Abstract
The deletion of a gene coding for a histidine kinase (sll0750, Hik8) in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 resulted in a conditional lethal phenotype with a pleiotropic effect on the expression of genes involved in glucose metabolism. This mutant had comparable doubling times to wild type (WT) in continuous-light-grown photoautotrophic and mixotrophic cultures, whereas it grew poorly under mixotrophic conditions with different light and dark cycles. Growth was completely stopped, and cells eventually died, when the light duration was less than 6 h on a 24-h regimen. Northern blot analysis demonstrated that steady-state transcript levels of genes encoding key enzymes of glycolysis, gluconeogenesis, the oxidative pentose phosphate pathway, and glycogen metabolism were significantly altered in a strain with mutant hik8 (Δhik8) grown with or without glucose. In some cases, differential expression was dependent on growth conditions (photoautotrophic versus mixotrophic). The enzyme activities of glucose-6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, and phosphofructokinase were significantly reduced in Δhik8 compared to WT. Glycogen determination indicated that Δhik8 accumulated glycogen under mixotrophic conditions but was unable to utilize these reserves for heterotrophic growth. The results suggest that the loss of gap1 transcription in the absence of Hik8 was the key factor that rendered cells unable to catabolize glucose and grow heterotrophically. Additionally, the transcript levels of the phytochrome gene (cph1) and its cotranscribed response regulator gene (rcp1) were significantly reduced and its dark inducibility was lost in Δhik8. The results demonstrated that Hik8 plays an important role in glucose metabolism and is necessary for heterotrophic growth.
Cyanobacteria are a morphologically divergent, but physiologically cohesive, group of organisms that exhibit oxygenic photosynthesis similar to that of higher plants. In the natural habitat, cyanobacteria face diurnal cycles of light and dark (LD). In the light, cyanobacteria assimilate CO2 via the Calvin cycle using ATP and NADPH generated by photosynthesis. The fixed carbon enters the glycolytic pathway and can be utilized to generate reducing power, cofactors, and building blocks for the biosynthetic pathway or can be assimilated in the form of glycogen. In the dark, glucose residues derived from glycogen are catabolized via the oxidative pentose phosphate (OPP) pathway, the lower portion of glycolysis, and an incomplete tricarboxylic acid (TCA) cycle, leading to the production of NADPH and biosynthetic intermediates for maintenance and growth (10, 28).
Genome sequencing of several cyanobacteria has revealed the presence of all genes required for carbohydrate metabolism, but the elucidation of biochemical evidence for the functional role of the gene products has just begun (10, 30). The glycolytic genes studied to date in some detail include glucose-6-phosphate dehydrogenase (G6PD) (21, 22), 6-phosphogluconate dehydrogenase (6PGD) (2), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5, 12, 29), pyruvate kinase (11), phosphoenolpyruvate carboxylase (15), fructose-1,6-bisphosphate aldolase (FBA) (17), and phosphofructokinase (PFK) (18, 28). Furthermore, regulation of dark carbon metabolism and its coordinated control in cyanobacteria are still poorly understood. It is important to note that the Calvin cycle and the glycolysis and gluconeogenesis pathways function in the same metabolic compartment. In addition, cyanobacterial thylakoid membranes harbor both photosynthetic and respiratory electron transport chains, and some components are shared by both processes (23). Therefore, one would expect that functionally equivalent reactions in anabolic and catabolic pathways of cyanobacteria would be controlled in order to prevent any futile cycles.
The unicellular, transformable cyanobacterium Synechocystis sp. strain PCC 6803 has been an important model organism, in part because of its versatile growth characteristics (i.e., photoautotrophic [PA], mixotrophic [MT], or heterotrophic [HT] growth). The availability of its complete genome (Cyanobase; http://www.kazusa.or.jp/cyano/cyano.html) has enabled us to construct a DNA microarray (19). High-throughput analysis of gene expression using DNA microarrays under a variety of environmental conditions is a powerful tool for the elucidation of gene function and regulation. In addition, the precise understanding of metabolic and physiological responses to individual gene deletions can provide deeper insights into an organism's control and regulation of central metabolism. We have used the Synechocystis sp. strain PCC 6803 microarray to analyze global gene expression in response to nutrient alterations and environmental stresses (14, 26).
We have performed transcriptional profiling with these arrays in a Synechocystis sp. strain PCC 6803 hik8 deletion strain. Hik8 is one of over 40 histidine kinases known to be encoded by the Synechocystis sp. strain PCC 6803 genome, and little is known about their roles in the perception of environmental or intracellular stimuli (16). We previously found that this gene was differentially regulated in response to oxidative stress and to changing iron concentrations (14, 26). Hik8 has significant protein sequence similarity to SasA from Synechococcus sp. strain PCC 7942, a protein that interacts with KaiC (9); KaiC, along with KaiA and KaiB, is involved in the circadian control in Synechococcus sp. strain PCC 7942 (8). It has been shown that SasA is required to sustain robust circadian rhythms (9). However, impaired growth of a ΔsasA strain, but not that of a ΔkaiABC strain, under LD cycles suggested additional functions (9). Our results showed that a number of genes involved in glucose metabolism were differentially expressed in a strain with mutant hik8 (Δhik8). Furthermore, Hik8 seemed to be required for the expression of the cyanobacterial phytochrome gene cph1. In this report, we demonstrate that hik8 has a pleiotropic effect on the expression of genes involved in central glucose metabolism.
MATERIALS AND METHODS
Cyanobacterial strains and growth.
Wild-type (WT) and mutants of Synechocystis sp. strain PCC 6803 were grown in liquid BG-11 medium (with or without 5 mM glucose) at 30°C under a light intensity of 20 to 30 μE m−2 s−1 as described by Singh and Sherman (27). Growth of strains was followed under different growth conditions by monitoring the optical density at 730 nm. HT growth was monitored after 7 days in complete darkness.
RNA isolation and RNA blotting.
Total RNA from Synechocystis sp. strain PCC 6803 sampled at various time points was isolated using the procedure described by Singh and Sherman (27). Microarray experiments were carried out as described by Singh et al. (26). Five micrograms of total RNA was used for Northern blot analysis as described by Sambrook et al. (20). DNA probes were labeled with [32P]dCTP by using the Ready-to-go labeling kit (Amersham Pharmacia).
Construction of mutants.
