Abstract
The NfrA protein, a putative essential oxidoreductase in the soil bacterium Bacillus subtilis, is induced under heat shock and oxidative stress conditions. In order to characterize the function of an homologous NfrA protein in Staphylococcus aureus, an nfrA deletion strain was constructed, the protein was purified, the enzymatic activity was determined, and the transcriptional regulation was investigated. The experiments revealed that NfrA is not essential in S. aureus. The purified protein oxidized NADPH but not NADH, producing NADP in the presence of flavin mononucleotide, suggesting that NfrA is an NADPH oxidase in S. aureus. In addition, the NfrA enzyme showed nitroreductase activity and weak disulfide reductase activity. Transcription was strongly induced by ethanol, diamide, and nitrofurantoin. Hydrogen peroxide induced nfrA transcription only at high concentrations. The expression of nfrA was independent of the alternative sigma factor σB. Furthermore, the transcriptional start site was determined, which allowed identification of a PerR box homologous sequence upstream of the nfrA promoter. The observations presented here suggest that NfrA is a nonessential NADPH oxidoreductase which may play a role in the oxidative stress response of S. aureus, especially in keeping thiol-disulfide stress in balance.
Staphylococcus aureus is a widely distributed opportunistic pathogen that is often part of the normal flora of the upper respiratory tract, as well as the skin surface, of healthy individuals. Under opportune circumstances, however, S. aureus can cause a wide variety of human diseases, which can range from mild superficial skin infections to deep and systemic infections, such as endocarditis, osteomyelitis, and septicemia (20). In addition, S. aureus is a common food pathogen that produces various superantigenic enterotoxins (6). As S. aureus is a typical nosocomial pathogen that causes up to 17% of all hospital-related infections and antibiotic treatment is essential to cure patients, the recent spread of antibiotic-resistant strains has raised major concerns worldwide (35). Most alarmingly, high-level vanA vancomycin resistance has recently been transferred from Enterococcus faecalis to S. aureus in the hospital setting (39). This event has spawned the fear that in the near future superbugs that are refractory to any antibiotic treatment will develop. Therefore, there is an urgent need to find new target structures for treatment, preferably components of pathways or cellular structures which are not targeted by currently used antibiotics (33).
To conquer different environmental niches, bacteria must adapt quickly to the changing conditions by enhanced expression of a certain subset of genes. For example, during the oxidative stress caused by oxygen-derived radicals or the disulfide stress under thiol-oxidizing conditions, several oxidoreductive systems are activated (14, 16, 18, 26, 36). For Bacillus subtilis, DNA microarray analysis revealed that NfrA is induced by superoxide stress and H2O2 stress. These results indicate that the putative essential NADPH-dependent oxidoreductase NfrA (YwcG) might play an important role in the oxidative stress response in B. subtilis (28). Transcription of the nfrA gene in this organism is growth phase dependent. The highest transcription rates were observed in the early stationary phase and were dependent on the activity of the alternative sigma factor σD, which is usually necessary to transcribe genes involved in motility and cell division (23). Homologous NfrA proteins are present in several gram-negative and gram-positive pathogens, including Escherichia coli, Shigella flexneri, Salmonella enterica serovar Typhimurium, Vibrio cholerae, Listeria monocytogenes, E. faecalis, and Bacillus anthracis (The Institute for Genome Research TIGR Microbial Database [http://www.tigr.org/tdb/mdb/mdbcomplete.html]). These proteins are probably part of the oxidative stress response systems of the bacteria. Because of their wide distribution among bacterial pathogens and possible essential role, homologous NfrA proteins may serve as putative targets for new anti-infectives.
In this study, the homologous NfrA (SA0367) protein of S. aureus was characterized. We constructed an nfrA deletion strain which showed that the protein is not essential in S. aureus. Further biochemical analysis of the NfrA protein revealed an NADPH oxidoreductase activity. In addition, transcriptional regulation of the nfrA gene was investigated under different stress conditions. Our results provide evidence that S. aureus NfrA is involved in maintenance of the cellular redox state and the disulfide stress response.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli and S. aureus were grown in Luria-Bertani (LB) medium, and liquid cultures were shaken at 200 rpm. The bacteria were grown at 37°C, unless indicated otherwise. Antibiotics were used at the following concentrations: 100 μg of ampicillin ml−1 and 10 μg of chloramphenicol ml−1 for E. coli and 10 μg of erythromycin ml−1 and 10 μg of chloramphenicol ml−1 for S. aureus. Growth curve analysis was done without addition of antibiotics.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or phenotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F−endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1(A) Δ(argF-lac)U169φ80dlacZΔM15 | BRL |
| BL21(DE3)/pLysS | F−ompT hsdSB (rB− mB−) gal dcm (DE3)/pLysS (Camr); strain for overexpression of proteins | Invitrogen |
| S. aureus strains | ||
| 8325 | NCTC 8325 (wild type, with 11-bp deletion in rsbU) | Laboratory stock |
| RN4220 | NCTC 8325-4-r (restriction mutant, with 11-bp deletion in rsbU) | Laboratory stock |
| MA12 | Clinical nasal isolate | 34 |
| MA12.2 | sigB deletion strain of MA12 | 34 |
| 8325 ΔnfrA | nfrA deletion strain of 8325 | This study |
| Plasmids | ||
| pBT2 | Shuttle vector, Apr in E. coli, Cmr in S. aureus | 2 |
| pKSt2 | Deletion vector for nfrA, erythromycin cassette flanked by fragments upstream and downstream of nfrA in pBT2; Apr in E. coli, Emr and Cmr in S. aureus | This study |
| pGEM-T | Apr | Promega |
| pGEM-TnfrAPEx | Apr, 832 bp of 5′ nfrA and upstream region for primer extension sequencing reaction | This study |
| pGKSt1 | Apr, 837-bp EcoRI-Xbal fragment upstream of nfrA in pGEM-T | This study |
| pGKSt2 | Apr, 874-bp BamHI-PstI fragment downstream of nfrA in pGEM-T | This study |
| pEC1 | Apr, EmrermB fragment in pUC18 | 2 |
| pGKSt11 | Apr, Emr EcoRI-PstI fragment of ermB in pGEM-T | This study |
| pCRT7/CT-TOPO | Apr, protein expression vector | Invitrogen |
| pCRT7/CT-TOPO-nfrA | Apr, protein expression vector for expression of NfrA | This study |
Construction of S. aureus nfrA deletion strain.
