Abstract
The unusual and complex cell wall of pathogenic mycobacteria plays a major role in pathogenesis, with specific complex lipids acting as defensive, offensive, or adaptive effectors of virulence. The phthiocerol and phthiodiolone dimycocerosate esters (PDIMs) comprise one such category of virulence-enhancing lipids. Recent work in several laboratories has established that the Mycobacterium tuberculosis fadD26-mmpL7 (Rv2930-Rv2942) locus plays a major role in PDIM biosynthesis and secretion and that PDIM is required for virulence. Here we describe two independent transposon mutants (WAg533 and WAg537) of Mycobacterium bovis, both of which carry an insertion in Mb0100 (= M. tuberculosis Rv0097) to reveal a new locus involved in PDIM biosynthesis. The mutations have a polar effect on expression of the downstream genes Mb0101, Mb0102 (fadD10), Mb0103, and Mb0104 (nrp), and Mb0100 is shown to be in an operon comprising these genes and Mb0099. Reverse transcription-PCR analysis shows elevated transcription of genes in the operon upstream from the transposon insertion sites in both mutants. Both mutants have altered colony morphology and do not synthesize PDIMs or glycosylphenol-PDIM. Both mutants are avirulent in a guinea pig model of tuberculosis, and when tested as a vaccine, WAg533 conferred protective immunity against M. bovis infection at least equal to that afforded by M. bovis bacillus Calmette-Guérin.
Tuberculosis, caused by closely related members of the Mycobacterium tuberculosis complex, continues to have a major impact on human and animal health worldwide and is responsible for the death of approximately two million people each year, primarily in developing nations (14). Mycobacterium bovis, the pathogen responsible for bovine tuberculosis, is a broad-host-range member of the M. tuberculosis complex, and its transmission to humans is probably responsible for some 5% of human tuberculosis deaths (15). The current tuberculosis vaccine, M. bovis bacillus Calmette-Guérin (BCG), has shown highly variable efficacy, and a significantly better vaccine is urgently required.
In New Zealand, traditional test and slaughter approaches to eradication of bovine tuberculosis from domestic livestock have been frustrated by the presence of introduced wildlife, particularly the Australian brushtail possum (Trichosurus vulpecula), which maintains a reservoir of infection (23). Extensive wildlife culling operations over many years have failed to eliminate infected possums from many parts of the country, and vaccination of wildlife against tuberculosis is being investigated. Any vaccine developed for tuberculosis control in the New Zealand environment must be compatible with large-scale vaccination of animals, and this requirement has directed research towards the development of rationally attenuated strains of M. bovis with vaccine efficacy (9, 11, 12). More-detailed investigation of the attenuation of some of these strains (10, 38) is also contributing to our understanding of the molecular determinants required for tuberculosis pathogenesis.
Among the known determinants required for virulence in pathogenic mycobacteria are complex lipid components of the mycobacterial cell wall that act as defensive, offensive, or adaptive effectors of virulence. The phthiocerol and phthiodiolone dimycocerosate esters (PDIMs) comprise one such category of virulence-enhancing lipids produced by members of the M. tuberculosis complex and closely related species (17). PDIMs are built upon polyketide scaffolds and comprise multimethyl-branched long-chain mycocerosic acids diesterified with long-chain phthiocerol or phthiodiolone diols (28) (Fig. 1). Additional PDIM variants include the phenol- and glycosylphenol-PDIMs (Fig. 1). Recent work in several laboratories has established that proteins encoded by genes at the M. tuberculosis fadD26-mmpL7 locus (fadD26, ppsA to ppsE, drrA to drrC, papA5, mas, fadD28, and mmpL7) play major roles in PDIM biosynthesis and secretion and that PDIM is required for virulence (7, 16, 33, 35). PpsA to PpsE and Mas are multifunctional polyketide synthases responsible for fatty-acid-chain extension utilizing either malonyl CoA (PpsA to PpsC) or methylmalonyl coenzyme A (CoA) (PpsD, PpsE and Mas) and leading to phthiocerol (PpsA to PpsE) and mycocerasate (Mas) biosynthesis. Onwueme et al. (29) have recently proposed that PapA5 is required for diesterification of phthiocerol with mycocerosate to produce PDIM. FadD26 belongs to a family of long-chain fatty acyl-AMP ligases that activate long-chain fatty acids as acyl-adenylates for subsequent transfer to their cognate multifunctional polyketide synthases, and in the case of FadD26, transfer is to PpsA (39). Several additional proteins involved in PDIM and glycosylphenol-PDIM biosynthesis are encoded by genes located immediately downstream from the fadD26-mmpL7 locus (13, 31, 32).
FIG. 1.
PDIM (3) biosynthesis involves diesterification of mycocerosic acid (2) with phthiocerol (1). R1 is -CH3 for phthiocerols, phthiodiolone, and PDIMs or methyl-d-rhamnose-phenol-CH2 -for glycosylphenol PDIM. R2 is -OCH3 for phthiocerol and glycosylphenol PDIM and =O for phthiodiolone. R3 is -CH3 or -H. R4 denotes a thioester intermediate.
Despite our increasing knowledge of PDIM and glycosylphenol-PDIM biosynthesis, considerable gaps still remain. Here, we describe the characterization of two avirulent transposon mutants, WAg533 and WAg537, derived from the virulent M. bovis wild-type strains WAg200 and WAg201, respectively. While the two mutants were independently identified, they each carry a disruption in Mb0100 (= M. tuberculosis Rv0097) and show the same defect in PDIM and glycosylphenol-PDIM biosynthesis. When WAg533 was tested as a tuberculosis vaccine in guinea pigs, it was at least as effective as BCG.
MATERIALS AND METHODS
Bacterial strains, growth media, and culture conditions.
Plasmids were propagated at 37°C in Escherichia coli XL1 Blue MR in Luria-Bertani broth or Luria-Bertani agar (Difco) supplemented with kanamycin (50 μg/ml). M. bovis strains examined were the following: WAg200, a virulent wild-type isolate from New Zealand cattle, and its transposon mutant, WAg533; WAg201, also a virulent wild-type isolate from New Zealand cattle, and its transposon mutant, WAg537; and a signature tag mutant derived from WAg201, WAg579, that carries a disruption in pks15/1 (11). M. bovis wild-type and transposon mutant strains were cultured at 37°C in Middlebrook 7H9 (Difco) liquid medium and on Middlebrook 7H11 (Difco) solid medium as previously described (12). For 96-well microtiter plates, mutants were cultured in Tween-albumin broth (Tween 80, Dubos broth base [Difco], oleic acid-albumin-dextrose complex [Difco]) (25). Transposon and signature tag mutants were selected and cultured on solid or liquid medium supplemented with 20 μg of kanamycin/ml. M. bovis was cultured from guinea pig spleens (half the spleen) and lungs (left apical lobe) at the conclusion of vaccine efficacy trials as previously described (12).
Transposon mutagenesis.
