demonstrated in a central venous catheter infection model of a rat and in a subcutaneous foreign-body infection model in mice (43, 44). In addition, the ica operon is one of the few genetic markers of S. epidermidis that differs between invasive strains and strains from the skin flora (15, 16).
PIA/PNAG production and biofilm formation in S. epidermidis are regulated by a variety of environmental factors, including high osmolarity (3% NaCl), ethanol (4%), glucose, growth in anaerobic conditions, high temperature, and subinhibitory concentrations of certain antibiotics (7, 10, 12, 27, 28). Genetic regulation involving icaR and the alternative sigma factor σB has been reported. The gene icaR, located adjacent to the ica operon, encodes a transcriptional repressor involved in the environmental regulation of icaADBC operon expression in S. epidermidis and S. aureus (8, 24, 25). A transcriptional analysis revealed that icaADBC transcription was strongly repressed in mutants with defective σB, whereas icaR was upregulated, a finding suggesting that σB controls transcription of the icaADBC operon by an icaR-dependent pathway in S. epidermidis (27, 28). However, the alternative transcription factor σB plays contradictory roles in controlling biofilm formation of S. aureus. While an initial work by Rachid and coworkers implied that σB was a regulator of the biofilm formation process of S. aureus (42), our results have demonstrated that biofilm and PIA/PNAG production were not affected in a S. aureus σB mutant compared with its wild-type strain (47). Moreover, ica operon expression in S. epidermidis can be turned on and off by the insertion and excision of the insertion sequence IS256 at specific hot spots in the icaA and icaC genes (51). By this mechanism, PIA/PNAG production and biofilm formation phenotypes may be phase variable.
By screening a library of Tn917 insertions in a clinical S. aureus strain, we identified SarA as being essential for biofilm development by S. aureus (47). Nonpolar mutations of sarA in genetically unrelated S. aureus strains decreased ica transcription and PIA/PNAG production and completely prevented biofilm development. In S. epidermidis, a SarA protein has been described (14). This protein is highly related (84%) to the S. aureus SarA protein, suggesting that it could be involved in the control of virulence determinants. However, no data on sar regulation exist, and some evidence suggests the existence of particularities for each species. Thus, the order of the three promoters that control sarA expression is not the same in S. epidermidis as in S. aureus, a distinction which might reflect differences in regulation (14).
In this study, we have examined the role of SarA in the regulation of icaADBC operon expression, PIA/PNAG production, and biofilm formation in two genetically unrelated biofilm-positive clinical isolates of S. epidermidis. Our results demonstrate that the sarA mutants show a severe defect in biofilm formation through a transcriptional downregulation of icaADBC operon expression and PIA/PNAG production by an IcaR-independent pathway.
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
The biofilm-forming strain S. epidermidis O-47 was isolated from a patient at the Institut für Medizinische Mikrobiologie und Hygiene, Universität zu Köln, Cologne, Germany (22). S. epidermidis CH845 was isolated from a patient with infection in a joint prosthesis (16). Both strains were selected because of their strong biofilm production phenotype, antibiotic susceptibility profile, and ability to accept recombinant DNA by protoplast transformation.
The most relevant bacterial strains and plasmids used and constructed in this study are listed in Table 1. Escherichia coli DH5α cells were grown in Luria-Bertani (LB) broth or on LB agar (Pronadisa) with appropriate antibiotics. Staphylococcal strains were cultured on Trypticase soy agar (TSA), in Trypticase soy broth supplemented with glucose (0.25% wt/vol; TSB-gluc), and in Congo red agar (11). Media were supplemented with appropriate antibiotics in the following concentrations: erythromycin, 2.5 μg/ml; ampicillin, 100 μg/ml; and chloramphenicol, 20 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Species | Relevant properties | Source or reference |
|---|---|---|---|
| Strains | |||
| RN4220 | S. aureus | Restriction-deficient mutant of 8325-4 | 29 |
| 15981 | S. aureus | Clinical isolate, biofilm-positive | 47 |
| 15981 Δica | S. aureus | Derivative of 15981; icaADBC-negative, biofilm-negative strain | 47 |
| 15981 ΔsarA | S. aureus | Derivative of 15981; sarA-negative, biofilm-negative strain | 47 |
| ISP479c | S. aureus | Derivative of 8325; biofilm-positive strain | 40 |
| ISP479c sarA- | S. aureus | Derivative of ISP479c; sarA-negative; biofilm-negative strain | 47 |
| O-47 | S. epidermidis | Clinical strain; icaADBC-positive; biofilm-positive strain | 22 |
| O-47 Δica::tet | S. epidermidis | Derivative of O-47; icaADBC mutant; biofilm-negative strain | 10 |
| CH845 | S. epidermidis | Clinical strain BM94314; icaADBC-positive; biofilm-positive strain | 16 |
| JP54 | S. aureus | 15981 ΔsarA(pJP19) | This study |
| JP55 | S. aureus | ISP479c sarA(pJP19) | This study |
| JP56 | S. epidermidis | Derivative of O-47; sarA mutant | This study |
| JP57 | S. epidermidis | Derivative of CH845; sarA mutant | This study |
| JP58 | S. epidermidis | JP56(pJP19) | This study |
| JP59 | S. epidermidis | JP57(pJP19) | This study |
| Plasmids | |||
| pCU1 | Shuttle plasmid | 2 | |
| pMAD | Shuttle vector with a termosensitive origin of replication for gram-positive bacteria | 1 | |
| pJP18 | Derivative of pMAD used to construct the deletions in the sarA gene | This study | |
| pJP19 | Vector for complementation experiments; a 1.1-kb PCR fragment containing sarA from S. epidermidis RP62A cloned in pCU1 | This study |
DNA manipulations.