DNA fragments comprising hik8 (sll0750) (5′-AGACTGGGACAAATTATTTAC-3′ and 5′-TCAACGGTAAACAGGCAACG-3′) and cph1-rcp1 (slr0473 and slr0474) (5′-ATGACCACCGTACAACTC-3′ and 5′-CGCCAAACGCTTTACGGCATC-3′) were amplified by PCR from genomic DNA of Synechocystis sp. strain PCC 6803. The amplified PCR fragments consisting of either hik8 (1,465 nucleotides [nt]) or cph1-rcp1 (2,889 nt) were cloned in the pGEM-T vector (Promega). A deletion mutation in hik8 was constructed by replacing 627 nt of the coding region (codons 8 to 217), following Bbs1 digestion, with a 2.0-kb spectinomycin resistance cassette (from plasmid pRL453). Similarly, a deletion mutation in cph1-rcp1 was constructed by replacing 1,837 nt of the coding region (codons 237 to 748 for cph1 and codons 1 to 97 for rcp1), following Hpa1 digestion, with a 2.0-kb spectinomycin resistance cassette (from plasmid pRL453). WT Synechocystis sp. strain PCC 6803 was transformed with the various constructs, and transformants were selected on plates containing 40 μg of spectinomycin ml−1. After the mutants were streaked multiple times over 6 weeks, complete segregation was confirmed by PCR and Southern blotting (data not shown). For complementation of the mutant strain with the WT gene, Δhik8 was transformed with the hik8 gene. Following 24 h of incubation in the light in the presence of glucose, cells were plated on BG-11 plates containing 5 mM glucose and incubated in continuous darkness. After a 10-day incubation, several colonies were obtained. Colonies were grown in liquid cultures, and PCR was used to confirm the integration of hik8 (data not shown).
Glycogen determination.
The glycogen content in Synechocystis sp. strain PCC 6803 was quantified by using the anthrone reagent as described by Schneegurt et al. (24) with some modifications. The crude extracts of Synechocystis sp. strain PCC 6803, sampled at various time points, were isolated using a Braun homogenizer and clarified by centrifugation at 1,000 × g. The glycogen granules were extracted using trichloroacetic acid, precipitated by ethanol, and resuspended in 200 μl of water. One milliliter of anthrone reagent (0.05% anthrone in 72% sulfuric acid) was added, and the solution was boiled for 20 min at 100°C in a water bath. The tubes were cooled, and the absorbance was measured at 620 nm. The concentration of glycogen was determined using a glucose standard curve and was represented per milligram of chlorophyll.
Enzyme assays.
Enzymatic activities of G6PD and 6PGD in cell extracts were measured spectrophotometrically by monitoring the glucose-6-phosphate- and 6-gluconate phosphate-dependent generation of NADPH formation at 340 nm, respectively, as described by Schaeffer and Stanier (22). Cyanobacterial cells were washed twice with 50 mM Tris-maleate, pH 6.5, and extracts were isolated using a Braun homogenizer. The crude extracts were clarified by centrifugation at 12,000 × g and 4°C for 5 min. GAPDH activity was measured spectrophotometrically by monitoring the glyceraldehyde-3-phosphate-dependent generation of NADH and NADPH formation at 340 nm as described by Valverde et al. (29). Cell extracts were prepared in 50 mM Tris-Cl buffer, pH 7.5, 1 mM EDTA, and 10 mM 2-mercaptoethanol. Both phosphorylating and nonphosphorylating enzymatic activities were determined in the presence of 10 mM Na2HAsO4. PFK activity was determined by measuring NADH oxidation at 340 nm in the presence and absence of ATP and fructose-6-phosphate as described by Kotlarz and Buc (13). The extract was prepared with the same buffer used for GAPDH activity determinations but included 2 mM fructose-6-phosphate. Protein concentration was estimated by the Bradford technique (1).
Electron microscopy.
The preparation of cyanobacterial cells for electron microscopy was based on the procedure described by Giberson et al. (7). Samples were prepared using a scientific microwave (Ted Pella, Inc., Redding, Calif.) equipped with a variable wattage control and a PELCO Coldspot water recirculator to maintain constant temperature in the oven. Cells were spun down and fixed in 2% paraformaldehyde plus 2% glutaraldehyde in 0.1 M potassium phosphate buffer, pH 6.8, containing 750 mM NaCl for two cycles of 40 s at P1 (180 W) followed by a 3-min hold (no power) under a 5-mm Hg vacuum. After being washed twice with buffer for 40 s at P1, cells were postfixed in reduced osmium tetroxide [1% OsO4 plus 1.5% K2Fe(CN)6 in H2O] for one cycle of 40 s (P1) followed by a 3-min hold under a 5-mm Hg vacuum. Samples were spun down in 1.5% agarose (Sigma type VII low temperature gelling), cut into 1-mm blocks, and dehydrated three times in an ethanol series (30, 50, 70, 90, and 100%) for 40 s each at P1. Initial infiltration in propylene oxide (PO) plus Spurr's resin (3:1 and 1:1 mixtures) was done in the microwave (40 s each at P2 [300 W] and 5 mm Hg vacuum). Infiltration was continued at room temperature on a rotator (1 PO:3 resin overnight followed by 100% resin for 4 h). Blocks were embedded in 100% Spurr's resin in capsules and polymerized for 48 h at 60°C. Samples were imaged on an FEI Philips CM-10 biotwin electron microscope operated at 80 kV. The identification of glycogen granules was carried out as described by Sherman and Sherman (25).
RESULTS
Growth characteristics of strain Δhik8.
We generated two strains of Synechocystis sp. strain PCC 6803 by replacing most of the coding regions of the hik8 (codons 8 to 217 are missing) and the cph1-rcp1 (codons 237 to 748 for cph1 and codons 1 to 97 for rcp1 are missing) genes with a spectinomycin cassette. The parental strain and strains with hik8 or cph1-rcp1 deleted are referred to as WT, Δhik8, and Δcph1rcp1, respectively. Table 1 shows the growth characteristics of the WT and mutants under PA, MT, and HT conditions. The mutants grew similarly to WT under PA and MT conditions in continuous light (LL) with comparable doubling times. When the mutants were grown under PA conditions under various LD regimens, their doubling times increased significantly; however, they were similar to that of WT. Cell growth for all three strains virtually stopped (with a doubling time of about 130 h) with a light duration of 6 h on a 24-h regimen, suggesting that, under growth conditions used in the present study, light duration of less than 6 h was not sufficient for PA growth. In addition, loss of viability in WT and mutants, determined by the CFU on BG-11 plates under PA conditions, was similar up to 144 h of dark incubation (data not shown). However, Δhik8 grew poorly compared to WT and Δcph1rcp1 under MT conditions with various LD regimens and under HT conditions (Table 1). The doubling time of Δhik8 increased significantly as the light period decreased. Growth was completely stopped and cells eventually died with less than 6 h of light on a 24-h regimen. Additionally, transfer of actively growing MT cultures of Δhik8 into the dark for prolonged periods led to growth cessation and to eventual death (data not shown). The complementation of the Δhik8 strain with hik8 resulted in heterotrophic growth with a doubling time similar to that of WT, suggesting that the growth phenotype of Δhik8 was due to deletion of hik8 (data not shown). Thus, the loss of Hik8 reflected a deficiency in glucose utilization and/or glucose intolerance in the absence of light and suggested a role for this histidine kinase in the metabolism of glucose.