The suicide plasmid vector, pYUB553.1, containing transposon Tn5367 (26), which incorporates a kanamycin resistance gene as a selectable marker, was electroporated (41) into the wild-type, virulent M. bovis strains WAg200 and WAg201. In the case of WAg200, electroporated cells were plated to obtain single colonies, and insertional mutants were screened by eye to detect colonies with morphology differences. In the case of WAg201, electroporated cells were plated and single colonies were subcultured in 96-well microtiter plates and transferred in duplicate to multiple Omnitrays containing solid medium with and without cycloserine (2 μg/ml).
To determine the site of insertion of the kanamycin resistance gene in the chromosome of the transposon mutants derived from WAg200 and WAg201, chromosomal DNA (40) was digested with EcoRI (which does not cut within Tn5367), ligated into the EcoRI site of the cloning vector pBluescript II KS, electroporated into E. coli XL1-Blue MR, and selected with kanamycin. Following isolation of the insertional construct, the junction regions of the transposon-containing fragment and mycobacterial chromosome were sequenced with primers DMC213 (5′-ACGTTCCGACCAGCGTC-3′) and DMC214 (5′-TCAGGGTCGAGGCGTTG-3′).
To confirm that a single transposition event had occurred in each mutant, a Southern blot of KpnI-digested genomic DNA that had been extracted (40) from liquid cultures of the parental and transposon mutant strains was hybridized with a 1,171-bp probe to Mb0100 made by incorporating [32P]dCTP in a PCR using primers DMC638 (5′-AGCACTACAGTCCCGTTGCT-3′) and DMC376 (Table 1).
TABLE 1.
Oligonucleotide primers used for RT-PCR analysis to define the Mb0099-Mb0104 operon
| RT-PCR product, gene boundary examined | Plus-strand primer and sequencea | Minus-strand primer and sequencea |
|---|---|---|
| 1, Mb0098-Mb0099 | DMC371 | DMC372 |
| AAGCGGTCCAGGTCGGCATC | CTCAAGCCAGGCCAGATAGG | |
| 2, Mb0099-Mb0100 | DMC373 | DMC613 |
| GAAAGCGGCGTCGGGATACG | ACCGAGTCCCTCGCCTTTGA | |
| 3, Mb0100-Mb0101 | DMC375 | DMC376 |
| GCACCACCAAGATCGAGGAC | GTTCGACCGCGTTGAAGTGC | |
| 4, Mb0101-Mb0102 | DMC377 | DMC378 |
| TGCTTCGCGGCTGGTCGATC | AGCTTTGCACACGCCAGCAC | |
| 5, Mb0102-Mb0103 | DMC379 | DMC380 |
| GGTCGCATCGGCAGAGCTTG | ACAGCCTCCAGCTCGCGAAG | |
| 6, Mb0103-Mb0104 | DMC381 | DMC382 |
| CGCCGTCTGCGACGTGTTGT | CAGGTGCTCATCGGCTGACC | |
| 7, Mb0102-Mb0104 | DMC379 | DMC382 |
| GGTCGCATCGGCAGAGCTTG | CAGGTGCTCATCGGCTGACC | |
| 8, Mb0104-Mb0105 | DMC383 | DMC384 |
| GGTTGCAGCGGTTTGAGGCC | CCAGCCGTACCATCGCCATG | |
| 9, Mb0105-Mb0106 | DMC385 | DMC386 |
| TGGCGCTGGTTACCCAATGG | TCTGGGCGTTCGGGTACAAC |
Gene-specific primers expected to prime reverse transcription based upon gene annotation are shown in Table 1 as minus-strand primers; gene-specific primers not expected to prime reverse transcription based upon gene annotation are shown in Table 1 as plus-strand primers, except for RT-PCR product 1 (Mb0098-Mb0099), where both primers are expected to prime reverse transcription but not yield a RT-PCR product, and RT-PCR product 9 (Mb0105-Mb0106), where neither primer is expected to prime reverse transcription.
Determination of virulence and determination of vaccine efficacy.
All animal work was approved by the institution's Animal Ethics Committee. Virulence was tested by inoculating approximately 106 CFU of each strain subcutaneously into the flanks of three Dunkin-Hartley guinea pigs. After 8 weeks, the animals were sacrificed and autopsied, and virulence was assessed by determining the numbers of macroscopic spleen, lung, and liver lesions.
Vaccine efficacy trials were performed as previously described (12). Briefly, groups of six Dunkin-Hartley guinea pigs were vaccinated subcutaneously with 105 CFU of WAg533 or BCG. A control group of six animals was not vaccinated. Eight weeks after vaccination, all animals were challenged by aerosol with 2 to 10 CFU of wild-type M. bovis (WAg201). Five weeks after challenge, the animals were sacrificed and autopsied. The numbers of macroscopic spleen lesions were recorded, and samples of spleen and lung were subjected to mycobacterial culture and the numbers of CFU were recorded. Statistical analyses by analysis of variance were performed on log10 transformations of spleen and lung CFU and on numbers of macroscopic lesions in the spleen.
RNA extraction and RT-PCR.
RNA was prepared by adding four volumes of guanidinium thiocyanate solution directly to broth cultures (37), harvesting the cells by centrifugation (approximately 108 cells/extraction), and disrupting the cells in Lysing Matrix B tubes (Qbiogene), using a FastPrep cell disrupter (Qbiogene) in the presence of 1 ml of Trizol (Invitrogen). After Trizol extraction, residual DNA was removed by incubation in DNase I (Invitrogen), and the RNA was further purified by using an RNeasy minikit (QIAGEN). At least three independent RNA samples were prepared for each strain. Reverse transcription (RT) using Transcriptor reverse transcriptase (Roche) was performed according to the manufacturer's instructions, using random primers or, for some experiments detailed in the Results section, gene-specific primers (Table 1). PCR was performed in a GeneAmp PCR System 9700 (Applied Biosystems) instrument, using the following conditions: 94°C for 5 min (94°C, 45 s; 60°C, 30 s; 72°C, 90 s) 31 times and 72°C for 7 min at 4°C. PCR primers are shown in Table 1. Extensive control reactions utilizing DNaseI-treated RNA without reverse transcription were performed for all PCR primer-sample combinations. In all cases, these control reactions did not yield PCR products (data not shown). All PCR primer pairs produced the expected PCR product with genomic DNA as a template (data not shown).
Lipid analysis.
The production of PDIMs and glycosylphenol-PDIM by the parental and mutant strains was analyzed by two-dimensional thin-layer chromatography (TLC). Cultures were harvested during late exponential phase (optical density at 600 nm = 1.0) for each wild-type and mutant strain, heat killed, and dried under nitrogen. The dried bacterial pellet (50 to 100 mg) was subjected to biphasic extraction as described previously (3) and finally redissolved in hexane. Extracted mycobacterial lipids were separated by TLC on precoated, aluminum-backed, 10- by 10-cm ALGRAM Nano-Sil G/UV254 TLC plates (Macherey Nagel), using two two-dimensional solvent systems to reveal PDIMs (solvent system A) or glycosylphenol-PDIM (solvent system C) (18). Solvent system A was as follows: first dimension, petroleum ether/ethyl acetate, 98:2, three runs; second dimension, petroleum ether/acetone, 98:2, one run. Solvent system C was as follows: first dimension, chloroform/methanol, 96:4; second dimension, toluene/acetone, 80:20. TLC plates were dipped in 4% ethanolic phosphomolybdic acid (solvent system A) or 2.5% α-naphthol-10% sulfuric acid (or 10% sulfuric acid alone) (solvent system C) and heated at 100 to 120°C to reveal PDIMs and glycosylphenol-PDIM, respectively.