Routine DNA manipulations were performed with standard procedures (45) unless otherwise stated. Plasmid DNA from E. coli and staphylococci were purified with a Genelute plasmid miniprep kit (Sigma) according to the manufacturer's protocol, except that the staphylococcal bacterial cells were lysed by lysostaphin (Sigma; 12.5 μg/ml) at 37°C for 1 h before plasmid purification. Plasmids were introduced into the staphylococci by electroporation or by protoplast transformation with previously described methods (11, 19, 20). Restriction enzymes were purchased from Roche and used according to the manufacturer's instructions. Oligonucleotides were obtained from Invitrogen (Table 2).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) |
|---|---|
| Sarepi-1mX | TGCTCTAGAATGACTAAAGGGAGGTGCC |
| Sarepi-2c | GGCTAGGGAGTAAAACAGATATTTC |
| Sarepi-3m | ATCTGTTTTACTCCCTAGCCGCAATGTAGCATTTGCTATATTC |
| Sarepi-4cB | CGCGGATCCTAAATTAACTTCTAAAACAGAAG |
| Sarepi-5c | TGGATATGATATAAATAGGGAGG |
| Sarepi-6cX | TGCTCTAGAGGACATGCACCACATATCGAGG |
| Sarepi-7mB | CGCGGATCCGGTATATTAATATAACTAAAGGC |
| Sarepi-8c | TCTGTGATACGGTTGTTTACTCG |
| icaA-epi-1 | AACAAGTTGAAGGCATCTCC |
| icaA-epi-2 | GATGCTTGTTTGATTCCCT |
| icaR-epi-1 | GGTAAAGTCCGTCAATGGAA |
| icaR-epi-2 | CGCAATAACCTTATTTTCCG |
| icaR-aur-1c | ATTGCGTTATCAATAATCTTATC |
| icaA-aur-3c | TTGCAATTTCTTTACCTACCTTTCG |
| icaR-epi-7c | ATTGCGTTATCAATAATCTTATC |
| icaA-epi-3 | CATGCATTTTTTCACCTACCTTTCG |
| gyrB-epi-1 | TTATGGTGCTGGACAGATACA |
| gyrB-epi-2 | CACCGTGAAGACCGCCAGATA |
Staphylococcal chromosomal DNA was extracted with a Genelute bacterial genomic DNA kit (Sigma) according to the manufacturer's protocol, except that the bacterial cells were lysed by lysostaphin (Sigma; 12.5 μg/ml) at 37°C for 1 h before DNA purification. For Southern blot hybridization, the chromosomal DNA digested with HindIII was analyzed by agarose gel electrophoresis. Gels were blotted onto nylon membranes (Hybond-N 0.45-mm-pore-size filters, Amersham Life Science) with standard methods (3, 45). The PCR product of the amplified sarA gene with oligonucleotides Sarepi-5c and Sarepi-8c was used as a DNA probe. Labeling of the probe and DNA hybridization were performed according to the protocol supplied with the PCR-DIG DNA-labeling and chemiluminescent detection kit (Roche).
Allelic exchange of chromosomal genes.
To construct the deletions in sarA, we amplified by PCR two fragments of approximately 1,000 bp that flanked the left (oligonucleotides Sarepi-4cB and Sarepi-3m) and the right (oligonucleotides Sarepi-2c and Sarepi-1mX) of the sequence targeted for deletion (Table 2). Oligonucleotides Sarepi-2c and Sarepi-3m have a 20-base complementary region (underlined in the oligonucleotide sequence) to allow the products of the first PCR to anneal at their overlapping region. A second PCR was performed with primers Sarepi-1mX and Sarepi-4cB to obtain a single fragment. Specifically, 1 μl of each of the products of the first PCR was mixed with 10 pM of the outside primers and PCR amplified. The fusion products were purified and cloned into the BamHI and XbaI sites of plasmid pSC20 (F. Götz). The fragment was then cloned into the SalI and EcoRI sites of the shuttle plasmid pMAD (1), and the resulting plasmid, pJP18, was transformed into S. epidermidis by protoplast transformation. Plasmid pMAD contains a temperature-sensitive origin of replication and an erythromycin resistance gene. The plasmid was integrated into the chromosome through homologous recombination at the nonpermissive temperature (43.5°C). From the 43.5°C plate, one to five colonies were picked into 10 ml of TSB-gluc and incubated for 24 h at 30°C. Tenfold serial dilutions of this culture in sterile TSB-gluc were plated on TSA containing 5-bromo-4-chloro-3-indolyl-B-d-galactopyranoside at 150 μg ml−1). White colonies, which no longer contained the pMAD plasmid, were tested to confirm the replacement by PCR with oligonucleotides Sarepi-1mX and Sarepi-5c (Table 2) and by Southern blotting.