TABLE 1.
Growth characteristics of WT, Δhik8, and Δcph1rcp1 under various trophic conditions
| Growth condition | Light condition (L:D [h]) | Doubling time (h)a
|
||
|---|---|---|---|---|
| WT | Δhik8 | Δcph1rcp1 | ||
| PA | 24:0 | 15 ± 1 | 15 ± 2 | 14 ± 3 |
| 12:12 | 42 ± 2 | 44 ± 3 | 41 ± 3 | |
| 9:15 | 48 ± 3 | 48 ± 2 | 47 ± 2 | |
| 6:18 | 130 ± 6 | 130 ± 5 | 117 ± 15 | |
| MT | 24:0 | 9 ± 1 | 9 ± 1 | 9 ± 1 |
| 12:12 | 17 ± 3 | 24 ± 2 | 18 ± 2 | |
| 9:15 | 18 ± 1 | 55 ± 3 | 17 ± 2 | |
| 6:18 | 20 ± 2 | NGb | 17 ± 2 | |
| HT | 0:24 | Growth | NGc | Growth |
Doubling time for strains are means ± standard errors for n ≥ 3.
NG, no growth.
Complementation of Δhik8 with hik8 resulted in heterotrophic growth.
Differential expression of genes in strain Δhik8 in LD.
A DNA microarray experiment with PA-grown WT and Δhik8 under LL and then 1 h in the dark was used as described by Singh et al. (26) to profile the expression of genes in Δhik8 compared to WT. Preliminary data analysis showed that two categories of genes were particularly affected by the absence of a functional Hik8 (data not shown): (i) genes involved in glucose metabolism, and (ii) genes coding for ribosomal proteins, especially during dark growth. Additionally, the operon encoding phytochrome (cph1) and its cognate response regulator (rcp1) was significantly reduced in the Δhik8 strain irrespective of growth conditions. Therefore, Δcph1rcp1 was included in the study to determine if the differential expression of various genes by Hik8 relied on Cph1-Rcp1. To test the working hypothesis that Hik8 plays a role in glucose metabolism, we measured selected enzyme activities and used Northern blotting to determine the steady-state transcript levels of several key genes.
The genes included in the present study were those encoding key enzymes of glycolysis, gluconeogenesis, glycogen metabolism, and the OPP pathway (Fig. 1 and Table 2). Total RNA was isolated from exponentially growing WT and mutants under either PA or MT conditions in LL and after transfer to the dark for 0.25, 1, and 3 h followed by subsequent reillumination for 1 and 3 h. Several genes involved in glucose metabolism (with either low or high steady-state transcript levels) showed similar patterns during the LD transition in all three strains, whereas others had interesting changes depending on the growth conditions. The former category included genes encoding glucokinase, phosphoenolpyruvate carboxylase, malic enzyme, malate dehydrogenase, isocitrate dehydrogenase, transketolase, and pyruvate kinase (data not shown). Western blot analysis of some of these enzymes also showed no difference in protein levels in the three strains (data not shown). In contrast, several genes with key functions in glucose metabolism were differentially expressed in Δhik8 compared to WT and Δcph1rcp1.
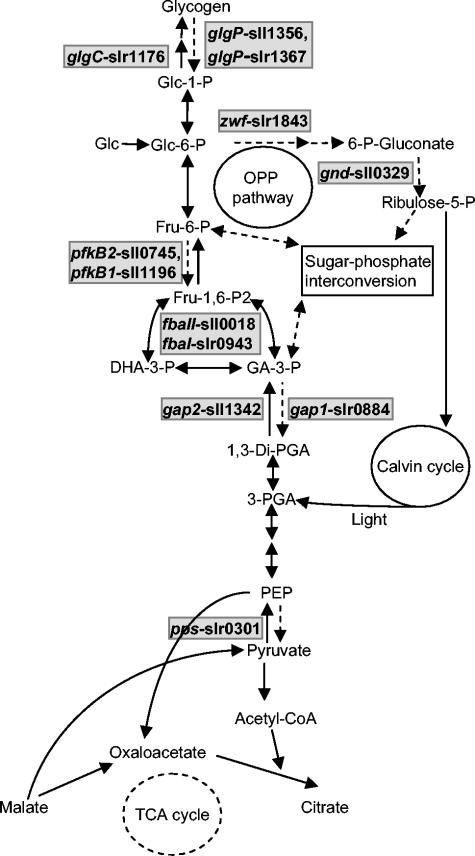
FIG. 1.
Simplified scheme depicting the glycolytic, OPP, and gluconeogenic pathways in Synechocystis sp. strain PCC 6803. The key enzymes that were examined in the present study are shown with possible redundant genes, based on the Kazusa annotation. The dotted, solid single, and double arrows indicate the direction of glucose (Glc) metabolism under either HT or PA growth conditions or both, respectively.
TABLE 2.
Open reading frame numbers, genes, and products of selected enzymes used in the present study involved in glucose metabolism in Synechocystis sp. strain PCC 6803
| ORFa | Gene | Product | Abbreviation or other name | EC no. |
|---|---|---|---|---|
| sll1196 | pfkB1 | Phosphofructokinase | PFK | 2.7.1.11 |
| sll0745 | pfkB2 | Phosphofructokinase | PFK | 2.7.1.11 |
| slr0884 | gap1 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH1, Gap1 | 1.2.1.12 |
| sll1342 | gap2 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH2, Gap2 | 1.2.159 |
| sll0018 | fball | Fructose-1,6-bisphosphatase | Cll-FBA | 4.1.2.13 |
| slr0943 | fbal | Fructose-1,6-bisphosphatase | Cl-FBA | 4.1.2.13 |
| slr1843 | zwf | Glucose-6-phosphate dehydrogenase | G6PD | 1.1.149 |
| sll0329 | gnd | 6-Phosphogluconate dehydrogenase | 6PGD | 1.1.1.44 |
| slr0301 | pps | Phosphoenolpyruvate synthase | Pps | 2.7.9.2 |
| slr1176 | glgC | ADP-gluose pyrophosphorylase | GlgC | 2.7.7.27 |
| slr1367 | glgP | Glycogen phosphorylase | GlgP | 2.4.1.1 |
| sll1356 | glgP | Glycogen phosphorylase | GlgP | 2.4.1.1 |
| slr0473 | cph1 | Cyanobacterial phytochrome | Cph1 | |
| slr0474 | rcp1 | Response regulator for cph1 | Rcp1 | |
| sll1127 | menB | 1,4-Dihydroxy-2-naphthoate synthase | MenB | 4.1.3.36 |
ORF, open reading frame.