In order to fractionate PDIMs from other nonpolar lipids, a cartridge purification system was developed. The petroleum-ether, nonpolar extract (3) was applied directly to a prewashed 500-mg normal-phase silica gel cartridge (Alltech), using 200 μl of petroleum ether. The sample was then eluted using 0.5% increments of diethyl ether in petroleum ether (0 to 5%) over 200 ml and collected in 20-ml fractions in test tubes. Fractions were dried by using a stream of argon, redissolved in petroleum ether, and analyzed by one-dimensional TLC by using direction 2 of solvent system A. The purified PDIMs were then reanalyzed by using solvent system A as described above, and a matrix-assisted laser desorption ionization-time-of-flight spectrum was recorded for the purified PDIM in comparison to M. tuberculosis PDIM standard.
Bioinformatics.
DNA and amino acid sequences for analysis were retrieved from the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), the Bovilist web server (http://genolist.pasteur.fr/BoviList/), the Tuberculist web server (http://genolist.pasteur.fr/TubercuList/), or the MycoBD web server (http://myco.bham.ac.uk//) as appropriate. BLAST searches were performed using BLAST resources available at the National Centre for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/ and http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). General sequence storage, analysis, and annotation were performed by using VectorNTI (Informax/Invitrogen).
RESULTS
Isolation of transposon mutants.
The virulent M. bovis wild-type strains WAg200 and WAg201 were both electroporated with the suicide plasmid vector, pYUB553.1, containing the transposon Tn5367. In the case of WAg200, more than 5,000 insertional mutants were screened by eye to detect colonies with morphology differences from the parent. One strain, WAg533, which consistently showed such differences on reculturing (Fig. 2), was found to be avirulent in guinea pigs, with no macroscopic spleen, lung, or liver lesions observed. By comparison, more than 100 macroscopic spleen lesions are typically observed in guinea pigs inoculated with the wild-type parental strains WAg200 and WAg201. In the case of WAg201, more than 2,000 insertional mutants were screened, and one strain was identified that was sensitive to cycloserine and was avirulent in guinea pigs, with no macroscopic spleen, liver, or lung lesions observed. Subsequent examination on solid medium showed that like WAg533, WAg537 had an altered morphology compared to its parent strain, WAg201 (Fig. 2). Thus, WAg200 and WAg201 colonies are typically flat, while colonies of WAg533 produce lumpy structures rising above the surface of the agar plate, and colonies of WAg537 are less lumpy than WAg533 colonies but develop a pronounced striated appearance (Fig. 2).
FIG. 2.
Colony morphologies of transposon mutants WAg533 and WAg537 compared to those of their respective parental wild-type strains, WAg200 and WAg201.
Sequencing out into the M. bovis genome from each end of the transposon inserted into each of mutants WAg533 and WAg537 demonstrated that an identical gene, Mb0100, was disrupted in each case. Figure 3A shows the gene annotations in the region of Mb0100 (21) (http://genolist.pasteur.fr/BoviList/) and the relative positions where transposon Tn5367 was inserted into WAg533 and WAg537.
FIG. 3.
Disruption of Mb0100 in transposon mutants WAg533 and WAg537. (A) Gene organization around Mb0100 showing annotated gene designations, the positions of RT-PCR gene boundary products 1 to 9, the relative locations of transposon insertion in WAg533 and WAg537, and the position of KpnI sites (descending arrowheads) flanking Mb0100. (B) Autoradiogram of a Southern blot of KpnI-digested genomic DNA from parental (WAg200 and WAg201) and transposon mutant (WAg533 and WAg537) strains hybridized with a Mb0100-derived probe.
Examination of the genome sequence in the region of Mb0100 in the sequenced M. bovis strain (AF2122/97) (http://genolist.pasteur.fr/BoviList/) and comparison with the M. tuberculosis H37Rv genome sequence (http://genolist.pasteur.fr/TubercuList/) identified KpnI sites flanking the insertion site (Fig. 3A) (KpnI does not cut within Tn5367). The existence of a single transposon insertion into Mb0100 in each mutant was demonstrated by hybridizing a Southern blot of KpnI-digested genomic DNA from each parent and mutant strain with an Mb0100-derived probe. Both parental strains displayed a 1,684-bp wild-type fragment, and as expected for a single transposition event, both mutants displayed a single fragment of 5.1 kb corresponding to the 1,684-bp wild-type KpnI fragment carrying a 3.4-kb Tn5367 insertion (Fig. 3B).
Vaccination studies.
WAg533 (WAg537 not tested) was examined for its efficacy as a vaccine against bovine tuberculosis. Vaccination with both WAg533 and BCG resulted in highly significant protection against virulent M. bovis for all measures examined (Table 2). WAg533 was better than BCG for all measures of protection examined, but the differences were not statistically significant (Table 2).
TABLE 2.
Vaccination of guinea pigs with WAg533 protects against aerosol challenge with M. bovisa
| M. bovis vaccination strain | Spleen lesions (mean no. ± SE) | M. bovis in spleen (log10 CFU ± SE) | M. bovis in lung (log10 CFU ± SE) |
|---|---|---|---|
| None | 32 ± 8.0A | 4.97 ± 0.17A | 4.22 ± 0.25A |
| BCG | 0.3 ± 0.33B | 1.53 ± 0.53B | 2.36 ± 0.36B |
| WAg533 | 0.0 ± 0.0B | 1.00 ± 0.00B | 2.30 ± 0.44B |
Values within the same column with the same superscript letter are not significantly different.
Transposon insertion into Mb0100 has a polar effect, disrupting expression of Mb0101 and Mb0102 and reducing expression of Mb0103 and Mb0104.
The annotated M. bovis AF2122/97 genome (21) (http://genolist.pasteur.fr/BoviList/) suggested that Mb0100 was located within an operon comprising Mb0099 (ppe1), Mb0100, Mb0101, Mb0102 (fadD10), Mb0103, and Mb0104 (nrp). Furthermore, each of the equivalent genes in M. tuberculosis is strongly induced in a senX3-regX3 mutant, suggesting that these genes are at least coregulated (30). If Mb0100 is in an operon with Mb0099, Mb0101, Mb0102, Mb0103, and Mb0104, then transposon insertion into Mb0100 in mutants WAg533 and WAg537 would be expected to have a polar effect on expression of the other, downstream genes. Consequently, a series of PCR primer pairs (Table 1) was designed to amplify across gene boundaries (1 to 9 in Fig. 3A) so that, coupled to reverse transcription, they would define the extent of the operon containing Mb0100 and the extent of the polar effect of the mutations.