Complementation of the mutants.
The sarA gene from S. epidermidis RP62A was amplified with high-fidelity thermophilic DNA polymerase (Dynazyme Ext, Finnzymes) with primers Sarepi-7mB and Sarepi-6cX (Table 2). The PCR product was cloned into the BamHI and XbaI sites of the pCU1 plasmid (2), and the resulting plasmid, pJP19, was transformed by electroporation or protoplast transformation into S. aureus RN4220 or S. epidermidis strains. Phage 80α was used to transduce pJP19 from RN4220 to strains 15981 and ISP479c (39).
Biofilm formation assays.
The quantification of the biofilm formation on abiotic surfaces was assessed basically as described elsewhere (11, 22). Macroscopic observation of biofilm on glass was performed as previously described (11). Briefly, cells were grown in 50 ml of B2 at 37°C, with a glass container, without shaking, for 1 day, and the walls of the container were visually (macroscopically) examined for the presence or absence of a white biofilm layer.
To analyze biofilm formation under flow conditions, we used 60-ml microfermenters (Pasteur Institute's Laboratory of Fermentation) with a continuous flow of 40 ml h−1 of TSB-gluc and constant aeration with sterile compressed air (0.3 bar). Submerged Pyrex slides served as the growth substratum. Approximately 108 bacteria from an overnight preculture of each strain grown in TSB-gluc were used to inoculate the microfermenters and cultivated 24 h at 37°C. Biofilm development was recorded with a Nikon Coolpix 950 digital camera.
PNAG detection.
PNAG production in the S. epidermidis strains was detected as described elsewhere (9), with an anti-S. aureus PNAG antibody diluted 1:10,000 (34). Bound antibodies were detected with a peroxidase-conjugated goat anti-rabbit immunoglobulin G antibody (Santa Cruz) diluted 1:5,000, and the Western blotting Luminol reagent (Santa Cruz Biotechnology).
Real-time quantitative PCR.
Total S. epidermidis RNA was prepared with the Fast RNA-Blue kit (Bio 101) according to the manufacturer's instructions. Two micrograms of each RNA was subjected, in duplicate, to DNase I (Invitrogen) treatment for 30 min at 37°C. The enzyme was inactivated at 65°C in the presence of EDTA. To verify the absence of genomic DNA in every sample, the RNA duplicates were reverse transcribed in the presence and absence of Moloney murine leukemia virus reverse transcriptase (Invitrogen). All preparations were purified with the QIAquick PCR purification kit (Qiagen); 25 ng of each reaction product was used for a real-time quantitative PCR with the iCycler machine (Bio-Rad) and the LC-DNA Master SYBR Green I mix (Bio-Rad). The icaA and icaR transcripts were amplified with primers icaA-epi-1/icaA-epi-2 and icaR-epi-1/icaR-epi-2, respectively (Table 2). The gyrB transcripts that are constitutively expressed were amplified as an endogenous control with the primers gyrB-epi-1 and gyrB-epi-2 (Table 2). The level of expression of icaA was normalized with respect to gyrB expression. Only samples with no amplification of gyrB in the minus-reverse transcriptase aliquot were included in the study.
To monitor the specificity, the final PCR products were analyzed by melting curves and electrophoresis. In each experiment, all the reactions were performed in triplicate. The relative transcriptional levels within distinct experiments were determined with the 2−ΔΔCT method (31). The results show the average ± standard error of the mean of at least four independent experiments.
Purification of SarA protein.
The cloning and purification of the His6-tagged SarA fusion protein were described earlier (6). The purified His6-tagged SarA protein was found to be more than 98% pure in a sodium dodecyl sulfate-12% polyacrylamide gel. The concentration of the purified protein was determined by the Bradford protein assay (Bio-Rad, Hercules, Calif.), with bovine serum albumin as the standard.
Electrophoretic mobility shift.
To determine if the recombinant SarA protein from S. aureus binds to the icaA promoter region, a 200-bp PCR-amplified fragment, representing the icaRA intergenic region from S. aureus (oligonucleotides icaR-aur-1c and icaA-aur-3c) or S. epidermidis (oligonucleotides icaR-epi-7c and icaA-epi-3) was end labeled with [γ-32P]ATP with T4 polynucleotide kinase. Labeled fragment (0.1 ng or 0.5 fmol) was incubated at room temperature for 20 min with various amounts of purified SarA protein in 25 μl of binding buffer (25 mM Tris-Cl [pH 7.5], 0.1 mM EDTA, 75 mM NaCl, 1 mM dithiothreitol, and 10% glycerol) containing 0.5 μg of calf thymus DNA (Amersham Pharmacia Biotech). The reaction mixtures were analyzed in an 8.0% nondenaturing polyacrylamide gel. The band shifts were detected by exposing dried gels to X-ray films.