Expression of glycolytic genes.
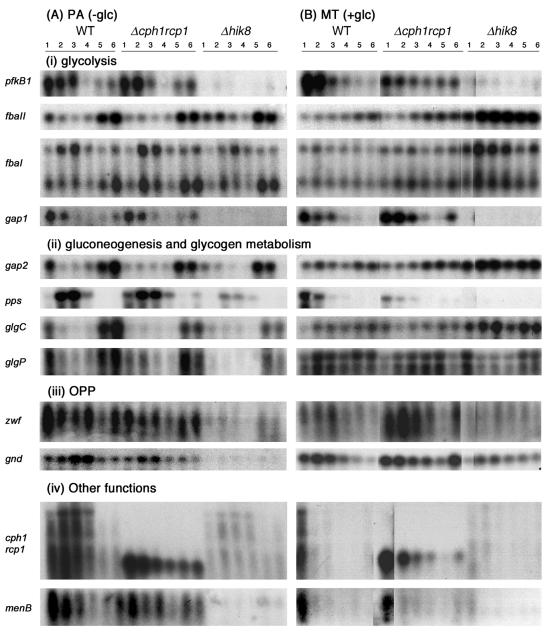
Synechocystis sp. strain PCC 6803 contains all the putative genes for glycolysis, and multiple genes are present for some enzymes. A recent metabolic flux study has shown that Synechocystis sp. strain PCC 6803 has an active glycolysis pathway (31), but few studies have been performed to substantiate the functional significance of the various glycolytic genes (10). Northern blot analysis showed that genes encoding PFK (pfkB1), GAPDH (gap1), and FBA (fbaII and fbaI) were differentially expressed in strain Δhik8. Synechocystis sp. strain PCC 6803 has two genes coding for PFK, pfkB1 and pfkB2 (Table 2), but their metabolic roles have not been clearly delineated. Comparative analysis of microbial genomes suggested that both pfk genes belong to the PFK-A family (4), whereas they were characterized biochemically as the PFK-B type (10). The transcript levels of pfkB1 in both WT and Δcph1rcp1 grown under PA and MT conditions gradually decreased after the cultures were transferred into the dark (Fig. 2) and increased slowly in both strains upon illumination under PA conditions, but not under MT conditions. In contrast, the transcript of pfkB1 was undetectable in Δhik8 under all growth conditions. The transcript level of pfkB2 was very low and did not change in any of the strains (data not shown). Enzyme activity of PFK in crude extracts of PA-grown Δhik8 was low and similar to that of WT (Table 3). However, the PFK activity in 3-h-dark-incubated, MT-grown Δhik8 was less than half that of the WT.
FIG. 2.
Northern blots of Synechocystis sp. strain PCC 6803 genes encoding proteins involved in (i) glycolysis, (ii) gluconeogenesis and glycogen metabolism, (iii) OPP, and (iv) other functions. Strains monitored were WT, Δhik8, and Δcph1rcp1 grown under PA (A) or MT (B) conditions in LL (lane 1) or after transfer to dark for 0.25 (lane 2), 1 (lane 3), and 3 h (lane 4) and subsequently reillumination for 1 (lane 5) or 3 h (lane 6). Five micrograms of total RNA isolated from these samples was electrophoresed on a 1.2% agarose gel, transferred to a nylon membrane, and hybridized with the respective gene probes. The data are representative of three separate biological replicates. The autoradiograms of these genes were exposed for different time periods for the sake of clarity.
TABLE 3.
Specific activities of G6PD, 6PGD, GADPH, and PFK in WT and Δhik8 cells grown under PA and MT conditions under continuous light or 3-h dark incubation
| Enzyme | Growth condition | Sp acta
|
|||||
|---|---|---|---|---|---|---|---|
| Light
|
3 h dark
|
||||||
| WT | Δhik8 | % Control | WT | Δhik8 | % Control | ||
| G6PD | PA | 69.8 ± 7.8 | 41.1 ± 3.8 | 59 | 66.9 ± 8.2 | 38.1 ± 5.1 | 57 |
| MT | 86.5 ± 4.6 | 75.0 ± 0.6 | 87 | 79.1 ± 8.3 | 69.6 ± 2.6 | 88 | |
| 6PGD | PA | 75.3 ± 7.3 | 32.1 ± 6.2 | 43 | 73.6 ± 6.6 | 29.2 ± 5.9 | 40 |
| MT | 97.7 ± 3.3 | 59.6 ± 2.6 | 61 | 91.0 ± 8.9 | 53.5 ± 2.2 | 59 | |
| GAPDH | PA | 203 ± 18 | 176 ± 21 | 87 | 211 ± 25 | 192 ± 19 | 91 |
| MT | 379 ± 32 | 273 ± 29 | 72 | 391 ± 45 | 335 ± 27 | 86 | |
| PFK | PA | 9.2 ± 1.0 | 8.0 ± 0.4 | 87 | 8.2 ± 0.4 | 5.9 ± 0.6 | 72 |
| MT | 16.7 ± 0.8 | 12.1 ± 0.1 | 72 | 24.4 ± 1.7 | 10.4 ± 1.1 | 43 | |
Specific activities are means ± standard errors for n = 3 and are represented as nanomoles of NADPH (for G6PD and 6PGD) or NADH (for GAPDH) produced per minute per milligram of protein; activities for PFK are reported as nanomoles of NADH oxidized per minute per milligram of protein.