Results presented in Fig. 4A, B, and C (which present results from typical experiments) show that Mb0099 to Mb0104 are cotranscribed in the wild-type strains WAg200 and WAg201, since all the intergene boundary RT-PCR products were observed, while expected negative controls (RT-PCR products 1 and 9) were not. The Mb0099-Mb0100 boundary RT-PCR product (RT-PCR product 2) was invariably weak compared to RT-PCR products 3, 4, 5, 6, and 7. Indeed, on several occasions, as shown for WAg200 and WAg201 in Fig. 4A, this product was not visible after 31 PCR cycles but could be detected by increasing the number of cycles. We suggest that the mRNA transcript is less stable in the region of the Mb0099-Mb0100 boundary than in the other gene boundaries examined. In contrast, disruption of Mb0100 resulted in a dramatic increase in expression across this gene boundary (compare RT-PCR products 2 in WAg533 and WAg537 with their parents), confirming that Mb0099 and Mb0100 are cotranscribed and suggesting the possible involvement of a feedback regulatory system controlling expression of the Mb0099-Mb0104 operon. Serial dilution RT-PCR experiments were performed to establish that transcription across the Mb0099-Mb0100 (RT-PCR product 2) gene boundary increased by a minimum of 8- to 16-fold in WAg533 compared to results for WAg200 and increased by a minimum of 8-fold in WAg537 compared to results for WAg201. A typical result is shown in Fig. 4C.
FIG. 4.
Disruption of Mb0100 in transposon mutants WAg533 and WAg537 results in altered expression of both upstream boundary RT-PCR products and downstream boundary RT-PCR products. (A) Expression analysis of RT-PCR gene boundary products 1 to 9 in parental (WAg200 and WAg201) and transposon mutant (WAg533 and WAg537) strains following reverse transcription with random primers. (B) Expression analysis of RT-PCR gene boundary products 5 to 7 in parental (WAg200 and WAg201) and transposon mutant (WAg533 and WAg537) strains following reverse transcription with gene-specific primers that were expected (left arrow) or not expected (right arrow) to prime reverse transcription. (C) Dilution RT-PCR analysis of RT-PCR gene boundary product 2 (Mb0099-Mb0100) in parental (WAg200 and WAg201) and transposon mutant (WAg533 and WAg537) strains.
Results presented in Fig. 4A, B, and C further show that in transposon mutants WAg533 and WAg537, transposon insertion into Mb0100 disrupted transcription across the Mb0100-Mb0101 (RT-PCR product 3) and Mb0101-Mb0102 (RT-PCR product 4) gene boundaries (compare the absence of RT-PCR products 3 and 4 in WAg533 and WAg537 with their presence in the parental strains). However, transposon insertion into Mb0100 did not disrupt transcription across the Mb0102-Mb0103 (RT-PCR product 5), Mb0103-Mb0104 (RT-PCR product 6), or Mb0102-Mb0104 (RT-PCR product 7) gene boundaries but resulted in decreased expression. Results shown in Fig. 4A were obtained by using random primers for reverse transcription. To confirm reduced expression across the Mb0102-Mb0103 (RT-PCR product 5), Mb0103-Mb0104 (RT-PCR product 6), and Mb0102-Mb0104 (RT-PCR product 7) gene boundaries, we used gene-specific primers for reverse transcription to again show that transcription across all three gene boundaries was reduced in WAg533 and WAg537 compared to their expression levels in WAg200 and WAg201 (Fig. 4B). Unexpectedly, we also observed transcription from the opposite strand (Fig. 4B), likely accounting for some of the expression observed across the Mb0102-Mb0103 (RT-PCR product 5), Mb0103-Mb0104 (RT-PCR product 6), and Mb0102-Mb0104 (RT-PCR product 7) gene boundaries obtained by using random primers for reverse transcription (Fig. 4A). Two partially overlapping open reading frames (with no similarity to any other genes detected in BLAST searches) are present in this region (data not shown), and their transcription may account for our results.
Results presented in Fig. 4A, B, and C show that in the wild-type strains, Mb0099 to Mb0104 are cotranscribed, since the relevant intergene boundary RT-PCR products are observed. However, RT-PCR results for WAg533 and WAg537 suggest that Mb0102-Mb0104 can be cotranscribed separately from Mb0099, Mb0100, and Mb0101. This indicates that there is an alternative promoter located immediately upstream from Mb0102. Consistent with this proposal, a possible mycobacterial promoter sequence (TCGAACN23TCCAATGGGAGGAAAGAAGTTTCAAGCTATG) with typical −10 and −35 regions (bold italic) (24) is found immediately upstream from, and partially overlapping, the annotated Mb0102 ATG start codon (underlined above). If so, this would imply that a more downstream start codon may be used, the first of which is highlighted in bold above.
We occasionally observed an RT-PCR product (RT-PCR product 8) indicative of transcription across the Mb0104-Mb0105 gene boundary (such an instance is shown for WAg201 in Fig. 4A). While detection of the Mb0104-Mb0105 boundary RT-PCR product clearly shows that the Mb0104-Mb0105 boundary can be cotranscribed, this product was detected in only a few samples. This, combined with the 175-bp separation between Mb0104 and Mb0105 and the observations by Parish et al. (30) that the M. tuberculosis genes equivalent to Mb0099, Mb0100, Mb0101, Mb0102, Mb0103, and Mb0104 are strongly induced in a senX3-regX3 mutant, while the M. tuberculosis gene equivalent to Mb0105 is not, together suggest that our detection of a Mb0104-Mb0105 boundary RT-PCR product in only a few samples most likely represents an occasional failure of transcription to terminate by the position of PCR primer DMC384.
Disruption of Mb0100 blocks biosynthesis of PDIMs and glycosylphenol-PDIM.
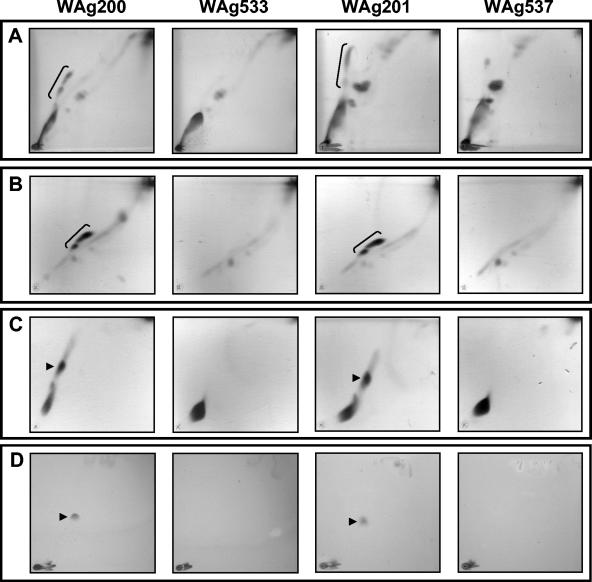
The differences in colony morphology between the two transposon mutants and their parental strains suggested that the mutants might be defective in some aspect of cell wall lipid biosynthesis, prompting an examination of the lipid profiles of the parental and mutant strains by TLC. Examination of the extracted lipid profiles under a range of TLC solvent systems designed to systematically profile a wide range of mycobacterial lipids (18) demonstrated that WAg533 and WAg537 both failed to produce PDIMs (Fig. 5A) or glycosylphenol-PDIM (Fig. 5D) but otherwise had normal lipid profiles. While an unidentified lipid is present in the PDIM region for WAg537 (Fig. 5A), additional cartridge fractionation and TLC analysis of lipids from the parental strains and transposon mutants showed that no PDIMs are produced (Fig. 5B and C). Additional lipids have been previously observed in the PDIM region (18).