The gene of interest (nfrA) was deleted by replacing the coding sequence of the gene with the coding sequence of the erythromycin resistance cassette (ermB) by a double-crossover event, as described by Brückner (2). Fragments upstream and downstream of the target gene were amplified by PCR, and restriction sites were added to the primers to facilitate cloning. For the upstream fragment EcoRI and XbaI restriction sites (underlined) were used (ywcGEcoRI [5′-CGG AAT TCC AGT TTC TCA CTC ATT ACC-3′] and ywcGXbaI [5′-TGC TCT AGA TTA CGA ACA TAT TGA GCT GC-3′]). The length of the fragment was 837 bp, and the fragment started 140 bp before the start codon of nfrA. For the downstream fragment PstI and BamHI restriction sites were added (ywcGPstI [5′-AAC TGC AGT ACA GCG ATA GCA AGA TAC C-3′] and ywcGBamHI [5′-CGG GAT CCC ATG GTG AAT TGC TTA AGC G-3′]). The length of the downstream fragment was 873 bp, and the fragment contained 11 bp of the 3′ end of the open reading frame (ORF) of nfrA. Between these two fragments the erythromycin resistance cassette (ermB) was cloned. The ermB gene was amplified from the pEC1 vector (2), and EcoRI and PstI restriction sites were added to the primers (ermBEcoRI [5′-CGG AAT TCG GTG ACA TCT CTC TAT TGT G-3′] and ermBPstI [5′-AAC TGC AGG GAA GCT GTC AGT AGT ATA CC-3′]). The length of the fragment was 1,364 bp. The fragments were first cloned into the pGEM-T vector (Promega, Mannheim, Germany). Then the fragments were cut out with the corresponding restriction enzymes and cloned into the temperature-sensitive shuttle vector pBT2 (2). Construction of this deletion vector was carried out in E. coli DH5α. The vector construct was introduced into S. aureus strain RN4220 by electroporation. After this the vector was introduced into S. aureus strain 8325 by transduction with phage Φ85. In this strain gene inactivation was carried out as described by Brückner (2).
RNA techniques.
RNA was isolated from cells at different times during bacterial growth, as indicated below for the different experiments. At least 4 ml of a bacterial culture was harvested and mixed with 2 volumes of RNA-protect (Qiagen, Hilden, Germany). The cells were centrifuged for 8 min at 3,000 × g, and the bacterial pellet was resuspended in 1 ml of RLT buffer and mechanically disrupted with glass beads (2 ml of lysing matrix E; Bio 101, Inc., San Diego, Calif.) in a Fast Prep shaker (Bio 101, Inc.). The cell lysate was centrifuged for 10 min at 16,000 × g, and the supernatant was used for RNA isolation. RNA was isolated with an RNeasy Mini kit (QIAGEN) used according to the instructions of the manufacturer. To remove the DNA template, RNA was treated with RNase-free DNase I (Roche, Penzberg, Germany). Two micrograms of RNA was reverse transcribed at 50°C for 1 h by using Superscript III reverse transcriptase (Invitrogen, Karlsruhe, Germany). For the reverse transcription 200-ng portions of random hexamer primers (Amersham, Freiburg, Germany) were used in each reaction. The reaction mixture was inactivated by incubating it at 70°C for 15 min. For amplification of the nfrA cDNA, the following primers were used: ywcGSo1 (5′-AAT GCT GCA TCG ATT GCA CC-3′) and ywcGSo2 (5′-GTA AAG TAT CGA GGA GTG GG-3′). As a control, 16S rRNA primers (16SrRNAup [5′-TTG CTT CTC TGA TGT TAG CG-3′] and 16SrRNAdown [5′-TCT AAT CCT GTT TGA TCC CC-3′]) were used. For relative quantification of nfrA expression, real-time PCR (RT-PCR) was performed by using the MyiQ device (Bio-Rad, Munich, Germany). For the amplification reaction, the iQ SYBER Green supermixture (Bio-Rad) was used. Relative quantification of nfrA transcription was performed as described by Pfaffl (32).
For Northern blot analyses 12 μg of total RNA was loaded into each lane of a 1.2% (wt/vol) denaturing agarose gel. The RNA gel was blotted on a nylon membrane (Schleicher and Schuell, Dassel, Germany) overnight by capillary blotting. DNA probes for detection of the nfrA or 16S rRNA transcripts were synthesized by PCR with the same primers that were used for the RT-PCR (see above). DNA probes were labeled by using a nonradioactive enhanced chemiluminescence detection kit (ECL; Amersham) as described by the manufacturer.
Primer extension.
RNA for primer extension assays was isolated from ethanol (6.5%)-induced S. aureus cells with an RNeasy mini kit (QIAGEN) used according to the instructions of the manufacturer. Primer extension analyses were carried out with infrared dye-labeled primer ywcGPEx (5′-ACA ACG TCT TCA CTT AAA GG-3′) with an automated DNA sequencer (LI-COR-DNA 4000; MWG-Biotech, Munich, Germany). The primer (4 pmol) was annealed in a thermocycler to 15 μg of RNA in 15 μl of H2O (RNase-free) by heating at 90°C for 2 min and subsequent cooling to 30°C within 30 min. Extension was performed in a 30-μl (total volume) mixture at 52°C for 90 min. For this procedure, Superscript III reverse transcriptase (Invitrogen) was used. Nucleic acids were precipitated with ethanol. The pellet was dissolved in 1 μl of H2O and 3 μl of loading buffer, and 0.5 or 1.5 μl of the sample was applied to a sequencing gel. Sequencing reactions were performed by using the same primer that was used for the primer extension analysis and a LI-COR sequencing kit as described by the manufacturer (Epicentre Technologies, Madison, Wis.). For the sequencing reaction the 5′ region of the nfrA gene containing 323 bp of the 5′ nfrA ORF and 539 bp of the upstream sequence was cloned into the pGEM-T vector (Promega). The primer sequences used for cloning the fragments were as follows: nfrAupPEx1, 5′-AGC AAA CCTTCC GTT GAA CC-3′; and nfrAupPEx2, 5′-AAG CTT GCG CTG TAA ATG G-3′.