Two genes coding for GAPDH in Synechocystis sp. strain PCC 6803, gap1 and gap2 (Table 2), were shown to function in the Calvin cycle, gluconeogenesis, and glycolysis (12, 29). The gap1 transcript levels responded similarly to those of pfkB1; the transcripts were undetectable in Δhik8, whereas there was a similar pattern in WT and Δcph1rcp1 during the LD transition. The gap2 transcripts decreased rapidly in all three PA cultures after transfer to the dark but increased quickly during reillumination. In contrast, gap2 transcripts were present under all conditions in MT cells (Fig. 2). The reduction of gap1 transcripts in Δhik8 had no effect on the enzyme activity of GAPDH, as levels were similar in both Δhik8 and WT measured in the presence (Table 3) or absence (data not shown) of arsenate. This was not surprising, as Valverde et al. (29) have shown that GAPDH2 shows both anabolic and catabolic activities and have concluded that only this enzyme is functional in Synechocystis sp. strain PCC 6803. However, a separate study by Koksharova et al. (12) has shown that, although GAPDH2 is the dominant activity, GAPDH1 is also present in Synechocystis sp. strain PCC 6803.
Synechocystis sp. strain PCC 6803 has two putative genes for FBA, fbaI and fbaII (Table 2). Both enzymes encoded by these genes have similar FBP-dependent activities, although fbaI was suggested to be functionally redundant (17). Both genes were differentially expressed depending on the growth conditions. The transcript levels of fbaII decreased when PA-grown WT was transferred to the dark but reappeared upon reillumination. A similar pattern was also observed in Δhik8, although a transient increase after 15 min of dark could be observed. The steady-state transcript of fbaII in MT-grown Δhik8 increased and remained quite high when cells were transferred to the dark. The transcript level of fbaI (Fig. 2, upper band) had a pattern similar to that of fbaII in MT-grown cells, whereas the transcripts initially increased in the dark in all PA-grown cells. Thus, increased transcript levels of fba genes in MT-grown Δhik8 suggested increased carbon flow towards gluconeogenesis, since Δhik8 was unable to catabolize glucose due to the loss of gap1 transcript (see Discussion).
Expression of OPP pathway genes.
The OPP pathway was suggested to be the major route of glucose catabolism in cyanobacteria (18, 22). G6PD (zwf) and 6PGD (gnd) control the carbon flow into the OPP pathway (Fig. 1). Both genes were differentially expressed in the Δhik8 strain compared to WT, with expression dependent on growth under PA or MT conditions. In PA-grown Δhik8, transcript levels of both zwf and gnd were greatly reduced. The gnd transcripts increased as cultures of WT and Δcph1rcp1 were transferred into the dark and were subsequently reduced upon reillumination. In MT-grown Δhik8, transcripts of both zwf and gnd were relatively less than those present in WT, but significantly higher than those present in PA-grown Δhik8.
Enzyme activities of G6PD and 6PGD were measured in WT and Δhik8 extracts isolated from PA- and MT-grown cells after 3 h in the dark. The PA-grown Δhik8 had only 59% G6PD activity compared to the WT, which changed little after dark incubation (Table 3). However, when cells were grown under MT conditions, G6PD activity in Δhik8 was 87% that of WT. Similarly, 6PGD activity was approximately 43 and 61% in Δhik8 compared to WT in PA- and MT-grown cells, respectively, and dark incubation had no effect on the enzyme activity (Table 3). Thus, enzyme activity and transcript levels suggested that Hik8 affected 6PGD, independent of growth conditions, whereas its effect on G6PD was dependent on growth conditions.
Expression of genes involved in gluconeogenesis and glycogen metabolism.
Phosphoenolpyruvate synthase (pps) controls the first committed step involved in the gluconeogenic process. Its transcriptional pattern was dependent on the presence or absence of glucose, and the pattern in WT was similar to that in Δcph1rcp1, but different from that in Δhik8. Its transcript levels decreased rapidly as MT-grown WT and Δcph1rcp1 cultures were transferred to the dark and did not reappear upon reillumination. In PA-grown WT and mutants, the pps transcript levels increased rapidly as cultures were transferred to the dark, eventually dropping by 3 h of dark, with patterns similar in WT and Δcph1rcp1. The transcript levels were much reduced in Δhik8 in both PA- and MT-grown cells compared to those in the other two strains.
The key regulatory enzymes involved in the assimilation (ADP-glucose pyrophosphorylase, glgC) and dissimilation (glycogen phosphorylase, glgP) of glycogen were differentially expressed in Δhik8. In PA-grown cultures of all three strains, the transcript patterns of both genes were similar; transcripts rapidly decreased as cultures were transferred to the dark but reappeared upon reillumination. In MT-grown cultures, the transcripts increased as WT and Δcph1rcp1 were transferred to the dark. Interestingly, transcript levels of glgC were higher in Δhik8 than in WT and Δcph1rcp1, whereas in Δhik8, the glgP transcript level decreased rapidly as cells were transferred to the dark. The results suggested that glycogen storage, but not degradation, was favored in MT-grown Δhik8.
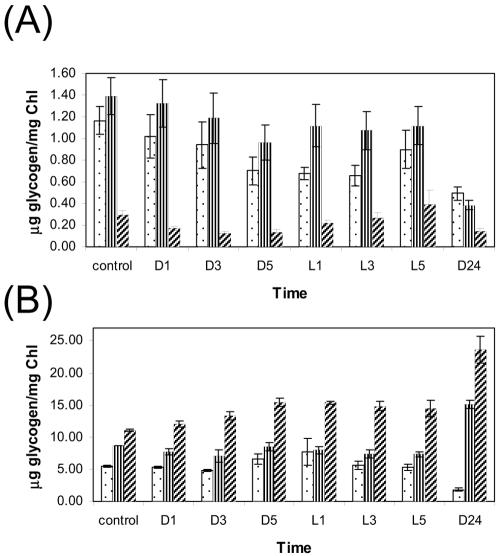
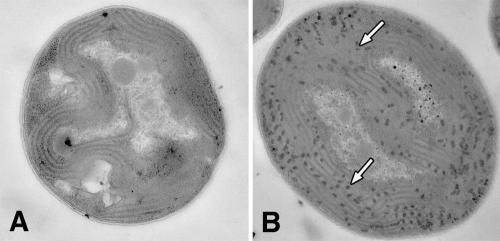
The glycogen content in the Δhik8 and WT cells was measured during growth under PA and MT conditions. Samples from exponentially growing cultures were collected over 34 h, which involved various LD transitions. As shown in Fig. 3A, Δhik8 had only approximately 25% of the glycogen level compared to WT and Δcph1rcp1. As expected, the glycogen content showed a typical response during LD transitions: the amount decreased during dark growth and increased upon reillumination (Fig. 3A). We also analyzed morphological changes in PA- and MT-grown cultures of Δhik8 and WT by electron microscopy and showed that glycogen granules in PA-grown WT (incubated for 4 h in dark) were far more numerous than in PA-grown Δhik8 (Fig. 4). Cellular morphology corresponded nicely with the biochemical determination of glycogen, as shown in Fig. 3. In contrast, glycogen accumulation and dissipation in the MT-grown cultures had quite different patterns; the glycogen content was almost twice as high in light-grown Δhik8 compared to WT (Fig. 3B). As expected, the glycogen content decreased in WT upon dark incubation, whereas the amount of glycogen increased in the dark in Δhik8.