FIG. 5.
Biosynthesis of PDIMs and glycosylphenol-PDIM is disrupted in transposon mutants WAg533 and WAg537. (A, B, and C) TLC showing the absence of PDIM biosynthesis in transposon mutants WAg533 and WAg537. The PDIM region is bracketed for the wild-type strains and comprises PDIMs A, B, and C (3). (B and C) TLC of the PDIM-containing fractions after cartridge fractionation. The PDIM region is bracketed for the wild-type strains in the fraction analyzed in panel B and comprises PDIMs A, B, and C, while the fraction analyzed in panel C contains only PDIM C (arrows) and triacylglycerol, which is present in all four strains, closer to the origin. (D) Absence of glycosylphenol-PDIM (arrows for the wild-type strains) biosynthesis in transposon mutants WAg533 and WAg537.
A matrix-assisted laser desorption ionization-time-of-flight spectrum of the purified PDIM from both M. bovis WAg200 and WAg201 afforded a series of molecular ions (m/z 1,304 to 1,390) consistent with a C86-C92 composition of PDIM (data not shown). These molecular species of M. bovis PDIM were slightly smaller than M. tuberculosis PDIM (C92-C100) and were consistent with previous observations of smaller mycocerosic fatty acids in PDIM from M. bovis in comparison to those from M. tuberculosis (6, 27).
To provide additional verification that glycosylphenol-PDIM was the lipid observed in Fig. 5D for both wild-type strains but was not present in either of the transposon mutant strains, we utilized a pks15/1-disrupted M. bovis mutant (WAg579) derived from WAg201 by selection for reduced virulence in a guinea pig model of bovine tuberculosis (11). We expected this mutant to produce PDIMs but not produce glycosylphenol-PDIM, based upon results from a similar M. bovis BCG pks15/1-disrupted mutant (13). As expected, WAg579 produced PDIMs (Fig. 6A) but did not produce glycosylphenol-PDIM (Fig. 6B).
FIG. 6.
WAg579, a pks15/1-disrupted mutant, produces PDIMs but does not produce glycosylphenol-PDIM. (A) PDIMs are produced by both WAg579 and its parental, wild-type strain, WAg201 (the PDIM region is bracketed). (B) Glycosylphenol-PDIM (arrow) is produced by WAg201 but not by WAg579.
Sequence analysis of the predicted proteins encoded by Mb0099-Mb0104.
We found that gene order at the Mb0099-Mb0104 locus is highly conserved among slow-growing pathogenic mycobacteria and is also substantially conserved in the related actinomycete Streptomyces avermitilis (Fig. 7). The predicted proteins encoded by these genes in M. bovis are examined in detail below.
FIG. 7.
Colocalization of genes (not drawn to scale) in the Mb0099-Mb0104 (ppe1-nrp) operon in M. bovis is highly conserved in the M. tuberculosis, M. leprae, M. marinum, and S. avermitilis genomes.
The predicted protein encoded by Mb0099 (PPE1) is a member of the PPE multigene family (21) but is of unknown function. The predicted protein encoded by Mb0100 shows greatest similarity to members of the alpha-ketoglutarate-dependent dioxygenase superfamily, particularly members of the taurine dioxygenase subfamily, and most likely functions as a dioxygenase or more generally as an oxidoreductase. Members of the superfamily are characterized by a requirement for Fe(II), which is bound by a conserved three-amino-acid His-X-Asp/Glu-Xn-His motif. Mb0100 retains this motif as His97-Ile-Asp99-X160-His260. The predicted protein encoded by Mb0101 shows no similarity to any other proteins, with the exception of orthologs in mycobacteria and S. avermitilis (Fig. 7), and its function is unknown. The predicted protein encoded by Mb0102 (FadD10), like other mycobacterial FadD proteins, is similar to acyl-CoA synthetases that convert free fatty acids into acyl-coenzyme A thioesters as a prelude to fatty acid degradation. FadD10 belongs to the larger of two M. tuberculosis FadD clusters (39) and, like other members of the large cluster, shows relatively low similarity to other members of the cluster. The predicted protein encoded by Mb0103 shows limited similarity to acyl carrier protein domains involved in polyketide biosynthesis. The characteristic acyl carrier protein Asp-Ser-Leu (DSL) motif that serves as a 4′-phosphopantetheine attachment site is conserved as DSV in Mb0103.
The predicted protein encoded by Mb0104 (Nrp) shows significant similarity to a wide range of nonribosomal peptide synthetases (NRPSs) from a broad range of species. NRPSs are organized as iterative modules, one for each amino acid to be incorporated into the peptide product. A minimal elongation module consists of a 55-kDa adenylation (A) domain responsible for substrate selection and activation, a 10-kDa downstream peptidyl carrier protein (PCP) domain, and a 50-kDa condensation (C) domain, located N-terminal to the A domain, that catalyses peptide bond formation. Two modules are present in Nrp and the identical gene in M. tuberculosis, but only one module is present in the orthologous NRPSs in Mycobacterium leprae, Mycobacterium marinum, and S. avermitilis (Fig. 8).
FIG. 8.
Comparative domain relationships for the Nrp homologs from M. bovis, M. tuberculosis, M. leprae, M. marinum, and S. avermitilis shown in Fig. 7. Each condensation (C) domain, adenylation (A) domain, peptidyl carrier protein (PCP) domain, or dehydrogenase (D) domain within each protein is shown as a circle (not to scale). One module (comprising single, sequential C, A, and PCP domains) is present in the M. leprae, M. marinum and S. avermitilis Nrps, while two modules (A1, C1, PCP1 [module 1] and A2, C2, PCP2 [module 2]) are present in the M. bovis and M. tuberculosis Nrps. The lines between domains show the similarity of the M. leprae and M. marinum C, A, and PCP domains to the module 1 domains from M. bovis and M. tuberculosis and the weaker similarity of the S. avermitilis C, A, PCP, and D domains to the module 2 domains and D domain from M. bovis and M. tuberculosis.