Purification and characterization of the NfrA protein.
The NfrA protein was expressed as a His-tagged fusion protein by using a pCRT7TOPO TA expression kit (Invitrogen). The NfrA protein was amplified with primers TrywcGup (5′-ATG TCA GAA CAT GTA TAT AAT CTT GTG-3′) and ywcGdown (5′-TCG CTG TAT TAA GCC TGA TT-3′). The PCR product was amplified from chromosomal DNA by using the TRIPL Master system (Eppendorf, Hamburg, Germany). The PCR product was cloned into the pCRT7/CT-TOPO-expression vector (Invitrogen). For protein expression, the plasmid was transformed into E. coli BL21(DE3). Cells were grown in LB medium to an optical density at 600 nm (OD600) of 0.5. Isopropyl-β-d-thiogalactoside (IPTG) was added to a final concentration of 1 mM. Five hours after induction, cells were harvested and resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole; pH 8.0). The same volume of glass beads (diameter, 0.25 to 0.5 mm; Roth, Karlsruhe, Germany) and 10 U of benzonase (Merck, Darmstadt, Germany) were added to the sample, and the cells were lysed by vortexing the mixture vigorously for 10 min at 4°C. The sample was centrifuged twice for 10 min at 6,000 rpm at 4°C. The supernatants were subjected to treatment with an Ni-nitrilotriacetic acid resin spin column used according to the manufacturer's instructions. The column was washed three times with 40 mM imidazole and subsequently eluted five times with imidazole at a concentration of 250 mM. Only the last two eluted fractions were used for enzymatic analysis of the protein. The protein was dialyzed against 0.1 M NaPO4 buffer with 50 mM NaCl. Enzymatic assays were performed at 21°C in 0.1 M NaPO4 buffer containing 50 mM NaCl at pH 6.8; 100 μM flavin mononucleotide (FMN) (or 100 μM flavin adenine dinucleotide [FAD]) was added as an electron acceptor. The change in the NADPH (or NADH) concentration was inferred from the change in absorption at 340 nm (absorption coefficient for NADPH at 340 nm [ɛ], 6.22 M−1 cm−1). The initial velocity of the enzymatic reaction was determined for 10, 15, 20, 50, 100, and 200 μM NADPH. Km values were determined by using a Lineweaver-Burk plot. The reductase activity of NfrA was also analyzed with nitrofurazone, nitrofurantoin, ferricyanide, and 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) in the presence of 100 μM NADPH and 20 μM FMN. The reaction was initiated by addition of FMN. The initial velocity was determined by monitoring the change in absorbance of the reaction mixture every 8 s for the first 1.5 min. The following molar absorption coefficients were used: 12.96 M−1 cm−1 (for nitrofurazone at 400 nm), 1.0 M−1 cm−1 (for ferricyanide at 420 nm), 12.0 M−1 cm−1 (for nitrofurantoin at 420 nm), and 13.6 M−1 cm−1 (for DTNB at 412 nm).
Antibiotic susceptibility tests.
The MIC was determined by the microdilution method performed according to the suggestions of the National Center for Clinical Laboratory Standards. The MIC was determined in 96-well microtiter plates by using a 200-μl (final volume) mixture without agitation. The initial inoculum was 2 × 103 bacterial cells per well. The plates were incubated for 18 h at 30°C.
Sequence analysis and database search.
An analysis of the S. aureus strain 8325 sequence was performed with the database of the University of Oklahoma Advanced Center for Genome Technology (http://www.genome.ou.edu/). The sequences of S. aureus strains N315, Mu50, and MW2 are available at the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov). The sequence of S. aureus strain COL was obtained from The Institute for Genomic Research website (http://www.tigr.org). The sequences of S. aureus strains MRSA252 and MSSA476 are available at the Sanger Institute website (http://www.sanger.ac.uk). BLAST searches of the nucleotide and protein sequences were done by using the database of the NCBI. The search for ORFs was performed with the Vector NTI Suite program (InforMax, North Bethesda, Md.) and the ORF finder program of NCBI.
RESULTS
Chromosomal location of the nfrA gene.
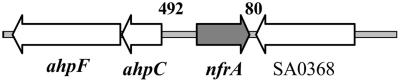
The organization of the nfrA gene is the same in all seven S. aureus strains sequenced so far (S. aureus N315, Mu50, MW2, MRSA252, MSSA476, 8325, and COL). The nfrA gene is transcribed monocistronically. The length of the transcript was estimated by Northern blot analyses to be about 900 bp (data not shown). The ORF consists of 756 bp which, when expressed, gives rise to a 251-amino-acid protein. Next to the nfrA gene is the ahpCF operon, which codes for the small and large subunits of the alkyl hydroperoxide reductase (Fig. 1).
FIG. 1.
Chromosomal localization of the nfrA gene in S. aureus strain N315. The chromosomal organization of the nfrA gene is the same for all seven S. aureus strains sequenced so far. The nfrA gene gives rise to a monocistronic transcript that is about 900 bp long. The length of the transcript was estimated by Northern blot analysis. The protein consists of 251 amino acids. The adjacent genes are transcribed from the DNA minus strand. The ahpC and ahpF genes form an operon and encode the small and large subunits of the alkyl hydroperoxide reductase, respectively. SA0368 encodes a hypothetical protein which is similar to proton/sodium glutamate symport proteins. The numbers above the intergenic region indicate the numbers of nucleotides between adjacent ORFs.