FIG. 3.
Glycogen content in WT (dotted bars), Δcph1rcp1 (line bars), and Δhik8 (cross line bars). Exponentially grown cells under PA (A) or MT (B) conditions were transferred to the dark (1, 3, and 5 h) and then reilluminated with light (1, 3, and 5 h) followed by 24 h of growth in the dark. The glycogen content was measured as described in Materials and Methods. The values for PA and MT are means ± standard errors for five and three separate experiments, respectively. Chl, chlorophyll.
FIG. 4.
Electron micrographs of Synechocystis sp. strain PCC 6803 PA-grown WT (A) and Δhik8 (B) cells. Cells were grown to mid-logarithmic phase under PA conditions, transferred to the dark for 4 h, and prepared for microscopy as described in Materials and Methods. Arrows indicate glycogen granules.
Expression of cph1-rcp1 and menB.
Two genes that were differentially expressed in Δhik8, but not related to glucose metabolism, were cph1 and its cognate response regulator, rcp1, and the 1,4-dihydroxy-2-naphthoate synthase gene (menB). As shown in Fig. 2, cph1-rcp1 transcript levels transiently increased in the PA-grown WT culture after transfer to the dark before decreasing to the level found in light-grown cells. A small, truncated transcript of the expected size (∼0.8 kb) was also detected in strain Δcph1rcp1. This transcript represented the synthesis of mRNA that originated upstream of cph1 until the site of the spectinomycin resistance cassette insertion at codon 237 (see Materials and Methods for details). In contrast, the cph1-rcp1 transcripts were severely reduced in the Δhik8 stain (Fig. 2). When MT-grown WT cells were transferred to the dark, the level of cph1-rcp1 transcript decreased rapidly. Again, cph1-rcp1 transcripts were undetectable in Δhik8. The transcript level of menB decreased as WT and Δcph1rcp1 grown under PA conditions were transferred to the dark and reappeared upon reillumination. In contrast, the transcript was undetectable in Δhik8 under all conditions.
DISCUSSION
In this study, we provide evidence that a histidine kinase, Hik8, plays a significant role in the glucose metabolism in Synechocystis sp. strain PCC 6803. Hik8 is critical for the HT growth of Synechocystis sp. strain PCC 6803, as strain Δhik8 does not grow heterotrophically or if given less than 6 h of light on a 24-h regimen; in addition, those cells grown under MT growth conditions die when transferred to the dark. The transcript levels of two important genes involved in glucose catabolism (pfkB1 and gap1) were nondetectable in Δhik8, and both the transcript levels and the enzymatic activities of enzymes involved in the OPP pathway (zwf and gnd) were also reduced. Interestingly, glycogen levels increased significantly in Δhik8 when MT-grown cells were transferred to the dark as opposed to the expected decrease that occurred in WT. This physiological result was mirrored by the differential expression of the genes involved in glycogen metabolism in Δhik8 compared to WT. Hik8 is also required for the expression of cyanobacterial phytochrome and its response regulator.
The transcript level of pfkB1 was greatly reduced in Δhik8. PFK is a major controlling step in glycolysis, and the loss of PFK activity would greatly impact glucose catabolism (3). The importance of PFK in HT growth of Synechocystis sp. strain PCC 6803 has been demonstrated recently by metabolic flux determination (31). It was shown that, in contrast to insignificant flux through PFK in MT, the flux through PFK was approximately 59% of the glucose in HT-grown cultures. However, it is unlikely that the absence of PFK would prevent glucose catabolism in HT-grown cells, as glucose can be efficiently catabolized through the OPP pathway. Ribulose-5-phosphate generated by the OPP pathway can easily be recycled into glyceraldehyde-3-phosphate and fructose-6-phosphate by transketolase and transaldolase (Fig. 1). Glyceraldehyde-3-phosphate can then be further catabolized into glycolytic products, whereas fructose-6-phosphate can be isomerized to glucose-6-phosphate that will feed the OPP pathway again, thus bypassing the involvement of PFK. Since the transcript patterns of transketolase and transaldolase were similar in the mutants compared to WT (data not shown), we concluded that the pathway involved in the regeneration of glyceraldehyde-3-phosphate and fructose-6-phosphate was active in Δhik8.
The transcript levels of zwf and gnd and the enzymatic activities of their products were significantly reduced in Δhik8. These two genes control the entry of glucose into the OPP pathway and are regulated by many factors, including LD transitions (22). Indeed, a recent metabolic study showed that over 90% of the glucose was catabolized through the OPP pathway during HT growth (31). Thus, the reduced G6PD and 6PGD activities in Δhik8 will significantly alter glucose degradation by the OPP pathway, although its contribution to the lethal phenotype of Δhik8 during HT growth remains to be determined. Several studies have shown that deletion of both zwf and gnd has no dramatic effect on the survival of cells in the dark (2, 21). Interestingly, the deletion of gnd resulted in a conditional lethal phenotype of Synechococcus sp. strain PCC 7942, apparently due to the accumulation of 6-phosphogluconate (2). It is important to note that, in MT-grown Δhik8, 6GPD activity was more affected (61% of WT) than G6PD activity (87% of WT). Such differences in enzyme activities could lead to the accumulation of more 6-phosphogluconate compared to its physiological level vis-a-vis glucose-6-phosphate, a situation similar to that for the conditional lethal gnd mutant strain.
An interesting transcriptional pattern in Δhik8 was observed for the two genes encoding GAPDH. The two gap genes encode distinct enzymes that participate in at least three metabolic processes, i.e., the Calvin cycle, glycolysis, and gluconeogenesis (12, 29). There is unequivocal evidence that GAPDH2 operates in the Calvin cycle and gluconeogenesis. In contrast, the role of GAPDH in catabolic glucose degradation is not very clear. Valverde et al. (29) suggested that GAPDH2 was involved in catabolic glucose degradation, whereas Koksharova et al. (12) concluded that GAPDH1 operated exclusively in catabolic glucose degradation. Synechocystis sp. strain PCC 6803 has an incomplete TCA cycle, and cells constantly need to replenish the TCA cycle intermediates that are utilized for biosynthetic reactions. Our results demonstrated that levels of gap2 transcription were similar in both WT and mutants, suggesting that the Calvin cycle and gluconeogenesis were active in all three strains. Thus, intermediates of the Calvin cycle fed the TCA cycle under PA and MT conditions, resulting in similar levels of growth of Δhik8 and WT (Fig. 1). We suggest that the loss of gap1 transcription in the absence of a functional Hik8 renders cells unable to catabolize glucose and thus their inability to grow under HT conditions. It is interesting that a Δgap1 mutant was unable to grow in low light in the presence of glucose, although it grew like WT under PA conditions (12).