Statistical analysis.
The data indicating gene expression were compared with the Kruskal-Wallis and the Mann-Whitney tests. All the tests were two-sided, and the significance level was 5%. The statistical analysis was performed with the SPSS program.
RESULTS
sarA gene of S. epidermidis restored the biofilm formation capacity of the S. aureus ΔsarA mutants.
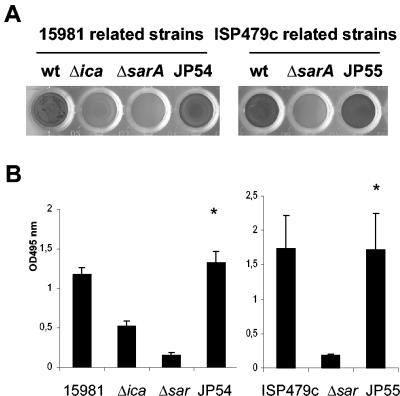
Considering the similarity of the SarA proteins of S. aureus and S. epidermidis, we investigated the functional relationship of the sarA gene in both species. The S. aureus sarA mutant clones 15981 ΔsarA and ISP479c sarA mutant (47) complemented with plasmid pJP19 (carrying a PCR-amplified 1,109-bp fragment containing the sarA gene from S. epidermidis under the control of its own promoter) were analyzed for their capacity to form a biofilm. As shown in Fig. 1, we found that the complemented strains JP54 and JP55 regained their capacity to form biofilms compared with their respective sarA mutants. Evidently, the SarA protein of S. epidermidis is functional in S. aureus.
FIG. 1.
Biofilm formation phenotype of two unrelated S. aureus sarA mutant clones carrying pJP19, a shuttle plasmid containing the sarA gene from S. epidermidis. Biofilm formation capacity differences correspond to 24-h biofilm formed on polystyrene microtiter plates after staining with 0.1% safranin. The microtiter plates and mean optical density values obtained (A495) are shown. (A) Left: wells corresponding to wild-type 15981, 15981 Δica (negative control), 15981 ΔsarA, and JP54 (15981 ΔsarA carrying pJP19). Right: wells corresponding to wild-type ISP479c, ISP479c ΔsarA, and JP55 (ISP479c ΔsarA carrying pJP19). (B) Mean optical density values. Bars represent the mean values, and error bars represent the standard error of the mean. Significant differences in adherence were noted between complemented and noncomplemented sarA mutant strains (*, P < 0.01).
Deletion of sarA in S. epidermidis resulted in a reduced capacity to form a biofilm in vitro.
To assess the role of SarA in S. epidermidis, we constructed nonpolar deletions of the sarA gene in two unrelated S. epidermidis clinical strains by allelic exchange with the pMAD plasmid (see Materials and Methods). As shown in Fig. 2A and B, the deletion mutants JP56 (S. epidermidis O-47 ΔsarA) and JP57 (S. epidermidis CH845 ΔsarA) were reduced in their capacity to form a biofilm on polystyrene microtiter plates compared to the wild-type parent strains O-47 and CH845, respectively. When the JP56 strain was complemented with plasmid pJP19, carrying the wild-type sarA gene, biofilm formation was restored (Fig. 2A and B). In contrast, although strain JP59 (JP57 carrying plasmid pJP19) formed large clumps in broth cultures, it was unable to form a biofilm on microtiter plates (data not shown). However, macroscopic examination of biofilm formation in a glass container revealed that upon 1 day of culture, the wild-type (O-47 and CH845) and the complemented (JP58 and JP59) strains formed an obvious biofilm on the glass surface, whereas the sarA mutants JP56 and JP57 did not (Fig. 2C). These results strongly suggest that the sarA gene is required for biofilm formation in S. epidermidis.
FIG. 2.
Loss of biofilm formation in two genetically unrelated S. epidermidis sarA mutants. (A) Biofilm formation capacity of S. epidermidis O-47 and CH845 (wild-type strains), their corresponding sarA mutants JP56 and JP57, the JP56 strain complemented with plasmid pJP19 (JP58), and O-47 Δica::tet as a negative control on polystyrene microtiter plates after 24 h in TSB-gluc medium at 37°C. The bacterial cells were stained with safranin and quantified by determining the absorbance at 495 nm. (B) Significant differences in adherence were noted between wild-type strains and their isogenic sarA mutants as well as between the complemented versus noncomplemented JP56 (O-47 sarA mutant) strain (*, P < 0.05). (C) Phenotypic differences in the capacity to form a 24-h biofilm on the surface of a glass container (visual observation) between wild-type strains O-47 and CH845, their corresponding sarA mutants (JP56 and JP57, respectively), and their sarA mutants complemented with plasmid pJP19 (JP58 and JP59, respectively).
Biofilm formation of the sarA mutants in continuous-flow culture microfermenters.