The nfrA gene is transcribed throughout the life cycle of S. aureus.
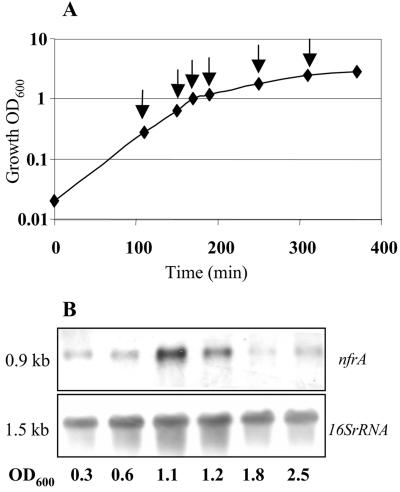
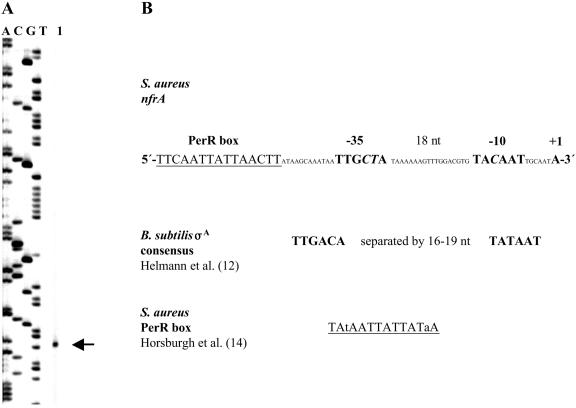
Transcription analysis of nfrA expression was performed by RT-PCR and Northern blot analysis. These experiments revealed that the nfrA gene is transcribed throughout the life cycle of S. aureus. Transcription was slightly increased during late-exponential growth (at OD600 between 1.0 and 1.2), during which there were twofold to threefold increases compared to mid-log or stationary-phase cells (Fig. 2). The promoter region of the nfrA gene was determined by primer extension analysis of S. aureus strain 8325 (Fig. 3). Transcription starts at an adenine nucleotide. The 5′ end of the transcript is 44 nucleotides upstream of the putative GTG translation start codon. The −10 and −35 sequences upstream of the transcriptional start site correlate well with the consensus sequence of σA-dependent promoters (12) (Fig. 3). In addition, a putative PerR box was identified 12 nucleotides upstream of the −35 promoter region (Fig. 3).
FIG. 2.
(A) Growth curve for S. aureus MA12 in LB medium at 37°C under aerobic conditions. Samples for RNA isolation were taken in different growth phases, as indicated by the arrows. (B) Northern blot analysis of RNA isolated at the early and mid-exponential and stationary phases. Northern blots were hybridized to nfrA and 16S rRNA probes.
FIG. 3.
(A) Primer extension analysis of the nfrA transcript. Total RNA was isolated from ethanol-induced, mid-log-phase cells of S. aureus 8325. The product of the primer extension reaction was separated by electrophoresis under denaturing conditions alongside sequencing reaction mixtures with the same primer. The position of the transcription start site is indicated by an arrow. The transcript starts at an adenine. (B) Nucleotide sequence of the nfrA promoter. The sequence is compared to the B. subtilis σA promoter consensus sequence. Putative promoter elements are indicated by large boldface letters. Nucleotides deviating from the consensus sequence are indicated by italics. Transcription of the nfrA transcript starts after a σA-dependent promoter. The sequence of a putative PerR box 12 nucleotides upstream of the promoter region is underlined. nt, nucleotides.
Effects of different stress stimuli on the expression of nfrA.
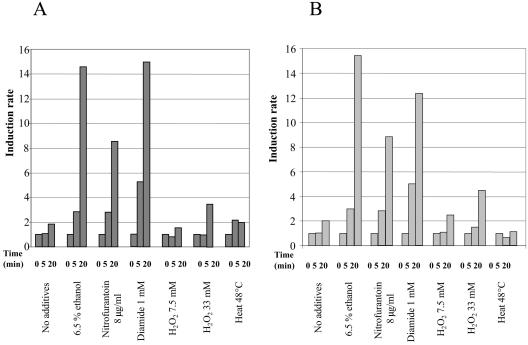
It was previously determined that expression of the nfrA gene of B. subtilis was induced upon heat shock (24). In a DNA microarray experiment with B. subtilis it was recently shown that nfrA was also induced upon oxidative stress (28). We examined the growth of S. aureus 8325 (a laboratory strain) and S. aureus MA12 (a clinical isolate) under different stress conditions. The bacteria were grown in LB medium at 37°C until the OD600 was between 0.5 and 0.7, and then stress was applied to a portion of the culture. For heat stress the culture was shifted to 48°C, for salt stress NaCl was added to a final concentration of 0.7 M, and for ethanol stress ethanol was added to a final concentration of 6.5%. RNA was isolated from the stressed and unstressed bacteria and was subjected to Northern blot analyses and RT-PCR. In contrast to nfrA expression in B. subtilis (24), no induction of nfrA was observed in response to heat in S. aureus (Fig. 4). There was also no induction of nfrA in response to salt stress (data not shown). However, transcription of nfrA was induced 3-fold with ethanol stress for 5 min and 16-fold with ethanol stress for 20 min (Fig. 4). A concentration of ethanol of 10% led to approximately 25-fold induction of nfrA mRNA after 20 min of incubation. Isopropanol induced nfrA transcription to the same extent as ethanol (data not shown). Induction of nfrA transcription was still observed when ethanol was added to a culture in the stationary phase at an OD600 of 2.8 (data not shown).
FIG. 4.