Nrp condensation domain 2 (C2) (Fig. 8) retains the conserved HHX3DG motif found in NRPS condensation domains and the more extended HX3DX14Y motif that is also found in the conserved Pap proteins (5). N-terminal to the first A domain (Fig. 8) is a second putative condensation domain (C1) not detected by BLAST searches. This domain retains the HHX3DG motif and a modified HX3DX13Y motif. The specific A domain amino acid residues responsible for substrate selectivity have been defined (8, 36). In Nrp domain A2, the corresponding residues are DAWTVAAICK, showing that A2 is likely specific for phenylalanine. In contrast, Nrp domain A1 shows comparatively low similarity to typical NRPS A domains, and it is not possible to predict the cognate amino acid substrate. The Nrp orthologs from M. leprae and M. marinum share only the A1 domain. In contrast, the Nrp ortholog from S. avermitilis shares an A2-like domain. While Nrp PCP domain 1 (Fig. 8) shows similarity only to the equivalent domain in the Nrp orthologs in M. leprae and M. marinum, PCP2 shows high similarity to PCP domains in NRPSs from a wide range of species. PCP2 retains the typical phosphopantetheine attachment site DSL motif, while PCP1 carries a modified GSL motif. The most carboxy-terminal region of Nrp contains an extended region with similarity to putative NRPS dehydrogenase/reductase domains. Overlapping the N-terminal half of the dehydrogenase/reductase domain is an epimerase domain that retains the conserved cofactor-binding motifs GxxGxxG and YxxxK (as GATGFLG and YGTSK) of the larger, and diverse, short-chain dehydrogenase/reductase family. The M. bovis and M. tuberculosis dehydrogenase/reductase domains show high similarity to the C-terminal regions in NRPSs involved in the biosynthesis of glycopeptidolipids in Mycobacterium avium, Mycobacterium smegmatis, and M. avium subsp. paratuberculosis and high similarity to the C-terminal region of FadD9. Lower similarity to a range of enzymes including other NRPSs, polyketide synthases, and fatty acid synthases is also observed. In contrast, the M. leprae and M. marinum dehydrogenase domains (Fig. 8), while carrying the GxxGxxG and YxxxK dehydrogenase motifs, show no significant similarity to any other proteins.
DISCUSSION
The cell wall of pathogenic mycobacteria has long attracted attention as a major determinant of mycobacterial pathogenesis, with specific complex lipids mediating the organism's interaction with its host. A defining feature of the cell wall in strains of the M. tuberculosis complex and closely related species is the presence of several classes of long-chain multimethyl-branched fatty acids, a broad category that includes the phthiocerol- and phthiodiolone-dimycocerosate esters. Two independent signature-tagged mutagenesis studies have identified several attenuated mutants that carry disruptions in genes within the primary PDIM biosynthetic locus (7, 16). This locus comprises 13 genes, fadD26-mmpL7, transcribed in three operons (6) that encode proteins required for the synthesis or transport of PDIMs (1, 2, 6, 7, 16, 20). More recently, Pinto et al. (33) have shown that an M. tuberculosis strain disrupted in drrC, a PDIM biosynthetic locus gene required for translocation of PDIMs, provided better protective immunity than BCG in mice.
Here we have identified an additional locus, Mb0099-Mb0104 (ppe1-nrp), involved in the synthesis of PDIMs and glycosylphenol-PDIM. We have shown that disruption of Mb0100 in strains WAg533 and WAg537 disrupts PDIM and glycosylphenol-PDIM biosynthesis, causing a loss of virulence. When one of the resulting avirulent strains (WAg533) was tested for vaccine efficacy, all measures of protection examined were better than BCG, but the differences were not statistically significant. This may point to a limited ability of our vaccine protocol to resolve differences between BCG and potentially better vaccine candidates. Whether or not a modified protocol can be devised that has sufficient resolving power awaits further experimentation. In addition, it is possible that a double-knockout mutant comprising a Mb0100 deletion and a deletion within another gene (11, 12) may produce a vaccine strain that can be shown to be significantly better than BCG even using our present protocol.
We found that WAg533 and WAg537 produce neither PDIMs nor glycosylphenol-PDIM, indicating that disruption of Mb0100 affects a common branch of the PDIM/glycosylphenol-PDIM biosynthetic pathway rather than a glycosylphenol-PDIM-specific branch, such as phenol or sugar residue addition. Consequently, it was unclear if the avirulent phenotype of WAg533 and WAg537 in guinea pigs was the result of their failure to specifically produce glycosylphenol-PDIM or their failure to produce glycosylphenol-PDIM and PDIMs. Clarification was provided by comparison of the virulence properties and lipid profiles of WAg533 and WAg537 with those of WAg579, a mutant disrupted in pks15/1 (11) that, like a similar M. bovis BCG pks15/1-disrupted mutant (13), produces PDIMs but does not produce glycosylphenol-PDIM. The parental strains WAg200 (parent of WAg533) and WAg201 (parent of WAg537 and WAg579) are highly virulent in guinea pigs, typically causing more than 100 macroscopic spleen lesions per animal and multiple macroscopic lesions in the lung and liver. In contrast, no macroscopic spleen lesions, nor macroscopic liver or lung lesions, were observed after inoculating guinea pigs with WAg533 or WAg537. By comparison, WAg579 caused an average of 12 to 13 macroscopic spleen lesions per animal but no macroscopic lung or liver lesions (11). This difference in the virulence properties of WAg533 and WAg537 (avirulent) versus WAg579 (reduced virulence) shows that the absence of glycosylphenol-PDIM production by WAg533 and WAg537 cannot account for their avirulence. Thus, the avirulent phenotype of WAg533 and WAg537 requires disruption of the common branch of the PDIM/glycosylphenol-PDIM biosynthetic pathway.
Our results show that while the absence of glycosylphenol-PDIM production by WAg533 and WAg537 cannot account for their avirulence, glycosylphenol-PDIM nevertheless plays an important role in the pathogenicity of M. bovis in guinea pigs. Interestingly, most strains of M. tuberculosis have a frameshift mutation in pks15/1 and do not produce glycosylphenol-PDIM, showing that they do not require glycosylphenol-PDIM for virulence. While expression of M. bovis pks15/1 in M. tuberculosis strain H37Rv “restored” glycosylphenol-PDIM biosynthesis, the virulence of this strain was not examined (13). Conversely, some hypervirulent strains of M. tuberculosis do produce glycosylphenol-PDIM, and disruption of pks15/1 in one such strain resulted in reduction in its virulence in mice to equal the virulence of M. tuberculosis strain H37Rv (34).
RT-PCR experiments with RNA from the two wild-type strains used in this study demonstrated that Mb0099 to Mb0104 are cotranscribed in an operon. This observation is supported by gene annotation and by results arising from a senX3-regX3 deletion mutant of M. tuberculosis in which the genes equivalent to Mb0099-Mb0104 are strongly coinduced (30). Consequently, our finding that transcription of Mb0102-Mb0104 was detected in both mutant strains was unexpected. A potential alternative promoter region is present immediately upstream from Mb0102, and future studies will examine its functionality. We also observed increased transcription of the Mb0099-Mb0100 boundary in both WAg533 and WAg537. Since at least one of the genes in the Mb0099-Mb0104 operon represents a potential target for antituberculosis drug therapy, and the promoter of the Mb0099-Mb0104 operon is apparently up-regulated when Mb0100 is disrupted, there is potential to examine use of the Mb0099-Mb0104 promoter in a high-throughput promoter-based assay to identify candidate drugs for antituberculosis therapy.