Extracellular proteases, including extracellular cysteine (Ecp) and serine (Esp) proteases, have been described in S. epidermidis (13). The first enzyme shows extended sequence similarity to the S. aureus cysteine protease (staphopain), and the second resembles the serine protease produced by that species. In S. aureus, the expression of the extracellular proteases is repressed by SarA, since their production is upregulated in sarA mutants (5). In S. epidermidis, nothing is known about their regulation, but it is likely that a similar control by SarA occurs. To test that, the proteolytic activity of the sarA mutant JP56 and JP57 strains and their parental strains was analyzed on 1.5% skimmed milk agar plates. As shown in Fig. 3A, the larger proteolytic halo around the sarA mutant colonies of JP56 and JP57 compared with that in the wild-type strains indicated an enhanced capacity of these strains to produce extracellular proteases and suggested that SarA is a repressor of protease production in S. epidermidis.
FIG. 3.
(A) Increased protease production by S. epidermidis sarA mutants. S. epidermidis wild-type strains O-47 and CH845 and their corresponding sarA mutants were grown in skimmed milk agar plates. (B) Biofilm formation in continuous-flow culture microfermenters of S. epidermidis O-47 and its derivative sarA mutant JP56. Biofilm development in microfermenters (upper) or on the corresponding Pyrex slides removed from the microfermenters (lower) after 24 h of growing in TSB-gluc at 37°C is shown.
Thus, we wondered whether the decreased biofilm formation by the sarA mutant strains could be the consequence of the accumulation of extracellular proteases in the microtiter plates and the degradation of a surface protein required for biofilm formation. To examine this possibility, we used microfermenters, where the medium is continuously replenished (17). As shown in Fig. 3B, the wild-type strain O-47 adhered abundantly to the submerged Pyrex spatula and, after 24 h, formed a thick biofilm. In contrast, JP56 only formed microcolonies on the surface of the slide and developed little biofilm thereafter.
Deletion of the sarA gene in S. epidermidis eliminates PIA/PNAG production.
The impact of the SarA regulator on the production of PIA/PNAG, the product of the IcaADBC proteins, was initially investigated by a dot blot with a specific anti-PNAG polyclonal antiserum (gift from G. Pier). The specificity of the polyclonal antiserum was confirmed by the absence of signal with the S. epidermidis O-47 Δica::tet strain. Our results showed that the JP56 and JP57 sarA mutants produced decreased amounts of PNAG, whereas PNAG production was restored in the complemented mutants JP58 and JP59 (Fig. 4).
FIG. 4.
Loss of PNAG production in S. epidermidis sarA mutant strains. Cell surface extracts from overnight cultures of S. epidermidis wild-type strains O-47 and CH845, their corresponding sarA mutants JP56 and JP57, respectively, the JP56 and JP57 strains complemented with plasmid pJP19 (strains JP58 and JP 59, respectively), and the O-47 Δica::tet as a negative control, treated as described in Materials and Methods, were spotted onto nitrocellulose filters. PNAG production was detected with an anti-PNAG polyclonal antibody. The sarA mutants produced lower levels of PNAG product.
SarA upregulates icaADBC expression.
The results discussed above suggested that sarA might control biofilm formation through icaADBC expression. To investigate whether the decrease in PIA/PNAG production observed in the sarA mutants was caused by a reduction of the icaADBC operon expression, we used real-time quantitative PCR. Although the extent of the decrease in the icaA RNA levels was different in both mutants, the results showed that the sarA mutation resulted in a significant (P < 0.05) decrease of icaADBC operon transcription compared to that of the wild-type strains at the mid-log exponential growth phase (OD650 = 1; Fig. 5). Similar results were obtained with RNA purified at the early stationary phase (OD650 = 2) (data not shown). These results indicate that SarA is a transcriptional activator of the icaADBC operon. However, it is worth noting the existence of considerable residual transcription of the icaADBC operon in the SarA mutant strain, a finding suggesting that SarA activity alone cannot account for the total icaADBC operon expression.
FIG. 5.
Real time quantification of ica expression on S. epidermidis wild-type strains and their corresponding sarA mutants. Asterisks denote significance (P < 0.05).
Recombinant SarA binds to the icaA promoter region.
As the level of transcript of icaA was decreased in sarA mutants, we speculated that SarA may bind to the icaA promoter region to modulate icaADBC expression. To verify this, we employed a 200-bp icaA promoter (from S. aureus or S. epidermidis) for DNA binding assays. The DNA fragments were end labeled with [γ-32P]ATP and used in gel shift assays with various amounts of purified SarA protein (Fig. 6A). The retarded protein-DNA complex could be detected with as little as 0.1 μg of SarA (≈3.3 nM). As the concentrations of the SarA protein increased, the retarded protein-DNA complex became the predominant band, with complete conversion at ≈0.2 to 0.3 μg of SarA. The presence of shifted bands with different sizes suggests the existence of several SarA-binding sites into the icaADBC promoter (Fig. 6B). Overall, these results support that SarA can bind to the icaADBC promoter and act as an activator of icaADBC transcription.