Results of the RT-PCR experiments. Various stresses were applied to exponentially growing cells of S. aureus 8325 (A) and MA12 (B), as indicated at the bottom. Total RNA of the bacteria was isolated before and 5 and 20 min after exposure to the stress. Equal amounts of RNA (2 μg) were reverse transcribed to cDNA with random hexamer primers, and then the cDNA was subjected to PCR with gene-specific primers as described in Materials and Methods. For normalization of the samples 16S rRNA was used. The increase in expression of the nfrA mRNA at 5 and 20 min was calculated relative to the expression at zero time before stress was applied. Relative quantification of transcription was performed as described by Pfaffl (32). The data are the averages of two independent experiments.
We also investigated transcription of nfrA in response to oxidative stress. A portion of a culture was incubated with either 7 or 33 mM H2O2 at the mid-log phase. Addition of 7 mM H2O2 had no significant effect on transcription of nfrA. At the higher concentration of H2O2 (33 mM) a three- to fourfold increase in the nfrA transcript was observed (Fig. 4). Both S. aureus strains were surprisingly resistant to H2O2; after addition of 33 mM H2O2 a growth arrest of about 20 min occurred, and then the cells resumed growth. This could have been due to high levels of catalase activity, which in some S. aureus strains quickly detoxifies H2O2 (5, 38).
Further investigations of nfrA expression were performed with diamide and nitrofurantoin. Diamide is a specific oxidant for thiols and causes disulfide stress in B. subtilis (18). The transcription of nfrA was induced about fivefold 5 min after addition of 1 mM diamide to mid-log-phase cells. After 20 min the induction was about 12- to 15-fold (Fig. 4).
Addition of nitrofurantoin to a bacterial culture can also lead to oxidative stress (22, 40). Nitrofurantoin was added to mid-log-phase growing cells at a concentration of 8 μg/ml (twice the MIC), and ninefold induction of nfrA was observed 20 min after addition of the compound (Fig. 4). Since the nitrofurantoin was dissolved in dimethyl sulfoxide, the impact of the solvent on nfrA transcription was also tested. This experiment revealed no influence of the solvent dimethyl sulfoxide on nfrA transcription (data not shown). All induction experiments were analyzed by RT-PCR (Fig. 4) and Northern blot hybridization (data not shown).
Role of the alternative sigma factor σB in transcription of nfrA in response to different stress stimuli.
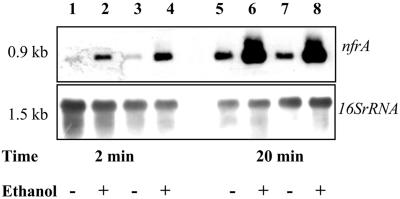
In S. aureus the alternative sigma factor σB is involved in the H2O2- or heat-induced stress response (3, 9). S. aureus strain 8325 is deficient in the RsbU phosphatase (8), which is necessary to activate σB (10, 29). Therefore, we investigated whether S. aureus strain MA12, which is fully competent for the σB pathway (34), responded differently to oxidative stress caused by H2O2, nitrofurantoin, diamide, or heat stress. The experiments revealed no differences in transcription of nfrA between S. aureus MA12 and strain 8325 (Fig. 4). To rule out the possibility that σB plays an indirect role in stress-induced transcription of nfrA, the induction of nfrA in a σB deletion strain (MA12.2) was also investigated. We performed induction experiments with ethanol using S. aureus strain MA12 and an isogenic mutant of this strain, MA12.2 (34). The same extent of transcription of the nfrA gene in response to ethanol was observed in both strains (Fig. 5). These results clearly demonstrated that σB is not involved in nfrA transcriptional regulation.
FIG. 5.
Northern blot analysis of nfrA transcription in S. aureus strain MA12 (lanes 1, 2, 5, and 6) and in the isogenic σB deletion strain MA12.2 (lanes 3, 4, 7, and 8). Bacteria were treated with 6.5% ethanol in the mid-log phase. RNA was isolated at different times after addition of ethanol. The expression of nfrA was compared to the expression in samples to which no ethanol was added, as indicated at the bottom.
Biochemical characterization of the NfrA protein revealed that it is an NADPH-dependent FMN-containing oxidoreductase.
For biochemical characterization of the NfrA protein, NfrA was expressed as a His-tagged fusion protein, as described in Materials and Methods. The calculated molecular mass of the NfrA protein is 28.6 kDa. Analyses of the NfrA protein on denaturing protein gels revealed that the molecular mass of the protein was about 32 kDa. The reason for the difference is the addition of the His tag to the protein. The C-terminal tag accounted for 3.3 kDa.
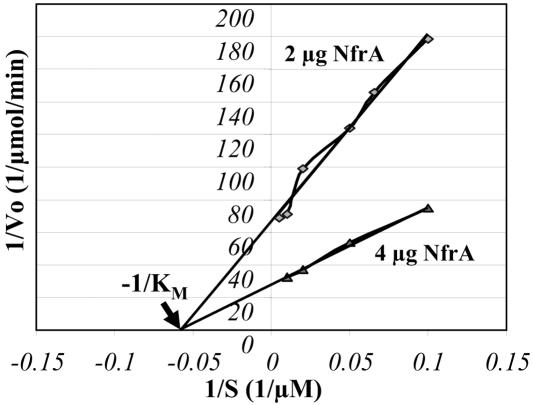
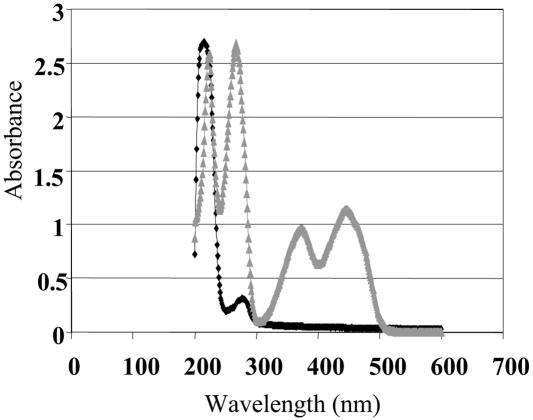
The homology of NfrA to bacterial oxidoreductases suggests that the protein has a similar function in S. aureus. To analyze the enzymatic activity of NfrA, the purified protein was added to samples containing NADH or NADPH as the substrate and 0.1 M FMN as the electron acceptor. The Km was determined with a Lineweaver-Burk plot. The Km for NADPH was estimated to be 17 μM and was determined for two different concentrations of the enzyme (Fig. 6). There was no enzymatic activity when NADH was used as the substrate. Heat inactivation of the enzyme at 70°C for 10 min also abolished the enzymatic reaction. Furthermore, the enzymatic reaction was analyzed with 0.1 M (constant concentration) FAD as the electron acceptor instead of FMN. No enzymatic reaction was observed with FAD as the electron acceptor, either with NADPH or with NADH. There was also no enzymatic reaction when an electron acceptor like FMN was omitted. The absorption spectrum of the purified NfrA protein had peaks at 214 and 280 nm, which is typical for solutions of pure protein. In contrast, the spectrum of a solution of the free cofactor FMN had peaks at 224, 268, 375, and 448 nm (Fig. 7). These results suggest that the cofactor FMN was only loosely bound to the enzyme and was lost during purification of the protein. In addition, we analyzed the reductase activity of NfrA with different electron acceptors. The enzyme was active with organic nitro compounds, like nitrofuran and nitrofurantoin. The NfrA enzyme also catalyzed the reduction of the disulfide DTNB to the thiol 5-thio-2-nitrobenzoic acid, although the observed disulfide reductase activity was low (Table 2). All enzymatic activities were dependent on addition of FMN.