While we have shown that the Mb0099-Mb0104 (ppe1-nrp) locus is involved in PDIM and glycosylphenol-PDIM biosynthesis and general functions for several of the encoded proteins can be proposed, their specific functions are unknown. Nevertheless, an analysis of likely function allows diagrammatic construction of a possible reaction mechanism involving some of the encoded proteins, as shown in Fig. 9 and as described in the accompanying figure legend. Future experiments will test the validity of this model. Of the three genes in the Mb0099-Mb0104 operon not included in our proposed reaction mechanism (Fig. 9), some support for a potential role in lipid biosynthesis exists for Mb0100, since other Fe(II)/αKG-dependent dioxygenases are known to play several roles in lipid metabolism (22).
FIG. 9.
Putative sequential reaction mechanism involving FadD10, Mb0103, and Nrp in the activation and transfer of a fatty acid to produce a lipid carrier molecule. In this model, FadD10 activates a long-chain fatty acid precursor of phthiocerol and/or mycocerosic acid biosynthesis as a CoA thioester (no. 1). The activated fatty acid is covalently attached to the 4′-phosphopanthetheine prosthetic (PP) group within the acyl carrier protein domain of Mb0103 (no. 2 and 3) and from there transferred (possibly by the action of Nrp condensation domain C1) to an unknown amino acid selected and activated by Nrp adenylation domain A1 (no. 4). Condensation domain C2 catalyzes peptide bond formation between this acylated amino acid and phenylalanine (selected and activated by adenylation domain A2) to create the mature acylated dipeptide thioester (no. 5).
Cotranscription of Mb0104 (nrp) with Mb0102 (fadD10) and Mb0103 suggests the surprising possibility that Nrp is involved in PDIM and glycosylphenol-PDIM biosynthesis. Overall, Nrp shows greatest similarity (contiguous from domain A2 to the C terminus) to Mps, an NRPS involved in the biosynthesis of glycopeptidolipids in M. smegmatis (4). Glycopeptidolipids are major secreted constituents of the cell envelopes of many species of mycobacteria, including M. smegmatis, members of the M. avium-intracellulare complex, M. avium subsp. paratuberculosis, and Mycobacterium scrofulaceum. Disruption of glycopeptidolipid biosynthesis affects colony morphology (19). However, M. bovis does not produce secreted glycopeptidolipids. Instead, if Nrp is involved directly in PDIM and glycosylphenol-PDIM biosynthesis, we suggest that it may be involved in the biosynthesis of an intracellular peptidolipid carrier molecule that serves as the donor of the long-chain fatty acid(s) entering the specific pathway to phthiocerol/glycosylphenol-phthiocerol and/or mycocerosic acid biosynthesis (Fig. 9). Buglino et al. (5) have recently determined the crystal structure of PapA5, an acyltransferase encoded within the PDIM biosynthetic cluster that Onwueme et al. (29) propose as the catalyst for the diesterification of phthiocerol and phthiodiolone with mycocerosate. PapA5 shares structural and sequence motifs characteristic of CoA-dependent acyltransferases, including the condensation domain from VibH, an NRPS (5). In our model the novel condensation domain (C1) of Nrp may act to esterify a long-chain fatty acid with the carboxy acid selected by adenylation domain A1.
Finally, while we have identified a new locus, Mb0099-Mb0104 (ppe1-nrp), that encodes proteins with roles in the common PDIM/glycosylphenol-PDIM biosynthetic pathway, further studies are required to identify the specific roles for each protein and to determine if all six genes in the operon are required.
Acknowledgments
This work was supported by the New Zealand Foundation for Research, Science and Technology. G.S.B., Lister Jenner Research Fellow, acknowledges support from the Medical Research Council and the Lister Institute of Preventive Medicine. P.R.W. acknowledges support from Department for Environment, Food and Rural Affairs (United Kingdom).
REFERENCES
- 1.Azad, A. K., T. D. Sirakova, N. D. Fernandes, and P. E. Kolattukudy. 1997. Gene knockout reveals a novel gene cluster for the synthesis of a class of cell wall lipids unique to pathogenic mycobacteria. J. Biol. Chem. 272:16741-16745. [DOI] [PubMed] [Google Scholar]
- 2.Azad, A. K., T. D. Sirakova, L. M. Rogers, and P. E. Kolattukudy. 1996. Targeted replacement of the mycocerosic acid synthase gene in Mycobacterium bovis BCG produces a mutant that lacks mycosides. Proc. Natl. Acad. Sci. USA 93:4787-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besra, G. S. 1998. Preparation of cell-wall fractions from mycobacteria. Methods Mol. Biol. 101:91-107. [DOI] [PubMed] [Google Scholar]
- 4.Billman-Jacobe, H., M. J. McConville, R. E. Haites, S. Kovacevic, and R. L. Coppel. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33:1244-1253. [DOI] [PubMed] [Google Scholar]
- 5.Buglino, J., K. C. Onwueme, J. A. Ferreras, L. E. N. Quadri, and C. D. Lima. 2004. Crystal structure of PapA5, a phthiocerol dimycocerosyl transferase from Mycobacterium tuberculosis. J. Biol. Chem. 279:30634-30642. [DOI] [PubMed] [Google Scholar]
- 6.Camacho, L. R., P. Constant, C. Raynaud, M.-A. Lanéelle, J. A. Triccas, B. Gicquel, M. Daffé, and C. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 7.Camacho, L. R., D. Ensergueix, E. Perez, B. Gicquel, and C. Guilhot. 1999. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol. Microbiol. 34:257-267. [DOI] [PubMed] [Google Scholar]
- 8.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 9.Collins, D. M. 2000. New tuberculosis vaccines based on attenuated strains of the Mycobacterium tuberculosis complex. Immunol. Cell. Biol. 78:342-348. [DOI] [PubMed] [Google Scholar]
- 10.Collins, D. M., R. P. Kawakami, B. M. Buddle, B. J. Wards, and G. W. de Lisle. 2003. Different susceptibility of two animal species infected with isogenic mutants of Mycobacterium bovis enabled the identification of phoT with roles in tuberculosis virulence and phosphate transport. Microbiology 149:3203-3212. [DOI] [PubMed] [Google Scholar]
- 11.Collins, D. M., B. Skou, S. White, S. Bassett, L. Collins, R. For, K. Hurr, G. Hotter, and G. W. de Lisle. Generation of attenuated Mycobacterium bovis strains by signature-tagged mutagenesis in search of novel vaccine candidates. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 12.Collins, D. M., T. Wilson, S. Campbell, B. M. Buddle, B. J. Wards, G. Hotter, and G. W. de Lisle. 2002. Production of avirulent mutants of Mycobacterium bovis with vaccine properties by the use of illegitimate recombination and screening of stationary-phase cultures. Microbiology 148:3019-3027. [DOI] [PubMed] [Google Scholar]
- 13.Constant, P., E. Perez, W. Malaga, M.-A. Lanéelle, O. Saurel, M. Daffé, and C. Guilhot. 2002. Role of the pks15/1 gene in the biosynthesis of phenolglycolipids in the Mycobacterium tuberculosis complex. Evidence that all strains synthesize glycosylated p-hydroxybenzoic methyl esters and that strains devoid of phenolglycolipids harbor a frameshift mutation in the pks15/1 gene. J. Biol. Chem. 277:38148-38158. [DOI] [PubMed] [Google Scholar]
- 14.Corbett, E. L., C. J. Watt, N. Walker, D. Maher, B. G. Williams, M. C. Raviglione, and C. Dye. 2003. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 163:1009-1021. [DOI] [PubMed] [Google Scholar]
- 15.Cosivi, O., J. M. Grange, C. J. Daborn, M. C. Raviglione, T. Fujikura, T. Cousins, D. R. A. Robinson, H. F. Huchzermeyer, I. de Kantor, and F. X. Meslin. 1998. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerg. Infect. Dis. 4:59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 17.Daffe, M., and M. A. Laneelle. 1988. Distribution of pthiocerol diester, phenolic mycosides and related compounds in mycobacteria. J. Gen. Microbiol. 134:2049-2055. [DOI] [PubMed] [Google Scholar]
- 18.Dobson, G., D. E. Minnikin, S. M. Minnikin, J. H. Pharlett, M. Goodfellow, M. Ridell, and M. Magnusson. 1985. Systemic analysis of complex mycobacterial lipids, p. 237-265. In M. Goodfellow and D. E. Minnikin (ed.), Chemical methods in bacterial systematics. Academic Press, London, United Kingdom.