FIG. 6.
Autoradiogram of a nondenaturing 8% polyacrylamide gel with purified SarA protein and a 198-bp γ-32P-radiolabeled DNA fragment containing the intergenic promoter region of the icaRA genes. A. Lanes 1 to 6, mobility of the 198-bp radiolabeled DNA fragment (≈3 ng) of the S. aureus icaRA promoters region in the presence of 0, 50, 100, 200, 300, and 500 ng of purified protein, respectively; lanes 7 and 8, mobility of the same fragment with 300 ng of SarA, but in the presence of a 100-fold excess (molar ratio) of the unlabeled 198-bp fragment as the specific competitor (lane 7) and a 100-fold excess of unlabeled 148-bp intergenic sarUT promoter fragment (36) as the nonspecific competitor (lane 8). Lanes 9 to 16 are similar to lanes 1 to 8 except that the 198-bp fragment was from the ica promoter region of S. epidermidis. B. The nucleotide sequence of the 198-bp fragment containing the ica intergenic region of S. aureus (AF086783) and S. epidermidis (U43366) is shown and marked with the putative binding regions for SarA protein as determined based on the SarA consensus binding site (6).
SarA does not affect icaR RNA levels.
It has been reported that mutation of either σB or rsbU, an activator of σB, results in complete abolition of biofilm formation and a drastic decrease in S. epidermidis ica transcription (27, 28). A transcriptional analysis revealed that icaR is upregulated in these mutants lacking σB functions; apparently, a σB-dependent regulatory intermediate negatively regulates IcaR. It has also been shown that one of the sarA promoters (P1) is σB dependent (14, 27). We therefore speculated that σB might modulate sarA expression and the ensuing ica transcription. If such an association existed, expression of the icaR gene should be increased in the sarA mutants. An analysis of the transcriptional activity of the icaR gene by real-time PCR showed that the levels of icaR transcripts were not increased in the JP56 and JP57 sarA mutant strains (data not shown), a result suggesting control of the biofilm formation process by σB- and icaR-independent pathways.
DISCUSSION
Although the genetic arsenal responsible for pathogenesis differs between S. aureus and S. epidermidis, both species are capable of forming biofilms in a PIA/PNAG-dependent manner. In this context, a thorough comprehension of the mechanism by which PIA/PNAG is regulated in S. epidermidis and S. aureus is an important prerequisite for understanding biofilm formation and could ultimately lead to the development of methods to repress the expression of this important virulence factor. It is becoming increasingly apparent, however, that the transcriptional regulation of the genes involved in PIA/PNAG synthesis, the ica genes, is complex. This study provides experimental evidence to demonstrate that SarA regulates the expression of the ica operon in clinical isolates of S. epidermidis.
In S. aureus, the role of the SarA protein in pathogenesis has been extensively analyzed with regard to its involvement in the expression of extracellular and cell wall-associated virulence determinants. In addition, we and others have recently reported that SarA is essential for biofilm development in S. aureus (4, 47). However, little is known about the regulation of virulence by SarA in S. epidermidis. Without the typical virulence determinants of S. aureus, S. epidermidis is a common skin resident, the regulation of whose virulence determinants is still largely unknown. The existence of a SarA homolog in S. epidermidis encouraged us to examine whether SarA controls biofilm formation in coagulase-negative staphylococci. The discovery that SarA controls this process, intimately related to the persistence and antibiotic resistance of S. epidermidis infections, is a significant step in our initial approach toward understanding gene regulation in this pathogen.
Recently, in describing the role of σB in S. epidermidis biofilm formation, Knobloch and coworkers presented a model of the transcriptional and posttranscriptional regulation of PIA/PNAG synthesis and accumulation in that organism (27). This model revealed a complex regulation of PIA/PNAG synthesis, involving at least three different regulatory pathways. Two of these pathways act through the transcriptional regulation of the negative regulator IcaR, and the third pathway is a glucose-dependent proteinaceous factor of PIA/PNAG synthesis (12). Based on our results, we proposed two additional pathways: one depending on the global regulator SarA and the other depending on the δ-toxin, as previously described by Vuong and coworkers (48) (Fig. 7).
FIG. 7.
Model of regulation of PIA/PNAG synthesis in S. epidermidis.
Two lines of evidences indicated that the pathway used by SarA in the control of icaADBC transcription is different from that utilized by σB. First, our results demonstrated that SarA binds to the icaA promoter and that icaR transcription is not increased in the SarA-defective strains in comparison to wild-type strains. In contrast, a σB deficiency provokes an upregulation of icaR transcription. Second, Knobloch and coworkers observed that supplementation of growth media with ethanol decreased icaR transcription in a σB mutant, a change leading to increased icaA transcription and a biofilm-positive phenotype. However, in these growth conditions, the σB-dependent sarA transcript was absent in the σB mutant, a finding suggesting that SarA did not influence the IcaR-dependent regulation of PIA/PNAG synthesis (27). Furthermore, SarA influences the regulation of biofilm formation via an agr-dependent pathway.