FIG. 6.
Lineweaver-Burk plot and deduced Km value. The plot is a double-reciprocal plot of initial velocity versus concentration of NADPH. The Km was determined for two different amounts of enzyme, as indicated on the graph, and with 100 μM (constant concentration) FMN as the electron acceptor. The Km was 17 μM for NADPH at 21°C and pH 6.8.
FIG. 7.
Absorption spectra of the purified NfrA protein and free FMN. The black symbols show the spectrum of NfrA at a concentration of 0.3 mg/ml (9.4 μM), and the grey symbols show the spectrum at 10 μM FMN in 0.1 M NaPO4 buffer (pH 6.8) containing 50 mM NaCl. The NfrA spectrum has peaks at 214 and 280 nm, whereas the peaks for free FMN are at 224, 268, 375, and 448 nm.
TABLE 2.
Electron acceptor specificities of NfrA and NfsAa
| Electron acceptor | Reductase activity (μmol/min/mg of protein)
|
|
|---|---|---|
| NfrA | NfsA | |
| FMN | 16 | 1 |
| Nitrofurazone | 20 | 73 |
| Nitrofurantoin | 15 | 82 |
| Ferricyanide | 97 | 251 |
| DTNB | 0.12 | NDb |
Data from reference 42.
ND, not determined.
Deletion of the nfrA gene.
Attempts by Moch et al. (23) to inactivate the homologous nfrA gene of B. subtilis were not successful. Therefore, it was assumed that the nfrA gene is essential for growth of B. subtilis. We deleted the nfrA gene of S. aureus strain 8325 by replacing the coding sequence of the nfrA gene with the coding sequence of the erythromycin resistance cassette ermB, as described in Materials and Methods. The change was confirmed by Southern blot analyses (data not shown). The absence of the nfrA transcript was also confirmed by Northern blot analyses and by RT-PCR (data not shown). The nfrA gene is clearly not essential in S. aureus. The nfrA deletion strain did not show any growth defect in LB medium at 30, 37, or 42°C compared to the wild-type strain (data not shown). The susceptibility to hydrogen peroxide and diamide was not altered in the nfrA mutant strain compared to the wild type. Furthermore, the nfrA mutant was not attenuated in a mouse sepsis model (data not shown).
DISCUSSION
Determinations of the complete genome sequences of bacterial pathogens are accelerating the exploration of virulence mechanisms and also provide the basis for a thorough investigation of the physiology of microorganisms. Comparative genome analyses along with functional genomic studies help identify particular genes or pathways in a given organism. It is a great challenge in the postgenome era to unravel the functions of unknown proteins, which account for approximately one-third of all proteins. In this report, we characterized a novel oxidoreductase (SA0367) in S. aureus. Searching putative new targets for anti-infectives, we found that S. aureus produces a protein with a high level of homology to the oxygen-insensitive NADPH nitroreductase NfsA of E. coli, which is the major nitroreductase in this organism, and to NfrA (YwcG) of B. subtilis (23, 41). NfrA is widely distributed among bacterial species. For example, homologous counterparts were found by BLAST searches in L. monocytogenes, Listeria innocua, and E. faecalis and in different Bacillus species, including B. subtilis, B. cereus, and B. halodurans, and B. anthracis, as well as in several gram-negative bacteria, such as E. coli, S. flexneri, S. enterica serovar Typhimurium, and V. cholerae. In the NfrA protein of S. aureus and the NfrA protein of B. subtilis 42% of the amino acids are identical.