The importance of glycogen synthesized as the storage material during the active photosynthetic process and its subsequent catabolism during dark periods has been well documented (24). Indeed, our results indicated that exponential-phase, PA-grown cells had lower glycogen levels and that glycogen dissipation began soon after cells were transferred to the dark. Yet, Δhik8 had less glycogen under the same conditions. Clearly, addition of glucose to the culture medium had an impact on the glycogen content; nonetheless, the WT glycogen content decreased as cells were transferred to the dark, whereas the Δhik8 glycogen content increased after cells were transferred into the dark. The increased glycogen content in Δhik8 could have resulted from its inability to catabolize glucose due to the absence of GAPDH1. Since glucokinase expression levels were similar in both WT and Δhik8 (data not shown), it would suggest that glucose enters the glycolytic pathway in Δhik8 similar to WT. However, the inability to catabolize glucose in Δhik8 resulted in rerouting the glycolytic intermediates towards glycogen accumulation. Accordingly, the genes involved in the assimilation and dissimilation of glycogen, particularly those encoding allosterically regulated ADP-glucose pyrophosphorylase and glycogen phosphorylase, were differentially expressed in Δhik8 compared to WT in MT-grown cells. Therefore, glycogen accumulation was favored in Δhik8 in the presence of glucose.
Another important aspect of our study is the requirement of Hik8 for the expression of the cyanobacterial phytochrome Cph1 and its response regulator, Rcp1. Cph1 has been shown to mediate red-far-red reversible phosphorylation of Rcp1 (32), and a role for Cph1 has been suggested during LD transitions (6). As reported in this work and as shown previously (6), Cph1-Rcp1 transcript levels increased during transfer to the dark in WT. Garcia-Dominguez et al. (6) suggested the involvement of a photoreceptor in the regulation of Cph1-Rcp1 expression during LD transition. It is clear that this regulation was impaired in Δhik8, suggesting that Hik8 may be involved in mediating the expression of cph1-rcp1 during LD transition.
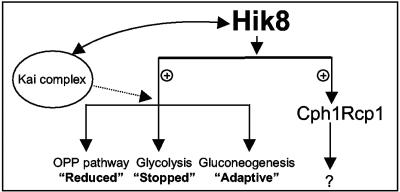
Figure 5 provides a summary model for the role of Hik8. The effect of Hik8 on the expression of the glycolytic genes can be categorized into three groups: (i) reduced, for genes whose transcript levels were significantly lower in Δhik8 compared to WT (e.g., the OPP pathway genes zwf and gnd); (ii) stopped, for genes whose transcript levels were undetectable in Δhik8 compared to WT (e.g., the glycolytic genes pfkB1 and gap1); and (iii) adaptive, for genes whose transcript levels changed depending on the growth conditions (e.g., glgC, glgP, fbaII, and several others). As described earlier, the growth phenotype of Δhik8 is unlikely due to the first category, although significantly reduced activities of these two key OPP enzymes (combined with the reduced activity of PFK) should slow the flux movement through glycolysis. We conclude that loss of gap1 transcripts in Δhik8 resulted in the observed growth phenotype.
FIG. 5.
Diagrammatic model for Hik8 function. Hik8 is required for the transcription of cph1-rcp1 and various genes involved in glucose metabolism. The activation may be direct or indirect and possibly involves an unidentified response regulator. A possible interaction between Hik8 and KaiABC is shown based on the report of Iwasaki et al. (9). The dotted line indicates the potential, but nonproven, involvement of a Hik8 and Kai complex in the regulation of glycolytic genes. The question mark indicates the unknown genes regulated by Cph1-Rcp1.
Our model also shows that Hik8 is required for the expression of Cph1. In general, sensory kinases function by autophosphorylation in response to changes in the environmental conditions, with subsequent phosphorylation of their cognate response regulators. Thus, the Hik8-mediated response should include at least one response regulator, but the identity of this activator is unknown. Unfortunately, preliminary analysis of microarray data did not identify any response regulators whose expression was overly changed in Δhik8 compared to WT (data not shown). Our results clearly demonstrated that the role of Hik8 in glucose metabolism is independent of cph1-rcp1. It also remains to be seen if expression of these genes by Hik8 involves interactions with other proteins. One possibility shown in Fig. 5 is its interaction with KaiABC. The Hik8 homolog has been shown to be associated with the KaiABC complex in Synechococcus sp. strain PCC 7942 (9). We are in the process of generating kaiABC mutants from Synechocystis sp. strain PCC 6803 to study its impact on expression of glycolytic genes during LD transitions.
Finally, cyanobacteria, like other organisms, can compensate for the loss of the majority of enzymes involved in glucose metabolism either by isoenzymes or by rerouting of carbon fluxes through alternative pathways. By coordinating the expression of genes encoding enzymes involved in glycolysis, in the OPP pathway, in gluconeogenesis, and in glycogen metabolism, Hik8 acts as a multifaceted regulator of glucose metabolism. In addition, by controlling the very core of glucose metabolism, Hik8 can influence many other cellular activities and thereby facilitate adaptation to different challenges or growth conditions. The link between metabolism and protein synthesis was manifested in the polysome profiles of WT and Δhik8. Our initial microarray results indicated that virtually all of the ribosomal protein genes were reduced in Δhik8 compared to the WT (data not shown). We later demonstrated that Δhik8 had a greater proportion of free ribosomes relative to polysomes compared to the WT regardless of growth conditions (i.e., PA versus MT) and that 4 h of dark incubation led to an even higher proportion of free ribosomes (data not shown). We will pursue this relationship of metabolism to protein synthesis, as well as the connection to circadian rhythms, in future experiments.
Acknowledgments
We thank Debra Sherman, Director of the Life Sciences Microscopy Facility, for the electron micrographs.