The gene for δ-toxin is encoded within the gene for RNAIII, and its expression is therefore directly linked to agr activity (49). The agr two-component system does not affect the level of icaADBC expression. However, probably because of its detergent-like physicochemical properties, the agr-encoded δ-toxin, when present, abolished biofilm-forming capacity in S. epidermidis (48). Interestingly, although the S. epidermidis O-47 strain used in this study has been described as a natural agr mutant (48), deletion of the sarA gene in this strain inhibited biofilm development. These data are consistent with the conclusion that SarA affects biofilm formation via an agr-independent pathway.
In addition, other potential regulators remain to be included in this complex regulatory system. Two additional regulators, tcaR and rbf, present in the genomes of the sequenced S. epidermidis strains, have recently been described as being involved in S. aureus biofilm development. The tcaR gene is a negative regulator of ica transcription, though deletion of tcaR alone did not induce any changes in PIA/PNAG production or in adherence to polystyrene (25). On the other hand, Rbf is involved in the regulation of the multicellular aggregation step of S. aureus biofilm formation in response to glucose and salt. This regulation is probably mediated through a still-unidentified protein of 190 kDa (30). Finally, it is important to note that several SarA homologs, involved directly or indirectly in gene regulation, have been described in S. aureus. One of these regulators, SarR, has been identified in S. epidermidis (50), although is not present in all of the S. epidermidis strains analyzed (35).
In S. aureus, SarR, a 115-residue polypeptide, represses SarA expression during the postexponential phase by binding to the sarA promoter region (35). If the function of the SarR homolog in S. epidermidis is similar to that assigned in S. aureus, it is tempting to speculate that, in a mature biofilm colonizing medical devices, the expression of SarR by some bacteria could turn off their biofilm formation capacities, so that individual S. epidermidis cells could leave the biofilm and colonize new surfaces.
Proteolytic enzymes are secreted by a large number of prokaryotic organisms. In most cases, they are involved in nutrient acquisition, but a growing body of evidence indicates that peptidases produced by pathogenic bacteria are important virulence factors. In S. aureus, the expression of proteinases is tightly regulated at the level of transcription by the global regulators agr, sarA, and σB (46). In addition, proteolytic activity is controlled at the posttranslational level by a cascade of activation of the secreted zymogens. Both systems work in concert to regulate the function of these enzymes, which are suggested to facilitate S. aureus dissemination from initial colonization sites (32). This process occurs via an elaborate modification of bacterial surface proteins (26, 37, 38), changing the bacterial phenotype from adhesive to invasive.
Similar to S. aureus, S. epidermidis ΔsarA overproduces proteases. Evidence supports the implication of proteases in the regulation of biofilm development. Thus, the inactivation of gelE, encoding a zinc-metalloprotease gelatinase, prevents, by a still-uncharacterized mechanism, the primary attachment of and biofilm development by Enterococcus faecalis (21). Furthermore, it has been demonstrated that the regulation of the Hms phenotype of Yersinia pestis, involved in biofilm formation, results from the degradation of the HmsH, HmsR (homolog to IcaA), and HmsT proteins at 37°C (41).
In a previous study, we showed that inhibition or deletion of the main proteases of S. aureus (aur and ssp) in a sarA-null background was unable to restore biofilm formation. In this study, we used microfermenters in which the continuous replenishment of medium might impair the accumulation of high quantities of proteases in the growing extracellular medium. However, we cannot rule out the accumulation of a protease in the vicinity of the bacteria, a protease which, either by its physical presence or by its enzymatic activity, could be responsible for the biofilm deficiency of both the S. epidermidis and S. aureus strains. The cleavage of a surface protein could affect the hydrophobicity of the bacterial surface and prevent its attaching to surfaces such as plastic or glass. Alternatively, the overproduction of proteases could result in the degradation of Ica proteins responsible for PIA/PNAG synthesis. This rationale could explain the dissociation between residual icaADBC transcription and the absence of PIA/PNAG production in sarA mutants. In this context, it is important to note that glucose-dependent posttranscriptional regulation of PIA/PNAG synthesis has been described (12).
S. aureus and S. epidermidis are the gram-positive bacteria most often associated with medical implant-related infections. We have shown that both species control the PIA/PNAG-dependent biofilm formation process via sarA and that deletion of the sarA genes reduces the ability to produce PIA/PNAG and form a biofilm in vitro. Due to the high level of morbidity associated with S. epidermidis and S. aureus infections as well as the high frequency of infection by both organisms, the sarA gene could represent an important potential clinical target for the prevention of chronic infections associated with prosthetic medical devices.
Acknowledgments
We express our gratitude to G. B. Pier for providing the polyclonal antibodies against PNAG; F. Götz for plasmids pCU1 and pSC20 and strains O-47 and O-47 Δica::tet; Névine El Solh for strain CH845 (BM94314); and M. Arnaud and M. Debarbouille for plasmid pMAD.