In B. subtilis nfrA is transcribed in an nfrA-ywcH operon, in which nfrA encodes an oxidoreductase and ywcH encodes a putative monooxygenase (24). The protein homologous to NfrA in E. coli, NfsA, shares 37% amino acids with the NfrA protein of S. aureus. This protein is transcribed in an operon in which ybjC, a small ORF whose function is unknown, is located upstream of nfsA (19, 30). In contrast to B. subtilis and E. coli, we found that the nfrA gene is transcribed monocistronically in S. aureus. Interestingly, we identified a symmetrical sequence that is 12 nucleotides upstream of the −35 box of the putative nfrA promoter which resembles the consensus sequence of the perR box described previously (14). In the putative perR box (TTCAATTATTAACTT) upstream of the nfrA promoter 9 of 14 nucleotides are identical to nucleotides in the consensus sequence (TATAATTATTATAA). PerR is a manganese-dependent transcriptional repressor which is involved in the regulation of oxidative stress-related components like KatA, AhpC, FtnA, and MrgA (14, 15, 27). Most, but not all, members of the PerR regulon are induced by H2O2 (14). Induction of nfrA following addition of hydrogen peroxide in this study, however, was observed only at high concentrations (up to a final concentration of 33 mM). Even with these high concentrations of hydrogen peroxide the induction of nfrA transcription was rather weak, and there was only a three- to fourfold increase after 20 min of cultivation in the presence of hydrogen peroxide. At an H2O2 concentration of 33 mM growth arrest of the bacteria was observed for about 20 min, and after this time the bacteria continued to grow, which indicates that the cells suffered stress under these conditions. Interestingly, Weber et al. (38) reported that the alkyl hydroperoxide reductase (AhpC), which belongs to the PerR regulon in S. aureus, was not induced in strain COL during the first 10 min of oxidative stress at very high concentrations of H2O2 (100 mM) and, importantly, the catalase (KatA) was even repressed during the first 30 min of H2O2 stress. Horsburgh et al. (14) observed slight induction of katA (twofold) and ahpC (threefold) after addition of 0.5 mM H2O2 in strain S. aureus 8325-4 using lacZ fusions. Overall, only a few proteins were significantly induced under H2O2 stress conditions, indicating that the regulation of the oxidative stress response due to H2O2 in S. aureus is different from the regulation of this response in B. subtilis (4, 7, 14, 28, 38).
The S. aureus strain 8325 used in this study was surprisingly resistant to hydrogen peroxide, which might have been due to the high catalase activity (14, 38). Probably, the high catalase activity contributed to the quick detoxification of hydrogen peroxide. Also, the thioredoxin (trxA) and thioredoxin reductase (trxB) genes, which are part of thiol-specific redox systems in S. aureus that play a major role in the protection of cells against toxic oxygen species, are not induced by H2O2 but are induced by the thiol-specific oxidant diamide and by τ-butyl hydroperoxide, an organic peroxide (37). TrxB has also been shown to belong to the PerR regulon in S. aureus (14). All these reports may indicate that the oxidative stress-related induction potential of hydrogen peroxide in S. aureus is low. Notably, it has also been reported that the perR gene of B. subtilis is not induced by H2O2 stress (7) but is induced by disulfide stress (18). This induction pattern is similar to that of the nfrA gene of S. aureus reported here, as transcription of the nfrA gene was strongly induced upon oxidative stress caused by diamide or nitrofurantoin. Diamide is a specific oxidant for thiols and induces disulfide stress in bacteria. Disulfide stress can be considered a subcategory of oxidative stress, as one major effect of reactive oxygen species is the oxidation of thiols, resulting in disulfide bond formation (18). Nitrofurantoin causes oxidative stress by generation of superoxide radicals. Nitrofurantoin is reduced by cellular enzymes to the free radical. Under aerobic conditions, the nitrofurantoin radical undergoes oxidation to regenerate the parent compound with concomitant production of superoxide (21, 31, 40). Interestingly, diamide and nitrofurantoin induce similar patterns of proteins, as recently demonstrated by a proteomic approach, suggesting that nitrofurantoin also causes disulfide stress (1).
In addition, transcription of nfrA was strongly induced in response to ethanol. In B. subtilis ethanol induces heat stress-specific proteins which are controlled by the alternative sigma factor σB, probably because ethanol treatment simulates heat stress by increasing the formation of nonnative proteins (11, 25). We demonstrated that induction of nfrA by ethanol, diamide, and nitrofurantoin is not under control of the alternative sigma factor σB, which has been shown to be involved in the oxidative stress response in S. aureus (9, 13, 17). The induction pattern was similar in the sigB wild-type-strain MA12, the corresponding sigB mutant MA12.2, and the rsbU deletion strain 8325, which has an 11-bp deletion in the rsbU gene that is necessary to fully activate SigB-dependent transcription (10, 17, 29). Furthermore, primer extension assays revealed that transcription of nfrA starts at promoter sequences typical for SigA-dependent transcription. The nfrA gene of B. subtilis is induced by heat stress (24), as well as by oxidative stress caused by hydrogen peroxide and paraquat (28). Regulation of nfrA expression by use of different sigma factors in B. subtilis, however, is still being investigated since Moch et al. have identified putative SigA-, SigB-, and SigD-dependent promoter sequences in front of the nfrA gene. Moreover, heat induction of nfrA was still observed in a SigB- and SigD-deficient strain and also in a CtsR- and HrcA-deficient strain (24).
The data presented here indicate that regulation of nfrA expression is different in S. aureus and B. subtilis. Finally, the exact function of NfrA in both organisms has to be defined. We found that the NfrA protein of S. aureus possesses NADPH-dependent oxidoreductase activity. The same enzymatic function has been determined for NfrA of B. subtilis and for NfsA of E. coli. It was previously shown that NfsA has broad electron acceptor specificity and reduces flavins, nitro compounds, and quinones (42). We showed that the NfrA protein of S. aureus can reduce FMN in the presence of NADPH. Moreover, we also demonstrated that the NfrA enzyme of S. aureus reduces organic nitro compounds like nitrofurantoin and nitrofurazone, like the NfsA enzyme of E. coli. Interestingly, NfrA also exhibited weak disulfide reductase activity. Thus, the NfrA enzyme of S. aureus might contribute electrons from NADPH to different oxidized or otherwise damaged proteins under different stress conditions. Further work has to be done to find out which molecules interact directly with the NfrA enzyme and to determine the exact role of nfrA in the responses to different kinds of oxidative stress in S. aureus.
NfrA was postulated to be an essential protein in B. subtilis (23). Therefore, we analyzed whether NfrA is also essential in S. aureus. Importantly, we were able to construct an nfrA deletion strain, suggesting that this protein is not essential in S. aureus. The nfrA deletion mutant showed the same pathogenicity as the wild type in a mouse sepsis model. This finding may reflect a less important role of NfrA during infection. Nevertheless, the enzyme could have a significant function in the bacterial stress response during phases of oxidative stress in other types of infections. Although NfrA is not essential in S. aureus, the work presented here emphasized the importance of the enzyme as part of the oxidative stress response. Further work, particularly work to gain a better understanding of the role of NfrA during the infection process, is necessary before a decision can be made about whether NfrA or homologous proteins may serve as new targets for anti-infective therapy.