The research was funded by grant DE-FG02-99ER20342 from the Department of Energy.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Broedel, S. E., Jr., and R. E. Wolf, Jr. 1990. Genetic tagging, cloning, and DNA sequence of the Synechococcus sp. strain PCC 7942 gene (gnd) encoding 6-phosphogluconate dehydrogenase. J. Bacteriol. 172:4023-4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daldal, F. 1983. Molecular cloning of the gene for phosphofructokinase-2 of Escherichia coli and the nature of a mutation, pfkB1, causing a high level of the enzyme. J. Mol. Biol. 168:285-305. [DOI] [PubMed] [Google Scholar]
- 4.Dandekar, T., S. Schuster, B. Snel, M. Huynen, and P. Bork. 1999. Pathway alignment: application to the comparative analysis of glycolytic enzymes. Biochem. J. 343:115-124. [PMC free article] [PubMed] [Google Scholar]
- 5.Figge, R. M., C. Cassier-Chauvat, F. Chauvat, and R. Cerff. 2000. The carbon metabolism-controlled Synechocystis gap2 gene harbours a conserved enhancer element and a gram-positive-like −16 promoter box retained in some chloroplast genes. Mol. Microbiol. 36:44-54. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Dominguez, M., M. I. Muro-Pastor, J. C. Reyes, and F. J. Florencio. 2000. Light-dependent regulation of cyanobacterial phytochrome expression. J. Bacteriol. 182:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giberson, R. T., R. L. Austin, J. Charlesworth, G. Adamson, and G. A. Herrera. 2003. Microwave and digital imaging technology reduce turnaround times for diagnostic electron microscopy. Ultrastruct. Pathol. 27:187-196. [DOI] [PubMed] [Google Scholar]
- 8.Golden, S. S., and S. R. Canales. 2003. Cyanobacterial circadian clocks—timing is everything. Nat. Rev. Microbiol. 1:191-199. [DOI] [PubMed] [Google Scholar]
- 9.Iwasaki, H., S. B. Williams, Y. Kitayama, M. Ishiura, S. S. Golden, and T. Kondo. 2000. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell 101:223-233. [DOI] [PubMed] [Google Scholar]
- 10.Knowles, V. L., and W. C. Plaxton. 2003. From genome to enzyme: analysis of key glycolytic and oxidative pentose-phosphate pathway enzymes in the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 44:758-763. [DOI] [PubMed] [Google Scholar]
- 11.Knowles, V. L., C. S. Smith, C. R. Smith, and W. C. Plaxton. 2001. Structural and regulatory properties of pyruvate kinase from the cyanobacterium Synechococcus PCC 6301. J. Biol. Chem. 276:20966-20972. [DOI] [PubMed] [Google Scholar]
- 12.Koksharova, O., M. Schubert, S. Shestakov, and R. Cerff. 1998. Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 36:183-194. [DOI] [PubMed] [Google Scholar]
- 13.Kotlarz, D., and H. Buc. 1982. Phosphofructokinases from Escherichia coli. Methods Enzymol. 90(Pt. E):60-70. [DOI] [PubMed] [Google Scholar]
- 14.Li, H., A. K. Singh, L. M. McIntyre, and L. A. Sherman. 2004. Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 186:3331-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luinenburg, I., and J. R. Coleman. 1990. A requirement for phosphoenolpyruvate carboxylase in the cyanobacterium Synechococcus PCC 7942. Arch. Microbiol. 154:471-474. [Google Scholar]
- 16.Marin, K., I. Suzuki, K. Yamaguchi, K. Ribbeck, H. Yamamoto, Y. Kanesaki, M. Hagemann, and N. Murata. 2003. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. USA 100:9061-9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakahara, K., H. Yamamoto, C. Miyake, and A. Yokota. 2003. Purification and characterization of class-I and class-II fructose-1,6-bisphosphate aldolases from the cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 44:326-333. [DOI] [PubMed] [Google Scholar]
- 18.Pelroy, R. A., G. A. Levine, and J. A. Bassham. 1976. Kinetics of light-dark CO2 fixation and glucose assimilation by Aphanocapsa 6714. J. Bacteriol. 128:633-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postier, B. L., H. L. Wang, A. Singh, L. Impson, H. L. Andrews, J. Klahn, H. Li, G. Risinger, D. Pesta, M. Deyholos, D. W. Galbraith, L. A. Sherman, and R. L. Burnap. 2003. The construction and use of bacterial DNA microarrays based on an optimized two-stage PCR strategy. BMC Genomics 4:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Scanlan, D. J., S. Sundaram, J. Newman, N. H. Mann, and N. G. Carr. 1995. Characterization of a zwf mutant of Synechococcus sp. strain PCC 7942. J. Bacteriol. 177:2550-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaeffer, F., and R. Y. Stanier. 1978. Glucose-6-phosphate dehydrogenase of Anabaena sp. Kinetic and molecular properties. Arch. Microbiol. 116:9-19. [DOI] [PubMed] [Google Scholar]
- 23.Schmetterer, G. 1994. Cyanobacterial respiration, p. 409-435. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer, Boston, Mass.
- 24.Schneegurt, M. A., D. M. Sherman, S. Nayar, and L. A. Sherman. 1994. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 176:1586-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherman, D. M., and L. A. Sherman. 1983. Effect of iron deficiency and iron restoration on ultrastructure of Anacystis nidulans. J. Bacteriol. 156:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, A. K., L. M. McIntyre, and L. A. Sherman. 2003. Microarray analysis of the genome-wide response to iron deficiency and iron reconstitution in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 132:1825-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh, A. K., and L. A. Sherman. 2000. Identification of iron-responsive, differential gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803 with a customized amplification library. J. Bacteriol. 182:3536-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stal, L. J., and R. Moezelaar. 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21:179-211. [Google Scholar]
- 29.Valverde, F., M. Losada, and A. Serrano. 1997. Functional complementation of an Escherichia coli gap mutant supports an amphibolic role for NAD(P)-dependent glyceraldehyde-3-phosphate dehydrogenase of Synechocystis sp. strain PCC 6803. J. Bacteriol. 179:4513-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, C., Q. Hua, and K. Shimizu. 2002. Integration of the information from gene expression and metabolic fluxes for the analysis of the regulatory mechanisms in Synechocystis. Appl. Microbiol. Biotechnol. 58:813-822. [DOI] [PubMed] [Google Scholar]
- 31.Yang, C., Q. Hua, and K. Shimizu. 2002. Metabolic flux analysis in Synechocystis using isotope distribution from 13C-labeled glucose. Metab. Eng. 4:202-216. [DOI] [PubMed] [Google Scholar]
- 32.Yeh, K. C., S. H. Wu, J. T. Murphy, and J. C. Lagarias. 1997. A cyanobacterial phytochrome two-component light sensory system. Science 277:1505-1508. [DOI] [PubMed] [Google Scholar]