This work was supported by grant BIO2002-04542-C02-01 from the Comisión Interministerial de Ciencia y Tecnología (C.I.C.Y.T.) and grants from the Cardenal Herrera-CEU University, from the Conselleria d'Agricultura, Pesca i Alimentació, and from the Generalitat Valenciana (CTIDIA/2002/62) to J.R.P. A.L.C. acknowledges financial support provided by the NIH (grant AI37142). Fellowship support for María Ángeles Tormo from the Conselleria de Cultura, Educación y Deporte and for Miguel Martí from the Conselleria d'Agricultura, Pesca i Alimentació is gratefully acknowledged.
REFERENCES
- 1.Arnaud, M., A. Chastanet, and M. Debarbouille. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Augustin, J., R. Rosenstein, B. Wieland, U. Schneider, N. Schnell, G. Engelke, K. D. Entian, and F. Götz. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149-1154. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien, Y. T., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 7.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216:171-177. [DOI] [PubMed] [Google Scholar]
- 9.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramton, S. E., M. Ulrich, F. Götz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:4079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penadés. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobinsky, S., K. Kiel, H. Rohde, K. Bartscht, J. K. Knobloch, M. A. Horstkotte, and D. Mack. 2003. Glucose-related dissociation between icaADBC transcription and biofilm expression by Staphylococcus epidermidis: evidence for an additional factor required for polysaccharide intercellular adhesin synthesis. J. Bacteriol. 185:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubin, G., D. Chmiel, P. Mak, M. Rakwalska, M. Rzychon, and A. Dubin. 2001. Molecular cloning and biochemical characterisation of proteases from Staphylococcus epidermidis. Biol. Chem. 382:1575-1582. [DOI] [PubMed] [Google Scholar]
- 14.Fluckiger, U., C. Wolz, and A. L. Cheung. 1998. Characterization of a sar homolog of Staphylococcus epidermidis. Infect. Immun. 66:2871-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frebourg, N. B., S. Lefebvre, S. Baert, and J. F. Lemeland. 2000. PCR-based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38:877-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galdbart, J. O., J. Allignet, H. S. Tung, C. Ryden, and N. El Solh. 2000. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182:351-355. [DOI] [PubMed] [Google Scholar]
- 17.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 18.Götz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 19.Götz, F., B. Kreutz, and K. H. Schleifer. 1983. Protoplast transformation of Staphylococcus carnosus by plasmid DNA. Mol. Gen. Genet. 189:340-342. [Google Scholar]
- 20.Götz, F., and B. Schumaker. 1987. Improvements of protoplast transformation in Staphylococcus carnosus. FEMS Microbiol. Lett. 40:285-288. [Google Scholar]
- 21.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heilmann, C., C. Gerke, F. Perdreau-Remington, and F. Götz. 1996. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64:277-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Götz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 24.Jefferson, K. K., S. E. Cramton, F. Götz, and G. B. Pier. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48:889-899. [DOI] [PubMed] [Google Scholar]
- 25.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to upregulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knobloch, J. K., S. Jager, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 72:3838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knobloch, J. K. M., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 30.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 32.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 33.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark, 3rd, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McAleese, F. M., E. J. Walsh, M. Sieprawska, J. Potempa, and T. J. Foster. 2001. Loss of clumping factor B fibrinogen binding activity by Staphylococcus aureus involves cessation of transcription, shedding and cleavage by metalloprotease. J. Biol. Chem. 276:29969-29978. [DOI] [PubMed] [Google Scholar]
- 38.McGavin, M. J., C. Zahradka, K. Rice, and J. E. Scott. 1997. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect. Immun. 65:2621-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 40.Pattee, P. A. 1981. Distribution of Tn551 insertion sites responsible for auxotrophy on the Staphylococcus aureus chromosome. J. Bacteriol. 145:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry, R. D., A. G. Bobrov, O. Kirillina, H. A. Jones, L. Pedersen, J. Abney, and J. D. Fetherston. 2004. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 186:1638-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rupp, M. E., P. D. Fey, C. Heilmann, and F. Götz. 2001. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 183:1038-1042. [DOI] [PubMed] [Google Scholar]
- 44.Rupp, M. E., J. S. Ulphani, P. D. Fey, K. Bartscht, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 46.Shaw, L., E. Golonka, J. Potempa, and S. J. Foster. 2004. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150:217-228. [DOI] [PubMed] [Google Scholar]
- 47.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 48.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 49.Vuong, C., F. Götz, and M. Otto. 2000. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect. Immun. 68:1048-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, Y. Q., S. X. Ren, H. L. Li, Y. X. Wang, G. Fu, J. Yang, Z. Q. Qin, Y. G. Miao, W. Y. Wang, R. S. Chen, Y. Shen, Z. Chen, Z. H. Yuan, G. P. Zhao, D. Qu, A. Danchin, and Y. M. Wen. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577-1593. [DOI] [PubMed] [Google Scholar]
- 51.Ziebuhr, W., V. Krimmer, S. Rachid, I. Löbner, F. Götz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]