Acknowledgments
This study was supported by a grant from the Bayerische Forschungsstiftung, by a grant from the Bundesministerium für Bildung und Forschung (BMBF) Network of Competence “Pathogenomics” Alliance “Gram-Positive Cocci,” and by the Deutsche Forschungsgemeinschaft (grant SFB479).
We thank Heike Bruhn and Klaus Heuner for technical advice and Susanne Engelmann for helpful discussions.
REFERENCES
- 1.Bandow, J. E., H. Brotz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brückner, R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1-8. [DOI] [PubMed] [Google Scholar]
- 3.Chan, P. F., S. J. Foster, E. Ingham, and M. O. Clements. 1998. The Staphylococcus aureus alternative sigma factor σB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model. J. Bacteriol. 180:6082-6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clements, M. O., and S. J. Foster. 1999. Stress resistance in Staphylococcus aureus. Trends Microbiol. 7:458-462. [DOI] [PubMed] [Google Scholar]
- 6.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuangthong, M., A. F. Herbig, N. Bsat, and J. D. Helmann. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184:3276-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 9.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hecker, M., and U. Volker. 1998. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the σB regulon. Mol. Microbiol. 29:1129-1136. [DOI] [PubMed] [Google Scholar]
- 12.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horsburgh, M. J., S. J. Wharton, A. G. Cox, E. Ingham, S. Peacock, and S. J. Foster. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44:1269-1286. [DOI] [PubMed] [Google Scholar]
- 16.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 17.Kullik, I. I., and P. Giachino. 1997. The alternative sigma factor σB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167:151-159. [DOI] [PubMed] [Google Scholar]
- 18.Leichert, L. I., C. Scharf, and M. Hecker. 2003. Global characterization of disulfide stress in Bacillus subtilis. J. Bacteriol. 185:1967-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liochev, S. I., A. Hausladen, and I. Fridovich. 1999. Nitroreductase A is regulated as a member of the soxRS regulon of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:3537-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 21.McOsker, C. C., and P. M. Fitzpatrick. 1994. Nitrofurantoin: mechanism of action and implications for resistance development in common uropathogens. J. Antimicrob. Chemother. 33(Suppl. A):23-30. [DOI] [PubMed] [Google Scholar]
- 22.Miller, C., L. K. Folkes, C. Mottley, P. Wardman, and R. P. Mason. 2002. Revisiting the interaction of the radical anion metabolite of nitrofurantoin with glutathione. Arch. Biochem. Biophys. 397:113-118. [DOI] [PubMed] [Google Scholar]
- 23.Moch, C., O. Schrogel, and R. Allmansberger. 1998. The σD-dependent transcription of the ywcG gene from Bacillus subtilis is dependent on an excess of glucose and glutamate. Mol. Microbiol. 27:889-898. [DOI] [PubMed] [Google Scholar]
- 24.Moch, C., O. Schrogel, and R. Allmansberger. 2000. Transcription of the nfrA-ywcH operon from Bacillus subtilis is specifically induced in response to heat. J. Bacteriol. 182:4384-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mogk, A., A. Völker, S. Engelmann, M. Hecker, W. Schumann, and U. Völker. 1998. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J. Bacteriol. 180:2895-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mongkolsuk, S., and J. D. Helmann. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45:9-15. [DOI] [PubMed] [Google Scholar]
- 27.Morrissey, J. A., A. Cockayne, K. Brummell, and P. Williams. 2004. The staphylococcal ferritins are differentially regulated in response to iron and manganese and via PerR and Fur. Infect. Immun. 72:972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 29.Palma, M., and A. L. Cheung. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson, E. S., S. E. Boucher, and I. B. Lambert. 2002. Regulation of the nfsA gene in Escherichia coli by SoxS. J. Bacteriol. 184:51-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson, F. J., R. P. Mason, J. Hovsepian, and J. L. Holtzman. 1979. Oxygen-sensitive and -insensitive nitroreduction by Escherichia coli and rat hepatic microsomes. J. Biol. Chem. 254:4009-4014. [PubMed] [Google Scholar]
- 32.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Projan, S. J. 2003. Why is big Pharma getting out of antibacterial drug discovery? Curr. Opin. Microbiol. 6:427-430. [DOI] [PubMed] [Google Scholar]
- 34.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richards, M. J., J. R. Edwards, D. H. Culver, and R. P. Gaynes. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21:510-515. [DOI] [PubMed] [Google Scholar]
- 36.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 37.Uziel, O., I. Borovok, R. Schreiber, G. Cohen, and Y. Aharonowitz. 2004. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weber, H., S. Engelmann, D. Becher, and M. Hecker. 2004. Oxidative stress triggers thiol oxidation in the glyceraldehyde-3-phosphate dehydrogenase of Staphylococcus aureus. Mol. Microbiol. 52:133-140. [DOI] [PubMed] [Google Scholar]
- 39.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 40.Youngman, R. J., W. F. Osswald, and E. F. Elstner. 1982. Mechanisms of oxygen activation by nitrofurantoin and relevance to its toxicity. Biochem. Pharmacol. 31:3723-3729. [DOI] [PubMed] [Google Scholar]
- 41.Zenno, S., T. Kobori, M. Tanokura, and K. Saigo. 1998. Purification and characterization of NfrA1, a Bacillus subtilis nitro/flavin reductase capable of interacting with the bacterial luciferase. Biosci. Biotechnol. Biochem. 62:1978-1987. [DOI] [PubMed] [Google Scholar]
- 42.Zenno, S., H. Koike, A. N. Kumar, R. Jayaraman, M. Tanokura, and K. Saigo. 1996. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J. Bacteriol. 178:4508-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]