- 19.Etienne, G., C. Villeneuve, H. Billman-Jacobe, C. Astarie-Dequeker, M.-A. Dupont, and M. Daffe. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis. Microbiology 148:3089-3100. [DOI] [PubMed] [Google Scholar]
- 20.Fitzmaurice, A. M., and P. E. Kolattukudy. 1998. An acyl-CoA synthase (acoas) gene adjacent to the mycocerosic acid synthase (mas) locus is necessary for mycocerosyl lipid synthesis in Mycobacterium tuberculosis var. bovis BCG. J. Biol. Chem. 273:8033-8039. [DOI] [PubMed] [Google Scholar]
- 21.Garnier, T., K. Eiglmeier, J.-C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hausinger, R. P. 2004. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit. Rev. Biochem. Mol. Biol. 39:21-68. [DOI] [PubMed] [Google Scholar]
- 23.Jackson, R., M. M. Cooke, J. D. Coleman, and R. S. Morris. 1995. Naturally occurring tuberculosis caused by Mycobacterium bovis in brushtail possums (Trichosurus vulpecula). I. An epidemiological analysis of lesion distribution. N. Z. Vet. J. 43:306-314. [DOI] [PubMed] [Google Scholar]
- 24.Kalate, R. N., S. S. Tambe, and B. D. Kulkarni. 2003. Artificial neural networks for prediction of mycobacterial promoter sequences. Comp. Biol. Chem. 27:555-564. [DOI] [PubMed] [Google Scholar]
- 25.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology, a guide for the level III laboratory. U.S. Department of Health and Human Services, Center for Disease Control, Atlanta, Ga.
- 26.McAdam, R. A., T. R. Weisbrod, J. Martin, J. D. Scuderi, A. M. Brown, J. D. Cirillo, B. R. Bloom, and W. R. Jacobs, Jr. 1995. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect. Immun. 63:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minnikin, D. E., G. Dobson, M. Goodfellow, M. Magnusson, and M. Ridell. 1985. Distribution of some mycobacterial waxes based on the phthiocerol family. J. Gen. Microbiol. 131:1375-1381. [DOI] [PubMed] [Google Scholar]
- 28.Minnikin, D. E., L. Kremer, L. G. Dover, and G. S. Besra. 2002. The methyl-branched fortifications of Mycobacterium tuberculosis. Chem. Biol. 9:545-553. [DOI] [PubMed] [Google Scholar]
- 29.Onwueme, K. C., J. A. Ferreras, J. Buglino, C. D. Lima, and L. E. N. Quadri. 2004. Mycobacterial polyketide-associated proteins are acyltransferases: proof of principle with Mycobacterium tuberculosis PapA5. Proc. Natl. Acad. Sci. USA 101:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parish, T., D. A. Smith, G. Roberts, J. Betts, and N. G. Stoker. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149:1423-1435. [DOI] [PubMed] [Google Scholar]
- 31.Perez, E., P. Constant, F. Laval, A. Lemassu, M.-A. Lanéelle, M. Daffé, and C. Guilhot. 2004. Molecular dissection of the role of two methyltransferases in the biosynthesis of phenolglycolipids and phthiocerol dimycocerosate in the Mycobacterium tuberculosis complex. J. Biol. Chem. 279:42584-42592. [DOI] [PubMed] [Google Scholar]
- 32.Perez, E., P. Constant, A. Lemassu, F. Laval, M. Daffé, and C. Guilhot. 2004. Characterization of three glycosyltransferases involved in the biosynthesis of the phenolic glycolipid antigens from the Mycobacterium tuberculosis complex. J. Biol. Chem. 279:42574-42583. [DOI] [PubMed] [Google Scholar]
- 33.Pinto, R., B. M. Saunders, L. R. Camacho, W. J. Britton, B. Gicquel, and J. A. Triccas. 2004. Mycobacterium tuberculosis defective in phthiocerol dimycocerosate translocation provides greater protective immunity against tuberculosis than the existing Bacille Calmette-Guérin vaccine. J. Infect. Dis. 189:105-112. [DOI] [PubMed] [Google Scholar]
- 34.Reed, M. B., P. Domenech, C. Manca, H. Su, A. K. Barczak, B. N. Kreiswirth, G. Kaplan, and C. E. Barry III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:184-187. [DOI] [PubMed] [Google Scholar]
- 35.Rousseau, C., N. Winter, E. Pivert, Y. Bordat, O. Neyrolles, P. Avé, M. Huerre, B. Gicquel, and M. Jackson. 2004. Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell. Microbiol. 6:277-287. [DOI] [PubMed] [Google Scholar]
- 36.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 37.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 38.Steyn, A. J. C., D. M. Collins, M. K. Hondalus, W. R. Jacobs, R. P. Kawakami, and B. R. Bloom. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. USA 99:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trivedi, O. A., P. Arora, V. Sridharan, R. Tickoo, D. Mohanty, and R. S. Gokhale. 2004. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 428:441-445. [DOI] [PubMed] [Google Scholar]
- 40.Van Soolingen, D., P. W. M. Hermans, P. E. W. de Haas, D. R. Soll, and J. D. A. van Embden. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 29:2578-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wards, B. J., and D. M. Collins. 1996. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol. Lett. 145:101-105. [DOI] [PubMed] [Google Scholar]